Introduction
Human cytomegalovirus (HCMV) is a widespread
opportunistic pathogen which is normally controlled by the immune
system. Primary infection is usually asymptomatic or may cause mild
symptoms in healthy individuals. However, accumulating evidence
supports the concept that periodic reactivation of HCMV can cause
severe medical conditions in immunocompromised patients such as
transplant, cancer or HIV-infected patients. Furthermore, vertical
transmission of HCMV during pregnancy is the leading cause of
congenital infection resulting in neurologic damage in neonates
[reviewed in (1,2)]. The exact mechanisms of
establishment, latency and reactivation of HCMV, although
fundamental for its successful persistence and dissemination,
remain poorly understood.
Lytic HCMV infection is temporally regulated and
viral gene expression takes place in three distinct phases, namely
the immediate-early (IE), early (E) and late (L) phases, with the
onset of each one being induced by the proteins expressed in the
previous one. The HCMV immediate-early promoter/enhancer (MIEP)
regulates the synthesis of the immediate-early protein 1 (IE1) and
immediate-early protein 2 (IE2) which are considered to control all
subsequent early and late events in HCMV replication, including
reactivation from latency, by antagonizing intrinsic and innate
immune responses (3). Both IE
proteins are abundantly transcribed and expressed as products of
alternative splicing and despite their relative homology, they
exert distinct functions; IE1 is essential for efficient viral
replication particularly at low multiplicities of infection
(4) whereas IE2 has a key role
during viral replication (5).
HCMV, as a successful opportunistic pathogen, takes control of
numerous host pathways such as metabolic and signalling pathways,
intrinsic and innate immune responses, apoptosis or even cell cycle
arrest strategies to prevent mitotic entry in order to ensure its
own maintenance and replication (6). HCMV interrupts the regular order of
steps in the cell division machinery by affecting key regulators of
cell cycle progression in normal fibroblasts in vitro
(7–9). HCMV infected cells have been found to
proceed to mitosis but this unscheduled mitotic entry is followed
by aberrant spindle formation, early separation of sister
chromatids and eventually cell death (10). Interestingly, the HCMV IE1 protein
has been known to be associated with condensed chromatin during
mitosis (11–20). The C-terminal (a.a. 476–491) of IE1
protein has been mapped as the responsible domain for this
intriguing association (17,21)
however, the molecular mechanism as well as the outcome of this
interaction between IE1 and chromatin remains to be determined.
Rho GTPases constitute a family of molecular
switches that control fundamental cellular processes such as signal
transduction pathways, cellular architecture and morphogenesis,
migration, innate and adaptive immunity while deregulation of Rho
GTPases have been associated with various human diseases and
disorders [reviewed in (22)].
Members of the Rho family, including the RhoA, RhoB and RhoC
isoforms have been shown to facilitate critical functions for a
productive HCMV infection such as deregulation of cytoskeleton at
early stages of infection, signal transduction, formation of the
viral assembly compartment or secretion of IL-11, both in normal
fibroblasts, the gold-standard cells for studying HCMV in
vitro (23–27) or even in a cancer background
(28). Combining the involvement
of RhoA isoform in the cell division procedure (29–31)
along with the mitotic defect in HCMV infected cells, we
investigated the role of RhoA GTPAse in the context of human
glioblastoma cells productively infected with HCMV. Our results
demonstrate that HCMV infected glioblastoma cells, despite the cell
cycle arrest observed in normal fibroblasts, are capable to
successfully complete all phases of mitosis and cytokinesis.
Furthermore, experiments depleting RhoA revealed the active role of
this Rho isoform in glioblastoma cell proliferation and mitosis
during HCMV lytic infection.
Materials and methods
Cells and viruses
Human Foreskin Fibroblasts (HFF) and the human
glioblastoma U373MG (Uppsala) cell line (kindly provided by
Professor Sunyoun Kim, Korea) were authenticated by short tandem
repeats DNA profiling and were grown in DMEM (Gibco BRL)
supplemented with 10% foetal bovine serum (Gibco BRL), 100 U/ml
penicillin and 100 µg/ml streptomycin under 5% CO2 in a
humidified incubator at 37°C. All cells were tested periodically
for mycoplasma contamination using the Mycoplasma Plus PCR primer
set (Stratagene) and found to be negative. The laboratory strain
HCMV AD169 strain as well as the the recombinant HCMV CR401 virus
expressing the IE1 fused to EGFP (12) were used in this study. The virus
stocks were propagated and titrated on HFF cells according to
standard protocols (32). For
viral infections, the cells were infected with HCMV at the
indicated MOI for 2 h and then the inoculum was removed and
replaced by fresh medium. The autofluoresecent construct
pHcRed1-H2A was transfected in U373MG cells to express histone H2A
fused to HcRed1 (12) using
Lipofectamine 3000 according to the manufacturer's instructions.
