Introduction
Nephrotic syndrome (NS) is one of the most common
causes of chronic kidney disease in the pediatric population
(1). NS is primarily characterized
by extensive proteinuria, hypoalbuminemia, hyperlipidemia and
edema, resulting from disruption of the glomerular filtration
barrier (2). A kidney biopsies
taken from children with NS are often associated with podocyte foot
process effacement, whereas kidney biopsies taken from healthy
children are not (3).
Previous studies have demonstrated that oxidized
low-density lipoprotein (oxLDL) may cause podocyte damage (4,5);
however, the underlying mechanism is not completely understood.
Evidence has suggested that oxLDL promotes the migration of
monocytes, smooth muscle cells and bone marrow-derived mesenchymal
stem cells (6–8); however, whether oxLDL results in
podocyte migration, as well as its associated mechanism, requires
further investigation.
CXC motif chemokine ligand 16 (CXCL16) is a
scavenger receptor that exists in two distinct forms: A
membrane-bound form and a soluble form (9). Membrane-bound CXCL16 can be released
as the soluble form upon digestion by ADAM metallopeptidase domain
(ADAM) proteins, specifically ADAM10 and ADAM17 (10). The membrane-bound form of CXCL16
binds to and internalizes oxLDL to promote the adhesion of cells
expressing CXC motif chemokine receptor 6 (CXCR6), the only known
receptor of CXCL16 (11). By
contrast, soluble CXCL16 functions as a chemoattractant to promote
the migration of CXCR6-expressing cells (12).
Previous studies have demonstrated that CXCL16 and
ADAM10 are expressed in the kidney (11,13,14),
and Gutwein et al (15)
reported that CXCL16 and ADAM10 are constitutively expressed in
podocytes. However, a limited number of studies have investigated
the role of CXCL16 and ADAM10 in proteinuria and the progression of
glomerular diseases.
Schramme et al (16) revealed that inhibition of ADAM10
and CXCL16 expression in mesangial cells results in a significant
reduction in cell proliferation and migration. Increasing evidence
has also demonstrated that soluble CXCL16 promotes cancer cell
migration in vitro (12).
Collectively, the results of the aforementioned studies suggest
that CXCL16 and ADAM10 may promote podocyte migration and may be
involved in the development of primary NS by mediating lipid
stimulation and inflammatory responses (17).
The present study investigated the expression of
ADAM10 and CXCL16 in an oxLDL-stimulated conditionally immortalized
mouse podocyte cell line (MPC5). Additionally, the effect of oxLDL
on cell migration was analyzed by performing functional assays. The
results suggested that oxLDL stimulation increased the expression
of ADAM10 and CXCL16, and promoted podocyte migration. Furthermore,
the effects of CXCL16 and ADAM10 overexpression and knockdown in
MPC5 cells were investigated, and the expression of actinin-α4
(ACTN4) was detected via western blotting.
Materials and methods
Cell culture
The conditionally immortalized mouse podocyte cell
line MPC5 was a gift from Professor Rong Wang (Department of
Nephrology, Shandong Provincial Hospital Affiliated to Shandong
University). Cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin, 100 µg/ml streptomycin and
10 U/ml mouse interferon-γ (IFN-γ; cat. no. C600059; Sangon Biotech
Co., Ltd.) at 33°C in a humidified incubator containing 5%
CO2 for 2–3 days (permissive condition). To induce
differentiation, cells were transferred to RPMI-1640 medium without
IFN-γ and incubated at 37°C in a humidified incubator with 5%
CO2 for 14 days (non-permissive condition).
Construction of recombinant short
hairpin (sh)RNA ADAM10 and shCXCL16 interference plasmids
Plasmids were constructed using the lentiviral
transferring plasmid, pLKO.1-TRC (pLKO.1; Jiangsu Laisen
Biotechnology Co., Ltd.). The sequences of the shADAM10 and
shCXCL16 were amplified and inserted into pLKO.1 to generate
recombinant pLKO.1-shADAM10 and pLKO.1-shCXCL16 for lentivirus
production. The sequences of the forward and reverse primers of the
shADAM10 and shCXCL16 are presented in Table I. Using synthetic shADAM10 and
shCXCL16 sequences as templates for PCR. PCR amplifications were
performed with Phanta Max Super-Fidelity DNA Polymerase (cat. no.
DC505; Vazyme Biotech Co., Ltd.), using the following thermocycling
conditions: 35 cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C
for 15 sec; extension at 72°C for 1 min; and maintained at 4°C. PCR
products were verified by 1% agarose gel electrophoresis and
recovered using a purification kit (cat. no. DC301; Vazyme Biotech
Co., Ltd.). The restriction enzymes AgeI and EcoRI
(New England BioLabs, Inc.) were used to perform a double digest of
the plasmid and the PCR gel purification products. The products
were separated by 1% agarose gel electrophoresis, purified and
incubated with T4 ligase overnight at 4°C. The products were
transformed into Escherichia coli, and a single colony was
randomly selected. The plasmid was extracted using a plasmid
extraction kit (cat. no. DC201; Vazyme Biotech Co., Ltd.). shADAM10
and shCXCL16 recombinant plasmids were verified by a double digest
and the products were detected by 1% agarose gel electrophoresis.
The plasmids were subsequently sent to Genscript for sequencing,
and the sequencing results were compared with the design
sequence.
 | Table I.shRNA sequences. |
Table I.
shRNA sequences.
| shRNA | Sequence (5→3) |
|---|
| shCXCL16-1-F |
CCGGGCAGGGTACTTTGGATCACATCTCGAGATGTGATCCAAAGTACCCTGCTTTTTGAATT |
| shCXCL16-1-R |
AATTCAAAAAGCAGGGTACTTTGGATCACATCTCGAGATGTGATCCAAAGTACCCTGC |
| shCXCL16-2-F |
CCGGGCTGGAAGTTGTTCTTGTGATCTCGAGATCACAAGAACAACTTCCAGCTTTTTGAATT |
| shCXCL16-2-R |
AATTCAAAAAGCTGGAAGTTGTTCTTGTGATCTCGAGATCACAAGAACAACTTCCAGC |
| shADAM10-1-F |
CCGGGCAGAGAGATACATTAAAGATCTCGAGATCTTTAATGTATCTCTCTGCTTTTTGAATT |
| shADAM10-1-R |
AATTCAAAAAGCAGAGAGATACATTAAAGATCTCGAGATCTTTAATGTATCTCTCTGC |
| shADAM10-2-F |
CCGGCCAGGAGAGTCTAAGAACTTACTCGAGTAAGTTCTTAGACTCTCCTGGTTTTTGAATT |
| shADAM10-2-R |
AATTCAAAAACCAGGAGAGTCTAAGAACTTACTCGAGTAAGTTCTTAGACTCTCCTGG |
Production of recombinant lentiviral
particles
293T cells (Conservation Genetics CAS Kunming Cell
Bank) were cultured in a 10 cm cell culture dish at 37°C with 5%
CO2 overnight. At 80% confluence, 293T cells were
co-transfected with 5.94 µg recombinant plasmid 5.94 µg
pLKO.1-shADAM10, 5.94 µg pLKO.1-shCXCL16 or 5.94 µg pLKO.1 (empty
vector), 4.86 µg psPAX2 and 9 µg pMD2.G (Jiangsu Laisen
Biotechnology Co., Ltd.) using 40 µl Lipofectamine® 2000
(cat. no. 11668019; Thermo Fisher Scientific, Inc.) to produce
lentivirus. Similarly, 293T cells were co-transfected with 5.94 µg
overexpression plasmid pCDH-CMV-ADAM10-Puro (pCDH-ADAM10; Vigene
Biosciences), 5.94 µg pCDH-CMV-CXCL16-Puro (pCDH-CXCL16; Vigene
Biosciences) or 5.94 µg pCDH (empty vector), 4.86 µg psPAX2 and 9
µg pMD2.G using 40 µl Lipofectamine® 2000 to produce
lentivirus. At 10 h post-transfection, the cell culture medium was
replaced with DMEM medium containing 10% FBS, 100 U/ml penicillin
and 100 µg/ml streptomycin. At 48 h post-transfection, the culture
medium was collected, filtered through a 0.45-µm filter and stored
at −80°C until further use.
