Introduction
During space flight, astronauts are exposed to space
radiation at 1 mSv.day−1, a dose rate ~1,000 times
higher than that on the Earth's surface (1). Space radiation consists of constant
low-dose, geomagnetic-trapped, high linear energy transfer (LET)
particles that are characterized by high relative biological
effectiveness (RBE) values (1).
Although the radiation dose during space station stays for 90 days
is <4.5 Gy.30 days−1, the lethal dose, chronic
high-LET radiation has been suggested to induce more severe DNA
damage compared with that of ground radiation (1). Human health should be protected from
such high-LET space radiation, thus, as a result, the RBE values of
space radiation have been extensively studied in the past decades
(2–4).
Nonetheless, the current knowledge of biological
effects of space radiation remains limited, as experiments in space
cannot be performed as frequently as on the ground. In addition,
reproducing the conditions of space flight on the Earth's surface
remains difficult. To evaluate the biological effects of space
radiation, conducting experiments in actual space are necessary.
Previous studies have investigated the biological responses of
prokaryotic and eukaryotic organisms to radiation during space
flight. In particular, D. melanogaster demonstrated a high
frequency of DNA mutations after space flight (5). In rat skin and muscle tissue, space
flight promoted p53 accumulation, a process known to be catalyzed
by radiation-induced DNA damage (6). In addition, Ohnishi et al
(7) reported the induction of
heat shock protein (HSP)72, a gene responsive to stresses
such as radiation and heat, in the muscle, skin and spleen of
goldfish, a non-model vertebrate, under space flight.
Melatonin is a hormone involved in the regulation of
the sleep-wake cycle, demonstrating protective properties against
ionizing radiation (8). The pineal
gland is a major source of melatonin; however, melatonin is also
synthesized in numerous extrapineal tissues, including the skin,
bone marrow and gastrointestinal tract, leading to high
concentrations of melatonin in peripheral tissues (9). Supporting the local production of
melatonin, N-acetylserotonine, a direct precursor of melatonin, is
also expressed in peripheral tissues (10). Although melatonin levels are
regulated by metabolism in the liver or peripheral organs (e.g.
kidney and uterus), the metabolites (e.g. 6-hydroxymelatonin and
5-methoxytryptamine) and melatonin function in both a
receptor-dependent and -independent manner (11,12).
The receptor-independent mechanism was identified to involve
melatonin serving as a mitochondrial-targeted antioxidant that
protected DNA and cells from damage (13). Melatonin has also been discovered
to attenuate the cytotoxicity of ionizing radiation, including DNA
damage and apoptosis, without exerting any cytotoxic effect on
normal organ function (8).
Therefore, melatonin may represent a promising compound to
alleviate the side effects of radiation, (e.g., as a radioprotector
during clinical radiotherapy).
Advances in next-generation sequencing (NGS) and
bioinformatics have permitted the determination of genome-wide
expression profiles, as well as their associated biological
functions and gene networks, even in non-model organisms with no
established databases (14–16).
Using NGS in goldfish, our previous study demonstrated that
melatonin suppressed the osteoclastic activity induced by
microgravity, indicating that melatonin may be a potential drug to
treat space flight-induced bone loss (17). However, it remains unknown whether
melatonin has an impact on biological responses induced by space
radiation.
The present study aimed to examine the effect of
radiation during space flight on gene expression levels in goldfish
scales, a model that was previously used to examine the biological
responses to various stimuli (17). In addition, to determine the
potential of melatonin as a radioprotective drug during space
flight, RNA-sequencing (RNA-seq) analysis with subsequent de
novo transcriptome assembly and computational gene expression
analysis were also performed.
Materials and methods
Fish scale isolation and culture
Equal numbers of male and female immature
Carassius auratus (goldfish) specimens, purchased from
Higashiyama Fish Farm, were grown until the body size reached 10–12
cm. A total of 16 goldfish were kept in an aquarium at 26°C under a
12 h light/dark cycle and fed every morning. Thereafter, the scales
were collected from the goldfish under anesthesia (64 scales were
collected from each goldfish). For anesthesia, the goldfish were
placed into a 300 mg/l (0.03%) ethyl 3-aminobenzoate and methane
sulfonic acid salt solution (MS-222; Nacalai Tesque, Inc.)
neutralized with 0.03% sodium bicarbonate. The fish scales were
isolated and cultured during space flight as previously described
(17). Briefly, the isolated fish
scales were regenerated for 14 days and packaged into a culture
chamber for space flight and ground (G) experiments. The fish
scales were stored at 4°C prior to microgravity culture and
incubated for 86 h at 22°C under in-flight microgravity (F-µG) or
in-flight artificial 1 gravity (F-1G; without microgravity) by
centrifugation at 1 × g and 22°C as previously reported (18). Fish scales under F-µG and G
conditions (F-1G excluded due to limited sample numbers) were also
treated with Leibovitz's L-15 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) containing 0.01% DMSO and 1 µM melatonin
(Sigma-Aldrich; Merck KGaA). F-µG + melatonin and G + melatonin,
respectively) and were maintained at 4°C just before space flight
(5). All procedures in the present
study were approved by the Institutional Animal Care and Use
Committee of Kanazawa University (Noto-cho, Japan) and performed in
accordance with the guidelines for animal experiments provided by
the Ethics Committee of Kanazawa University.
RNA isolation and sequencing
Total RNA was isolated from fish scales using the
RNeasy Mini kit (Qiagen, Inc.), according to the manufacturer's
protocol. RNA quality was analyzed using a Bioanalyzer 2100
instrument and the RNA6000 Nano LabChip® kit (both
Agilent Technologies, Inc.). RNA samples with RNA integrity number
values of >9.0 were used for library construction. RNA
concentration was determined by Nanodrop spectrometer (Thermo
Fisher Scientific, Inc.) and Bioanalyzer followed by library
construction. RNA-seq libraries for paired-end reads 100 bp in
length from biologically triplicated samples were constructed using
TruSeq RNA Sample Prep kit v2 (cat. no. RS-122; Illumina, Inc.).
10–20 pM pooled libraries were sequenced using HiSeq™ 2000
(Illumina, Inc.). The raw sequence reads were deposited at the DNA
Data Bank of Japan (DDBJ; http://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA008502)
under the DDBJ Sequence Read Archive (DRA) accession no.
DRA009118.
