Introduction
Diabetic nephropathy (DN) is becoming the most
pervasive complication for both types I and II diabetes, and is
also the major cause of end-stage renal disease in the majority of
developed and developing countries (1). Clinically, DN is characterized by
mesangial hypertrophy, mesangial cell (MC) proliferation,
albuminuria, extracellular matrix (ECM) accumulation and kidney
fibrosis (2,3). It has been recognized that the
glomerular MCs and ECM accumulation are involved in a variety of
biological events (4). A recent
study suggested that high concentration glucose-induced MCs
contribute to the initiation and development of kidney fibrosis
with an elevated level of fibroin and cytokines expression
(5). Thus, it is of utmost
importance to investigate the key targets and mechanisms of action
underlying the action of MCs in high glucose for the treatment and
prevention strategies of DN.
Studies have also begun to demonstrate the
participation and regulatory mechanism of non-coding RNAs (ncRNAs)
on DN development (6,7). Circular RNAs (circRNAs) are a novel
class of endogenous ncRNAs, which mediate diverse physiological and
pathological changes in the human body (8), including DN, and are characterized by
a covalently-closed loop formed by back-splicing between the 5′ to
3′ ends of the polyA tail (9,10).
Initially, circRNAs were incorrectly recognized as splicing errors
without any biological functions. There is an increasing evidence
indicating that some circRNAs play a significant role in regulating
various human diseases through a competing endogenous RNA (ceRNA)
mechanism, also known as a micro (mi)RNA sponge (5,11).
For example, Li et al (12)
revealed that overexpression of circRNA circ-0001785 promotes the
proliferative ability of osteosarcoma cells, acting as a ceRNA by
sponging miR-1200 to upregulate HOXB2. circ homeodomain interacting
protein kinase 3 (HIPK3) is upregulated in Ang II-induced cardiac
fibroblasts (CFs) and heart tissues, promoting the proliferation
and migration of CFs by acting as a miR-29b-3p sponge (13), suggesting that circRNAs may
accelerate the development of fibrosis by acting as an miRNA
sponge. Additionally, Chen et al (1) have shown that the circ LDL receptor
related protein 6 (LRP6) acts as a ‘sponge’ of miR-205 to promote
ECM-related protein synthesis in MCs by upregulating high mobility
group box 1 (HMGB1) and activating Toll-like receptor 4
(TLR4)/NF-κB pathway. Similarly, a study by Hu et al
(5) found that the circRNA_15698
/miR-185/TGF-β1 signaling pathway promoted ECM accumulation in DN
MCs. Although advances have been made into the roles of circRNAs in
human diseases, the role of more novel circRNAs on ECM accumulation
and fibrosis-associated proteins, as well as the detailed
mechanisms of actions that circRNAs play in MCs remains
unclear.
The present study demonstrated a high expression
level of mmu_circRNA_0000491 (chr13:94111710-94126034) in mice with
DN, as well as the MCs that received high concentration glucose
treatment. In addition, the present study also investigated the
involvement of the circRNA_0000491/miR-101b/TGFβ-receptor type I
(TGFβRI) axis on the ECM of DN tissue. It was hypothesized that
circRNA_0000491 worsens the accumulation of ECM of MCs through
negatively suppressing miR-101b by targeting TGFβRI.
Materials and methods
Animals
A total of three six-week-old spontaneous male
diabetic db/db mice (weight: ~18.3–20 g) and three
age-matched control littermate db/m mice (weight: ~17.6–21.4
g) were obtained from the Shanghai SLAC Laboratory Animal Co., Ltd.
All mice were housed in a pathogen-free facility under a controlled
temperature of 22±1°C, 60±5% humidity and a 12-h light and dark
cycle, with free access to water and a regular standard diet. All
the procedures were approved by the Animal Care and Welfare
Committee of Zhejiang Chinese Medical University (approval no.
ZSLL-2018-192).
circRNA sequencing
The db/m control and db/db mice were
euthanized by cervical dislocation, then the kidney cortex from the
renal tissues was isolated. Subsequently, total RNA was extracted
from the kidney cortex using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and RNA purity,
concentration and integrity were measured using a
NanoPhotometer® spectrophotometer (IMPLEN), a
Qubit® RNA assay kit in the Qubit® 2.0
Flurometer (Thermo Fisher Scientific, Inc.) and an RNA Nano 6000
Assay kit in the Bioanalyzer 2100 system (Agilent Technologies,
Inc.), respectively. circRNA microarray analysis was performed by
Novogene Co., Ltd. Briefly, a total of 5 µg RNA per sample was
applied as input material for the RNA sample preparations. Firstly,
ribosomal (r)RNA was removed using the Epicentre Ribo-zero™ rRNA
Removal kit (Epicentre; Illumina, Inc.) and rRNA free residue was
cleaned up by ethanol precipitation. Subsequently, the linear RNA
was digested with 3 units of RNase R (Epicentre; Illumina, Inc.)
