Introduction
Breast cancer (BC) is the most common cancer
occurring in women, and ~271,270 patients were newly diagnosed with
BC in 2019 in the United States (1). According to the National Center for
Health Statistics (NCHS), the mortality rate of BC in 2012–2016 was
20.6%, which ranked BC as the second most life-threatening disease
in the United States following lung/bronchus cancer. In addition,
the NCHS estimated that BC would cause ~42,260 deaths in 2019
(1). BC is typically categorized
as basal-like [estrogen receptor low/progesterone receptor
low/human epidermal growth factor receptor 2 (HER2) low], luminal
A-type (estrogen receptor high/HER2 low), luminal B-type (estrogen
receptor or progesterone receptor high, either HER2 high or low,
and Ki67 high) or HER2-type (estrogen receptor low/progesterone
receptor low/HER2 high) based on the cell types of origin. Notably,
the incidence of luminal A-type is the highest among all types of
BC (2). As numerous individuals
are likely to be affected by BC over the coming years, elucidating
the mechanisms underlying BC to identify a novel prognostic marker
could be important for BC therapy.
Cancer cells regularly exhibit altered chromosome
numbers; these alterations are collectively known as aneuploidy
(3,4). Aneuploidy, which is usually caused by
an abnormal number and size of the centrosome, may accelerate
tumorigenesis and carcinogenesis by causing chromosomes to separate
unequally during mitosis (4). It
has been reported that 65–90% of BC cells exhibit aneuploidy
(3). Centromere protein M (CENPM)
is an essential centromere component that is associated with
several other centromere proteins [including CENPA (5), CENPC (6), CENPI (7) and CENPH (8)], and is required for chromosome
separation. The CENPM protein interacts with other proteins to form
a vital complex that preserves kinetochore and spindle microtubule
attachment during metaphase (9).
Overexpression of CENPM leads to unequal numbers of chromosomes in
cells; these cells subsequently exit mitosis, survive and lead to
aneuploidy (10). In addition,
high CENPM expression has been reported to be associated with
primary melanoma (11), bladder
cancer (12), hepatocellular
carcinoma (13), and head and neck
squamous cell carcinoma (14).
Based on these studies, the present study hypothesized that
aberrant CENPM could function as an oncogene by intervening in the
progression of the cell cycle.
The present study demonstrated that CENPM mRNA
expression was upregulated in various types of cancer and that
elevated CENPM in BC was highly associated with low patient
survival probability. By studying CENPM-related genes, it was
identified that five key genes associated with CENPM could
accelerate the cell cycle in BC. In addition, in MCF7 cells, CENPM
overexpression significantly enhanced cell proliferation and
inhibited apoptosis in vitro. Therefore, the findings of the
present study indicated that CENPM may control tumor progression
and survival rate in patients with BC.
Materials and methods
Catalogue of Somatic Mutations in
Cancer (COSMIC) database analysis
The COSMIC (https://cancer.sanger.ac.uk/cosmic) is a database of
gene mutations associated with various types of human cancer, which
includes 1,420,135 samples and 26,878 papers (COSMIC v89). Using
this database, information about CENPM gene mutational signatures
according to cancer types was extracted, and the association
between CENPM mutations, including point mutations, insertions and
deletions, and the risk of BC tumorigenesis was summarized.
Detailed information about the mutational signature method is
provided in previous studies (15,16).
Oncomine database analysis
Oncomine v4.5 (https://www.oncomine.org/resource/login.html) is a
database of information levels, DNA copy numbers, drug sensitivity
and other parameters in normal samples and multiple types of cancer
samples. After a target gene is selected, the Oncomine database
returns a list of gene expression analyses that have been conducted
in different microarray studies, allowing easy exploration of the
diseases that are possibly caused by the selected gene. In the
present study, the analyses revealing elevated CENPM expression
were acquired to create a graph showing the cancer types that are
significantly associated with CENPM gene expression. The detailed
methods used for Oncomine database analysis are described in a
previous study (17).
Subsequently, Gene Expression Profiling Interactive Analysis
(GEPIA) v1 (http://gepia.cancer-pku.cn/) was applied to reveal
CENPM mRNA expression levels in several types of cancer samples vs.
normal samples. The detailed methods used for GEPIA are described
in a previous study (18).
cBioPortal database analysis
cBioPortal v3.4.4 (http://www.cbioportal.org/) is a database of
information on gene expression, genetic alterations, coexpression
and survival status that enables researchers to analyze and explore
topics associated with tumorigenesis. Upon selection of invasive
breast carcinoma [The Cancer Genome Atlas (TCGA), cell 2015]
(19), and the gene of interest
(CENPM), the database returned a list of genes highly associated
with CENPM. To visualize the protein-protein interaction network,
the Search Tool for the Retrieval of Interacting Genes/proteins
(STRING) database v11.0 (https://string-db.org/) was accessed with the gene
names provided by the cBioPortal database, the nodes and lines were
modified with Cytoscape (CytoHubba) v3.42 (https://cytoscape.org/), and protein expression was
examined by using the Human Protein Atlas v19 (HPA; www.proteinatlas.org). GeneCards Version 4.11
(https://www.genecards.org/) was used to
search for the functional information of the candidate genes. The
detailed methods used for gene coexpression analysis are described
in previous studies (20,21).
Kaplan-Meier plotter analysis
The UALCAN database (September 23, 2019 release;
http://ualcan.path.uab.edu), a
user-friendly platform that links to cancer omics databases TCGA
and MET500 (September 11, 2017 release; http://met500.path.med.umich.edu/), can be used to
evaluate gene expression vs. patient survival probability or tumor
grade for multiple types of cancer. To calculate the effect of the
target gene expression on patient survival, UALCAN was used to
create a Kaplan-Meier plot with sample data by using the ‘survival’
and ‘survminer’ packages. The detailed methods are provided in a
previous study (22). The
association between CENPM mRNA upregulation and patient survival
status in various types of cancer were identified. To estimate the
prognostic value of CENPM in specific BC types, including luminal
A-type, luminal B-type, HER2-positive-type and basal-like-type BC,
CENPM (Gene ID, 218741_at, JetSet best probe set) was queried on
the Kaplan-Meier Plotter database (August 1, 2019 release;
http://www.kmplot.com) that links to Gene
Expression Omnibus, European Genome-Phenome Archive and TCGA data,
and the results clearly demonstrated the potential effects of high
or low levels of CENPM on patient survival. The survival curves of
samples with high gene expression and low/medium gene expression
were compared by log rank test. The detailed methods are provided
in a previous study (23).
Reverse transcription-quantitative
(RT-q)PCR
MCF7 cells (American Type Culture Collection) were
cultured in DMEM medium (Sigma-Aldrich; Merck KGaA) containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
then received treatment in serum-free DMEM medium. Cellular mRNA
was extracted and purified from MCF7 cells (1×106/ml)
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.), chloroform, isopropanol and alcohol. The cDNA was
synthesized by using RevertAid RT Reverse Transcription Kit (Thermo
Fisher Scientific, Inc.), in accordance with the manufacturer's
protocols. Real time PCR was performed with ABI PRISM 7300 Sequence
Detection System (Applied Biosystems, USA). After RT, the target
gene mRNA expression was detected with the primers shown in
Table I and with FastStart
Universal SYBR Green Master (Rox; Roche Molecular Systems, Inc.),
in accordance with the manufacturer's protocols. The thermal
cycling conditions were: Denaturation 10 min at 95°C, followed by
45 cycles of denaturation at 95°C for 30 sec, annealing at 56°C for
30 sec and extension at 72°C for 30 sec, and then CENPM, ERCC6L,
SHCBP1, CKS2, RAD51, KIF4A, HMMR, SPAG5, CDC25C, and RACGAP1 mRNA
levels were quantified as previously reported (24).
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| PCNA |
CCTGCTGGGATATTAGCTCCA |
CAGACCCATTTACTTGTGTTGGA |
| GAPDH |
ACAACTTTGGTATCGTGGAAGG |
GCCATCACGCCACAGTTTC |
| KI67 |
ACGCCTGGTTACTATCAAAAGG |
CAGCGGTAGGTGTCGAAGC |
| CENPM |
GCGGACTCGATGCTCAAAGA |
TTCTGGAGACTGTATTTGCTGTG |
| ERCC6L |
CTCTGGCTTGCTACTTTATCGAG |
TGCATCAAACATACCGGAAAGG |
| SHCBP1 |
GCTACCGTGATAAACCAGGTTC |
AGGCTCTGAATCGCTCATAGA |
| CKS2 |
TTCGACGAACACTACGAGTACC |
GGACACCAAGTCTCCTCCAC |
| RAD51 |
CAACCCATTTCACGGTTAGAGC |
TTCTTTGGCGCATAGGCAACA |
| KIF4A |
TACTGCGGTGGAGCAAGAAG |
CATCTGCGCTTGACGGAGAG |
| HMMR |
ATGATGGCTAAGCAAGAAGGC |
TTTCCCTTGAGACTCTTCGAGA |
| SPAG5 |
TTGAGGCCCGTTTAGATACCA |
GCTTTCCTTGGAGCAATGTAGTT |
| CDC25C |
TCTACGGAACTCTTCTCATCCAC |
TCCAGGAGCAGGTTTAACATTTT |
| RACGAP |
ATGATGCTGAATGTGCGGAAT |
CGCCAACTGGATAAATTGGACTT |
Western blot analysis
Briefly, after protein was extracted from MCF7 cells
with RIPA buffer according to the manufacture's protocol (Cell
Signaling Technology, Inc.), the concentration of protein was
determined by Bradford protein assays kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocols, and 40 µg
proteins were loaded into each lane of a 12 or 15% SDS-PAGE gel.