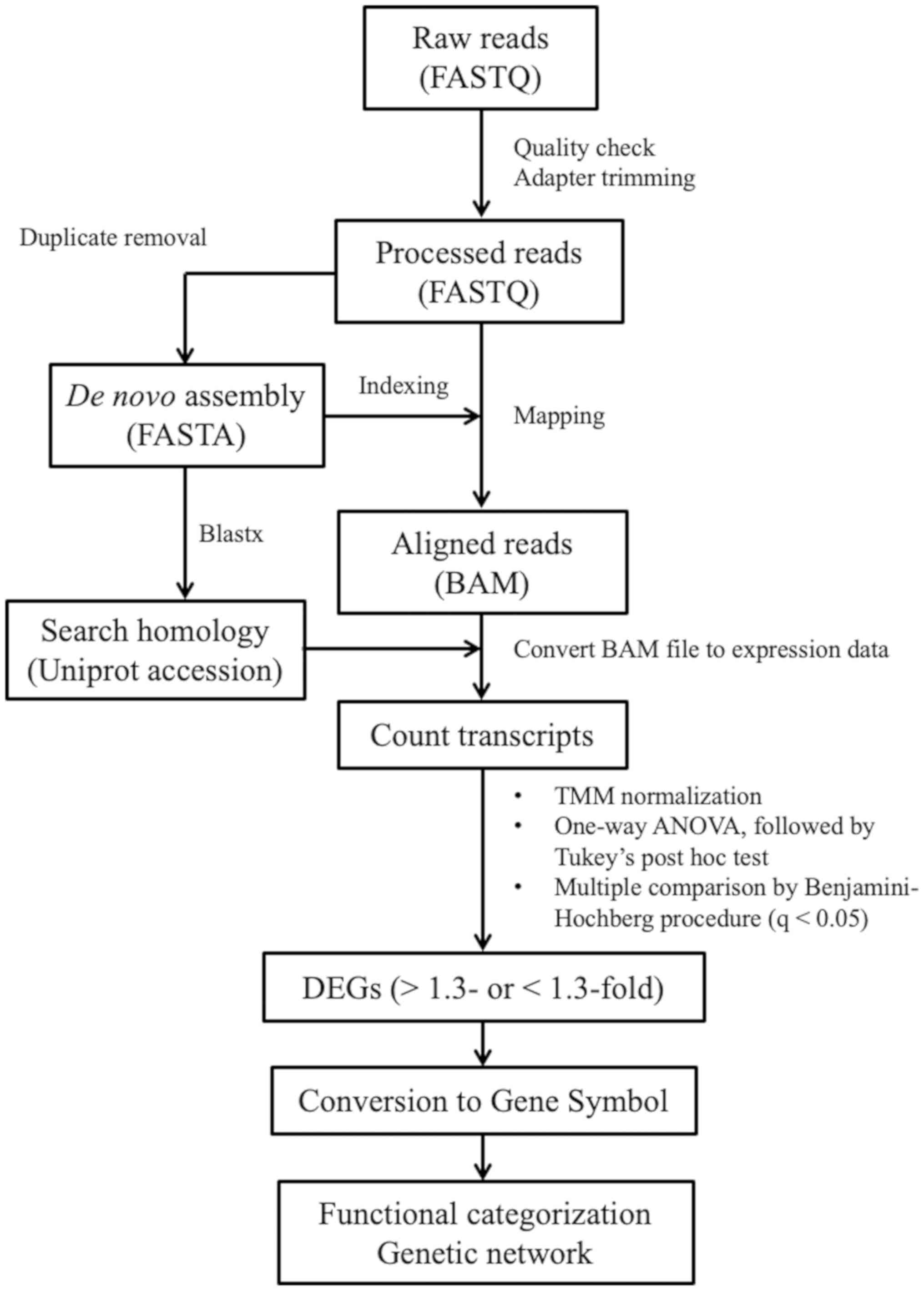
Quality control check and trimming of
reads
Quality check of raw read sequences was performed
using FastQC (v0.11.9;
bioinformatics.babraham.ac.uk/projects/fastqc). Adaptors and short,
low-quality reads (Q<20 and <36 bp) were trimmed using
Trimmomatic v0.39 (usadellab.org/cms/?page=trimmomatic) with default
parameters (19). The number of
raw and processed reads are presented in Table I.
 | Table I.Summary of the number of raw reads,
processed reads and mapping rate of transcriptome data. |
Table I.
Summary of the number of raw reads,
processed reads and mapping rate of transcriptome data.
| Treatment | Sample ID | Raw reads,
count | Processed reads,
count | Mapping rate,
% |
|---|
| F-1G | S001 | 21,382,556 | 20,946,002 | 92.46 |
| F-1G | S002 | 21,506,294 | 21,104,591 | 92.27 |
| F-1G | S003 | 28,155,809 | 27,650,984 | 92.39 |
| F-µG | S004 | 21,156,047 | 20,789,017 | 92.34 |
| F-µG | S005 | 21,655,997 | 21,255,721 | 92.10 |
| F-µG | S006 | 21,139,783 | 20,768,132 | 92.23 |
| G | S007 | 25,132,537 | 24,638,896 | 91.50 |
| G | S008 | 24,131,108 | 23,677,305 | 91.47 |
| G | S009 | 20,658,328 | 20,215,123 | 92.18 |
| F-µG +
melatonin | S010 | 23,724,796 | 23,497,407 | 91.89 |
| F-µG +
melatonin | S011 | 22,484,040 | 22,260,350 | 91.68 |
| F-µG +
melatonin | S012 | 22,737,402 | 22,469,979 | 91.66 |
| G + melatonin | S013 | 25,120,774 | 24,599,748 | 91.39 |
| G + melatonin | S014 | 24,027,734 | 23,539,894 | 91.93 |
| G + melatonin | S015 | 24,379,991 | 23,958,109 | 92.25 |
De novo transcriptome assembly and
read mapping
Following PCR duplicate removal using filterPCRdupl
v1.01 (20), the de novo
transcriptome assembly of processed reads was performed using
Trinity (version r2012-10-05) with default parameters to generate a
reference sequence of transcripts (21). Trinity was equipped with Management
and Analysis System for Enormous Reads (Maser; cell-innovation.nig.ac.jp/index_en.html), a graphical
user interface-based tool for NGS analysis, which was used to run
Trinity for de novo assembly. PCR duplicates were filtered
to remove the sequence noise and decrease the number of transcript
candidates. Sequence reads including PCR duplicates were mapped to
the reference using Bowtie2 v2.2.5 with strict parameters (−D 20-R
3-N 0-L 20-i S,1,0.50) (22) to
increase the rate of unique mapping. The mapping rates are
presented in Table I.
Transcriptome annotation using
Blastx
Blastx 2.2.26+ (blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE=BlastSearch&PROGRAM=blastx)
equipped with Maser was used to annotate the reference sequences
using default parameters and an e-value cutoff of
<10−10. The resulting top hit genes were listed, and
a total of 84,748 from 175,357 transcript candidates were
annotated. All UniProt accession numbers, subject IDs and sequence
identities are provided in Table
SI.
Transcript quantification and
identification of differentially expressed genes (DEGs)
Estimation of relative RNA expression levels from
mapped reads and principal component analysis (PCA) was performed
using Strand NGS v3.3 (strand-ngs.com) with the Trimmed Mean of M value (TMM)
normalization method (23). TMM
normalization is more reliable than using fragments per kilobase of
exon per million reads mapped (12). DEGs were identified using one-way
ANOVA followed by post-hoc Tukey's test and multiple testing
Benjamini-Hochberg correction to calculate false discovery rate and
adjust P-value. Genes were considered DEGS where q-value (adjusted
P-value) <0.05 and fold change >1.3 or <-1.3 for up- and
downregulated genes, respectively. Venn diagrams were drawn using
Strang NGS.