The initial HCMV CR401 infection of U373MG cells was subsequently
followed by transfection of the cells with the pHcRed1-H2A
construct.
RhoA knocking down
RhoA protein was knocked down using small
interfering RNA (siRNA) oligonucleotides against RhoA mRNA. The
siRNA sequence for RhoA was CGGAATGATGAGCACACAATT and the siRNA
sequence for the negative control was GAATAGACCCGTGATAGTACA. U373MG
cells were transfected with 2 µg of siRNA against RhoA mRNA or the
control siRNA using Lipofectamine 3000 according to the
manufacturer's recommended protocol. Western blot analysis was used
in order to determine the efficient knockdown of RhoA. After 48 h,
the culture medium was changed to 1% serum DMEM and the cells were
infected with HCMV AD169 wild type virus. All experiments were in
triplicates.
Western blot analysis
The protein lysates from cells were separated by 12%
Tris-glycine SDS-PAGE and electrophoretically transferred onto PVDF
membrane. The membrane was incubated in blocking solution at room
temperature for 1 h and subsequently with primary antibodies
against RhoA and β-actin overnight at 4°C. Visualization of
proteins was verified by a chemiluminesence detection method, using
the Image Lab Software 6.0.1 (Bio-Rad Laboratories, Inc.).
MTT assay
MTT assay was performed to determine the cell
proliferation rate of mock and HCMV infected U373MG after the
knockdown of RhoA protein. 1×105 cells were transfected
with the siRNA scrambled or the siRNA for RhoA and afterwards were
infected with HCMV AD169 wild type virus (MOI 2 pfu/cell) or mock
infected, in order to quantify the proliferation rate at 1, 2 and 3
days after the infection. The MTT reagent (3-(4,
5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) (Sigma
Aldrich) was used and the cells were incubated for 4 h at 37°C
followed by the addition of 150 µl MTT solvent (dimethyl
sulfoxide-DMSO) for 15 min and finally the measurement of the
absorbance at 590 nm with a reference filter of 620 nm.
Immunofluorescence analysis
For immunofluorescence, 1×105 U373MG
cells were transfected with the siRNA for RhoA or with the siRNA
scrambled on glass coverslips, were infected with the HCMV AD169
wild type virus at MOI 2 pfu/cell and after 1 day cells were fixed
with 4% PFA and permeabilized with 0.1% TritonX-100, for 10 min.
Cells were stained with the rabbit polyclonal antibody RhoA
(sc-179, Santa Cruz Biotechnology, Inc.), followed by a secondary
Cy5-labeled goat anti-rabbit antibody (Santa Cruz Biotechnology,
Inc.). The nuclei were stained with DAPI. The number of mitotic
cells were counted in randomly selected microscopy fields from each
condition and acquired with an epifluorescent Leica DMIRE2
microscope equipped with a Leica DFC300FX digital camera.
Statistical analysis
All data shown represent independent experiments
carried out in triplicate and are presented as the mean ± standard
deviation. Data were analyzed by one-way ANOVA followed by Tukey
post hoc test using GraphPad 8.3.1 software (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results and Discussion
Recruitment of IE1-72K onto metaphase
chromatin in live human fibroblasts and glioblastoma cells
Previous studies have shown that HCMV IE1 associates
with condensed chromatin of the host cell during mitosis and cells
engaged in viral replication enter into unscheduled mitosis
(10–20). Τhe recombinant HCMV CR401 virus
expressing IE1 fused to EGFP serves as an excellent tool for the
visualization of the IE1 protein in live infected cells, exhibiting
perfect colocalization with histone H2A in mitotic cells (12).
The direct interaction between HCMV IE1 and H2A-H2B
histones has been documented, precisely mapping the
chromatin-tethering domain (CTD) of IE1 and identifying that
several negatively charged H2A residues (E56, E61, E64, and D90)
composing the nucleosomal acidic pocket, but not acidic residues
outside the pocket, selectively direct the H2A interaction with the
IE1 CTD (21).