Following gel electrophoresis and transfer of the separated
proteins onto a nitrocellulose membrane, the membrane was treated
with 10 ml blocking buffer (Beyotime Institute of Biotechnology,
Inc.) containing 5% BSA for 1 h at room temperature. Subsequently,
the membrane was incubated with anti-cleaved caspase-3 (cat. no.
9661; 1:1,000; Cell Signaling Technology, Inc.), anti-Bax (cat. no.
5023; 1:1,000; Cell Signaling Technology, Inc.), anti-Bcl-2 (cat.
no. 15071; 1:1,000; Cell Signaling Technology, Inc.),
anti-cyclin-dependent kinase subunit 2 (CKS2; cat. no. ab155078;
1:1,000; Abcam, Inc.), anti-GAPDH (cat. no. 5174; 1:1,000; Cell
Signaling Technology, Inc.) or anti-β-actin (cat. no. 4970;
1:1,000; Cell Signaling Technology, Inc.) primary antibodies for 12
h at 4°C. Following incubation with anti-rabbit secondary
antibodies (cat. no. 7074; 1:5,000; Cell Signaling Technology,
Inc.), or anti-mouse secondary antibodies (cat. no. 7076; 1:5,000;
Cell Signaling Technology, Inc.) for 1 h at room temperature.
Protein bands were detected with Electro-chemiluminescence (ECL)
Plus reagents (Cytiva), according to the manufacturer's
instructions, the membrane was imaged with a GeneGnome XRQ
Chemiluminescence Imaging system (Gene Company Ltd.), and the
density was quantified with ImageJ software (v1.46; National
Institutes of Health).
Cell cycle analysis
To construct CENPM-overexpression vectors, CENPM
cDNA was amplified by polymerase chain reaction from the MCF7 cells
with the following primers: Forward,
5′-CATGCTAGCATGTCGGTGTTGAGGCCCCTGGA-3′; reverse:
5′-TTCAAGCTTTCACAGGTCCTCCAGGGAGGGGC-3′. The PCR product was
digested with NheI and HindIII, and then subcloned into an
expression vector pIRES2-EGFP (pIRES2-EGFP, Clontech Laboratories,
Inc.). The plasmid was termed the CENPM-overexpression vectors, and
then CENPM-overexpression vectors at a final concentration 0, 1.5
or 2 µM with or without control vector (pIRES2-EGFP) were
transfected into MCF7 cells (1×106) using the
Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. MCF7
cells were transfected for 12 h, grown in DMEM medium supplemented
with 5% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.) for 24 h at 37°C and then fixed in 70% ethanol for 12 h at
4°C. The ethanol-fixed samples were washed twice with PBS, digested
with RNase A (1 mg/ml; Sigma-Aldrich; Merck KGaA) for 30 min at
37°C, and then stained with propidium iodide solution (1 mg/ml;
Sigma-Aldrich; Merck KGaA) for 30 min at 4°C. The cell cycle was
examined by a BD FACSVerse flow cytometer (BD Biosciences, San
Jose, CA) at an excitation wavelength of 488 nm and the data were
analyzed with ModFit LT v4.0.5 Software (Verity Software House,
Inc.).
Cell Counting Kit-8 (CCK-8)-based
proliferation assay
CCK-8 assay was conducted to assess proliferation at
0, 12, 24 and 48 h and T-47D cells (American Type Culture
Collection) were cultured in RPMI-1640 medium (Sigma-Aldrich)
containing 10% fetal bovine serum at 37°C under a 5% CO2
atmosphere. Following transfection, the MCF7 and T-47D cells were
passaged at a density of 2,000 cells/well in 96-well plates, cells
in each well were incubated with 10 µl CCK-8 solution
(MedChemExpress) for 2 h at 37°C, and the proliferation rate was
calculated based on the absorbance at 450 nm, as determined using a
microplate reader (ELx800, BioTek Instruments).
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL) assay
For determination of apoptosis, control- or
CENPM-overexpressing MCF7 cells were stained using the One Step
TUNEL Apoptosis Assay Kit (Beyotime Institute of Biotechnology), as
described in the manufacturer's protocols, and immunofluorescence
was determined by fluorescence microscopy (Leica Microsystems
GmbH), with 5 randomly selected high-power fields to calculate the
ratio per slide.
Cell scratch assay
MCF7 cells (~90% confluence) were transfected with
control- or CENPM-overexpression vectors for 12 h, scratched with
200 µl pipette tips, washed with PBS, and grown in medium
supplemented with 1 or 5% FBS for 0 and 48 h (25). Following the indicated culture
times, the gaps were imaged with a fluorescence microscope (Leica
Microsystems GmbH) and the width of each gap was analyzed with
ImageJ (v1.46; National Institutes of Health).
RNA interference
Three CENPM-targeting short hairpin (sh)RNAs were
constructed; the sequences were as follows: shCEN #a,
5′-CCTGATCGTGTTTGTGGTTAA-3′; shCEN #b, 5′-GCTGACTCCATAAACATTCTC-3′;
shCEN #c, 5′-GCGGACTCGATGCTCAAAGAG-3′. The shRNAs specifically
targeting human CENPM were cloned into psi-LVRU6GP (GeneCopoeia,
Inc.) and the non-targeting sequence 5′-ACAGAAGCGATTGTTGATC-3′ was
cloned into the same vector and used as the shControl. A Lenti-Pac
HIV Expression Packaging kit (GeneCopoeia, Inc.) was used for
shRNA-encoding lentivirus packaging, according to the
manufacturer's protocols. The detailed methods used for lentivirus
infection and confirmation of CENPM knockdown are provided in a
previous study (26). Briefly,
293T cells (~70% confluence) were seeded in 6 well plate for 24 h,
transfected with 500 ng shControl, shCEN #a, shCEN #b, or shCEN #c,
250 ng pMD2.G, 250 ng psPAX2, and Lipofectamine® 2000
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h, then
the medium was refreshed using freshly prepared 5% DMEM. At 72 h
after transfection, the supernatants were harvested, the 400 µl
virus solution was added to the cells and the knockdown of the
target gene was confirmed by real time PCR.
Colony formation test
Transfected cells were plated in 6-well plates with
1,000 cells per well and then cultured for 10 days until colonies
were visible to the eye. Cells were washed twice with
phosphate-buffered saline (PBS), fixed with methanol for 30 min at
room temperature, and then stained with crystal violet for 30 min
at room temperature. The number of colonies was counted.
Statistical analysis
All experiments were conducted at least three times
independently, and the data were analyzed by two-tailed, unpaired
Student's t-test or one-way ANOVA followed by Tukey's or Dunnett's
multiple comparisons test. The data are presented as the means ±
standard error of the mean and P<0.05 was considered to indicate
a statistically significant difference. The survival curves of
samples with high gene expression and low/medium gene expression
were compared by log rank test. Data were analyzed using GraphPad
v5.01 (GraphPad Software, Inc.).
Results
Expression levels of CENPM in various
types of human cancers
By using the Oncomine database, the transcription
levels of CENPM in normal and cancerous human specimens were
examined (Fig. 1A). CENPM mRNA was
significantly upregulated in 14 of 20 types of cancer (Fig. 1A, fold change >2,
P<1×10−4, gene rank: Top 10%). The 43 datasets
associated with elevated CENPM mRNA expression included 4,697
samples, and the fold changes in expression ranged between 2.008
and 9.628. CENPM mRNA upregulation appeared most often in BC
(22.2%), cervical cancer (40%), liver cancer (23%), lung cancer
(22.2%), lymphoma (16%) and sarcoma (16%) (Fig. 1B). Subsequently, the mRNA
expression levels of CENPM were analyzed using an online database
(GEPIA) (18) to evaluate the
effect of CENPM on human cancer. The results demonstrated that
breast invasive carcinoma (BRCA), cervical squamous cell carcinoma
and endocervical adenocarcinoma (CESC), liver hepatocellular
carcinoma (LIHC), sarcoma (SARC), diffuse large B-cell lymphoma
(DLBC), lung adenocarcinoma (LUAD) and lung squamous cell carcinoma
(LUSC) tissues had significantly elevated levels of CENPM mRNA
(Fig. 1C and D).