Analysis of molecular functions and
gene networks
To determine the molecular functions of the DEGs and
gene networks involved, expression data were analyzed using
Ingenuity Pathway Analysis (IPA) tools (Qiagen, Inc.). In order to
import DEGs, the transcript candidates were converted to Gene IDs
using The Database for Annotation, Visualization and Integrated
Discovery 6.8 (24). IPA is a
web-delivered application that enables the identification,
visualization and exploration of molecular interaction networks in
gene expression data. The top five molecular functions were
identified, and their associated gene networks were visualized to
obtain information on the interactions of upregulated and
downregulated genes during melatonin treatment. The network is
displayed graphically as nodes (genes) and edges (biological
associations). Heatmaps of the expression values of genes on the
identified gene networks were constructed via Microsoft Excel 2019
software (Microsoft Corporation).
Results
Identification of differentially
expressed transcripts responsive to space radiation and melatonin
treatment
The effect of melatonin on gene expression
alterations in response to microgravity was determined in a
previous study by comparing fish scale DEGs on ground and in space
flight in the presence or absence of microgravity (17). In the present study, to identify
the candidate transcripts responsive to space irradiation, de
novo transcriptome analysis of fish scales was performed,
focusing on upregulated and downregulated genes during space flight
between fish scales with 1G or microgravity (F-1G and F-µG;
Fig. 1). After quantification and
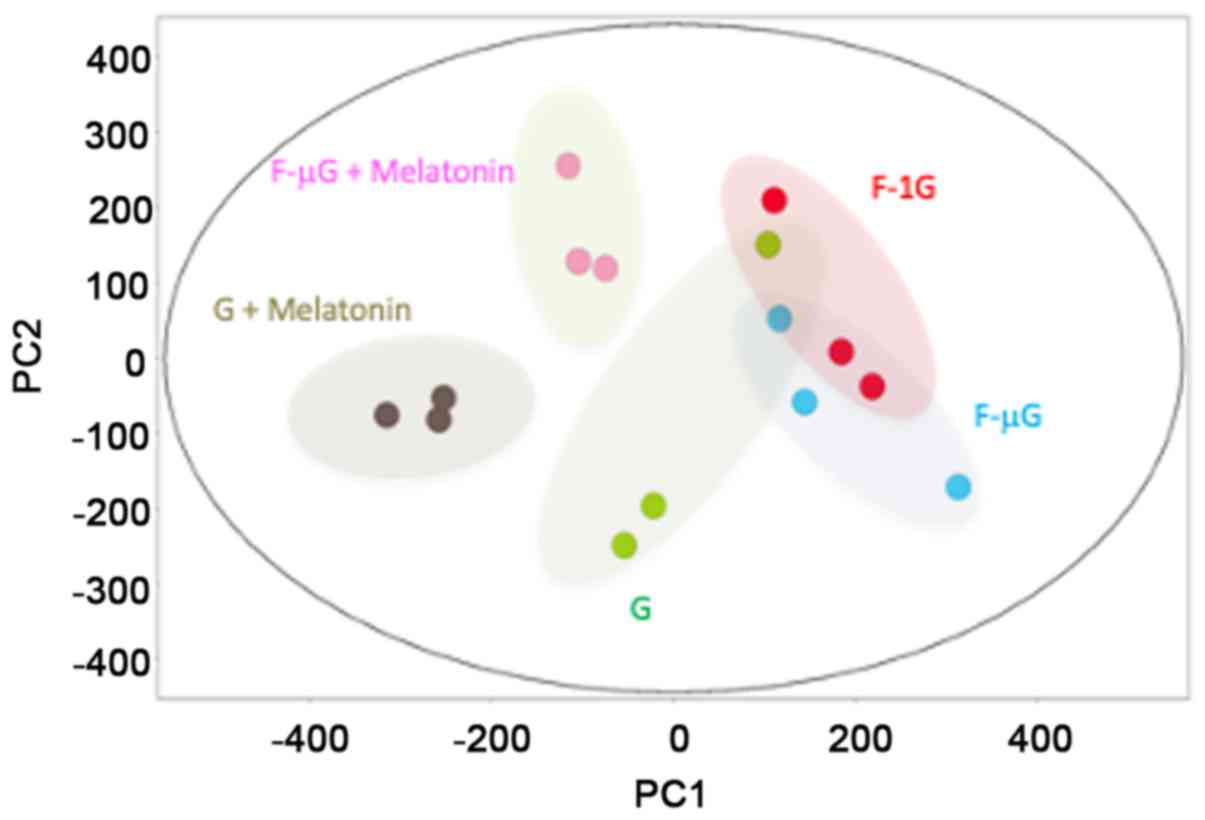
TMM normalization of mapped reads, principal component analysis
(PCA) was performed on the expression data (Fig. 2). The expression pattern of
transcripts in the G group (low-radiation control) was distinct
compared with melatonin treatment on ground (G + melatonin) or in
space flight (F-µG + melatonin). Moreover, the untreated space
flight groups F-1G and F-µG displayed similar expression patterns
to the G group, which were distinct from the melatonin-treated
groups.
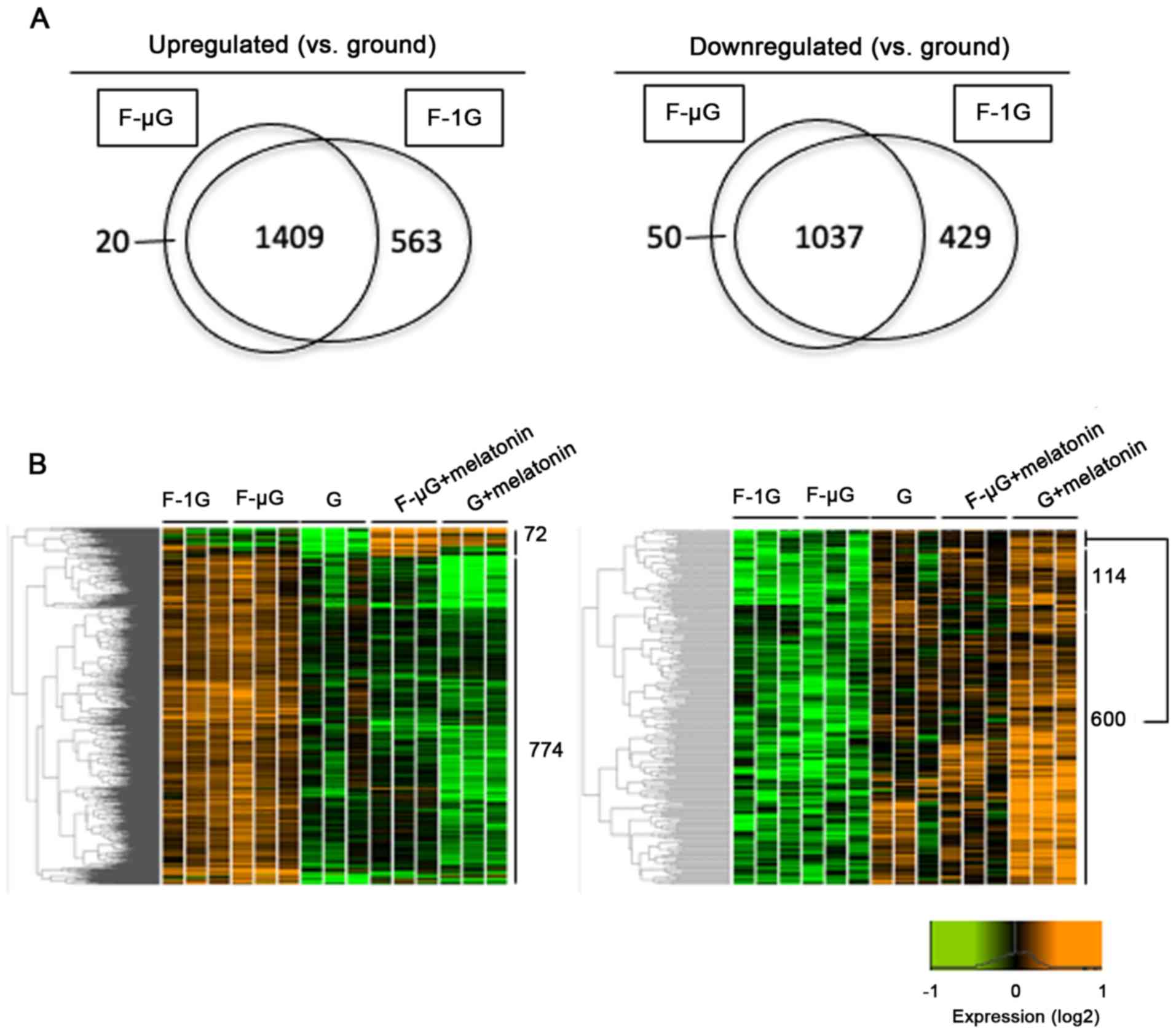
Among differentially expressed 2,446 transcripts
between the ground and space flight groups, 1,409 were upregulated
and 1,037 were downregulated during space flight, regardless of the
presence of microgravity (Fig.