Given that HCMV arrests cell cycle progression, we
attempted to identify the step in the mitotic process that is
impeded by timelapse microscopy. Single cells co-expressing CR401
EGFP-IE1 and HcRed1-H2A, presenting identical localization onto
condensed chromosomes, were monitored throughout the course of
mitosis. In contrast to the normal cultured fibroblasts, in which
mitosis lasts approximately for 1 h, HCMV infected fibroblasts
remained at several mitotic phases for prolonged times. The
IE1-EGFP protein evidently became associated with condensing
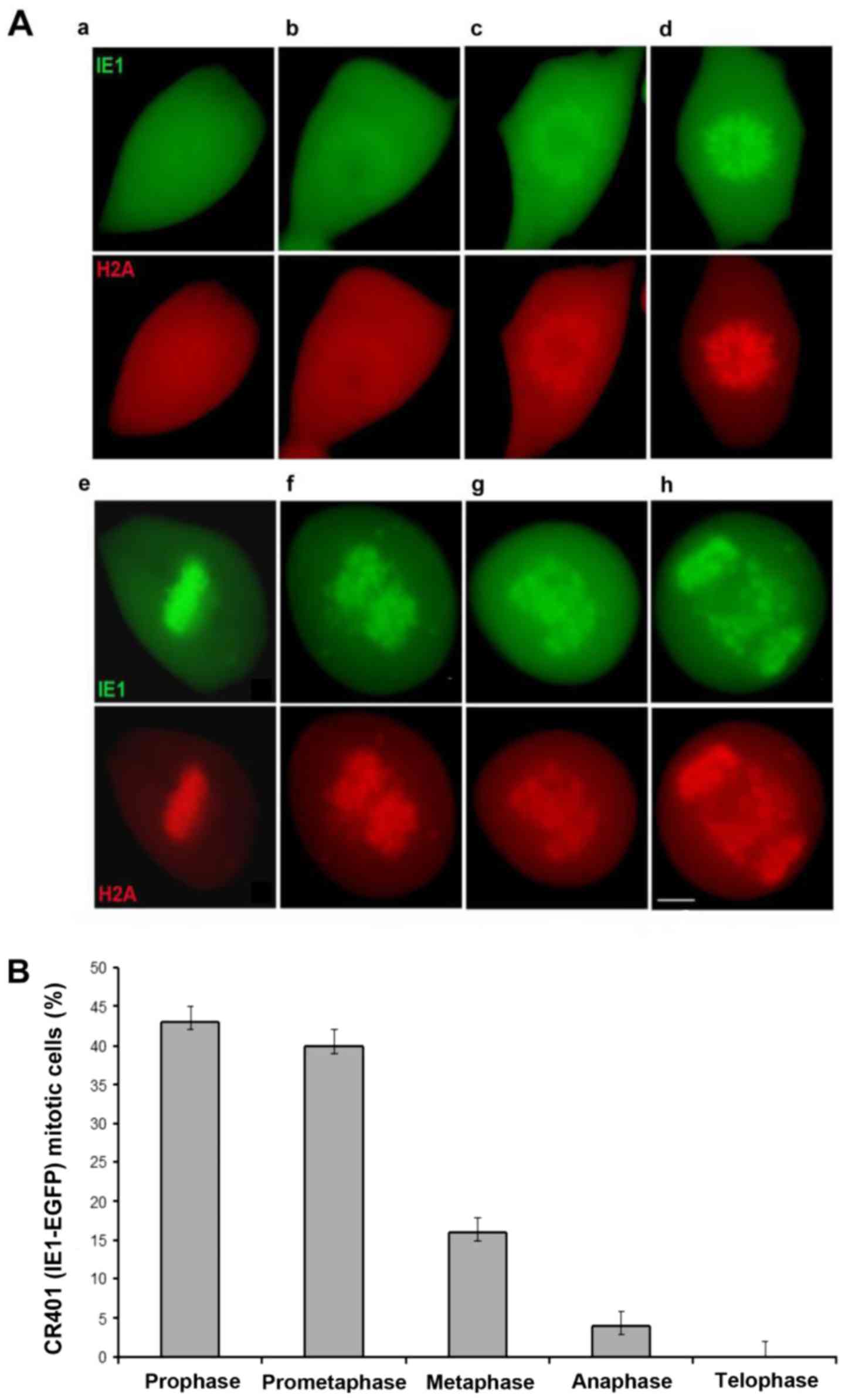
chromosomes altering its initial diffuse nuclear localization to a
compacted pattern alike H2A in prophase and prometaphase (Fig. 1Aa-d). IE1-EGFP mitotic chromosomes
were properly aligned in metaphase (Fig. 1Ae) however, a severe defect was
apparent in anaphase, revealing irregular chromosomal migration
which results in a significant number of lagging chromosomes
(Fig. 1Af-h) and consequently in
failure to complete the cell division. Detailed analysis examining
a large number of randomly selected HCMV CR401 EGFP-IE1 mitotic
cells, determined their distribution in relation to all mitotic
phases (Fig. 1B). The vast
majority of the mitotic cells were accumulated in prophase and
prometaphase followed by cells progressing to metaphase. A minimal
number of cells had advanced to an aberrant anaphase whilst
telophase cells were completely absent. Therefore, the progression
from metaphase to anaphase appears to be the critical transition
step, blocking the efficient cell division and rendering the cells
as non-productively infected cells.