CXCL16 and ADAM10 overexpression and
knockdown in MPC5 cells
CXCL16 and ADAM10 overexpression and knockdown
lentiviruses were used to infect MPC5 cells. MPC5 cells
(5×105) were infected with 200 µl lentivirus
(~2×107 viral particles) and 2 µl polybrene
(Sigma-Aldrich; Merck KGaA). At 48 h 37°C post-transfection,
puromycin (3 µg/ml) was used to screen the cells and surviving
cells were used for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using a Reverse
Transcription system (Vazyme Biotech Co., Ltd.) according to the
manufacturer's protocol. The temperature protocol used for reverse
transcription was as follows: 50°C for 20 min and 85°C for 2 min.
Subsequently, qPCR was performed. The sequences of the primers used
for qPCR are presented in Table
II. The following thermocycling conditions were used for qPCR:
95°C for 5 min; 45 cycles of 95°C for 10 sec and 60°C for 30 sec.
Relative gene expression was determined by the 2−ΔΔCq
method (18) and normalized to the
internal reference gene β-actin.
 | Table II.Primers used for reverse
transcription-quantitative PCR. |
Table II.
Primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence (5→3) |
|---|
| CXCL16- | F:
ACCCTTGTCTCTTGCGTTCT |
| Mus-1 | R:
GCTTTTGGACTGCAACTGGA |
| CXCL16- | F:
ACTCAGCACTCCACTCTTCC |
| Mus-2 | R:
ATGGCTGCAGTGAGGAAGAA |
| ADAM10- | F:
TTTGTTGAATCTGGCCAGCC |
| Mus-1 | R:
TCCCTTCCTTTGCACAGTCA |
| ADAM10- | F:
ATTGCTGAGTGGATTGTGGC |
| Mus-2 | R:
GATAACTCTCTCGGGGCCTC |
| β-actin-Mus | F:
TGGAATCCTGTGGCATCCATGAAAC |
|
| R:
TAAAACGCAGCTCAGTAACAGTCCG |
Western blotting
Cells were lysed with RIPA lysis buffer (Sangon
Biotech Co., Ltd.) supplemented with protease and phosphatase
inhibitors. Total protein was quantified using the BCA Protein
Assay kit (Beyotime Institute of Biotechnology). Equal quantities
of protein (30 µg/lane) were separated via 10% SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore). The membranes were
blocked with TBS-T (0.05% Tween-20) containing 5% non-fat milk at
room temperature for 1 h. Subsequently, the membranes were
incubated at 4°C overnight with primary antibodies targeted
against: β-actin (1:1,000; cat. no. 3700T; Cell Signaling
Technology), CXCL16 (1:1,000; cat. no. 60123-1-Ig; Proteintech),
ADAM10 (1:1,000; cat. no. BS60338; Bioworld Technology.) and ACTN4
(1:1,000; cat. no. BS90025; Bioworld Technology). Following primary
incubation, the membranes were incubated with horseradish
peroxidase-conjugated Affinipure Goat Anti-Rabbit IgG (H+L;
1:5,000; cat. no. SA00001-2; Proteintech) or Affinipure Goat
Anti-Mouse IgG (H+L; 1:5,000; cat. no. SA00001-1; Proteintech)
secondary antibodies for 1 h at room temperature. Protein bands
were visualized using Western Bright ECL (Thermo Fisher Scientific,
Inc.). Protein expression was quantified using Image Lab software
(version 4.1; Bio-Rad Laboratories) with β-actin as the loading
control.
Cell migration assay
Cells were treated with oxLDL (50, 80 or 100 µg/ml;
Beijing Solarbio Science & Technology Co., Ltd.) for 48 h at
37°C. Cell migration was evaluated by performing a Transwell assay
(Corning, Inc.). Cells were seeded (5×104 cells/well)
into the upper chamber with RPMI-1640 without FBS. RPMI-1640 medium
containing 10% FBS was added into the lower chamber. Following
incubation at 37°C for 12 h, cells on the surface of the Transwell
inserts were removed with a cotton swab, and cells on the underside
of the insert filter were fixed with 100% ethyl alcohol and stained
with 1% crystal violet at 24°C for 15 min. Stained cells were
visualized using a light microscope (magnification, ×200).
Oil red O staining
MPC5 cells were treated with oxLDL (50 or 100 µg/ml)
for 48 h at 37°C. Subsequently, Oil Red O was dissolved in a small
amount of 100% isopropyl alcohol to a volume of 100 ml and sealed
in a brown bottle at 4°C. Cells were rinsed with PBS, fixed with
10% neutral formalin for 30 min at 24°C, stained with Oil Red O for
10 min at 24°C, rinsed with 60% isopropyl alcohol and observed
under an optical microscope (magnification, ×400).
ELISA
The cell culture medium was collected and the levels
of released CXCL16 were measured using an ELISA kit (cat. no.
AD3057Mo; Andygene) according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard deviation.
All experiments were performed in triplicate. Statistical analyses
were performed using GraphPad Prism software (version 5.0; GraphPad
Software, Inc.). Comparisons among multiple groups were analyzed
using one-way ANOVA followed by Tukey's post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
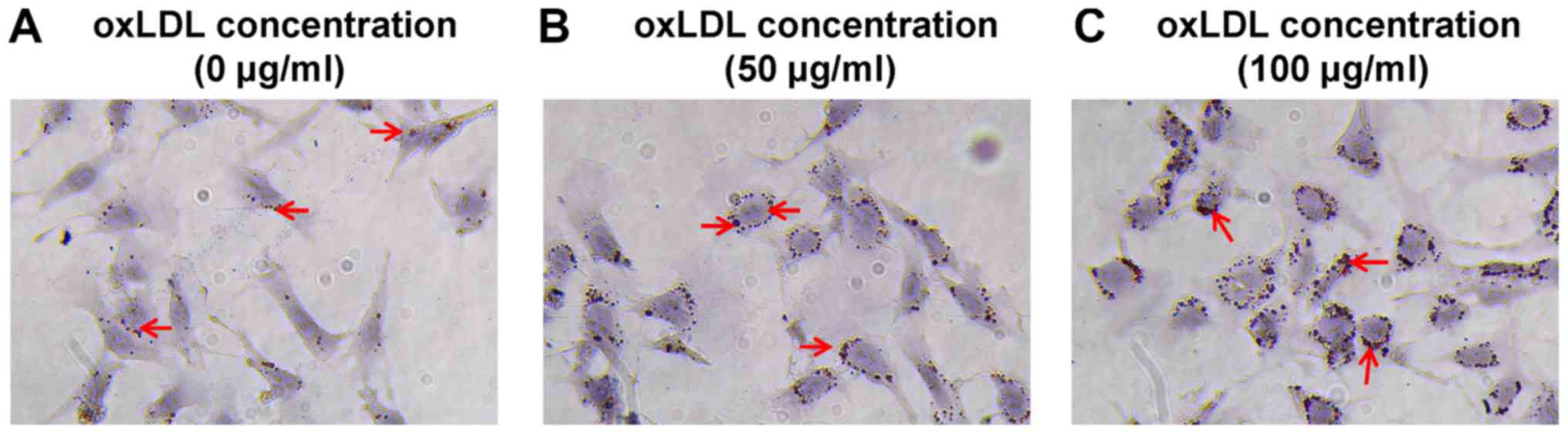
Lipid accumulation is increased in
oxLDL-treated cells
MPC5 cells were cultured in RPMI-1640 medium and
treated with 50 or 100 µg/ml oxLDL (Beijing Solarbio Science &
Technology Co., Ltd.) for 48 h. Control cells were treated with
DMSO. A previous study indicated that 80 µg/ml oxLDL was the most
suitable dose (17); therefore, a
high dose above the optimal dose (100 µg/ml) and a low dose below
the optimal dose (50 µg/ml) were used in the present study. Oil Red
O staining was performed to observe intracellular lipid
accumulation. The control group displayed a few visible lipid drops
(Fig. 1A), whereas a large number
of lipid drops were observed in oxLDL-treated cells oxLDL (50 and
100 µg/ml; Fig. 1B and C).