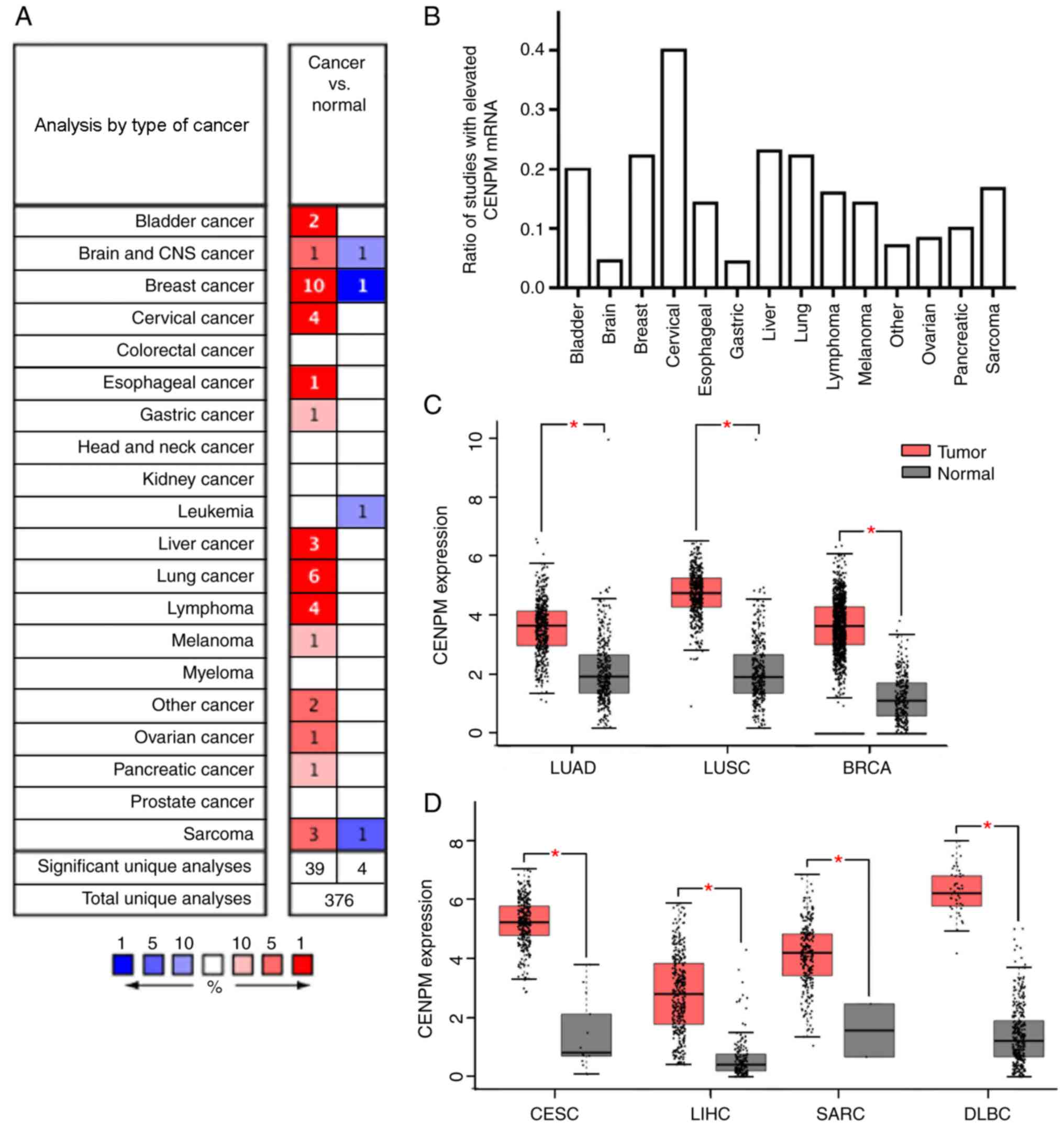 | Figure 1.Expression levels of CENPM in various
types of human cancer. (A) Analyses performed on data for various
cancer types from the Oncomine database revealed that CENPM mRNA
expression was increased in tumor samples compared with normal
samples (fold change >2, P<1×10−4, gene rank: Top
10%). (B) Number of studies revealing CENPM mRNA upregulation
relative to the total number of studies examined. The data were
collected from the Oncomine database and included data for various
cancer types (fold change >2, P<1×10−4, gene rank:
Top 10%). (C and D) mRNA expression levels of CENPM were
significantly higher in patients with LUAD (n=483) vs. normal
individuals (n=347), patients with LUSC (n=486) vs. normal
individuals (n=338), patients with BRCA (n=1085) vs. normal
individuals (n=291), patients with CESC (n=306) vs. normal
individuals (n=13), patients with LIHC (n=369) vs. normal
individuals (n=160), patients with SARC (n=262) vs. normal
individuals (n=2) and patients with DLBC (n=47) vs. normal
individuals (n=337) in Gene Expression Profiling Interactive
Analysis. *P<0.05. CENPM, centromere protein M; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; CESC, cervical
squamous cell carcinoma and endocervical adenocarcinoma; LIHC,
liver hepatocellular carcinoma; DLBC, diffuse large B-cell
lymphoma; BRCA, invasive breast carcinoma; SARC, sarcoma. |
The protein expression levels of CENPM were detected
using the Human Protein Atlas (HPA; www.proteinatlas.org). Since immunohistochemical (IHC)
staining data for CENPM in SARC or DLBC were not available in the
HPA database, the protein expression levels of CENPM in SARC or
DLBC were not investigated. As shown in Fig. 2A, the protein expression levels of
CENPM were slightly elevated in BRCA (n=2 for normal breast tissue
and n=2 for BRCA tissue) and CESC (n=2 for normal cervical tissue
and n=2 for CESC tissue), and no significant difference was
observed in LIHC (n=3 for normal liver tissue and n=1 for LIHC
tissue; P>0.05) and LUSC (n=2 for normal lung tissue and n=4 for
LUSC tissue). However, it is highly recommended that further IHC
studies be performed on normal and cancerous samples to verify
these findings. Furthermore, the IHC staining assay illustrated
that the CENPM protein was primarily expressed in the nuclei of
cells and its localization was associated with its function
(9).
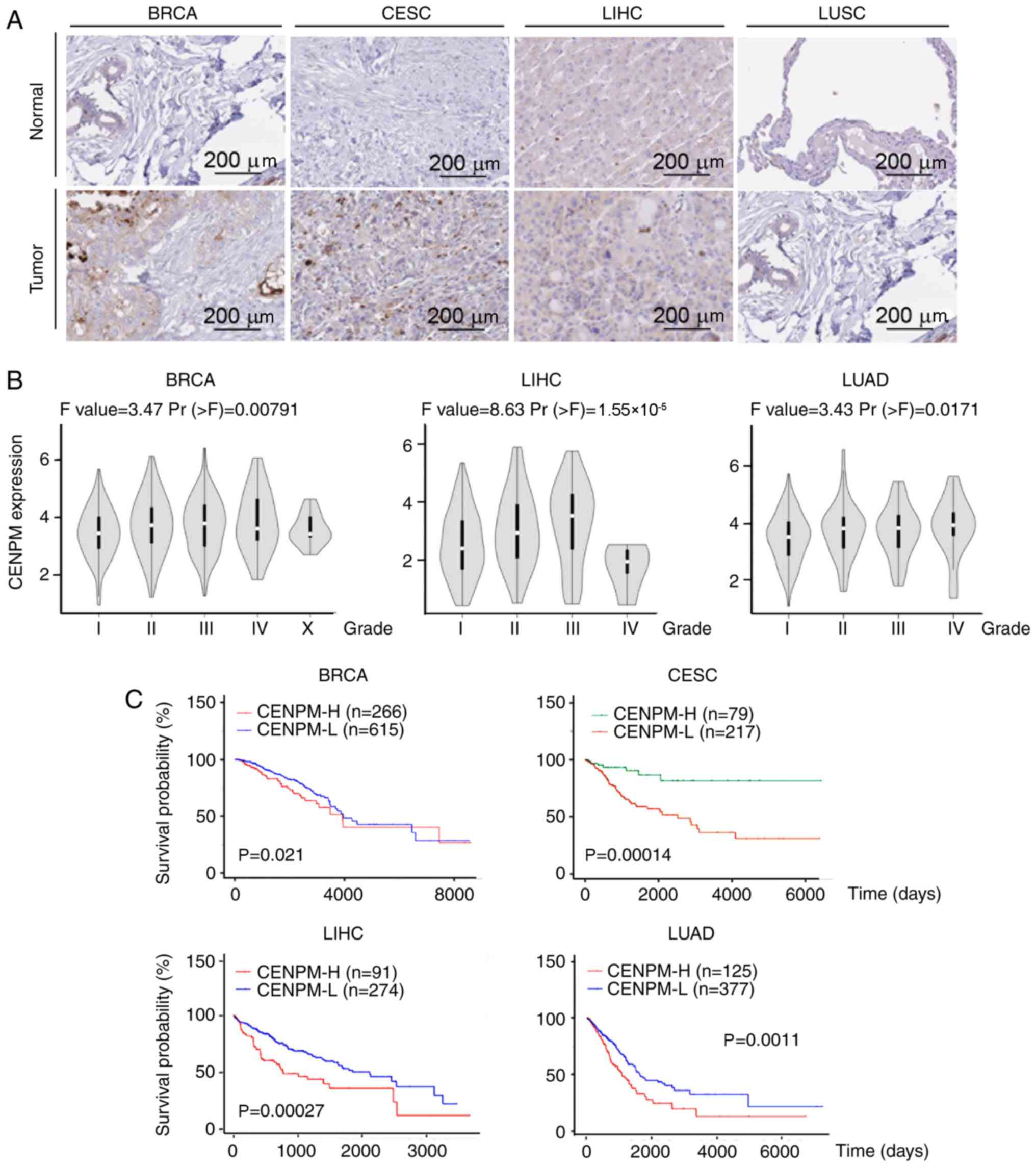 | Figure 2.Expression levels of CENPM in various
types of human cancer. (A) Immunohistochemical staining of CENPM
protein levels in breast (n=2 for normal breast tissue and n=2 for
BRCA tissue), cervix (n=2 for normal cervical tissue and n=2 for
CESC tissue), liver (n=3 for normal liver tissue and n=1 for LIHC
tissue) and lung (n=2 for normal lung tissue, and n=4 for LUSC
tissue) samples. The data for the samples, which were collected and
stained with a CENPM antibody, were obtained from the Human Protein
Atlas. Scale bars, 200 µm. (B) Data on CENPM mRNA expression levels
in BRCA, LIHC, and LUAD of different stages were downloaded from
the Gene Expression Profiling Interactive Analysis database. (C)
Kaplan-Meier plotter analysis of 881 patients with BRCA, 296
patients with CESC, 365 patients with LIHC and 502 patients with
LUAD who had high or low CENPM mRNA expression. CENPM, centromere
protein M; BRCA, invasive breast carcinoma; CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; LIHC, liver
hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; LUAD,
lung adenocarcinoma. |
Next, the association of CENPM gene expression with
various types of human cancer was examined, and the association
between CENPM mRNA expression and cancer grade was further examined
using the GEPIA database. As shown in Fig. 2B, elevated CENPM mRNA expression
was positively associated with tumor grade in BRCA, LIHC and LUAD,
but barely in CESC and DLBC (data not shown). The comparatively few
specimens from patients with SARC (Fig. 1D, n=2 for normal tissue) made the
results of the analysis of the association of CENPM gene expression
with tumor grade in SARC less meaningful (data not shown). Next, it
was examined whether CENPM gene expression in BRCA, CESC, LIHC,
SARC, DLBC, LUAD and LUSC was associated with overall survival
rates using the online UALCAN databases (22). As shown in Fig. 2C, lower survival rates in BRCA (881
patients, P=0.021), LIHC (365 patients, P=0.00027) and LUAD (502
patients, P=0.0011) were associated with higher CENPM mRNA levels.