The abortive infection observed in mitotic normal
fibroblasts prompted us to investigate the fate of cell division in
HCMV-infected glioblastoma cells. HCMV expression has been detected
in a high percentage of human glioblastomas, the most malignant
primary brain tumor [reviewed in (33)] while human glioblastoma cell lines
are permissive and support HCMV gene expression (34–36).
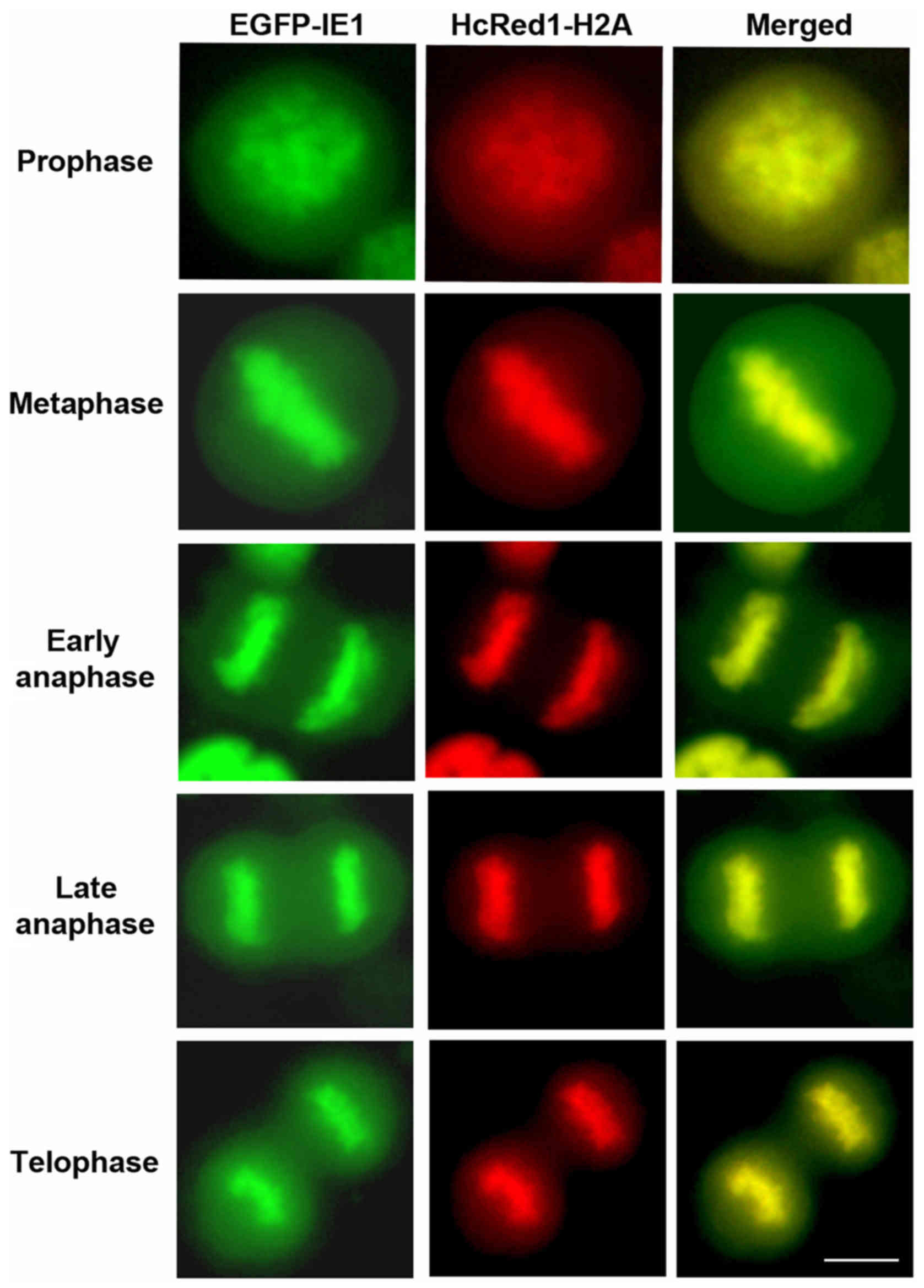
U373MG glioblastoma cells were infected with HCMV CR401 EGFP-IE1
virus while HcRed1-H2A in those cells served as a marker for
condensed chromosomes. Live cell imaging showed that glioblastoma
infected cells were capable of successfully completing mitosis. As
shown in Fig. 2, IE1-EGFP in tight
association with H2A was readily detectable in mitotic chromosomes,
following the strict sequential order of all phases of cell
division and cytokinesis, without encountering the defect observed
in fibroblasts. The above experiments demonstrate that there is an
apparent differentiation regarding mitotic progression, depending
on the cell line background, normal or cancerous.