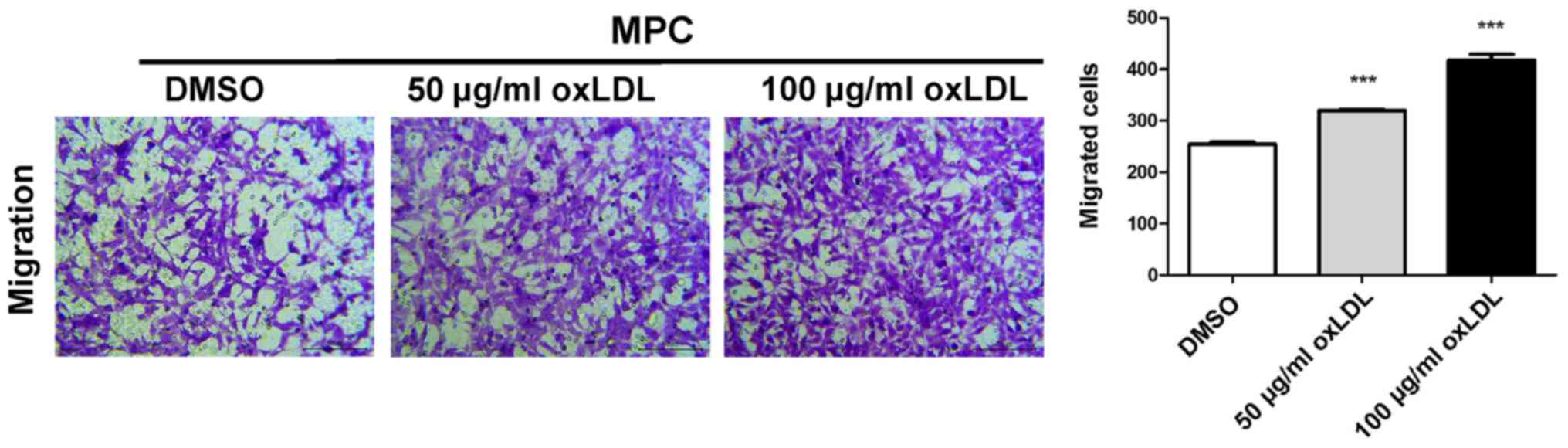
oxLDL stimulates podocyte
migration
The migratory ability of MPC5 cells was assessed by
performing a Transwell assay. The number of migratory cells was
significantly increased in a dose-dependent manner in the 50 and
100 µg/ml oxLDL groups compared with the control group (Fig. 2).
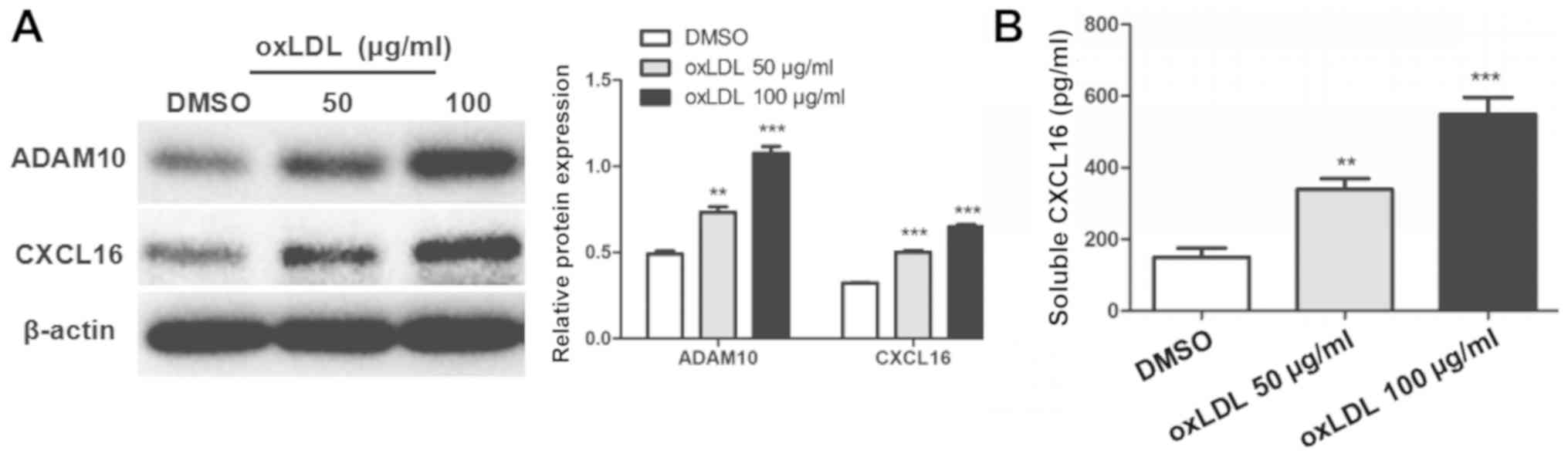
oxLDL stimulation increases ADAM10 and
CXCL16 expression
oxLDL stimulation significantly increased the
protein expression levels of ADAM10 and CXCL16 in MPC5 cells
compared with DMSO-treated cells (Fig.
3A). Moreover, the level of soluble CXCL16 in the culture
medium was significantly increased in the 50 and 100 µg/ml oxLDL
groups compared with the control group (Fig. 3B).
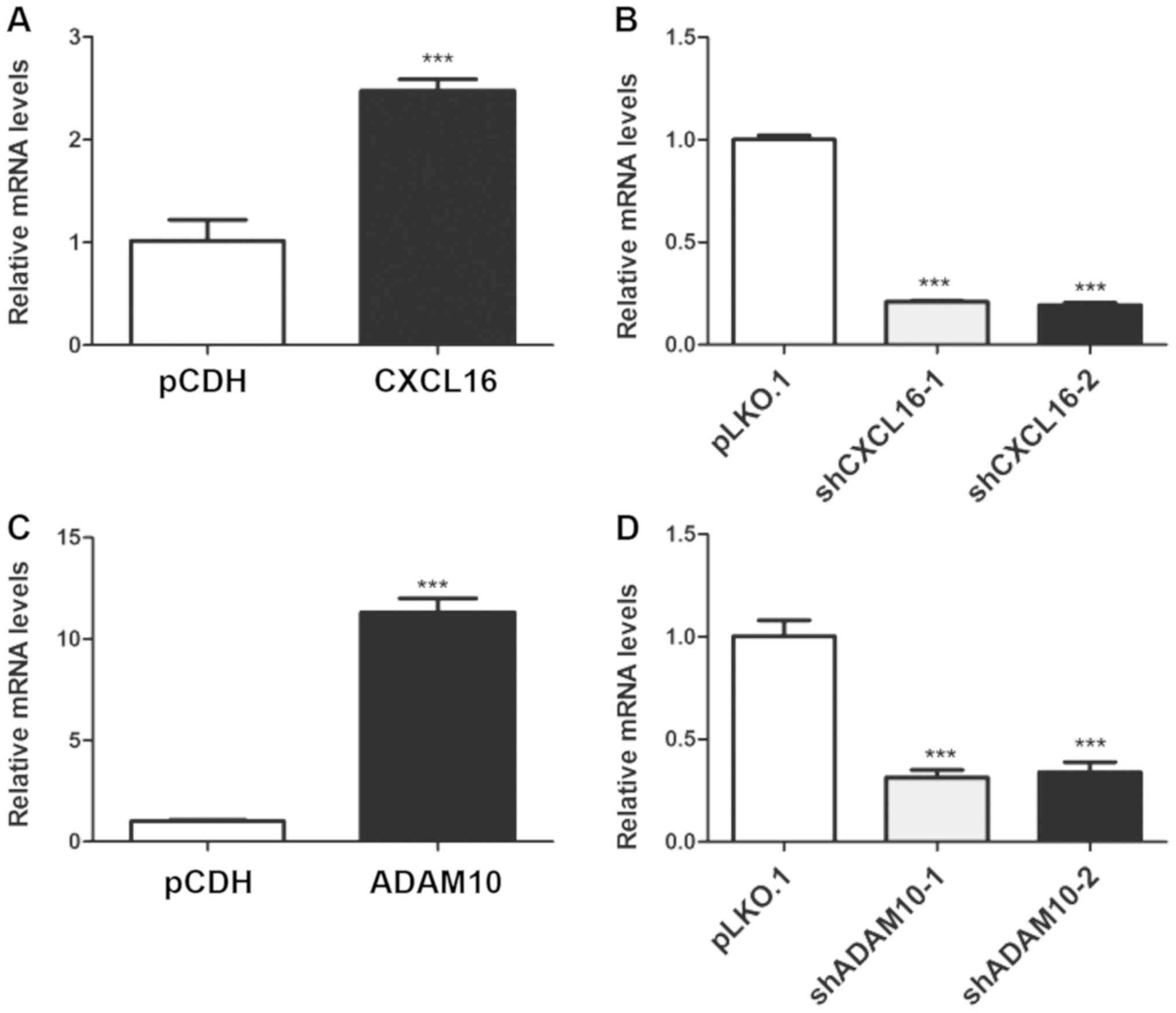
Generation of stable CXCL16- and
ADAM10-overexpression and knockdown podocyte cell lines
Transfection efficiency was assessed via RT-qPCR.