By contrast, elevated CENPM mRNA levels were associated with higher
survival rates in patients with CESC (296 patients, P=0.00014) but
were not associated with survival rates in SARC (259 patients,
P=0.29), DLBC (47 patients, P=0.039; too few patients to confirm)
or LUSC (494 patients, P=0.25) (data not shown). Thus, the results
regarding the proportion of studies indicating CENPM upregulation
(Fig. 1A and B), the mRNA and
protein expression levels of CENPM (Figs. 1C and D, and 2A) in tumor vs. normal samples, and the
association between CENPM mRNA expression and tumor stage (Fig. 2B) and patient survival time
(Fig. 2C) demonstrated that CENPM
was highly associated with BC among various types of cancer and
suggested that CENPM could be used as a prognostic marker for
BC.
CENPM modulates a biological network
including nine genes associated with the cell cycle in BC
Given the predictive function of CENPM, the
hypothesis that the molecule could also serve as a prognostic
marker in different types of BC was further tested using the
Kaplan-Meier Plotter database (27). As shown in Fig. 3A, higher CENPM mRNA expression
levels in patients with luminal A-type BC (339 patients, P=0.0052)
were highly associated with lower survival rates. However, the
expression levels of CENPM did not affect survival rate in luminal
B-type (Fig. 3B; 77 patients,
P=0.18), HER2-positive-type (Fig.
3C; 115 patients, P=0.33) or basal-like-type BC (Fig. 3D; 107 patients, P=0.71).
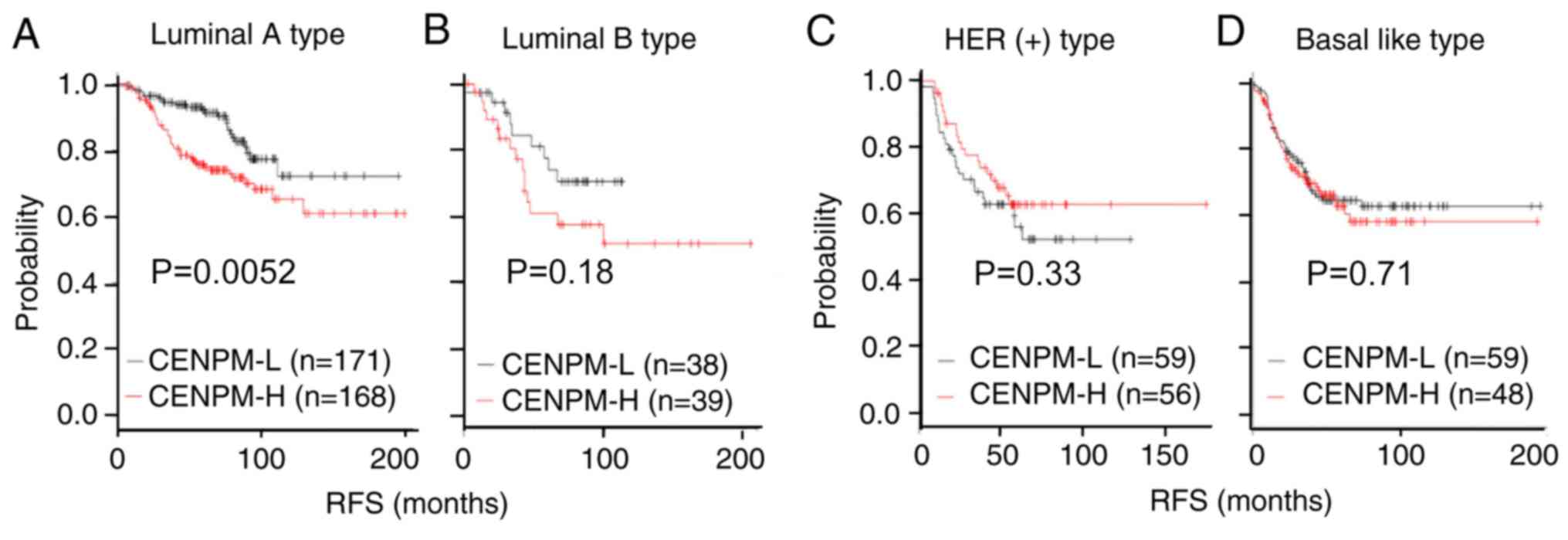 | Figure 3.CENPM mRNA expression is associated
with patient survival rate in breast cancer. Kaplan-Meier plotter
analysis of (A) 339 patients with ER-positive, PR-positive and
HER2-negative luminal A-type; (B) 77 patients with ER-positive,