3A). The impact of melatonin on DEGs was also assessed by
comparing the melatonin-treated samples. Among the 1,409
upregulated and 1,037 downregulated transcripts, the expression of
846 and 714 transcripts were altered by melatonin treatment
(Fig. 3B, left and right,
respectively). Among them, melatonin reversed the expression
patterns of 774 and 600 transcripts, respectively, that were
altered by space radiation (Fig.
3B). Furthermore, among 774 and 600 transcripts responsive to
melatonin, 576 and 219 transcripts had UniProt accession numbers
assigned by Blastx (Table SI).
Among the transcripts with UniProt accession numbers, 566
upregulated and 208 downregulated transcripts responsive to space
radiation had high sequence homology with genes identified in
Homo sapiens, Mus musculus, Danio rerio and other species
(Tables SII and SIII). The annotated transcripts were
designated as DEGs for subsequent computational analysis.
Functional analysis of genes
responsive to space radiation and melatonin
The molecular and cellular functions and
corresponding genetic networks of the DEGs were subsequently
identified in order to assess the impact of melatonin on gene
expression levels altered by space radiation. Analysis of canonical
pathways based on the IPA indicated that melatonin reduced the
number of DEGs associated with ‘Unfolded Protein Response’
consisting of HSP families that are upregulated under space
conditions (7), ‘NRF-2-mediated
Oxidative Stress Response’ consisting of superoxide dismutase
2, as well as ‘NER Pathway’ and ‘DNA Double-Strand Break Repair
by Non-Homologous End Joining’, consisting of DNA ligase (LIG)4,
excision repair cross-complementing (ERCC2) and protein kinase,
DNA-activated, catalytic subunit (PRKDC (Table II).
 | Table II.List of identified IPA canonical
pathways and associated genes that were differentially expressed in
response to space radiation and melatonin treatment. |
Table II.
List of identified IPA canonical
pathways and associated genes that were differentially expressed in
response to space radiation and melatonin treatment.
| Ingenuity Canonical
Pathways | −log (P-value) | Gene |
|---|
| Protein
Ubiquitination Pathway | 8.22 | ANAPC4, DNAJA1,
DNAJB1, DNAJB9, DNAJC3, HSP90AA1, HSP90B1, HSPA4, HSPD1, HSPE1,
PSMA3, PSMC1, PSMC2, PSMC4, PSMC5, PSMD10, PSMD6, PSMD8, UBE2D4,
UCHL1, USP22 |
| NRF2-Mediated
Oxidative Stress Response | 2.91 | CBR1, CLPP, DNAJA1,
DNAJA4, DNAJB1, DNAJB9, DNAJC3, GSR, PPIB, SOD2 |
| NER Pathway | 2.76 | ERCC2, GTF2H5,
LIG3, POLE3, POLK, RFC5, TCEA1 |
| Role of BRCA1 in
DNA Damage Response | 2.59 | BRIP1, CHEK2, MDC1,
RFC5, SMARCA4, SMARCD1 |
| DNA Double-Strand
Break Repair by Non-Homologous End Joining | 1.63 | LIG3, PRKDC |
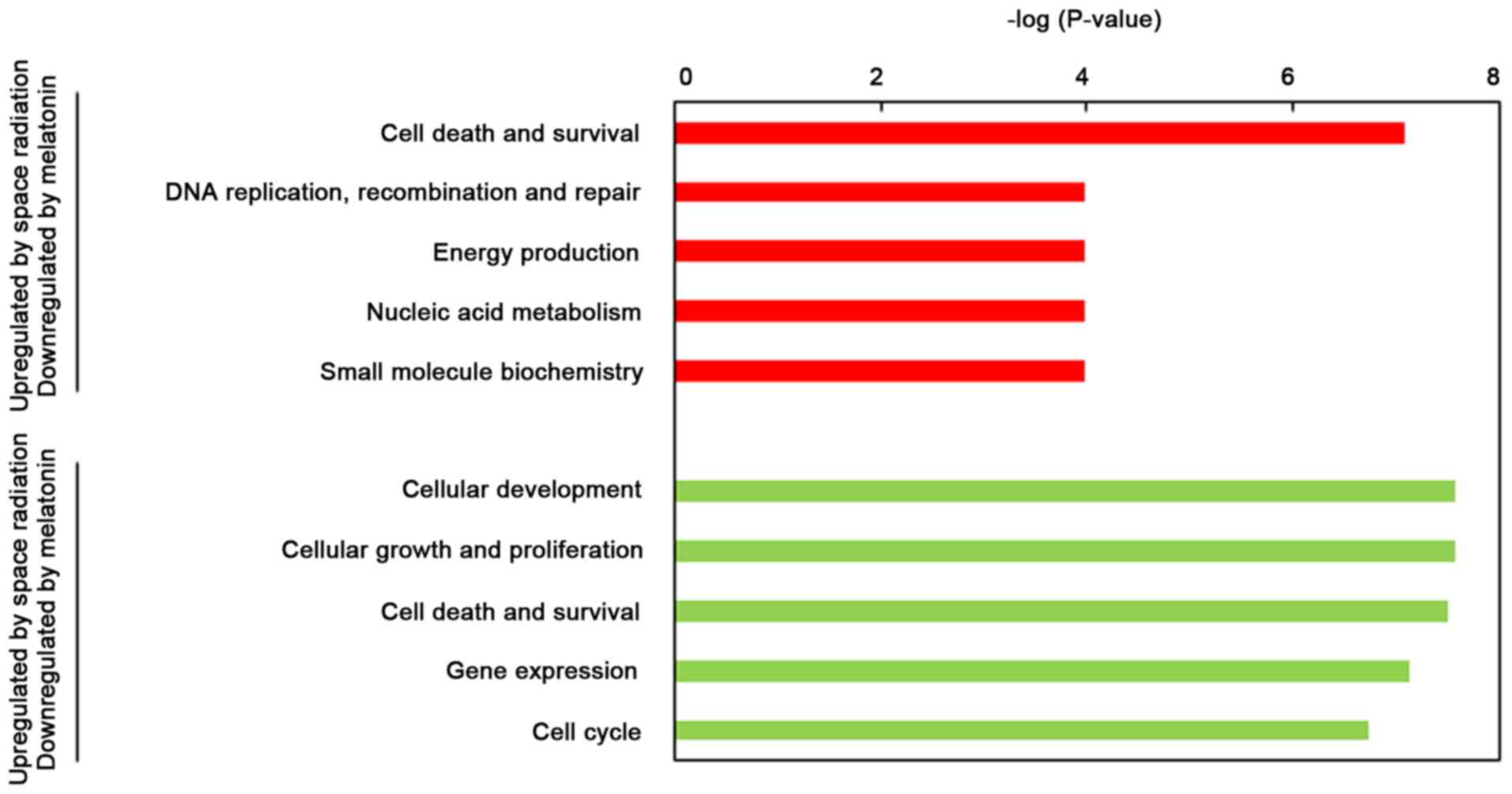
Functional analysis also suggested that the genes
downregulated by melatonin were involved in ‘Cell Death and
Survival’ and ‘DNA Replication, Recombination, and Repair’
(Fig. 4). Genes upregulated by
melatonin were also associated with ‘Cellular Development’, ‘Cell
Death and Survival’ and ‘Cellular Growth and Proliferation’
comprising Bcl family members and cyclin-dependent kinase 1B
(CDKN1B).