CXCL16 expression was significantly increased by 147.54% in the
overexpression group compared with the pCDH group (Fig. 4A). By contrast, CXCL16 expression
was significantly decreased by 79.11 and 80.68% in the shCXCL16-1
and shCXCL16-2 groups compared with the pLKO.1 group, respectively
(Fig. 4B). Similarly, ADAM10
expression was significantly increased by 1,012.07% in the
overexpression group compared with the pCDH group (Fig. 4C), whereas ADAM10 expression was
significantly decreased by 68.58 and 66.08% in the shADAM10-1 and
shADAM10-2 groups compared with the pLKO.1 group, respectively
(Fig. 4D).
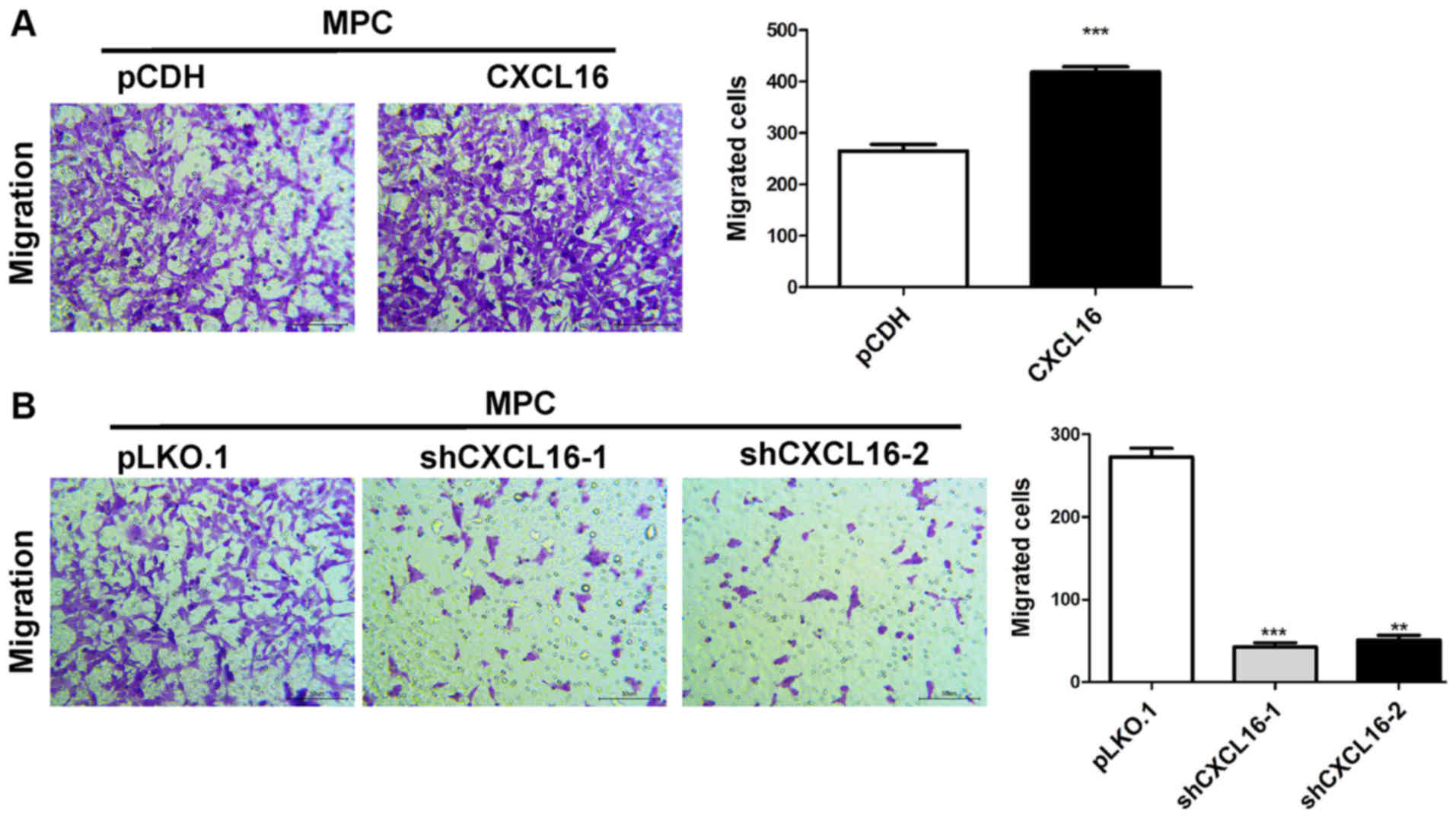
CXCL16 overexpression enhances
podocyte migration, whereas CXCL16 knockdown reduces podocyte
migration
The migratory ability of CXCL16-overexpression cells
was assessed using a Transwell assay. The results indicated that
CXCL16 overexpression significantly increased MPC5 cell migration
compared with the pCDH group (Fig.
5A). By contrast, CXCL16 knockdown significantly reduced MPC5
cell migration compared with the pLKO-1 group (Fig. 5B).