per µg of RNA. The sequencing libraries were generated with a
NEBNext® Ultra™ Directional RNA Library Prep kit for
Illumina® (New England BioLabs, Inc.) according to the
manufacturer's protocol. circRNA was detected and identified using
find_circ (14) and CIRI2
(15). The differentially
expressed circRNAs were identified by utilizing the edgeR R package
(v3.31.4; http://bioconductor.org) in R (v3.6.1;
http://www.r-project.org/) with the
threshold set at a P<0.01 and |log2(fold change)|>1. To
investigate the underlying function of candidate circRNAs, UCSC
Genome Browser (http://genome.ucsc.edu/) was used to illustrate their
composition (16).
Cell culture and transfection
MES13 cells from a SV40 transgenic mouse (SV40
MES13) were acquired from the Shanghai Academy of Life Sciences,
and maintained in DMEM (Sigma-Aldrich; Merck KGaA) supplemented
with 10% FBS (Thermo Fisher Scientific, Inc.) and 1% penicillin and
streptomycin (Welgene Inc.) in a 37°C incubator with 5%
CO2. MES13 cells were stimulated with 5.5 mM D-glucose
for the control group. For the model group, the MES13 cells were
stimulated with 30 mM glucose for 48 h in order to simulate the
condition of the DN cells. In addition, small interfering RNAs
(siRNAs), scrambled siRNA (si_Circ_Control), miR-101b
mimics/inhibitor and negative controls, used for cell
transfections, were purchased from RiboBio Co., Ltd. The sequences
used were as follows: CircRNA_0000491 si_Circ_1,
5′-ATATGGACCAAGAATACCAAA−3′; circRNA_0000491 si_Circ-2,
5′-TATATGGACCAAGAATACCAA−3′; si_Circ_Control,
5′-TTCTCCGAACGTGTCACGTTT−3′; miR-101b mimic,
5′-GUACAGUACUGUGAUAGCU-3′; miR-101b inhibitor,
5′-AGCUAUCACAGUACUGUAC-3′. All the specialized siRNAs were
transfected into MES13 cells using Lipofectamine® 3000
(Gibco; Thermo Fisher Scientific, Inc.). The subsequent experiments
were performed in MES13 cells following 48 h transfection.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was obtained from kidney samples and MES13
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA using a
PrimeScript RT reagent kit (Takara Bio, Inc.). The reverse
transcription conditions were 37°C for 15 min, and reverse
transcriptase inactivation at 85°C for 5 sec. For miRNA
quantification, cDNA synthesis was performed using the Mir-X™ miRNA
First-Strand Synthesis kit (Clontech Laboratories, Inc.).
Subsequently, qPCR was performed using a CFX96 Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc.) with the
amplification conditions: Pre-denaturation at 95°C for 30 sec,
followed by 40 cycles of denaturation at 95°C for 5 sec and
annealing at 60°C for 30 sec. The dissolution curve conditions were
65°C for 0.05 sec and 95°C for 0.5 sec. The primers used for
RT-qPCR were as follows: mmu_circ_0000491 forward,
5′-TCGGCTTGCAAAGGAGAAGTC−3′ and reverse,
5′-TGCATGCTTGCTGGTGGGTA-3′; E-cadherin (E-cad) forward,
5′-CTCAAGCTCGCGGATAACCA-3′ and reverse, 5′-AATCCTGCTGCCACGATTC-3′;
vimentin (Vim) forward, 5′-TGTGACAACTGCCGTAGACC-3′ and reverse,
5′-TAGCGGCATGAAGCACTCAA-3′; Fibronectin (FN) forward,
5′-AAGAAGGGCTCGTGTGACAG-3′ and reverse, 5′-GGAAGGGTTACCAGTTGGGG-3′;
α-SMA forward, 5′-AAGAGAGGGATCCTGACGCT-3′ and reverse,
5′-CTCCAGGGAGGAAGAGGAGG-3′; Type I collagen (Col. I) forward,
5′-CGTATCACCAAACTCAGAAGATGT−3′, and reverse,
5′-AGAGCCTCTAGCTCCTTGGG-3′; Type III collagen (Col. III) forward,
5′-ATGCCCACAGCCTTCTACAC-3′ and reverse, 5′-GGGTCACCATTTCTCCCAGG-3′;
Type IV collagen (Col. IV) forward, 5′-GGTGTTCCAGGAAGAGGCTT-3′ and
reverse, 5′-CATGCCTTGGAATCCTGGGT-3′; TGFβRI forward,
5′-AGCTCCTCATCGTGTTGGTG-3′ and reverse, 5′-AAACCGACCTTTGCCAATGC-3′;
miR-101b forward, 5′-GGGCTACTGTGATAGCTAAAA−3′; and
5′-CCAGTGCAGGGTCCGAGGTA-3′, as the reverse. In addition, the
transcriptional levels of mmu_circ_0000712, mmu_circ_0000898,
novel_circ_0001857 and novel_circ_0001778 were estimated in the
kidney samples of db/db mice. The sequences and primers of
mmu_circ_0000712, mmu_circ_0000898, novel_circ_0001857 and
novel_circ_0001778 are shown in Tables SI and SII. GAPDH was used as internal control
for the circRNA and mRNA. U6 acted as an internal standard of
miR-101b. Relative quantification of expression was performed
compared with internal standard with the 2−ΔΔCq method
(17).