Several viruses have evolved strategies to arrest
cycle progress and inhibit entry to mitosis, presumably in favour
of their own functions and overall successful replication and
propagation. HCMV represents a characteristic example of a virus
causing unscheduled cell division entry and distortion of mitotic
structures and subsequent mitotic blockade (10,12,20).
Remarkably, the HCMV-induced mitotic restrain observed in normal
fibroblasts is entirely abrogated in human glioblastoma cells which
unconditionally succeed to divide. In line with our finding,
efficient cell division has been observed in an additional
glioblastoma cell line permissive to HCMV, T98G cells, which
continue to divide, expressing all classes of viral genes. The
ability of HCMV to persist within U373MG cells may provide insights
into a mechanism for the sustained shedding of the virus.
Interestingly, the nuclear antigen 1 of Epstein-Barr virus and the
latency-associated nuclear antigen of the Kaposi sarcoma herpes
virus (HHV-8), both proteins expressed by gamma herpes viruses,
tether metaphase chromosomes, promoting viral genome maintenance
during latency and regulation of viral replication (37,38).
The specific role of IE1 binding onto condensed chromosomes remains
to be determined, however it is intriguing to suggest that the
chromatin-tethering activity of IE1 facilitates persistence of the
viral genomes in dividing cells. Furthermore, the chromosome
association properties of IE1 may have an additive role in the
uniform segregation of the HCMV genomes into daughter cells.
RhoA GTPase silencing reduces
proliferation and mitosis in human glioblastoma cells
RhoA GTPase has been shown to actively be involved
in several aspects of HCMV infection (23–28).
In addition, RhoA displays crucial roles in several stages of
mitosis such as cell cortex stiffening during cell rounding,
mitotic spindle formation [reviewed in (39)]. Combining the above, we tested the
implication of RhoA in cell division and proliferation of mock or
HCMV-infected U373MG cells by knocking down RhoA using the siRNA
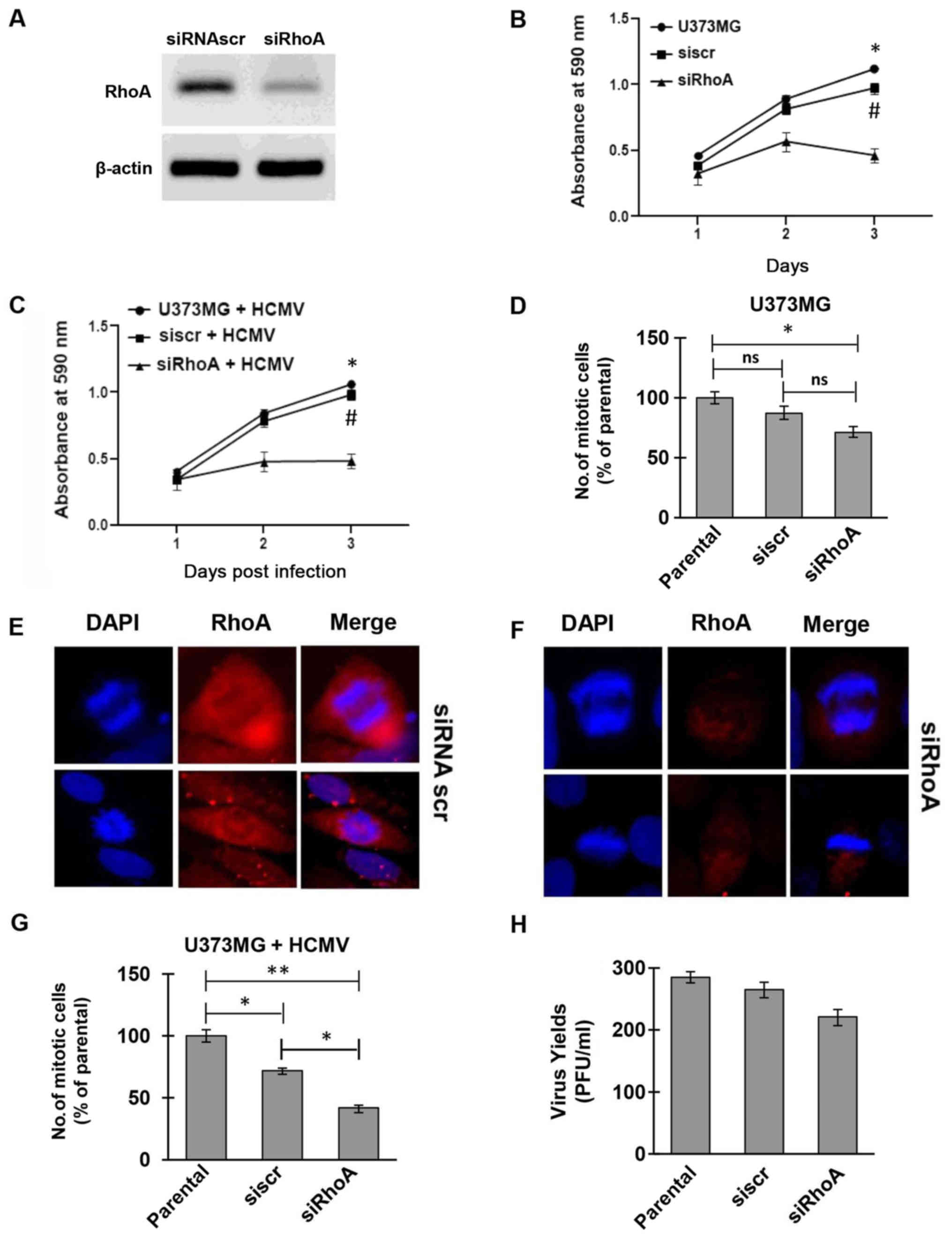
approach. The successful silencing of RhoA in U373MG cells used as
a cell model in the current study, was determined by western blot
analysis (Fig. 3A). The
proliferation rate of RhoA-depleted cells was significantly
decreased for a period of 3 days compared to both parental U373MG
and siRNA scramble-treated cells (Fig.