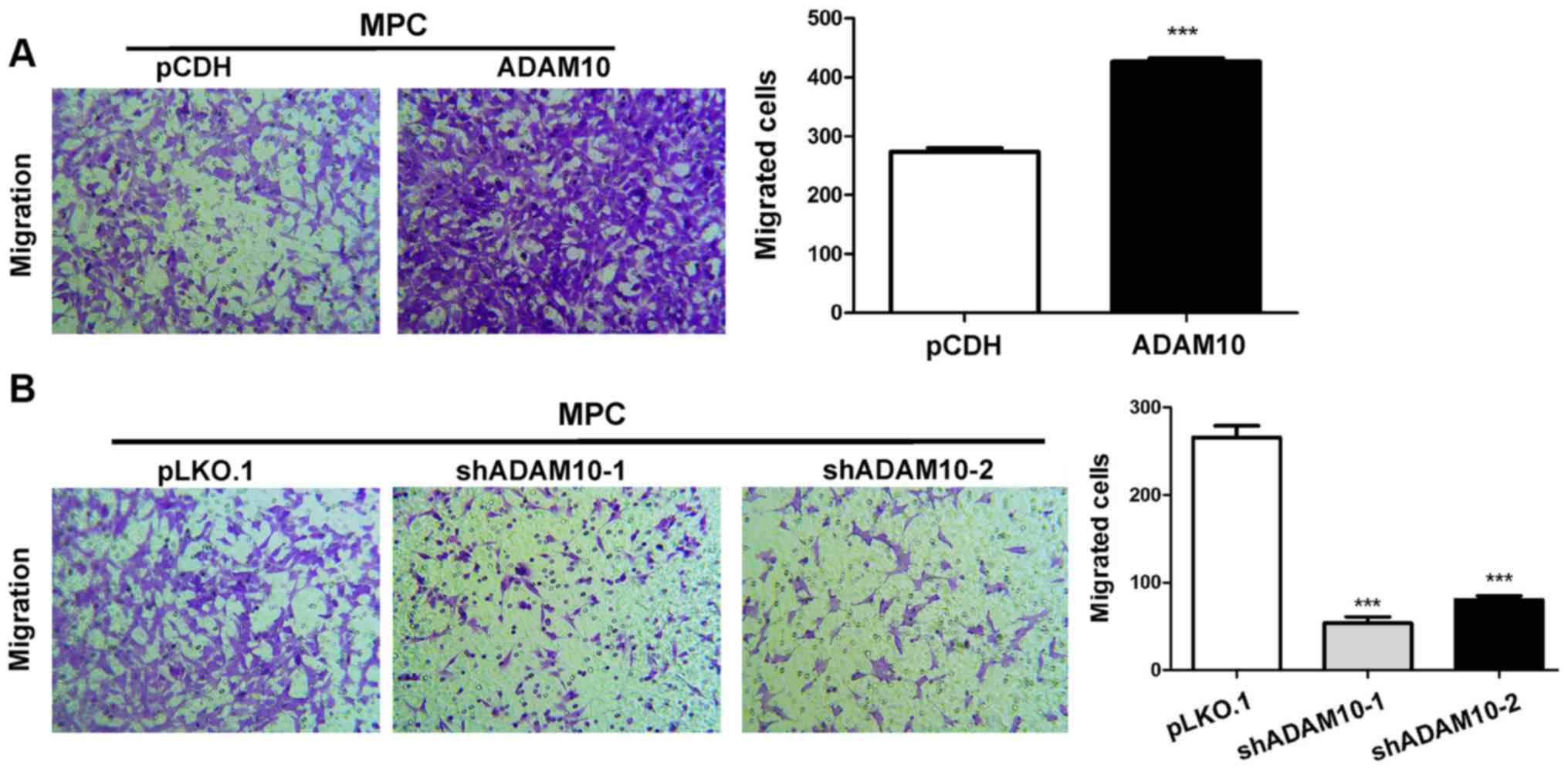
ADAM10 overexpression enhances
podocyte migration, whereas ADAM10 knockdown reduces podocyte
migration
The migratory ability of ADAM10-overexpression cells
was assessed using a Transwell assay. The results indicated that
ADAM10 overexpression significantly increased MPC5 cell migration
compared with the pCDH group (Fig.
6A), whereas ADAM10 knockdown significantly decreased MPC5 cell
migration compared with the pLKO.1 group (Fig. 6B).
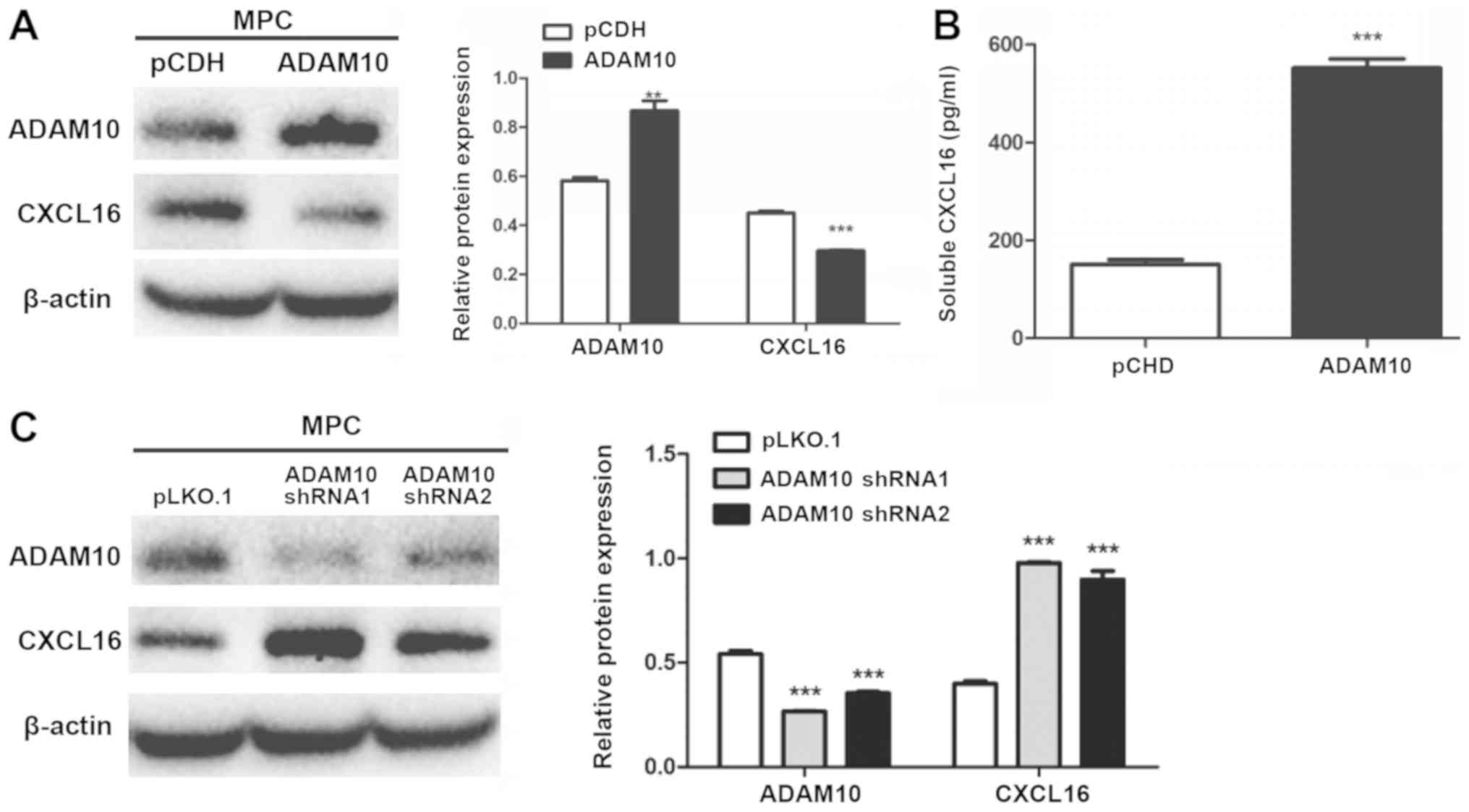
ADAM10 and CXCL16 protein expression
in MPC5 cells
ADAM10-overexpression and -knockdown cells were
subjected to western blotting. The expression level of
membrane-bound CXCL16 was significantly decreased in
ADAM10-overexpression cells compared with pCDH cells (Fig. 7A). Conversely, the expression level
of membrane-bound CXCL16 was significantly increased in
ADAM10-knockdown cells compared with pLKO.1 cells (Fig. 7C). Soluble CXCL16 release was
significantly increased in ADAM10-overexpression cells compared
with pCDH cells (Fig. 7B).