Gene networks responsive to space
radiation and melatonin
Gene network analysis by IPA was performed to
elucidate the interactions between DEGs responsive to space
radiation (Fig. 5).
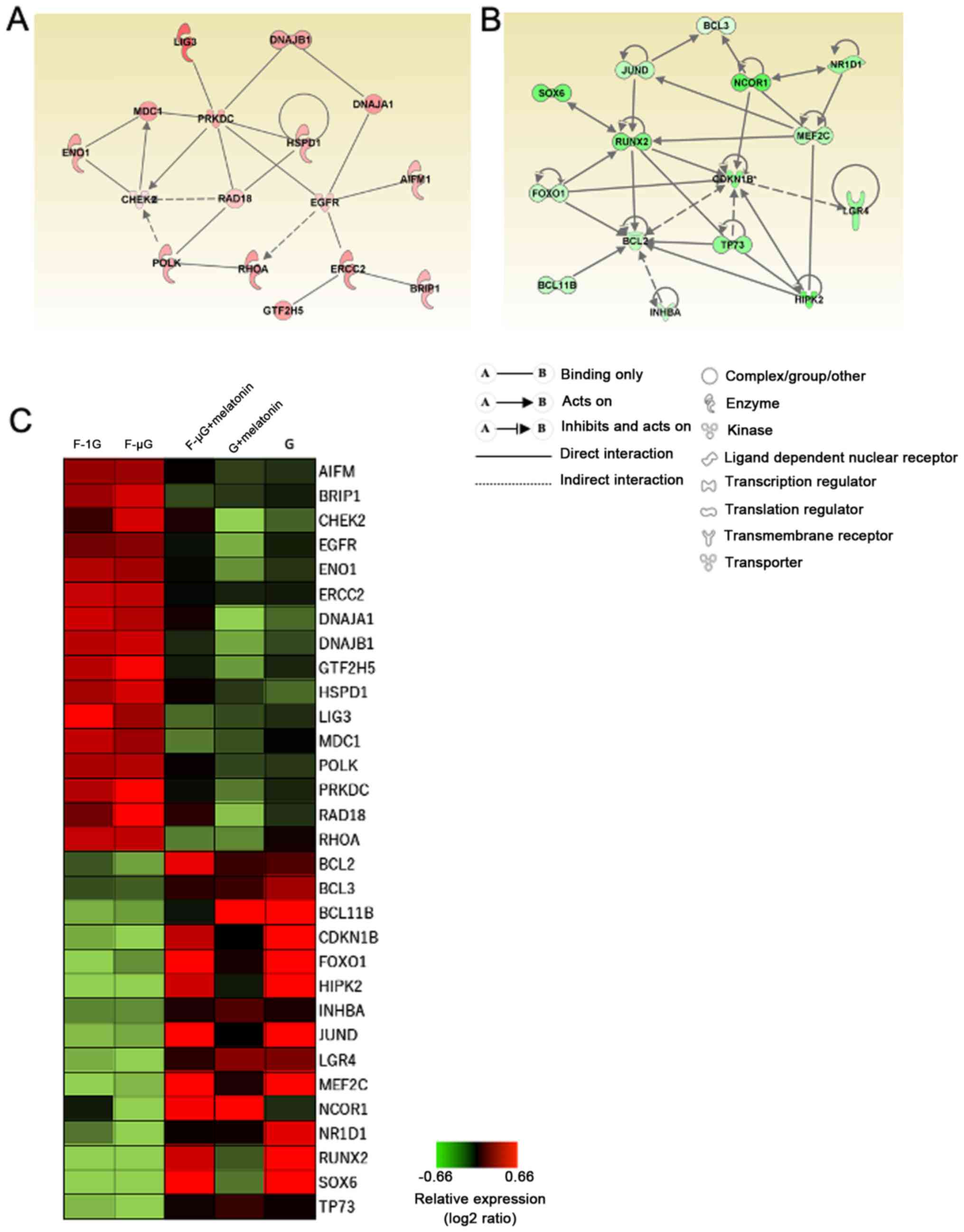 | Figure 5.Gene networks identified using
differentially expressed gene analysis. (A) Upregulated and (B)
downregulated genes following melatonin treatment were analyzed
using Ingenuity Pathway Analysis tools. Red nodes represent
upregulated genes, while the green nodes are for downregulated
genes. Solid lines and dashed lines indicate direct and indirect
interactions between molecules, respectively. (C) Heatmap of genes
in the identified networks based on relative expression values. The
green to red color spectrum represents the log2 ratios scaled
between-0.66 and 0.66 (low to high expression). G, ground samples;
F-µG, space flight samples with microgravity; F-1G space flight
samples with 1G. The expression value and gene names are also shown
in Table SII. EGFR, epidermal
growth factor receptor; ENO1, enolase 1; RHOA, Ras homologue family
member A; HIPK2, homeodomain-interacting protein kinase 2; INHBA,
inhibin subunit β A; LGR4, leucine-rich repeat-containing G
protein-coupled receptor 4; Sox6, SRY-box transcription factor
6. |
An upregulated gene network containing 16 genes was
identified, which included HSP family genes such as DNA J heat
shock protein member (DNAJ)A1, DNAJB1 and HSPD1. The network also
contained PRKDC, mediator of DNA damage checkpoint 1 (MDC1), LIG3,
Rad18 E3 ubiquitin ligase (RAD18), DNA polymerase κ (POLK) and
ERCC2, which are genes involved in DNA double-strand break (DSB)
repair and nucleotide excision repair (25,26)
(Fig. 5A and C). In addition,
apoptosis-inducing factor 1, mitochondria-associated (AIFM1) was
discovered to be upregulated by space radiation. AIFM1 encodes
apoptosis-inducing factor 1, which is released from the
mitochondria and involved in caspase-independent DNA fragmentation
during apoptosis (27). Checkpoint
kinase 2 (CHEK2) was also upregulated; this gene encodes CHK2, a
kinase involved in p53-dependent apoptosis induced by ionizing
radiation (28). Marker of
proliferation Ki67 (MKI67) and proliferating cell nuclear antigen
(PCNA) were also identified in the upregulated network.