Western blotting
Total cellular protein samples were acquired at 4°C
in RIPA buffer (Sigma-Aldrich; Merck KGaA), then the protein
concentration was estimated using a BCA kit (Beyotime Institute of
Biotechnology). Proteins (40 µg) were separated by SDS-PAGE (10–12%
gel) and transferred onto polyvinylidene fluoride membranes (GE
Healthcare). The membranes were blocked with 5% skim milk powder
for 2 h at room temperature, and incubated overnight at 4°C with
primary antibodies against E-cadherin (1:1,000, cat. no. 14472,
Cell Signaling Technology, Inc.), vimentin (1:1,000, cat. no. 5741,
Cell Signaling Technology, Inc.), FN (1:1,000, ab268020, Abcam),
α-SMA (1:1,000, cat. no. 68463, Cell Signaling Technology, Inc.),
Type I collagen (1:1,000, cat. no. 39952, Cell Signaling
Technology, Inc.), Type III collagen (1:1,000, ab184993, Abcam),
Type IV collagen (1:1,000, cat. no. 50273, Cell Signaling
Technology, Inc.), TGFβ1 (1:1,000, sc-130348, Santa Cruz
Biotechnology, Inc.), TGFβR1 (1:2,000, ab31013, Abcam),
phosphorylated (p-)Smad3 (1:2,000, ab52903, Abcam), and Smad3
(1:2,000, ab40854, Abcam), with GAPDH serving as an internal
control (1:1,000, D190090-0200, Sangon Biotech Co., Ltd.). After
exposure to horseradish peroxidase (HRP)-labeled goat anti-rabbit
(cat. no. A24531, 1:5,000, Invitrogen; Thermo Fisher Scientific,
Inc.) or goat anti-mouse secondary antibodies (cat. no. 31432,
1:5,000, Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 2 h, the bands were visualized with enhanced
chemiluminescence (Beyotime Institute of Biotechnology) and
analyzed using an Odyssey® IR scanner (LI-COR
Biosciences).
Luciferase reporter assay
Using the starBase v3.0 database (http://starbase.sysu.edu.cn/index.php),
the interactions between circ_0000491 and miR-101b were confirmed.
starBase is an open-source platform for studying the miRNA-ncRNA
interactions from CLIP-seq, degradome-seq and RNA-RNA interactome
data (18). In addition, the mRNAs
targeted by miR-101b were retrieved from the TargetScan (version
7.2, http://www.targetscan.org/vert_72/) (19), miRDB (http://mirdb.org/) (20) and PicTar(https://pictar.mdc-berlin.de/) databases. Then, the
luciferase reporter assay was applied in MES13 cells. Briefly,
MES13 cells were plated into 24-well plates at 60% confluence.
circ_0000491/TGFβRI reporter construct (mutant or wild-type) or the
empty reporter vector psiCHECK-2 (Promega Corporation) was
co-transfected with miR-101b mimics or control mimics using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
After 48 h of transfection, the luciferase activity was measured
with a Dual-Luciferase Reporter Assay System (Promega Corporation)
according to the manufacturer's protocol. The relative reporter
activity was normalized to the ratio of firefly luciferase to
Renilla luciferase.