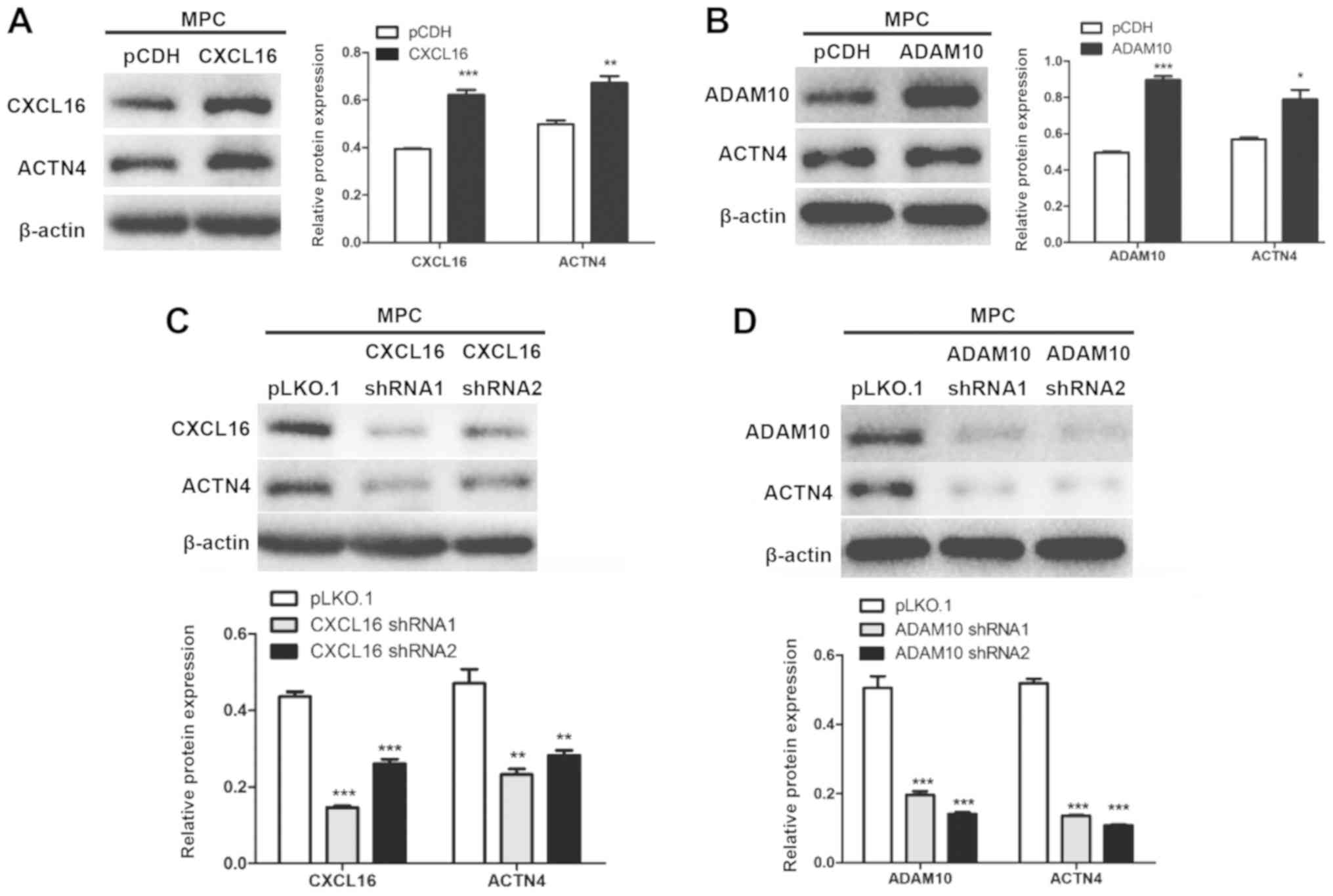
CXCL16 and ADAM10 modulate the
expression of ACTN4
CXCL16- and ADAM10-overexpression cells displayed
significantly increased ACTN4 protein expression levels compared
with pCDH cells (Fig. 8A and B).
By contrast, CXCL16- and ADAM10-knockdown cells displayed
significantly decreased ACTN4 expression levels compared with
pLKO.1 cells (Fig. 8C and D).
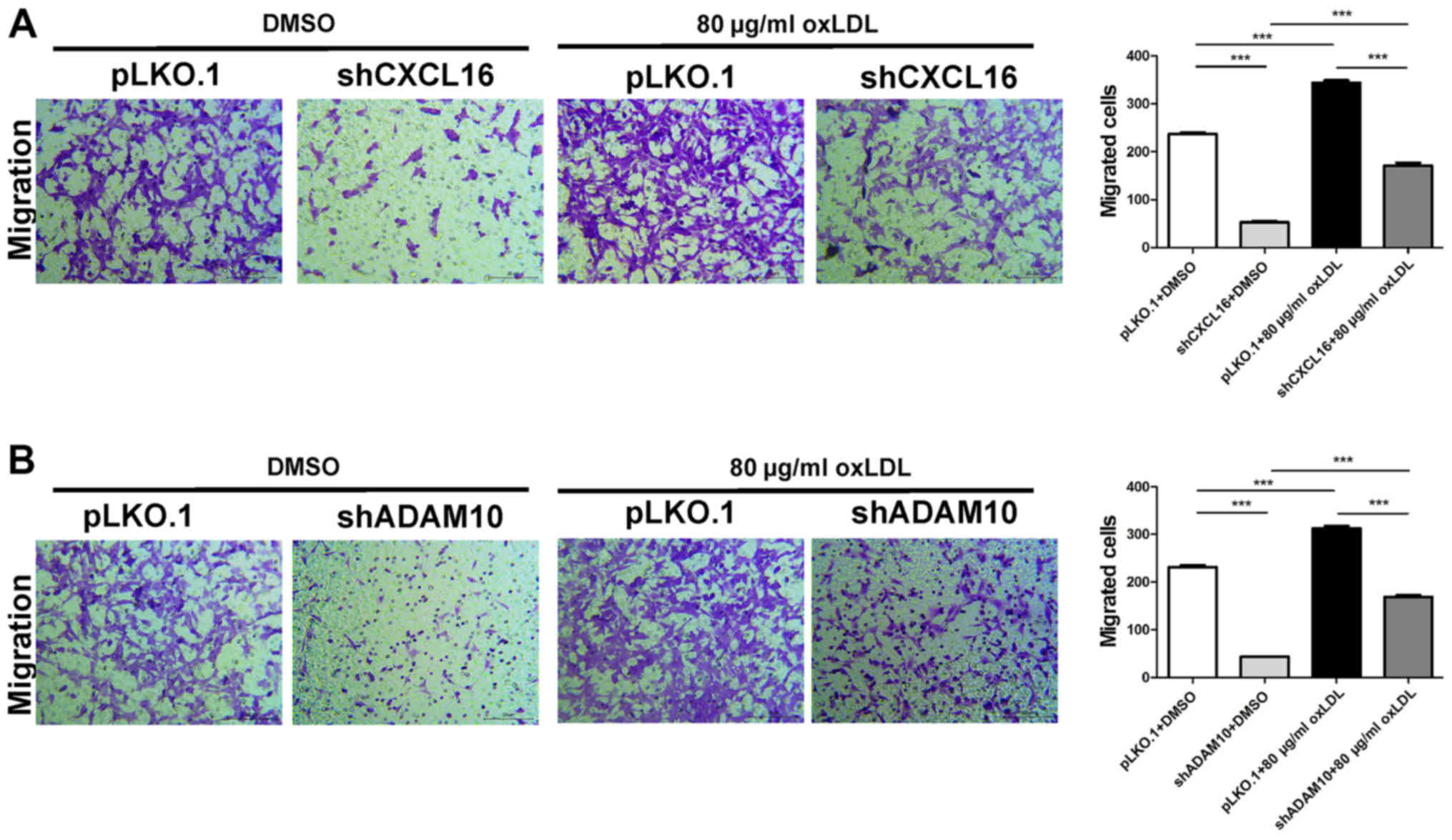
oxLDL stimulation enhances CXCL16- and
ADAM10 knockdown-mediated podocyte migration
Podocyte migration was assessed in CXCL16- and
ADAM10-knockdown cells following treatment with 80 µg/ml oxLDL. The
control group was treated with DMSO. The results indicated that the
number of migratory cells in the oxLDL-treated pLKO.1 group was
significantly increased compared with the oxLDL-treated shCXCL16
group. In addition, the number of migratory cells in the
DMSO-treated pKLO.1 group was significantly increased compared with
the DMSO-treated shCXCL16 group. Moreover, the oxLDL-treated pLKO.1
group displayed significantly increased cell migration compared
with the DMSO-treated pLKO.1 group. Similarly, the oxLDL-treated
shCXCL16 group displayed significantly increased cell migration
compared with the DMSO-treated shCXCL16 group (Fig. 9A). Consistently, the number of
migratory cells in the oxLDL-treated pLKO.1 group was significantly
increased compared with the oxLDL-treated shADAM10 group. The
number of migratory cells in the DMSO-treated pLKO.1 group was
significantly increased compared with the DMSO-treated shADAM10
group. Furthermore, the oxLDL-treated pLKO.1 group displayed
increased cell migration compared with the DMSO-treated pLKO.1
group and the oxLDL-treated shADAM10 group displayed significantly
increased cell migration compared with the DMSO-treated shADAM10
group (Fig. 9B). The results
indicated that oxLDL stimulation promoted the migration of CXCL16
and ADAM10-knockdown MPC5 cells.