3B). Super-infection of the same set of cells with HCMV AD169
further confirmed the successful replication rate of the
RhoA-expressing cells in contrast to the restricted cell division
observed in the virally infected RhoA-depleted cells (Fig. 3C).
Apart from the growth rate, we were also interested
in exploring the mitotic behavior of RhoA-expressing and silenced
HCMV infected cells. For that purpose, parental U373MG cells, siRNA
scramble and siRNA RhoA cells, were cultured for 2 days and
multiple random optical fields were monitored for the detection of
mitotic cells. Independently of the siRNA target (siscr or siRhoA),
all cells successfully accomplished cell division, with
RhoA-depleted cells presenting a significant reduction in the
number of mitotic cells compared to the parental and scramble cells
(Fig. 3D). The ability of both
siRNA scramble and siRNA targeting RhoA to successfully bring about
mitosis was also confirmed by immunofluorescence microscopy
(Fig. 3E and F). The mitotic rate
of the aforementioned siRNA silenced cells was further assessed in
the context of HCMV infection. In line with the proliferation
rates, all groups of infected cells (siscr or siRhoA) proceeded and
successfully completed mitosis and cytokinesis. However, the number
of mitotic cells with RhoA-depletion was statistically
significantly reduced compared to the parental and scramble cells
(Fig. 3G). The difference between
the HCMV-infected parental and siRNA scramble cells was of less
statistical significance, indicating a relative effect of the RNA
interference process on the mitotic cells. The aforementioned
observation that the siRNA scramble affected mitosis confirmed that
although non-targeting controls activate the RNAi machinery
allowing a baseline assessment of the effect of the introduction of
duplex RNA on gene expression, they can induce a stress response
within cells. In addition, considering that HCMV is known to
trigger multiple cellular stress responses, the difference in
number of mitotic cells between the parental and siRNA scramble
groups could be due to siRNA/HCMV-induced stress. Moreover, the
HCMV shedding was also determined by standard plaque assay, showing
an efficient egress of the virus from the glioblastoma cells,
presenting no statistically significant difference between siRNA
treated and non-treated cells (Fig.
3H). These findings provide a clear evidence for an active role
of RhoA GTPase during cell proliferation of uninfected but also
remarkably of HCMV infected glioblastoma cells which despite the
silencing of RhoA they are still capable of dividing properly.
Moreover, cellular DNA synthesis is ongoing, although restrained in
RhoA-knockdown cells, even in the presence of the HCMV genomes.
Previous studies have shown an active involvement of
RhoA GTPase during several aspects of HCMV cell cycle including
cytoskeleton reorganization, viral replication, assembly and egress
of the progeny virus (23–28). The current study provides evidence
for an additional implication of RhoA in cell proliferation and
mitosis of glioblastoma HCMV-infected cells. RhoA is a critical
regulator and ensures the proper formation of mitotic spindle and
inhibition of RhoA activity is known to affect distinct phases of
the mitotic progression (29–31).
We show that glioblastoma cells are cable in overcoming the cell
cycle arrest induced by HCMV and our findings on cell proliferation
and division after RhoA depletion further expands our knowledge
regarding the role of this particular Rho GTPase for viral genome
maintenance and shedding.
Acknowledgements
The authors would like thank Dr Melpomeni Tseliou
and Dr Panagiota Dimitropolou (Medical School, University of Crete,
Greece) for technical assistance.
Funding
This project was funded by the National Plan for
Science, Technology, and Innovation (MAARIFAH), King Abdulaziz City
for Science and Technology, Kingdom of Saudi Arabia (grant no.