Statistical analysis
All data are presented as the mean ± SD. Student's
unpaired t-tests (two tailed) were used for comparisons between two
groups. The statistical difference of the luciferase reporter assay
was evaluated using a one-way ANOVA followed by Tukey's post-hoc
tests. Statistical analysis was performed using SPSS 20.0 (IBM
Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
circRNA expression profile analysis in
DN mice
In order to investigate the differentially expressed
circRNAs in the kidney tissue, the present study employed mouse
circRNA sequencing in db/m and db/db mice with DN.
Heat maps (Fig. 1A) and volcano
plots (Fig. 1B) revealed that a
total of 40 circRNAs, including 18 upregulated circRNAs and 22
downregulated circRNAs, were differentially expressed, using
P<0.01 and |log2(fold change)|>1 as a significant cut off
criterion.
Overexpression of circRNA_0000491 in
kidney tissue and high glucose-treated mouse MES13 cells
RT-qPCR was used to evaluate mmu_circ_0000712,
mmu_circ_0000898, novel_circ_0001857, circRNA_0000491 and
novel_circ_0001778 levels in 3 paired kidney tissue samples
(Fig. S1). It was found that the
transcriptional levels of mmu_circ_0000712, novel_circ_0001857 and
novel_circ_0001778 were significantly increased in the kidney
tissue of db/db mice. Furthermore, according to the UCSC
Genome Browser, mmu_circ_0000491 is produced at the Homer1 gene
locus, containing exons 2–5 (Fig.
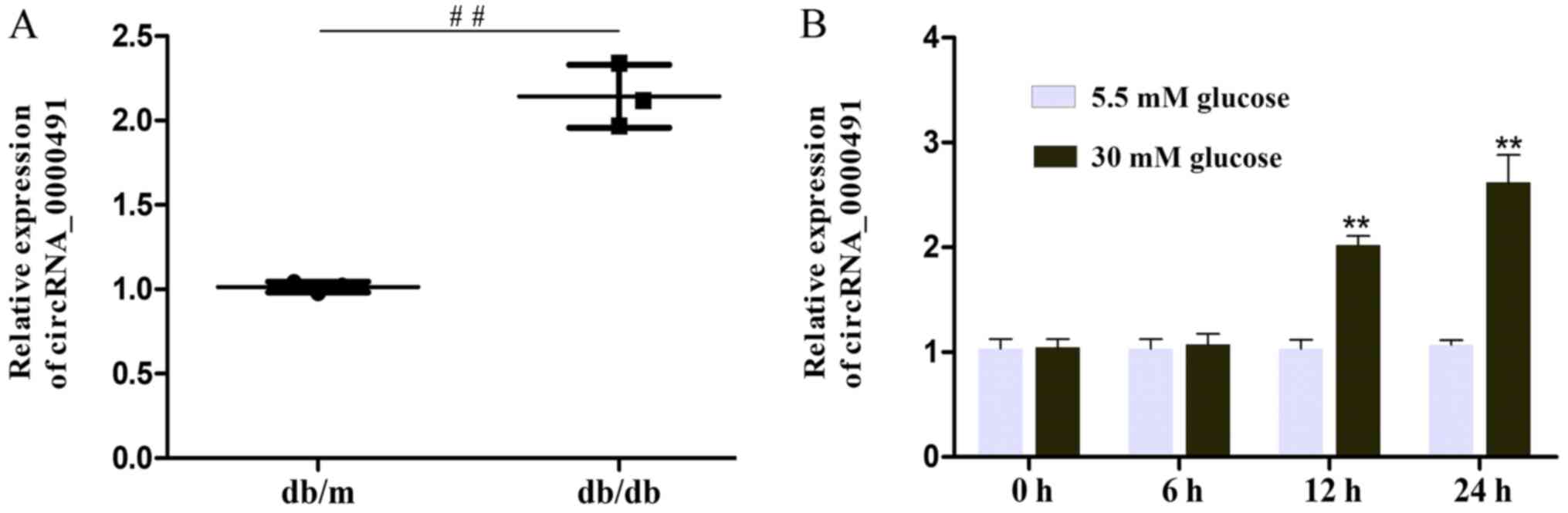
1C). The results also revealed that circRNA_0000491 expression
was markedly amplified in the db/db mice (Fig. 2A). Among the upregulated circRNAs,
it was noted that circRNA_0000491 was significantly increased;
therefore, this circRNA was selected for further experiments.
Additionally, it was observed that circ_0000491 expression was
markedly upregulated in MES13 cells exposed to high glucose for 12
and 24 h, respectively, when compared with the expression in
control cells (Fig. 2B). These
findings documented the high expression levels of circRNA_0000491
in db/db mouse kidney tissue.