Discussion
The present study investigated the effect of oxLDL
on podocyte lipid accumulation and migration. The results indicated
that oxLDL promoted podocyte lipid accumulation and enhanced MPC5
cell migration in a dose-dependent manner.
NS is a renal disease caused by disruption of the
glomerular filtration barrier that results in extensive
proteinuria, hypoalbuminemia, hyperlipidemia and edema (1). Based on kidney biopsies, minimal
change disease (MCD) is the most common cause of primary NS in
children, followed by focal segmental glomerulosclerosis (FSGS) and
membranoproliferative glomerulonephritis (2,3,19).
Renal biopsies of patients with proteinuria and kidney disease are
most often associated with podocyte foot process effacement, which
commonly occurs in NS (20).
The podocyte is a highly specialized, terminally
differentiated cell that constitutes a crucial component of the
glomerular filtration barrier (21–23).
Podocyte injury is a hallmark of proteinuria and glomerular
diseases (24,25), such as MCD, membranous
glomerulopathy, lupus nephritis and diabetic nephropathy (26–28).
Podocytes are a contractile and motile cell type,
and their motility must be finely regulated in order to maintain
the function of the glomerular filtration barrier (29). Previous studies have suggested that
podocytes may transition from a stationary state to a migratory
state in response to various stimuli, such as lipopolysaccharide or
puromycin aminonucleoside (30–32).
Podocyte migration integrates several functions related to adhesion
and rearrangement of the actin cytoskeleton (33–35),
and increased cell motility could serve as a potential mechanism
underlying foot process effacement and proteinuria (36,37).
The results of the present study suggested that oxLDL promoted
podocyte migration; however, the link between oxLDL-induced
migration, and foot process effacement and proteinuria requires
further investigation.
The present study also demonstrated that oxLDL
stimulation increased the expression of CXCL16 and ADAM10 in
podocytes compared with DMSO-treated cells. Furthermore, the levels
of both membrane-bound and soluble CXCL16 were increased in
oxLDL-stimulated cells compared with control cells. Schramme et
al (16) demonstrated that
inhibition of ADAM10 and CXCL16 in mesangial cells resulted in a
significant reduction in cell proliferation and migration.
Moreover, increasing evidence demonstrates that soluble CXCL16
promotes cancer cell migration in vitro (12).
The present study investigated the effect of CXCL16
and ADAM10 on podocyte migration. CXCL16 and ADAM10 overexpression
significantly increased podocyte migration compared with the
control groups. Furthermore, podocyte migration was significantly
decreased following CXCL16 and ADAM10 knockdown compared with the
control groups. CXCL16 exists in two forms in vivo, soluble
CXCL16 and membrane-bound CXCL16 (9). ADAM is a Zn2+ dependent
transmembrane and metalloproteinase superfamily, known as
‘molecular scissors’. ADAM10 is highly expressed in the kidney,
where it cleaves the extracellular domain of membrane-bound CXCL16
in a process known as ectodomain shedding, resulting in the release
of soluble CXCL16 into the circulation (10). ADAM10 overexpression is associated
with enhanced release of CXCL16 into culture supernatants and a
reduction of membrane-bound protein (10,15).
In the present study, membrane-bound CXCL16 expression was
decreased in ADAM10-overexpression cells, but was increased in
ADAM10-knockdown cells compared with control cells. Moreover,
soluble CXCL16 was upregulated in ADAM10-overexpression cells
compared with control cells.
In the present study, oxLDL was used to stimulate
CXCL16- and ADAM10-knockdown podocytes. oxLDL stimulation enhanced
CXCL16 and ADAM10 knockdown-mediated cell migration compared with
control cells, which suggested that oxLDL promoted podocyte
migration by regulating CXCL16 and ADAM10.
ACTN4 is a regulator of the actin cytoskeleton that
enables cell migration (38).
Several studies have demonstrated that podocyte-specific expression
of an FSGS-associated ACTN4 mutant (K256E) results in a reduction
in podocyte migration and causes proteinuria in mice (39,40).
Wang et al (41) reported
that nucleoporin 160 knockdown increased the expression of ACTN4
and enhanced podocyte migration. Therefore, to determine whether
CXCL16 and ADAM10 impacted the dynamic actin cytoskeleton
rearrangements of podocytes, the present study investigated the
expression of ACTN4 in CXCL16- and ADAM10-overexpression and
knockdown podocytes. The results revealed that ACTN4 was
upregulated in CXCL16- and ADAM10-overexpression cells, but was
downregulated in CXCL16- and ADAM10-knockdown cells compared with
control cells. Therefore, the regulation of CXCL16 and ADAM10
expression may influence ACTN4 expression, which may result in
podocyte migration; however, the link between CXCL16, ADAM10 and
ACTN4 requires further investigation.
The present study investigated the effects of oxLDL
on podocyte migration. The results revealed that oxLDL enhanced
podocyte migration, whereas CXCL16 and ADAM10 mediated the process.
More importantly, CXCL16 and ADAM10 overexpression and knockdown
significantly increased and decreased podocyte migration compared
with control cells, respectively. Furthermore, the results
indicated that CXCL16 and ADAM10 may regulate podocyte migration by
modulating the actin cytoskeleton. Collectively, the results
suggested that oxLDL promoted podocyte migration via regulation of
CXCL16 and ADAM10, and modulation of the actin cytoskeleton.
Therefore, future studies investigating CXCL16 and ADAM10 are
required, as the two proteins may serve as potential therapeutic
targets for NS in children.
Acknowledgements
The authors would like to thank Professor Rong Wang
(Shandong Provincial Hospital, Cheeloo College of Medicine,
Shandong University) for providing the podocyte cell line.
Funding
The present study was supported by the Natural
Science Foundation of Shandong Province (grant no. ZR2015HM009) and
the Shandong Key Research and Development Program (grant no.
2017GSF218005).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC and SS designed the present study. YC drafted the
manuscript. ZW, QL, LY, YZ and JW evaluated and interpreted the
data critically. SS revised and approved the final version of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
oxLDL
|
oxidized low-density lipoprotein
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
CXCL16
|
CXC motif chemokine ligand 16
|
|
MPC5
|
mouse podocyte clone-5
|
|
ACTN4
|
actinin-α4
|
|
CXCR6
|
CXC motif chemokine receptor 6
|
|
shRNA
|
short hairpin RNA
|
References
|
1
|
McCaffrey J, Lennon R and Webb NJ: The
non-immunosuppressive management of childhood nephrotic syndrome.
Pediatr Nephrol. 31:1383–1402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao X, Hwang DY and Kao HY: The role of
glucocorticoid receptors in podocytes and nephrotic syndrome. Nucl
Receptor Res. 5:52018. View Article : Google Scholar
|
|
3
|
Ranganathan S: Pathology of
podocytopathies causing nephrotic syndrome in children. Front
Pediatr. 4:322016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fornoni A, Merscher S and Kopp JB: Lipid
biology of the podocyte - new perspectives offer new opportunities.