The identified downregulated gene network contained
15 genes, including anti-apoptotic Bcl family genes such as BCL2,
BCL3 and BCL11B, that were downregulated by space radiation
(Fig. 5B and C). CDKN1B, which is
involved in cell-cycle progression, was also downregulated by space
radiation (29). Several
transcription factors, such as Forkhead box O1 (FOXO1),
runt-related transcription factor 2 (RUNX2), myocyte enhancer
factor 2C (MEF2C) and nuclear receptor corepressor 1 (NCOR1) were
also identified in the downregulated gene network. In both
networks, the treatment with melatonin recovered the expression
levels of DEGs altered by space radiation but did not markedly
affect gene expression in the control groups (Fig. 5C), indicating that melatonin may
reverse the changes in gene expression levels induced by space
radiation without affecting constitutive gene expression.
Discussion
The influence of low-dose rate radiation on human
health during space flight has been investigated in actual space
conditions. For instance, previous studies demonstrated that
HSP-encoding genes were upregulated by ionizing radiation, not only
on the ground but also during space flight in mammalian and
goldfish spleens (7,30). Similarly, the upregulation of
several HSP-encoding genes, such as DNAJA1, DNAJB1 and HSPD1, in
fish scales exposed to space radiation was also observed in the
present study, both in the absence and presence of microgravity.
Gene network analysis also suggested that the upregulated
HSP-coding genes may interact with other genes responsive to space
radiation. Among them, PRKDC is known to serve a pivotal role in
non-homologous end joining (NHEJ) of double-stranded DNA (31). DSBs, induced by direct ionization
of DNA or hydroxyl radical attacks, are the most harmful form of
DNA damage for cells, since even a single unrepaired DSB site can
cause cell death (32). In the
absence of sister chromatids in G0/G1-phase
cells, NHEJ is the major pathway for DSB repair (32). Although the upregulation of
PRKDC has not yet been demonstrated during space flight, it
may be a potential protective response to chronic low-dose high LET
space radiation. In the presence of sister chromatids in S and
G2/M-phase cells, the homologous recombination (HR)
pathway that is highly conserved among species, can lead to
accurate DSB repair (31). MDC1, a
component of the MDC1/RAD50 DSB repair protein/nibrin complex
(33), has been identified to bind
to damaged sites and recruit DNA damage sensor proteins, such as
ataxia telangiectasia, and DNA repair proteins, including tumor
suppressor 53 binding protein 1 to promote HR (34). In addition, MDC1 was discovered to
activate CHK2 in the mammalian DNA damage response pathway
(35). Protein-protein
interactions between MDC1 and CHK2 have not yet been reported in
goldfish. However, the simultaneous upregulation of these genes in
the present study suggested the possibility of coupled activation
in their corresponding proteins. In addition, RAD18 and POLK may
also potentially interact with CHK2. RAD18 is involved in
translesion synthesis, which prevents replication fork stalling at
damaged sites by interacting with POLK (36). CHK2 and RAD18 have been previously
identified to cooperatively maintain genome stability during
frequent lymphomagenesis in double knockout mice (37).
In the present study, melatonin treatment
counteracted the radiation-induced upregulation of DNA repair
genes. A previous study demonstrated that melatonin has the
potential to scavenge free radicals, such as reactive oxygen and
nitrogen species (38). Although
the detailed mechanism on how melatonin affects upregulated DNA
repair genes in response to space radiation has yet to be
determined, one possible explanation is that melatonin treatment
may serve as a radical scavenger that ameliorates DNA damage
(8), thus resulting in moderate
DNA repair.
Previous studies have reported that the Bcl-2
anti-apoptotic protein was downregulated by ionizing radiation,
thereby promoting apoptosis (39).
Melatonin has also been reported to protect cells from
radiation-induced apoptosis by recovering Bcl-2 expression
(40). Similarly, in the present
study, the expression of BCL-2 in goldfish scales was downregulated
by space radiation, which was subsequently recovered following
melatonin treatment. Furthermore, other anti-apoptotic Bcl family
genes, such as BCL3 and BCL11B (41,42),
were also downregulated by space radiation. The attenuation of BCL2
expression may be explained by the observed downregulation of
upstream transcription factors that bind the BCL2 promoter and
elicit BCL2 expression, such as FOXO1 and RUNX2 (43,44).
The expression of BCL family genes was also restored by melatonin
treatment, suggesting that melatonin may antagonize the
anti-apoptotic response elicited by space radiation.
The identified network contained BCL2-interacting
genes such as CDKN1B, RUNX2, FOXO1 and NCOR1. CDKN1B codes for
p27Kip1, a CDK inhibitor that promotes G1
cell cycle arrest by inhibiting CDK4 (45). In general, CDKN1B has been
found to be upregulated following DNA damage induced by acute
high-dose ionizing radiation (46). However, in the present study,
CDKN1B expression was downregulated by chronic low-dose space
radiation. Interestingly, in the present study, MKI67 and PCNA,
which are well-studied proliferation markers (47), were upregulated in the presence of
space radiation. Upregulation of proliferative genes may be
explained by the hormesis effect evoked by low-dose radiation
(48). The molecular mechanism
underlying hormesis effect remains unknown. However, the regulation
of CDKN1B expression by transcription factors RUNX2, FOXO1 and
NCOR1 may be one possible explanation (49–51).
MEF2C may also be a possible regulator, since MEF2C regulates RUNX2
expression by directly binding to its enhancer region (52). JunD proto-oncogene, AP-1
transcription factor subunit is also a candidate regulator because
its stable expression enhances the expression of RUNX2 (53).
Previous studies have demonstrated the
radioprotective effect of melatonin in a receptor-independent
manner (54), which was at least
partly mediated by radical scavenging and the activation of DNA
repair machinery (55,56). The present study demonstrated that
melatonin suppressed the expression levels of DNA repair genes that
were upregulated by radiation-induced DNA damage, supporting the
radioprotective effect of melatonin on DNA at the gene expression
level, and provided valuable information regarding the possible
application of melatonin as a radioprotector during space flight.
Melatonin has been suggested to exert a radioprotective effect
either directly or indirectly through its metabolite, as melatonin
can be rapidly metabolized in peripheral tissue (57,58).