Knockdown of circRNA_0000491
suppresses ECM accumulation of MES13 cells
circRNA microarray analyses identified a variety of
dysregulated circRNAs in the DN mouse kidney cortex (Fig. 1A). Subsequently, the present study
identified that circRNA_0000491 was expressed at significantly
higher levels in the kidney tissues of the db/db mice and it
was speculated that circRNA_0000491 participated in the
pathological process of ECM accumulation and renal fibrosis. In the
present study, ECM accumulation and fibrosis-associated proteins
levels, including E-cad, Vim, FN and α-SMA, as well as Col. Types
I, III and IV were investigated using RT-qPCR. The results
suggested that the mRNA expression levels of Vim, FN, α-SMA, Col.
I, III and IV were increased, while the levels of E-cad were
markedly decreased compared with control db/m mice and
normal glucose treated MES13 cells (Fig. 3A and B). The role of
circRNA_0000491 was further investigated in MES13 cells using a
specific siRNA. The knockdown efficiency of si_Circ_1 and si_Circ_2
in MES13 cells was determined by RT-qPCR analysis. The results
showed that the expression levels of circRNA_0000491 were
significantly decreased in the si_Circ_1 group compared with that
in the si_Circ_Control group. Subsequently, the present study
selected si_Circ_1 as it had a more effective knockdown efficiency
than si_Circ_2 for subsequent experiments (Fig. 3C). Moreover, the western blot
analysis revealed that the Vim, FN, α-SMA, Col. I, III and IV