PR-negative and HER2-negative luminal B-type; (C) 115 patients with
HER positive-type; and (D) 107 patients with basal-like-type breast
cancer, who had high or low CENPM mRNA expression. CENPM,
centromere protein M; ER, estrogen receptor; PR, progesterone
receptor; HER2, human epidermal growth factor receptor 2; -L, low;
-H, high; RFS, regression-free survival. |
To accurately determine the biological role of
CENPM, 1,199 genes determined to be associated with CENPM by
Spearman's rank correlation coefficient analysis (Spearman >0.4,
P<0.001) of 815 invasive breast carcinomas and one normal
specimen were selected from the cBioPortal database (19). A circular correlation network (data
not shown) with these genes was drawn that included 399 nodes and
11,495 edges according to the STRING database. Genes with degrees
of connectivity (as calculated with CytoHubba in Cytoscape) >100
were selected and it was identified that the mRNA expression levels
of RAD51 (Fig. 4A), KIF4A
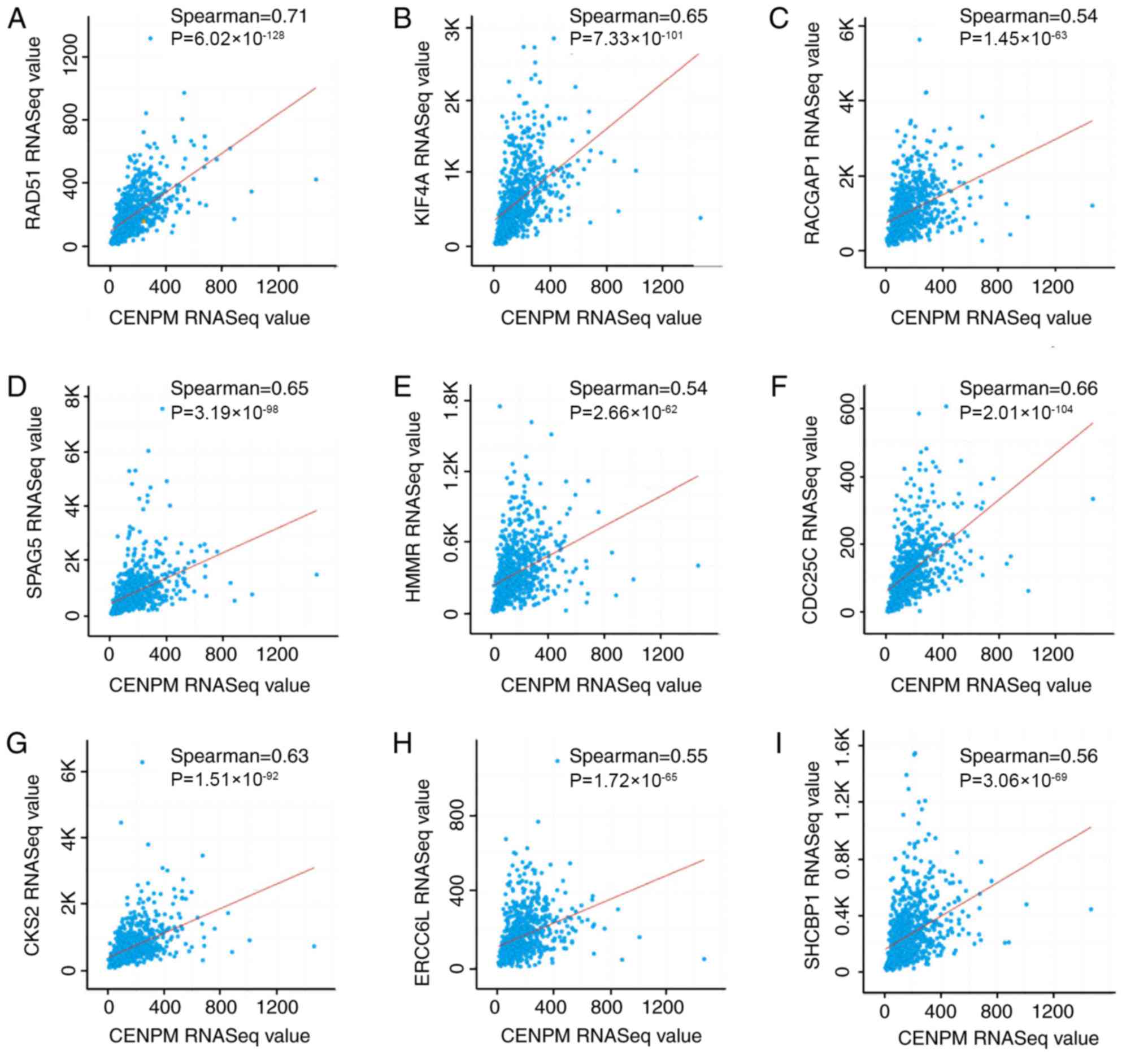
(Fig. 4B), RACGAP1 (Fig. 4C), SPAG5 (Fig. 4D), HMMR (Fig. 4E), CDC25C (Fig. 4F), CKS2 (Fig. 4G), ERCC6L (Fig. 4H) and SHCBP1 (Fig. 4I) were positively correlated with
those of CENPM in BRCA. In addition, these nine genes were
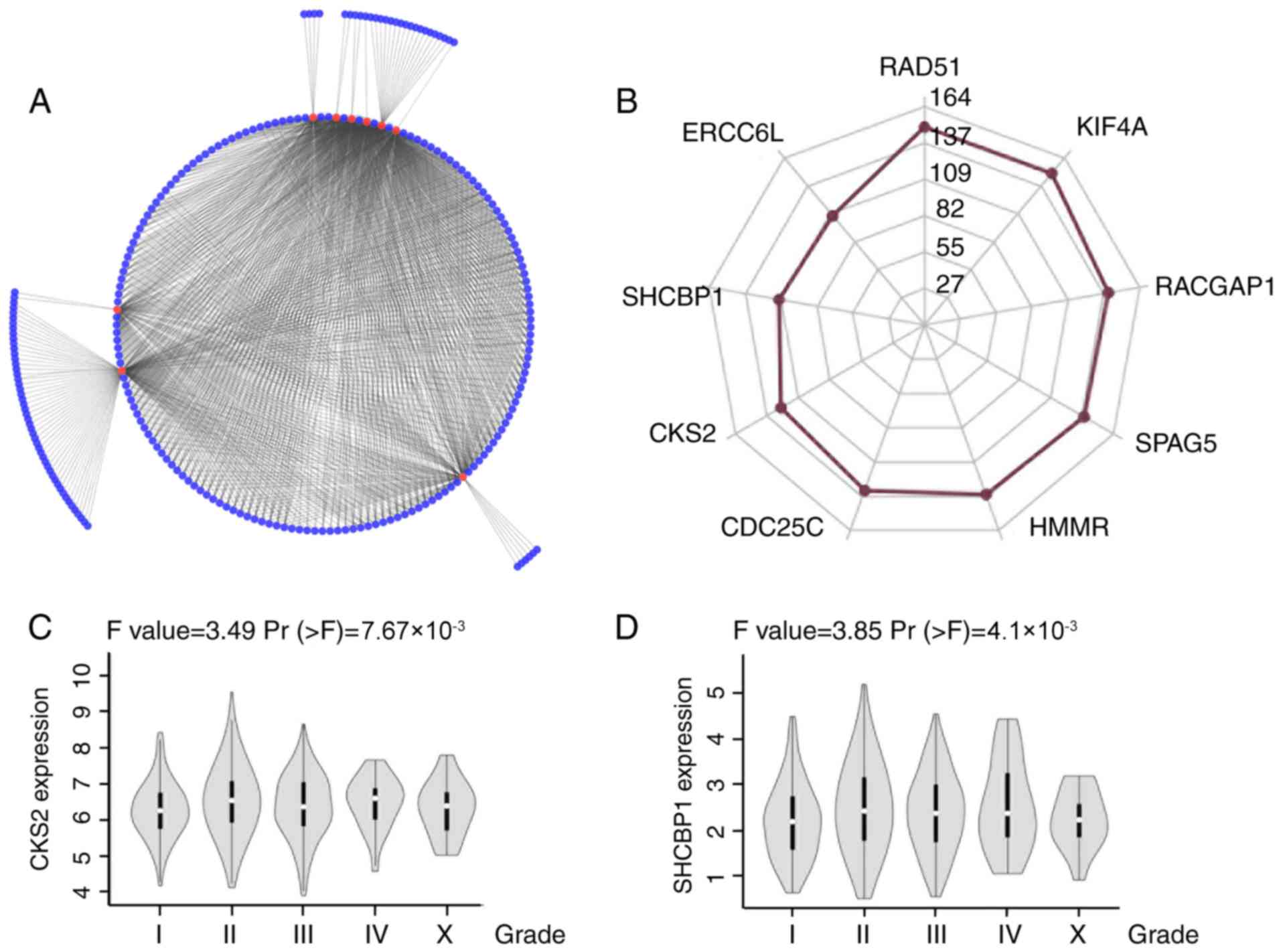
connected by 207 of 399 nodes (Fig. 5A
and B), suggesting the critical functions of these nine genes
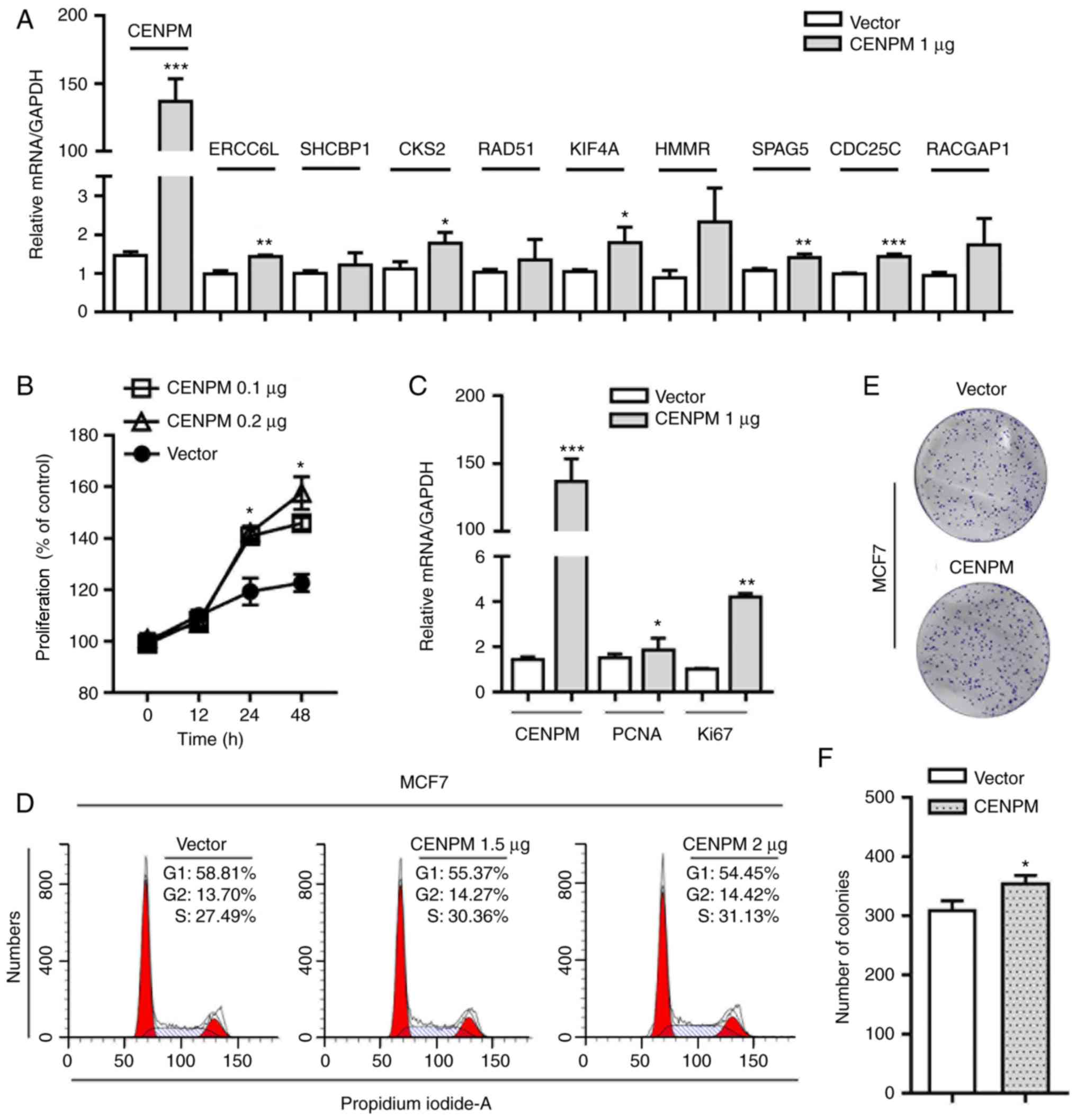
in the correlation network. Consistent with these findings, RT-qPCR
analysis revealed that ERCC6L, CKS2, KIF4A, SPAG5 and CDC25C mRNA
expression levels were significantly increased in MCF7 cells
transfected with a CENPM overexpression vector (Fig. 6A); the protein expression levels of
CKS2 were also identified to be increased in MCF7 cells with
elevated CENPM expression (Fig.