In conclusion, the findings of the present study
suggested that melatonin treatment may counteract the expression of
anti-apoptotic and proliferative genes attenuated by space
radiation. In addition, melatonin also counteracts the expression
levels of genes associated with DNA damage that were upregulated by
space radiation. Thus, melatonin may promote cell survival and
normal cell proliferation during space flight. Whether and how
melatonin affects the gene response of mammalian cells to space
radiation remains to be determined. Furthermore, comparisons
between transcriptome and proteome data should be performed to
confirm whether the genes responsive to space radiation are
functional in goldfish scales.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors wish to acknowledge the Japan Manned
Space System and Chiyoda Corporation for their technical support,
development of equipment and instruction on how to use the
equipment. The authors would also like to thank the Japanese
Astronaut Mr Soichi Noguchi for performing experiments at the
International Space Station.
Funding
The present study was supported partly by grants to
NS (Japan Aerospace Exploration Agency and Kanazawa University
Chozen Project), to AH (The Cooperative Research Program of The
Institute of Nature and Environmental Technology, Kanazawa
University; grant no. 17018) and to JH (The Cooperative Research
Program of The Institute of Nature and Environmental Technology,
Kanazawa University; grant no. 19024).
Availability of data and materials
The datasets are available in the DNA DataBank Japan
repository, https://ddbj.nig.ac.jp.
Authors' contributions
YF and YT analyzed and interpreted the data. YF
wrote the manuscript. AH and NS designed study. TY, JH and TS
supported data analysis and performed validation. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures in the present study were approved by
the Institutional Animal Care and Use Committee of Kanazawa
University (Noto-cho, Japan) and performed in accordance with the
guidelines for animal experiments provided by the Ethics Committee
of Kanazawa University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ohnishi K and Ohnishi T: The biological
effects of space radiation during long stays in space. Biol Sci
Space. 18:2627–205. 2004. View Article : Google Scholar
|
|
2
|
Ohnishi T and Nagaoka S: Emphasis of
biological research for space radiation. Biol Sci Space. 12:5–13.
1998.(In Japanese). View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi A and Ohnishi T: Space radiation
biology. Biol Sci Space. 15:40–46. 2001.(In Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hellweg CE, Dilruba S, Adrian A, Feles S,
Schmitz C, Berger T, Przybyla B, Briganti L, Franz M, Segerer J, et
al: Space experiment ‘Cellular Responses to Radiation in Space
(CellRad)’: Hardware and biological system tests. Life Sci Space
Res (Amst). 7:73–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ikenaga M, Yoshikawa I, Kojo M, Ayaki T,
Ryo H, Ishizaki K, Kato T, Yamamoto H and Hara R: Mutations induced
in Drosophila during space flight. Biol Sci Space. 11:346–350.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohnishi T, Inoue N, Matsumoto H, Omatsu T,
Ohira Y and Nagaoka S: Cellular content of p53 protein in rat skin
after exposure to the space environment. J Appl Physiol (1985).
81:183–185. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohnishi T, Tsuji K, Ohmura T, Matsumoto H,
Wang X, Takahashi A, Nagaoka S, Takabayashi A and Takahahsi A:
Accumulation of stress protein 72 (hsp72) in muscle and spleen of
goldfish taken into space. Adv Space Res. 21:1077–1080. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farhood B, Goradel NH, Mortezaee K,
Khanlarkhani N, Salehi E, Nashtaei MS, Mirtavoos-Mahyari H,
Motevaseli E, Shabeeb D, Musa AE and Najafi M: Melatonin as an
adjuvant in radiotherapy for radioprotection and
radiosensitization. Clin Transl Oncol. 21:268–279. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Slominski AT, Hardeland R, Zmijewski MA,
Slominski RM, Reiter RJ and Paus R: Melatonin: A cutaneous
perspective on its production, metabolism, and functions. J Invest
Dermatol. 138:490–499. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Slominski AT, Kim TK, Kleszczyński K,
Semak I, Janjetovic Z, Sweatman T, Skobowiat C, Steketee JD, Lin Z,
Postlethwaite A, et al: Characterization of serotonin and
N-acetylserotonin systems in the human epidermis and skin cells. J
Pineal Res. 68:e126262020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slominski RM, Reiter RJ,
Schlabritz-Loutsevitch N, Ostrom RS and Slominski AT: Melatonin
membrane receptors in peripheral tissues: Distribution and
functions. Mol Cell Endocrinol. 351:152–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slominski AT, Zmijewski MA, Semak I, Kim
TK, Janjetovic Z, Slominski RM and Zmijewski JW: Melatonin,
mitochondria, and the skin. Cell Mol Life Sci. 74:3913–3925. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan DX, Manchester LC, Qin L and Reiter
RJ: Melatonin: A mitochondrial targeting molecule involving
mitochondrial protection and dynamics. Int J Mol Sci. 17:21242016.
View Article : Google Scholar
|
|
14
|
Zerbino DR and Birney E: Velvet:
Algorithms for de novo short read assembly using de Bruijn graphs.
Genome Res. 18:821–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robertson G, Schein J, Chiu R, Corbett R,
Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al: De
novo assembly and analysis of RNA-seq data. Nat Methods. 7:909–912.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie Y, Wu G, Tang J, Luo R, Patterson J,
Liu S, Huang W, He G, Gu S, Li S, et al: SOAPdenovo-Trans: De novo
transcriptome assembly with short RNA-Seq reads. Bioinformatics.
30:1660–1666. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ikegame M, Hattori A, Tabata MJ, Kitamura
KI, Tabuchi Y, Furusawa Y, Maruyama Y, Yamamoto T, Sekiguchi T,
Matsuoka R, et al: Melatonin is a potential drug for the prevention
of bone loss during space flight. J Pineal Res. 67:e125942019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yano S, Masuda D, Kasahara H, Omori K,
Higashibata A, Asashima M, Ohnishi T, Yatagai F, Kamisaka S,
Furusawa T, et al: Excellent thermal control ability of cell
biology experiment facility (CBEF) for Ground-based experiments and
experiments onboard the Kibo Japanese Experiment Module of
International Space Station. Biol Sci Space. 26:12–20. 2012.
View Article : Google Scholar
|
|
19
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smeds L and Kunstner A: ConDeTri-a content
dependent read trimmer for Illumina data. PLoS One. 6:e263142011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haas BJ, Papanicolaou A, Yassour M,
Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber
M, et al: De novo transcript sequence reconstruction from RNA-seq
using the Trinity platform for reference generation and analysis.
Nat Protoc. 8:1494–1512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson MD and Oshlack A: A scaling
normalization method for differential expression analysis of
RNA-seq data. Genome Biol. 11:R252010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Winkler GS, Araujo SJ, Fieldler U,
Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT
and Weeda G: TFIIH With Inactive XPD helicase functions in
transcription initiation but is defective in DNA repair. J Biol
Chem. 275:4258–4266. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hirao A, Kong YY, Matsuoka S, Wakeham A,
Ruland J, Yoshida H, Liu D, Elledge SJ and Mak TW: DNA
damage-induced activation of p53 by the checkpoint kinase Chk2.