protein expression levels were suppressed and that the expression
of E-cad in the si_Circ_1 treated group was observably increased
compared with the si_Circ_NC group in HG-treated MES13 cells
(Fig. 3D-K). Taken together, these
findings confirmed that circRNA_0000491 knockdown suppressed the
epithelial-to-mesenchymal transition (EMT) process and
fibrosis-associated protein synthesis in MES13 cells.
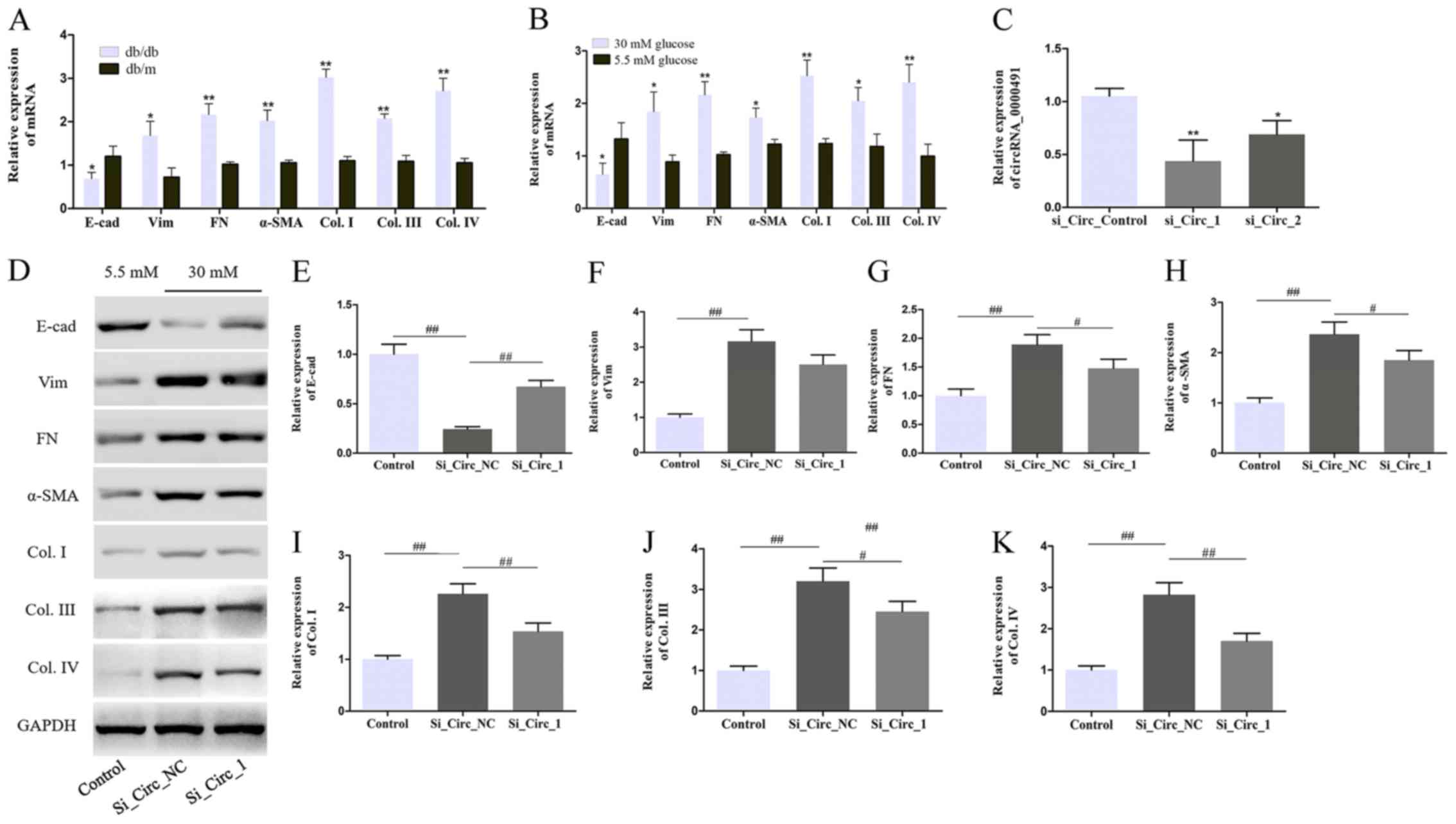 | Figure 3.Circ_0000491 silencing inhibits the
extracellular matrix accumulation and fibrosis-associated protein
synthesis in MES13 cells. RT-qPCR revealed the expression levels of
E-cad, Vim, FN, α-SMA, Col. I, Col. III and Col. IV in (A) diabetic
nephropathy mice and (B) MES13 cells treated with high glucose. (C)
The expression levels of circ_0000491 in MES13 cells transfected
using siRNA. (D) Western blotting showing the protein expression
levels of (E) E-cad, (F) Vim, (G) FN, (H) α-SMA, (I) Col. I, (J)
Col. III and (K) Col. IV in MES13 cells with high glucose treatment
and transfected with si_circ_0000491. *P<0.05, **P<0.01 vs.
the corresponding control group; #P<0.05,
##P<0.01 vs. the Si_Circ_NC group. Col., collagen;
circRNA, circular RNA; E-cad, E-cadherin; FN, fibronectin; si,
small interfering; SMA, smooth muscle actin; Vim, vimentin. |
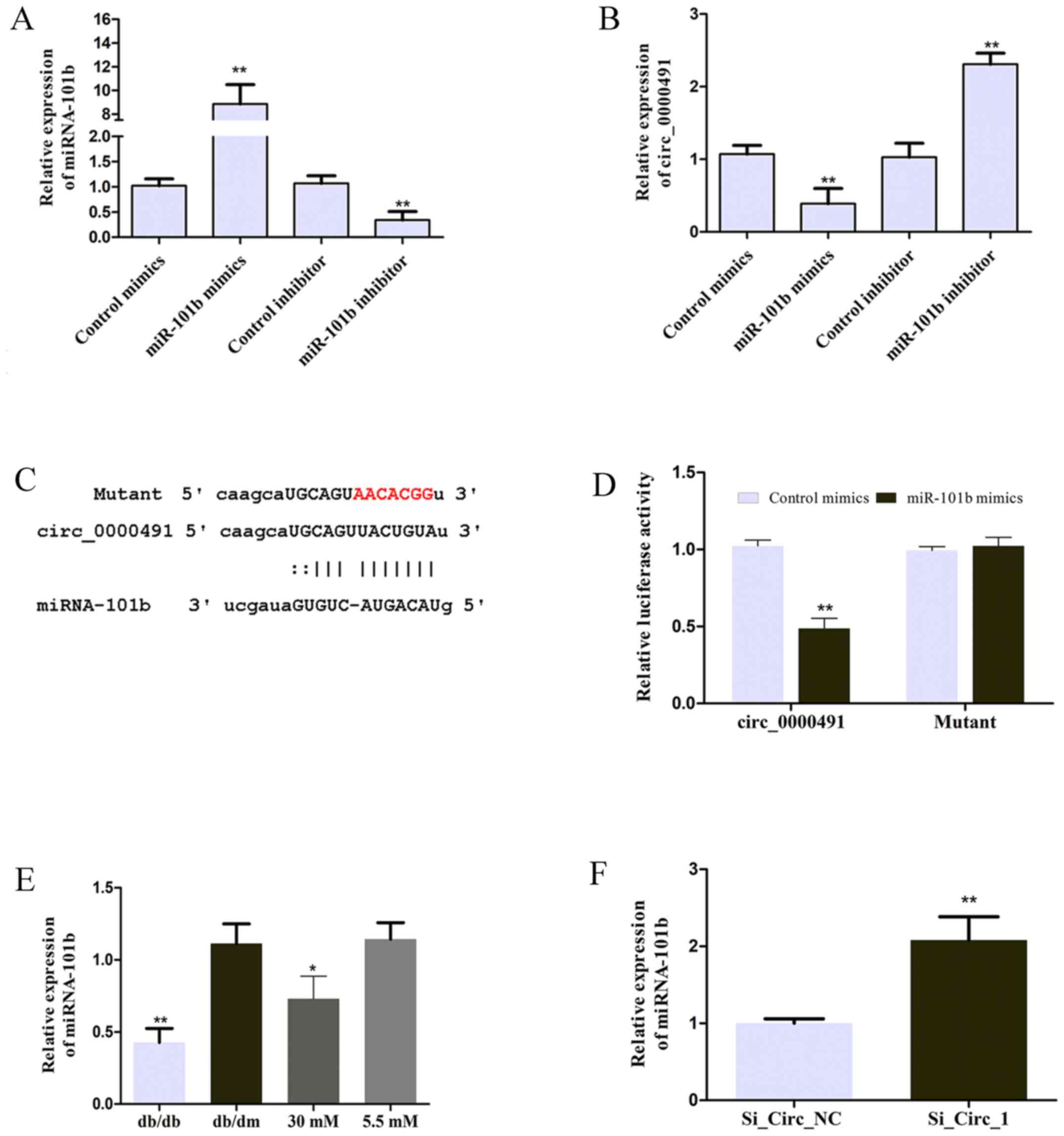
circRNA_0000491 sponges miR-101b in
MES13 cells
In the present study, MES13 cells were transfected
with miR-101b mimics or miR-101b inhibitor to achieve miR-101b
expression, as illustrated in Fig.