Nat Rev Nephrol. 10:379–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu M, Fan M, Zhen J, Lin J, Wang Q, Lv Z
and Wang R: FAK contributes to proteinuria in hypercholesterolaemic
rats and modulates podocyte F-actin re-organization via activating
p38 in response to ox-LDL. J Cell Mol Med. 21:552–567. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mertens A and Holvoet P: Oxidized LDL and
HDL: Antagonists in atherothrombosis. FASEB J. 15:2073–2084. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang F, Wang C, Wang H, Lu M, Li Y, Feng
H, Lin J, Yuan Z and Wang X: Ox-LDL promotes migration and adhesion
of bone marrow-derived mesenchymal stem cells via regulation of
MCP-1 expression. Mediators Inflamm. 2013:6910232013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rubina K, Talovskaya E, Cherenkov V,
Ivanov D, Stambolsky D, Storozhevykh T, Pinelis V, Shevelev A,
Parfyonova Y, Resink T, et al: LDL induces intracellular signalling
and cell migration via atypical LDL-binding protein T-cadherin. Mol
Cell Biochem. 273:33–41. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang H, Liao M, Zhao W, Zheng X, Xu F,
Wang H and Huang J: CXCL16/ROCK1 signaling pathway exacerbates
acute kidney injury induced by ischemia-reperfusion. Biomed
Pharmacother. 98:347–356. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kato T, Hagiyama M and Ito A: Renal ADAM10
and 17: Their physiological and medical meanings. Front Cell Dev
Biol. 6:1532018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu ZB, Chen Y, Gong YX, Gao M, Zhang Y,
Wang GH, Tang RN, Liu H, Liu BC and Ma KL: Activation of the
CXCL16/CXCR6 pathway by inflammation contributes to atherosclerosis
in patients with end-stage renal disease. Int J Med Sci.
13:858–867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang Y, Henderson FC Jr, Yi Q, Lei Q, Li Y
and Chen N: Chemokine CXCL16 expression suppresses migration and
invasiveness and induces apoptosis in breast cancer cells.
Mediators Inflamm. 2014:4786412014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garcia GE, Truong LD, Li P, Zhang P,
Johnson RJ, Wilson CB and Feng L: Inhibition of CXCL16 attenuates
inflammatory and progressive phases of anti-glomerular basement
membrane antibody-associated glomerulonephritis. Am J Pathol.
170:1485–1496. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schramme A, Abdel-Bakky MS, Gutwein P,
Obermüller N, Baer PC, Hauser IA, Ludwig A, Gauer S, Schäfer L,
Sobkowiak E, et al: Characterization of CXCL16 and ADAM10 in the
normal and transplanted kidney. Kidney Int. 74:328–338. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gutwein P, Abdel-Bakky MS, Schramme A,
Doberstein K, Kämpfer-Kolb N, Amann K, Hauser IA, Obermüller N,
Bartel C, Abdel-Aziz AA, et al: CXCL16 is expressed in podocytes
and acts as a scavenger receptor for oxidized low-density
lipoprotein. Am J Pathol. 174:2061–2072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schramme A, Abdel-Bakky MS, Kämpfer-Kolb
N, Pfeilschifter J and Gutwein P: The role of CXCL16 and its
processing metalloproteinases ADAM10 and ADAM17 in the
proliferation and migration of human mesangial cells. Biochem
Biophys Res Commun. 370:311–316. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Sun S, Zhou A, Yao X and Wang Y:
oxLDL-induced lipid accumulation in glomerular podocytes: Role of
IFN-γ, CXCL16, and ADAM10. Cell Biochem Biophys. 70:529–538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davin JC: The glomerular permeability
factors in idiopathic nephrotic syndrome. Pediatr Nephrol.
31:207–215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalluri R: Proteinuria with and without
renal glomerular podocyte effacement. J Am Soc Nephrol.
17:2383–2389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Assady S, Wanner N, Skorecki KL and Huber
TB: New insights into podocyte biology in glomerular health and
disease. J Am Soc Nephrol. 28:1707–1715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reiser J and Altintas M: Podocytes.
F1000Res. 5:1142016. View Article : Google Scholar
|
|
23
|
Tan X, Chen Y, Liang X, Yu C, Lai Y, Zhang
L, Zhao X, Zhang H, Lin T, Li R, et al: Lipopolysaccharide-induced
podocyte injury is mediated by suppression of autophagy. Mol Med
Rep. 14:811–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Greka A and Mundel P: Cell biology and
pathology of podocytes. Annu Rev Physiol. 74:299–323. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin T, Zhang L, Liu S, Chen Y, Zhang H,
Zhao X, Li R, Zhang Q, Liao R, Huang Z, et al: WWC1 promotes
podocyte survival via stabilizing slit diaphragm protein dendrin.
Mol Med Rep. 16:8685–8690. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Akchurin O and Reidy KJ: Genetic causes of
proteinuria and nephrotic syndrome: Impact on podocyte
pathobiology. Pediatr Nephrol. 30:221–233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu SM, Nissaisorakarn P, Husain I and Jim
B: Proteinuric kidney diseases: A Podocyte's slit diaphragm and
cytoskeleton approach. Front Med (Lausanne). 5:2212018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vallon V and Komers R: Pathophysiology of
the diabetic kidney. Compr Physiol. 1:1175–1232. 2011.PubMed/NCBI
|
|
29
|
Noris M and Remuzzi G: Non-muscle myosins
and the podocyte. Clin Kidney J. 5:94–101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin X, Wang W, Mao J, Shen H, Fu H, Wang
X, Gu W, Liu A, Yu H, Shu Q, et al: Overexpression of Myo1e in
mouse podocytes enhances cellular endocytosis, migration, and
adhesion. J Cell Biochem. 115:410–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kistler AD, Altintas MM and Reiser J:
Podocyte GTPases regulate kidney filter dynamics. Kidney Int.
81:1053–1055. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cechova S, Dong F, Chan F, Kelley MJ, Ruiz
P and Le TH: MYH9 E1841K mutation augments proteinuria and podocyte
injury and migration. J Am Soc Nephrol. 29:155–167. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shao H, Wang JH, Pollak MR and Wells A:
α-actinin-4 is essential for maintaining the spreading, motility
and contractility of fibroblasts. PLoS One. 5:e139212010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duning K, Schurek EM, Schlüter M, Bayer M,
Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem
MA, et al: KIBRA modulates directional migration of podocytes. J Am
Soc Nephrol. 19:1891–1903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fernández D, Horrillo A, Alquezar C,
González-Manchón C, Parrilla R and Ayuso MS: Control of cell
adhesion and migration by podocalyxin. Implication of Rac1 and
Cdc42. Biochem Biophys Res Commun. 432:302–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mundel P and Reiser J: Proteinuria: An
enzymatic disease of the podocyte? Kidney Int. 77:571–580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Z, Zhang L, Shi W, Chen Y, Zhang H, Liu
S, Liang X, Ling T, Yu C, Huang Z, et al: Spironolactone inhibits
podocyte motility via decreasing integrin β1 and increasing
integrin β3 in podocytes under high-glucose conditions. Mol Med
Rep. 12:6849–6854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shao H, Travers T, Camacho CJ and Wells A:
The carboxyl tail of alpha-actinin-4 regulates its susceptibility
to m-calpain and thus functions in cell migration and spreading.
Int J Biochem Cell Biol. 45:1051–1063. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Michaud JL, Chaisson KM, Parks RJ and
Kennedy CR: FSGS-associated alpha-actinin-4 (K256E) impairs
cytoskeletal dynamics in podocytes. Kidney Int. 70:1054–1061. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Feng D, DuMontier C and Pollak MR: The
role of alpha-actinin-4 in human kidney disease. Cell Biosci.
5:442015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang P, Zhao F, Nie X, Liu J and Yu Z:
Knockdown of NUP160 inhibits cell proliferation, induces apoptosis,
autophagy and cell migration, and alters the expression and
localization of podocyte associated molecules in mouse podocytes.
Gene. 664:12–21. 2018. View Article : Google Scholar : PubMed/NCBI
|