Science. 287:1824–1827. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shiloh Y and Ziv Y: The ATM protein
kinase: Regulating the cellular response to genotoxic stress, and
more. Nat Rev Mol Cell Biol. 14:197–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hartwell LH and Kastan MB: Cell cycle
control and cancer. Science. 266:1821–1828. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amundson SA, Grace MB, McLeland CB,
Epperly MW, Yeager A, Zhan Q, Greenberger JS and Fornace AJ Jr:
Human in vivo radiation-induced biomarkers: Gene expression changes
in radiotherapy patients. Cancer Res. 64:6368–6371. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bouwman P and Jonkers J: The effects of
deregulated DNA damage signalling on cancer chemotherapy response
and resistance. Nat Rev Cancer. 12:587–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takahashi A and Ohnishi T: Does gammaH2AX
foci formation depend on the presence of DNA double strand breaks?
Cancer Lett. 229:171–179. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van den Bosch M, Bree RT and Lowndes NF:
The MRN complex: Coordinating and mediating the response to broken
chromosomes. EMBO Rep. 4:844–849. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mochan TA, Venere M, DiTullio RA Jr and
Halazonetis TD: 53BP1 and NFBD1/MDC1-Nbs1 function in parallel
interacting pathways activating ataxia-telangiectasia mutated (ATM)
in response to DNA damage. Cancer Res. 63:8586–8591.
2003.PubMed/NCBI
|
|
35
|
Lou Z, Minter-Dykhouse K, Wu X and Chen J:
MDC1 is coupled to activated CHK2 in mammalian DNA damage response
pathways. Nature. 421:957–961. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Y, Gao Y, Zlatanou A, Tateishi S,
Yurchenko V, Rogozin IB and Vaziri C: Diverse roles of RAD18 and
Y-family DNA polymerases in tumorigenesis. Cell Cycle. 17:833–843.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanoue Y, Toyoda T, Sun J, Mustofa MK,
Tateishi C, Endo S, Motoyama N, Araki K, Wu D, Okuno Y, et al:
Differential roles of Rad18 and Chk2 in genome maintenance and skin
carcinogenesis following UV Exposure. J Invest Dermatol.
138:2550–2557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reiter RJ, Tan DX, Manchester LC and Qi W:
Biochemical reactivity of melatonin with reactive oxygen and
nitrogen species: A review of the evidence. Cell Biochem Biophys.
34:237–256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui YF, Ding YQ, Zhang Y, Xu H, Jin W, Liu
XL, Dong B, Mao JP and Mao BZ: Apoptotic characteristics of spleen
lymphocyte in mice irradiated by lethal dose and its relationship
to the expression of Bax and Bcl-XL proteins. Zhongguo Wei Zhong
Bing Ji Jiu Yi Xue. 17:109–112. 2005.(In Chinese). PubMed/NCBI
|
|
40
|
Mohseni M, Mihandoost E, Shirazi A,
Sepehrizadeh Z, Bazzaz JT and Ghazi-khansari M: Melatonin may play
a role in modulation of bax and bcl-2 expression levels to protect
rat peripheral blood lymphocytes from gamma irradiation-induced
apoptosis. Mutat Res. 738-739:19–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Garcia I, Cosio G, Lizarraga F,
Martínez-Ruiz G, Meléndez-Zajgla J, Ceballos G, Espinosa M, Pacheco
R and Maldonado V: Bcl-3 regulates UVB-induced apoptosis. Hum Cell.
26:47–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Karanam NK, Grabarczyk P, Hammer E, Scharf
C, Venz S, Gesell-Salazar M, Barthlen W, Przybylski GK, Schmidt CA
and Völker U: Proteome analysis reveals new mechanisms of
Bcl11b-loss driven apoptosis. J Proteome Res. 9:3799–3811. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Behl Y, Krothapalli P, Desta T, Roy S and
Graves DT: FOXO1 plays an important role in enhanced microvascular
cell apoptosis and microvascular cell loss in type 1 and type 2
diabetic rats. Diabetes. 58:917–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Browne G, Nesbitt H, Ming L, Stein GS,
Lian JB, McKeown SR and Worthington J: Bicalutamide-induced hypoxia
potentiates RUNX2-mediated Bcl-2 expression resulting in apoptosis
resistance. Br J Cancer. 107:1714–1721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kato JY, Matsuoka M, Polyak K, Massagué J
and Sherr CJ: Cyclic AMP-induced G1 phase arrest mediated by an
inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell.
79:487–496. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Savell J, Rao S, Pledger WJ and Wharton W:
Permanent growth arrest in irradiated human fibroblasts. Radiat
Res. 155:554–563. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Juríková M, Danihel Ľ, Polák Š and Varga
II: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Baldwin J and Grantham V: Radiation
Hormesis: Historical and current perspectives. J Nucl Med Technol.
43:242–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Komori T: Regulation of proliferation,
differentiation and functions of osteoblasts by Runx2. Int J Mol
Sci. 20:16942019. View Article : Google Scholar
|
|
50
|
Sakamaki J, Daitoku H, Yoshimochi K, Miwa
M and Fukamizu A: Regulation of FOXO1-mediated transcription and
cell proliferation by PARP-1. Biochem Biophys Res Commun.
382:497–502. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Malinen M, Saramäki A, Ropponen A,
Degenhardt T, Väisänen S and Carlberg C: Distinct HDACs regulate
the transcriptional response of human cyclin-dependent kinase
inhibitor genes to Trichostatin A and 1alpha, 25-dihydroxyvitamin
D3. Nucleic Acids Res. 36:121–132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kawane T, Komori H, Liu W, Moriishi T,
Miyazaki T, Mori M, Matsuo Y, Takada Y, Izumi S, Jiang Q, et al:
Dlx5 and mef2 regulate a novel runx2 enhancer for
osteoblast-specific expression. J Bone Miner Res. 29:1960–1969.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Naito J, Kaji H, Sowa H, Hendy GN,
Sugimoto T and Chihara K: Menin suppresses osteoblast
differentiation by antagonizing the AP-1 factor, JunD. J Biol Chem.
280:4785–4791. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Slominski AT, Zmijewski MA, Plonka PM,
Szaflarski JP and Paus R: How UV light touches the brain and
endocrine system through skin, and Why. Endocrinology.
159:1992–2007. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Skobowiat C, Brozyna AA, Janjetovic Z,
Jeayeng S, Oak ASW, Kim TK, Panich U, Reiter RJ and Slominski AT:
Melatonin and its derivatives counteract the ultraviolet B
radiation-induced damage in human and porcine skin ex vivo. J
Pineal Res. 65:e125012018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Janjetovic Z, Jarrett SG, Lee EF, Duprey
C, Reiter RJ and Slominski AT: Melatonin and its metabolites
protect human melanocytes against UVB-induced damage: Involvement
of NRF2-mediated pathways. Sci Rep. 7:12742017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim TK, Kleszczynski K, Janjetovic Z,
Sweatman T, Lin Z, Li W, Reiter RJ, Fischer TW and Slominski AT:
Metabolism of melatonin and biological activity of intermediates of
melatoninergic pathway in human skin cells. FASEB J. 27:2742–2755.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Slominski AT, Semak I, Fischer TW, Kim TK,
Kleszczyński K, Hardeland R and Reiter RJ: Metabolism of melatonin
in the skin: Why is it important? Exp Dermatol. 26:563–568. 2017.
View Article : Google Scholar : PubMed/NCBI
|