4A. The expression of circRNA_0000491 was negatively regulated
by miR-101b (Fig. 4B). The
suggestion that miR-101b may be sponged by circRNA_0000491 was
predicted using the starBase v3.0 platform and the seeding
sequences of circ_0000491 and miR-101b are presented in Fig. 4C. In order to validate whether
miR-101b is the direct target of circ_0000491, a luciferase
reporter assay was performed in the present study. The results
revealed that miR-101b mimics significantly inhibited the
luciferase activity mediated by wild-type circ_0000491-luciferase
reporter, which suggested an interaction between circ_0000491 and
miR-101b (Fig. 4D). In addition,
the results demonstrated that miR-101b expression was significantly
downregulated in DN mice and the high concentration glucose-treated
group (Fig. 4E) in comparison to
the control groups. Furthermore, miRNA-101b expression was
significantly increased in MES13 cells transfected with si_Circ_1
(Fig. 4F). These data suggested
that circ_0000491 negatively regulates miR-101b expression by
directly sponging miR-101b.
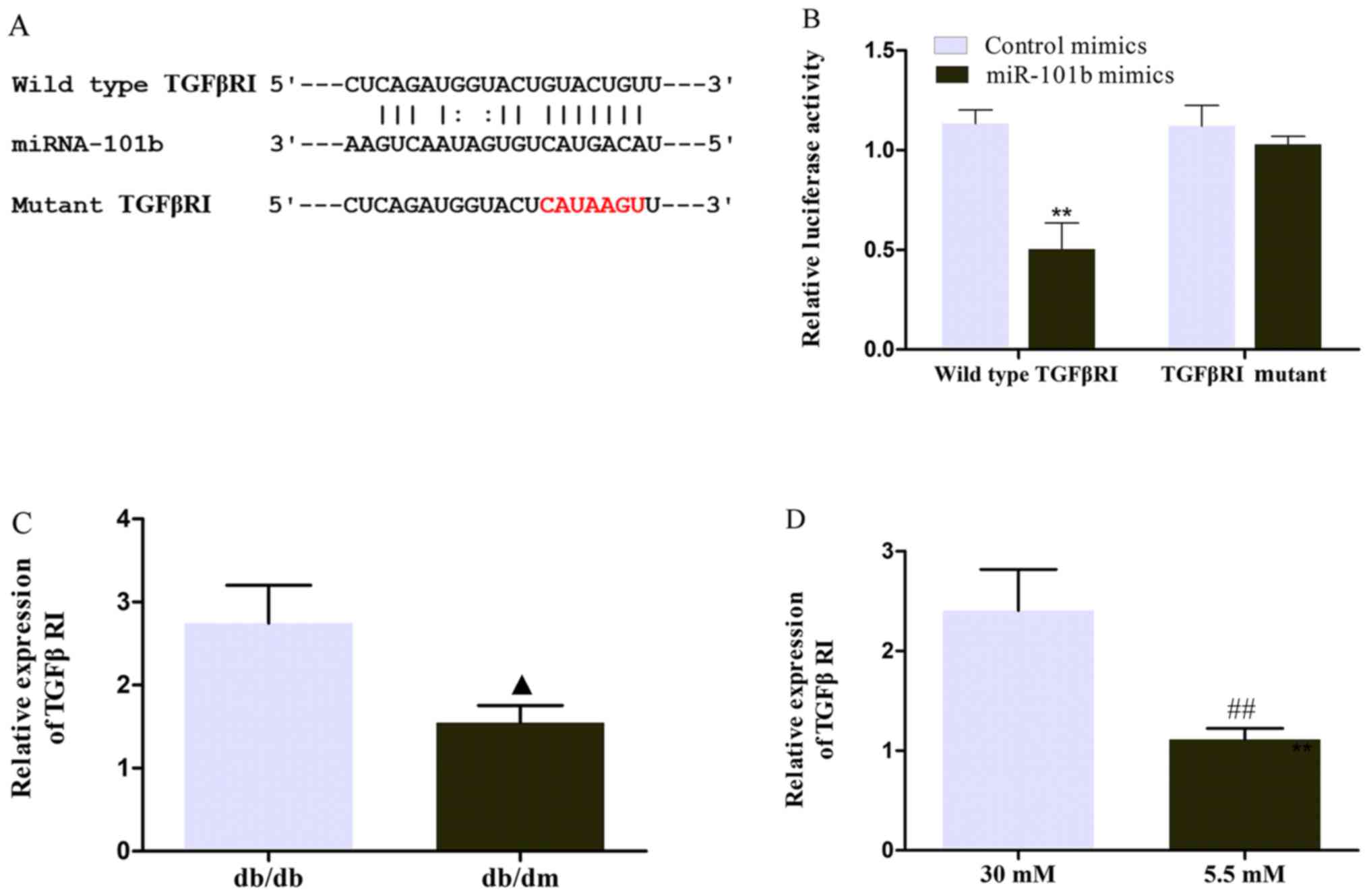
miR-101b sponges TGFβRI in MES13
cells
After confirming that circ_0000491 sponges miR-101b
in MES13 cells, it was necessary to further determine the
downstream target of the circRNA_0000491/miR-101b axis. According
to the prediction of the TargetScan, miRDB and PicTar databases,
TGFβRI may be a potential downstream target for miR-101b. The
underlying complementary binding sequence is presented in Fig. 5A. To further investigate the