12-MED2652-02).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SaudA and SaadA were involved in methodology, data
curation and writing the original draft of the manuscript. AAAQ was
involved in methodology, data curation and writing the manuscript.
CS and GS were involved in conceptualization of the study,
supervised the study, and wrote, reviewed and edited the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Griffiths P: The direct and indirect
consequences of cytomegalovirus infection and potential benefits of
vaccination. Antiviral Res. 176:30662020. View Article : Google Scholar
|
|
2
|
Mocarski ES Jr: Immunomodulation by
cytomegaloviruses: Manipulative strategies beyond evasion. Trends
Microbiol. 10:332–339. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Adamson CS and Nevels MM: Bright and
early: Inhibiting human cytomegalovirus by targeting major
immediate-early gene expression or protein function. Viruses.
12:1102020. View Article : Google Scholar
|
|
4
|
Greaves RF and Mocarski ES: Defective
growth correlates with reduced accumulation of a viral DNA
replication protein after low-multiplicity infection by a human
cytomegalovirus ie1 mutant. J Virol. 72:366–379. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchini A, Liu H and Zhu H: Human
cytomegalovirus with IE-2 (UL122) deleted fails to express early
lytic genes. J Virol. 75:1870–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanchez V and Spector DH: Subversion of
cell cycle regulatory pathways. Curr Top Microbiol Immunol.
325:243–262. 2008.PubMed/NCBI
|
|
7
|
Salvant BS, Fortunato EA and Spector DH:
Cell cycle dysregulation by human cytomegalovirus: Influence of the
cell cycle phase at the time of infection and effects on cyclin
transcription. J Virol. 72:3729–3741. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalejta RF and Shenk T:
Proteasome-dependent, ubiquitin-independent degradation of the Rb
family of tumor suppressors by the human cytomegalovirus pp71
protein. Proc Natl Acad Sci USA. 100:3263–3268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song YJ and Stinski MF: Effect of the
human cytomegalovirus IE86 protein on expression of E2F-responsive
genes: A DNA microarray analysis. Proc Natl Acad Sci USA.
99:2836–2841. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eifler M, Uecker R, Weisbach H, Bogdanow
B, Richter E, König L, Vetter B, Lenac-Rovis T, Jonjic S, Neitzel
H, et al: PUL21a-cyclin A2 interaction is required to protect human
cytomegalovirus-infected cells from the deleterious consequences of
mitotic entry. PLoS Pathog. 10:e10045142014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahn JH, Brignole EJ III and Hayward GS:
Disruption of PML subnuclear domains by the acidic IE1 protein of
human cytomegalovirus is mediated through interaction with PML and
may modulate a RING finger-dependent cryptic transactivator
function of PML. Mol Cell Biol. 18:4899–4913. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dimitropoulou P, Caswell R, McSharry BP,
Greaves RF, Spandidos DA, Wilkinson GW and Sourvinos G:
Differential relocation and stability of PML-body components during
productive human cytomegalovirus infection: Detailed
characterization by live-cell imaging. Eur J Cell Biol. 89:757–768.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huh YH, Kim YE, Kim ET, Park JJ, Song MJ,
Zhu H, Hayward GS and Ahn JH: Binding STAT2 by the acidic domain of
human cytomegalovirus IE1 promotes viral growth and is negatively
regulated by SUMO. J Virol. 82:10444–10454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krauss S, Kaps J, Czech N, Paulus C and
Nevels M: Physical requirements and functional consequences of
complex formation between the cytomegalovirus IE1 protein and human
STAT2. J Virol. 83:12854–12870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lafemina RL, Pizzorno MC, Mosca JD and
Hayward GS: Expression of the acidic nuclear immediate-early
protein (IE1) of human cytomegalovirus in stable cell lines and its
preferential association with metaphase chromosomes. Virology.