S1). However, the mRNA expression levels of SHCBP1, RAD51, HMMR
and RACGAP1 were only slightly and not significantly upregulated in
CENPM-overexpressing MCF7 cells (P>0.05; Fig. 6A).
The results of biological experiments and TCGA
database analyses confirmed that these nine genes were commonly
upregulated in BC vs. normal samples; in addition, they were
associated with tumor grade or mortality and involved in DNA
replication, DNA repair or chromosome segregation (Table II). Next, a positive association
was demonstrated between CKS2/SHCBP1 mRNA expression and tumor
grade, which, to the best of our knowledge, has not previously been
reported (Fig. 5C and D). These
findings indicated that CENPM could serve as a prognostic marker in
luminal A-type BC and may regulate tumorigenesis by altering cell
cycle-associated proteins.
 | Table II.The nine critical genes significantly
connected with centromere protein M. |
Table II.
The nine critical genes significantly
connected with centromere protein M.
| Gene | Degree | Kaplan-meier
survival P-value in BRCA | Function |
|---|
| RAD51 | 149 | 0.0045 | Participates in
homologous strand exchange, an important process in DNA repair |
| KIF4A | 149 | 0.036 | Translocates
transcription factor pcr1 to the plus ends of interdigitating
spindle microtubules during the metaphase to anaphase
transition |
| RACGAP1 | 140 | 0.00095 | Controls
cytokinesis by forming the central spindling complex and mediates
the rho-dependent signaling required for actomyosin contractile
ring assembly |
| SPAG5 | 138 | 0.019 | Essential component
of the mitotic spindle required for normal chromosome segregation
and progression into anaphase |
| HMMR | 135 | 0.027 | Forms a complex
with BRCA, which is a key gene associated with G2 to M transition
in breast cancer |
| CDC25C | 132 | 0.03 | Triggers mitosis in
the cell cycle |
| CKS2 | 124 | 0.012 | Promotes cell cycle
progression by triggering degradation of p27 |
| SHCBP1 | 111 | 0.031 | Serves a role in
signaling pathways to govern cellular proliferation |
| ERCC6L | 107 | 0.00071 | Acts as an
essential component of the spindle assembly checkpoint |
Upregulation of CENPM enhances
tumorigenesis in vitro
To study the precise effects of CENPM on BC, a CENPM
overexpression plasmid was constructed and CENPM mRNA expression
levels were detected in MCF7 cells transfected with a control
vector vs. the CENPM overexpression vector (Fig. 6A). MCF7 cells (Fig. 6B) and T-47D cells (Fig. S2) transfected with the CENPM
overexpression vector proliferated more quickly than the normal
control cells, as determined by CCK-8 assay. Conversely, MCF7 cells
transfected with CENPM shRNA proliferated more slowly than control
cells (Fig. S3A and B). Next
investigated were the proliferation-associated gene markers,
including Ki67 and proliferating cell nuclear antigen (PCNA), with
RT-qPCR assays, which demonstrated that the mRNA expression levels
of Ki67 and PCNA were elevated in CENPM-overexpressing MCF7 cells
(Fig. 6C). Considering that the
expression of numerous cell cycle-related genes was identified to
be positively associated with CENPM expression, it was hypothesized
that CENPM may enhance proliferation by affecting cells in mitosis.
The fluorescence-activated cell sorting results (Fig. 6D) demonstrated that the proportion
of MCF7 cells in S phase was elevated and that the proportion of
cells in G1 phase was decreased following transfection
with the CENPM overexpression vector, supporting the hypothesis
that CENPM accelerated the progression of cells through
G1 phase to promote tumor growth.
Since tumorigenesis and migratory ability are key
factors associated with tumor malignancy (28), a colony formation assay was
conducted to assess the tumorigenicity of CENPM. As shown in
Fig. 6E and 6F, CENPM-overexpressing MCF7 cells formed
significantly more colonies compared with control vector-expressing
MCF7 cells. However, the wound-healing assay demonstrated that
CENPM had little effect on the migration of MCF7 cells; and
migration rate was ~20% in both groups (Fig. S4).
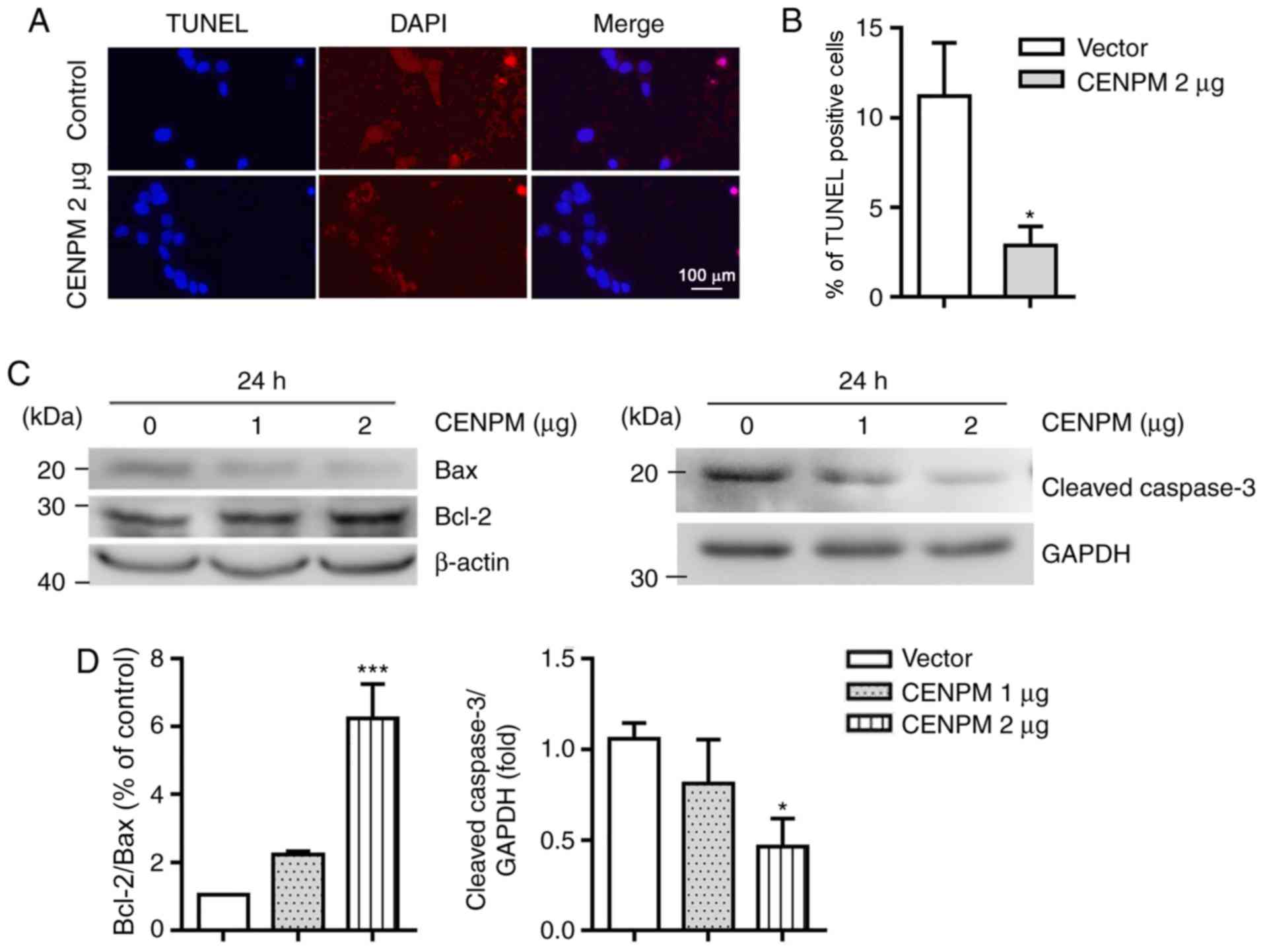
As imbalance between cell division and apoptosis
leads to tumorigenesis, it was hypothesized that investigation of
the role of CENPM in apoptosis could further elucidate the precise
function of CENPM in BC. To test this hypothesis, a TUNEL assay was
conducted and the results revealed that the amount of DNA damage
was decreased in CENPM-overexpressing MCF7 cells (Fig. 7A and B). In addition, western blot
analysis with anti-Bcl-2, anti-Bax and anti-cleaved caspase-3
antibodies further demonstrated the inhibitory role of CENPM in
apoptosis (Fig. 7C and D).
Therefore, overexpression of CENPM in MCF7 cells accelerated the
development of a BC-like state by interfering with proliferation
and apoptosis in vitro.
CENPM gene mutations
The COSMIC database (v89), was used to evaluate
whether the CENPM gene was substantially mutated among various
types of human cancer. The expression of CENPM was examined in
47,818 patient tissues representing 39 distinct types of cancer
from inception to May 15, 2019. Of the 39 cancer types, 19 were
associated with CENPM gene mutations, primarily 37 point mutations
(total percentage of mutated, 0.077%), and two frameshift deletions
(total percentage of mutated, 0.004%), and the corresponding
specimens were further examined. No insertions or complex mutations
were identified in the samples (Table III). Among the CENPM gene point
mutations, 19 were missense substitutions, and 18 were synonymous
substitutions with no changes in the protein-coding sequence. Point
mutations occurred in the skin (gene mutation rate, 0.54%), large
intestine (gene mutation rate, 0.39%) and endometrium (gene
mutation rate, 0.42%). Overall, considering the low gene mutation
rates, CENPM gene sequence changes may not be the main reasons for
tumorigenesis.
 | Table III.Centromere protein M gene mutations
among 47,818 patients with cancer (Catalogue of Somatic Mutations
in Cancer database). |
Table III.
Centromere protein M gene mutations
among 47,818 patients with cancer (Catalogue of Somatic Mutations
in Cancer database).
| Mutation type | Number | Ratio (%) |
|---|
| Nonsense
substitution | 0 | 0 |
| Missense
substitution | 19 | 0.039 |
| Synonymous
substitution | 18 | 0.038 |
| Inframe
insertion | 0 | 0 |
| Frameshift
insertion | 0 | 0 |
| Inframe
deletion | 0 | 0 |
| Frameshift
deletion | 2 | 0.004 |
| Complex
mutation | 0 | 0 |
Discussion
CENPM gene expression has been reported to be
associated with tumorigenesis in primary melanoma, bladder cancer,
hepatocellular carcinoma, and head and neck squamous cell
carcinoma. The present study analyzed the association of 13 types
of cancer (bladder, brain, breast, cervical, esophageal, gastric,
liver, lung, ovarian and pancreatic cancer, and lymphoma, melanoma
and sarcoma) with ectopic CENPM mRNA expression, and identified
that CENPM protein expression was upregulated in patients with BRCA
and CESC. In addition, it was revealed that CENPM mRNA expression
levels were positively associated with tumor grade in BRCA, LIHC
and LUAD, and with mortality in BRCA, CESC, LIHC and LUAD. Based on
the BC cell types of origin, the critical role of CENPM was further
confirmed in luminal A-type BC mortality. In general, the present
study implied that exploration of CENPM expression may enhance the
diagnostic accuracy and survival probability in patients with
cancer, particularly in patients with BC.
The protein interaction network identified nine key
genes [RAD51 (29), KIF4A
(30), RACGAP1 (31), SPAG5 (32), HMMR (33), CDC25C (34), CKS2 (35), SHCBP1 (36) and ERCC6L (37)] that interacted strongly with CENPM
mRNA. Subsequently, the functions of these nine genes (38–46)
were profiled in various types of cancer and it was identified that
all of them have been reported to accelerate the progression of
cancer, including BC and hepatocellular carcinoma (29–37).
RT-qPCR demonstrated that the expression levels of five of the nine
genes, ERCC6L, CKS2, KIF4A, SPAG5 and CDC25C, were increased in
MCF7 cells with elevated CENPM expression, implying that CENPM may
promote tumor progression by controlling the expression of these
five genes. Notably, normal cells utilize CDC25C to remove the
inhibitory phosphates and trigger the G2-prophase
transition (47), and utilize
SPAG5 (32), KIF4A (48), ERCC6L (49) or CKS2 (50) to properly position chromosomes
during metaphase. Of the nine genes, four (SHCBP1, RAD51, HMMR and
RACGAP1), were identified as being associated with CENPM through
cBioPortal correlation analysis; however, the mRNA expression
levels of these genes remained unchanged in CENPM-overexpressing
MCF7 cells. Further exploration is required to elucidate the
mechanism by which CENPM can regulate the expression of ERCC6L,
CKS2, KIF4A, SPAG5 and CDC25C in mitosis, and to confirm whether
the CENPM protein interacts with the SHCBP1, RAD51, HMMR and
RACGAP1 proteins. In addition, the present study demonstrated that
CENPM-overexpression promoted cell cycle progression in human BC
cells, and decreased the apoptotic rate, as calculated by TUNEL
assay. Taken together, these results indicated that ectopic CENPM
expression may accelerate the development of a BC-like state and
control mitosis by regulating the expression of KIF4A, RECC6L,
CKS2, CDC25C and SPAG5, and affect apoptosis in BC cells.
In order to explain why CENPM expression was
upregulated in patients with BRCA, point mutations, insertions,
deletions or complex mutations of CENPM were identified in the
present study; calculating the CENPM gene mutation rates among
various types of cancer may be helpful for revealing the precise
function of CENPM. Based on the low CENPM mutation rates, the
present study suggested that CENPM gene sequence changes (point
mutations) may not be the main reason underlying its effects on
tumorigenesis. Although mutations in proto-oncogenes or tumor
suppressor genes may be major drivers of carcinogenesis, the
epigenetic modification (including DNA methylation, acetylation,
phosphorylation and ubiquitylation) of CENPM may still serve a
critical role in gene expression regulation, which is also
considered an important factor for tumorigenesis (51). Thus, it is suggested that further
experiments should be conducted to explain which pathway
contributes to the upregulation of CENPM in BC cells.
In conclusion, the findings of the present study
indicated that upregulation of CENPM may enhance BC progression via
numerous mechanisms, including increasing upregulation of genes
linked to cell proliferation and reducing cell apoptosis. The
present study revealed, for the first time to the best of our
knowledge, that CENPM may be a novel therapeutic target for BC
progression.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81960655) to Y Liu,
and the Guizhou Medical University start-up fund for doctoral
talent [grant no. J-(2018)014].
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YL and WY performed the experiments; YL wrote and
revised the manuscript; PR and TZ analyzed the data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CENPM
|
centromere protein M
|
|
BC
|
breast cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
BRCA
|
breast invasive carcinoma
|
|
CESC
|
cervical squamous cell carcinoma and
endocervical adenocarcinoma
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
SARC
|
sarcoma
|
|
DLBC
|
diffuse large B-cell lymphoma
|
|
LUAD
|
lung carcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
References
|
1
|
Smith RA, Andrews KS, Brooks D, Fedewa SA,
Manassaram-Baptiste D, Saslow D and Wender RC: Cancer screening in
the United States, 2019: A review of current American cancer
society guidelines and current issues in cancer screening. CA
Cancer J Clin. 69:184–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vuong D, Simpson PT, Green B, Cummings MC
and Lakhani SR: Molecular classification of breast cancer. Virchows
Arch. 465:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li G, Cottier M, Sabido O, Gentil-Perret
A, Lambert C, Passebosc-Faure K, Genin C and Tostain J: The in vivo
DNA aneuploidization during expansion of conventional renal cell
carcinoma. In vivo. 16:341–344. 2002.PubMed/NCBI
|
|
4
|
Wu Q, Li B, Liu L and Sun S and Sun S:
Centrosome dysfunction: A link between senescence and tumor
immunity. Signal Transduct Target Ther. 5:1072020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Howman EV, Fowler KJ, Newson AJ, Redward
S, MacDonald AC, Kalitsis P and Choo KH: Early disruption of
centromeric chromatin organization in centromere protein A (Cenpa)
null mice. Proc Natl Acad Sci USA. 97:1148–1153. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meluh PB and Koshland D: Evidence that the
MIF2 gene of saccharomyces cerevisiae encodes a centromere protein
with homology to the mammalian centromere protein CENP-C. Mol Biol
Cell. 6:793–807. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu ST, Hittle JC, Jablonski SA, Campbell
MS, Yoda K and Yen TJ: Human CENP-I specifies localization of
CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis.