172:584–600. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nevels M, Brune W and Shenk T: SUMOylation
of the human cytomegalovirus 72-kilodalton IE1 protein facilitates
expression of the 86-kilodalton IE2 protein and promotes viral
replication. J Virol. 78:7803–7812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reinhardt J, Smith GB, Himmelheber CT,
Azizkhan-Clifford J and Mocarski ES: The carboxyl-terminal region
of human cytomegalovirus IE1491aa contains an acidic domain that
plays a regulatory role and a chromatin-tethering domain that is
dispensable during viral replication. J Virol. 79:225–233. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin HJ, Kim YE, Kim ET and Ahn JH: The
chromatin-tethering domain of human cytomegalovirus immediate-early
(IE) 1 mediates associations of IE1, PML and STAT2 with mitotic
chromosomes, but is not essential for viral replication. J Gen
Virol. 93:716–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilkinson GW, Kelly C, Sinclair JH and
Rickards C: Disruption of PML-associated nuclear bodies mediated by
the human cytomegalovirus major immediate early gene product. J Gen
Virol. 79:1233–1245. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Q, Chen P, Wang M, Fang J, Yang N, Li
G and Xu RM: Human cytomegalovirus IE1 protein alters the
higher-order chromatin structure by targeting the acidic patch of
the nucleosome. Elife. 5:e119112016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mücke K, Paulus C, Bernhardt K, Gerrer K,
Schön K, Fink A, Sauer EM, Asbach-Nitzsche A, Harwardt T, Kieninger
B, et al: Human cytomegalovirus major immediate early 1 protein
targets host chromosomes by docking to the acidic pocket on the
nucleosome surface. J Virol. 88:1228–1248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hall A: Rho family GTPases. Biochem Soc
Trans. 40:1378–1382. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Huang DY, Huong SM and Huang ES:
Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat
Med. 11:515–521. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seo JY, Yaneva R, Hinson ER and Cresswell
P: Human cytomegalovirus directly induces the antiviral protein
viperin to enhance infectivity. Science. 332:1093–1097. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Poncet D, Pauleau AL, Szabadkai G, Vozza
A, Scholz SR, Le Bras M, Brière JJ, Jalil A, Le Moigne R, Brenner
C, et al: Cytopathic effects of the cytomegalovirus-encoded
apoptosis inhibitory protein vMIA. J Cell Biol. 174:985–996. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goulidaki N, Alarifi S, Alkahtani SH,
Al-Qahtani A, Spandidos DA, Stournaras C and Sourvinos G: RhoB is a
component of the human cytomegalovirus assembly complex and is
required for efficient viral production. Cell Cycle. 14:2748–2763.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alarifi S, Alkahtani S, Al-Qahtani AA,
Stournaras C and Sourvinos G: Induction of interleukin-11 mediated
by RhoA GTPase during human cytomegalovirus lytic infection. Cell
Signal. 70:1095992020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tseliou M, Al-Qahtani A, Alarifi S,
Alkahtani SH, Stournaras C and Sourvinos G: The role of RhoA, RhoB
and RhoC GTPases in cell morphology, proliferation and migration in
human cytomegalovirus (HCMV) infected glioblastoma cells. Cell
Physiol Biochem. 38:94–109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Derksen PWB and van de Ven RAH: Shared
mechanisms regulate spatiotemporal RhoA-dependent actomyosin
contractility during adhesion and cell division. Small GTPases.
11:113–121. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakayama Y, Saito Y, Soeda S, Iwamoto E,
Ogawa S, Yamagishi N, Kuga T and Yamaguchi N: Genistein induces
cytokinesis failure through RhoA delocalization and anaphase
chromosome bridging. J Cell Biochem. 115:763–771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lian G, Wong T, Lu J, Hu J, Zhang J and
Sheen V: Cytoskeletal associated filamin A and RhoA affect neural
progenitor specification during mitosis. Cereb Cortex.
29:1280–1290. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Filippakis H, Dimitropoulou P, Eliopoulos
AG, Spandidos DA and Sourvinos G: The enhanced host-cell
permissiveness of human cytomegalovirus is mediated by the Ras
signaling pathway. Biochim Biophys Acta. 1813:1872–1882. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cobbs CS: Cytomegalovirus and brain tumor:
Epidemiology, biology and therapeutic aspects. Curr Opin Oncol.
25:682–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Awasthi S, Isler JA and Alwine JC:
Analysis of splice variants of the immediate-early 1 region of
human cytomegalovirus. J Virol. 78:8191–8200. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee K, Jeon K, Kim JM, Kim VN, Choi DH,
Kim SU and Kim S: Downregulation of GFAP, TSP-1, and p53 in human
glioblastoma cell line, U373MG, by IE1 protein from human
cytomegalovirus. Glia. 51:1–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo MH and Fortunato EA: Long-term
infection and shedding of human cytomegalovirus in T98G
glioblastoma cells. J Virol. 81:10424–10436. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marechal V, Dehee A, Chikhi-Brachet R,
Piolot T, Coppey-Moisan M and Nicolas JC: Mapping EBNA-1 domains
involved in binding to metaphase chromosomes. J Virol.
73:4385–4392. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Piolot T, Tramier M, Coppey M, Nicolas JC
and Marechal V: Close but distinct regions of human herpesvirus 8
latency-associated nuclear antigen 1 are responsible for nuclear
targeting and binding to human mitotic chromosomes. J Virol.
75:3948–3959. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chircop M: Rho GTPases as regulators of
mitosis and cytokinesis in mammalian cells. Small GTPases.
5:e297702014. View Article : Google Scholar : PubMed/NCBI
|