Nat Cell Biol. 5:341–345. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukagawa T, Mikami Y, Nishihashi A,
Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W
and Ikemura T: CENP-H, a constitutive centromere component, is
required for centromere targeting of CENP-C in vertebrate cells.
EMBO J. 20:4603–4617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Basilico F, Maffini S, Weir JR, Prumbaum
D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van
Gerwen S, et al: The pseudo GTPase CENP-M drives human kinetochore
assembly. eLife. 3:e029782014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Foltz DR, Jansen LE, Black BE, Bailey AO,
Yates JR III and Cleveland DW: The human CENP-A centromeric
nucleosome- associated complex. Nat Cell Biol. 8:458–469. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Wu F, Shi Y, Yang D, Xu M, Lai Y
and Liu Y: Identification of key candidate genes involved in
melanoma metastasis. Mol Med Rep. 20:903–914. 2019.PubMed/NCBI
|
|
12
|
Kim WT, Seo SP, Byun YJ, Kang HW, Kim YJ,
Lee SC, Jeong P, Song HJ, Choe SY, Kim DJ, et al: The anticancer
effects of garlic extracts on bladder cancer compared to cisplatin:
A common mechanism of action via centromere protein M. Am J Chin
Med. 46:689–705. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Z, Wang R, Chen F, Wang J and Huang X:
Five novel oncogenic signatures could be utilized as AFP-related
diagnostic biomarkers for hepatocellular carcinoma based on
next-generation sequencing. Dig Dis Sci. 63:945–957. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prystowsky MB, Adomako A, Smith RV,
Kawachi N, McKimpson W, Atadja P, Chen Q, Schlecht NF, Parish JL,
Childs G and Belbin TJ: The histone deacetylase inhibitor LBH589
inhibits expression of mitotic genes causing G2/M arrest and cell
death in head and neck squamous cell carcinoma cell lines. J
Pathol. 218:467–477. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang PJ, Chiu LY, Lee CC, Yeh YM, Huang
KY, Chiu CH and Tang P: mSignatureDB: A database for deciphering
mutational signatures in human cancers. Nucleic Acids Res.
46(D1):D964–D970. 2018. View Article : Google Scholar
|
|
16
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45(D1):D777–D783. 2017. View Article : Google Scholar
|
|
17
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45(W1):W98–W102. 2017. View Article : Google Scholar
|
|
19
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: Comprehensive molecular portraits of invasive lobular breast
cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and prognosis of topoisomerase isoforms in
non-small-cell lung cancer by using oncomine and kaplan-meier
plotter. PLoS One. 12:e01745152017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lackler KP, Cochran DL, Hoang AM, Takacs V
and Oates TW: Development of an in vitro wound healing model for
periodontal cells. J Periodontol. 71:226–237. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moore CB, Guthrie EH, Huang MT and Taxman
DJ: Short hairpin RNA (shRNA): Design, delivery, and assessment of
gene knockdown. Methods Mol Biol. 629:141–158. 2010.PubMed/NCBI
|
|
27
|
Gyorffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mierke CT: The matrix environmental and
cell mechanical properties regulate cell migration and contribute
to the invasive phenotype of cancer cells. Rep Prog Phys.
82:0646022019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ward A, Khanna KK and Wiegmans AP:
Targeting homologous recombination, new pre-clinical and clinical
therapeutic combinations inhibiting RAD51. Cancer Treat Rev.
41:35–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang
Y, Deng L, Lu Q and Luo S: FOXM1 promotes hepatocellular carcinoma
progression by regulating KIF4A expression. J Exp Clin Cancer Res.
38:1882019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang MY, Chen DP, Qi B, Li MY, Zhu YY, Yin
WJ, He L, Yu Y, Li ZY, Lin L, et al: Pseudogene RACGAP1P activates
RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to
promote hepatocellular carcinoma early recurrence. Cell Death Dis.
10:4262019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li M, Li A, Zhou S, Lv H and Yang W: SPAG5
upregulation contributes to enhanced c-MYC transcriptional activity
via interaction with c-MYC binding protein in triple-negative
breast cancer. J Hematol Oncol. 12:142019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai Y, Sheng Z, Chen Y and Wang J: LncRNA
HMMR-AS1 promotes proliferation and metastasis of lung
adenocarcinoma by regulating MiR-138/sirt6 axis. Aging.
11:3041–3054. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giono LE, Resnick-Silverman L, Carvajal
LA, St Clair S and Manfredi JJ: Mdm2 promotes Cdc25C protein
degradation and delays cell cycle progression through the G2/M
phase. Oncogene. 36:6762–6773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grey W, Ivey A, Milne TA, Haferlach T,
Grimwade D, Uhlmann F, Voisset E and Yu V: The Cks1/Cks2 axis
fine-tunes Mll1 expression and is crucial for MLL-rearranged
leukaemia cell viability. Biochim Biophys Acta Mol Cell Res.
1865:105–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Yang Y, Liu S, Tao T, Cai J, Wu J,
Guan H, Zhu X, He Z, Li J, et al: EGF-induced nuclear localization
of SHCBP1 activates β-catenin signaling and promotes cancer
progression. Oncogene. 38:747–764. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang G, Yu Z, Fu S, Lv C, Dong Q, Fu C,
Kong C and Zeng Y: ERCC6L that is up-regulated in high grade of
renal cell carcinoma enhances cell viability in vitro and promotes
tumor growth in vivo potentially through modulating MAPK signalling
pathway. Cancer Gene Ther. 26:323–333. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hannabuss J, Lera-Ramirez M, Cade NI,
Fourniol FJ, Nédélec F and Surrey T: Self-organization of minimal
anaphase spindle midzone bundles. Curr Biol. 29:2120–2130.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Breznau EB, Semack AC, Higashi T and
Miller AL: MgcRacGAP restricts active RhoA at the cytokinetic
furrow and both RhoA and Rac1 at cell-cell junctions in epithelial
cells. Mol Biol Cell. 26:2439–2455. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mack GJ and Compton DA: Analysis of
mitotic microtubule-associated proteins using mass spectrometry
identifies astrin, a spindle-associated protein. Proc Natl Acad Sci
USA. 98:14434–14439. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gruber J, Harborth J, Schnabel J, Weber K
and Hatzfeld M: The mitotic-spindle-associated protein astrin is
essential for progression through mitosis. J Cell Sci.
115:4053–4059. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maxwell CA, Benítez J, Gómez-Baldó L,
Osorio A, Bonifaci N, Fernández-Ramires R, Costes SV, Guinó E, Chen
H, Evans GJR, et al: Interplay between BRCA1 and RHAMM regulates
epithelial apicobasal polarization and may influence risk of breast
cancer. PLoS Biol. 9:e10011992011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cho YC, Park JE, Park BC, Kim JH, Jeong
DG, Park SG and Cho S: Cell cycle-dependent Cdc25C phosphatase
determines cell survival by regulating apoptosis signal-regulating
kinase 1. Cell Death Differ. 22:1605–1617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Frontini M, Kukalev A, Leo E, Ng YM,
Cervantes M, Cheng CW, Holic R, Dormann D, Tse E, Pommier Y and Yu
V: The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2
activity in maintaining replicative fidelity and neurodevelopment.
Dev Cell. 23:356–370. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schmandt R, Liu SK and McGlade CJ: Cloning
and characterization of mPAL, a novel Shc SH2 domain-binding
protein expressed in proliferating cells. Oncogene. 18:1867–1879.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Baumann C, Körner R, Hofmann K and Nigg
EA: PICH, a centromere-associated SNF2 family ATPase, is regulated
by Plk1 and required for the spindle checkpoint. Cell. 128:101–114.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shen Y, Sherman JW, Chen X and Wang R:
Phosphorylation of CDC25C by AMP-activated protein kinase mediates
a metabolic checkpoint during cell-cycle G2/M-phase transition. J
Biol Chem. 293:5185–5199. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Poser E, Caous R, Gruneberg U and Barr FA:
Aurora A promotes chromosome congression by activating the
condensin-dependent pool of KIF4A. J Cell Biol. 219:e2019051942019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yen TJ: Polo delivers a PICH to the
kinetochore. Cell. 128:20–21. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Spruck CH, de Miguel MP, Smith AP, Ryan A,
Stein P, Schultz RM, Lincoln AJ, Donovan PJ and Reed SI:
Requirement of Cks2 for the first metaphase/anaphase transition of
mammalian meiosis. Science. 300:647–650. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang T, Gong Y, Meng H, Li C and Xue L:
Symphony of epigenetic and metabolic regulation-interaction between
the histone methyltransferase EZH2 and metabolism of tumor. Clin
Epigenetics. 12:722020. View Article : Google Scholar : PubMed/NCBI
|