association between miR-101b and TGFβRI, a luciferase reporter
assay was performed. It was found that the luciferase activity was
significantly downregulated in wild-type TGFβRI cells
co-transfected with miR-101b mimics (Fig. 5B). Furthermore, RT-qPCR revealed
that the mRNA expression levels of TGFβRI were markedly increased
in DN mice and high glucose-treated MES13 cells compared with the
corresponding control groups (Fig. 5C
and D). These data verified that TGFβRI servers as the target
of miR-101b.
miR-101b rescues the effects of
circ_0000491 in MES13 cells by targeting TGFβRI
It was hypothesized that the miR-101b/TGFβRI axis is
one-way and that circ_0000491 promotes ECM accumulation and
fibrosis in DN cells. To test this hypothesis, the present study
performed rescue experiments in which circ_0000491-knockdown
cultures underwent transfection with an miR-101b inhibitor and
scrambled control. Western blotting revealed that the protein
expression of TGFβ1, TGFβRI, phosphorylated (p)Smad3, Vim, FN and
α-SMA, as well as Col. I, III and IV were all increased; however,
E-cad decreased in the miR-101b inhibitor transfected cells. In
addition, the cotransfection of miR-101 inhibitor and si_Circ_1
reversed the effect of circ_0000491 silencing (Fig. 6A-K). Collectively, these results
suggested that circ_0000491 acts as a sponge of miR-101b,
regulating high glucose-induced ECM accumulation and the expression
levels of fibrosis-associated proteins in MCs by upregulating
TGFβRI and suppressing miR-101b.
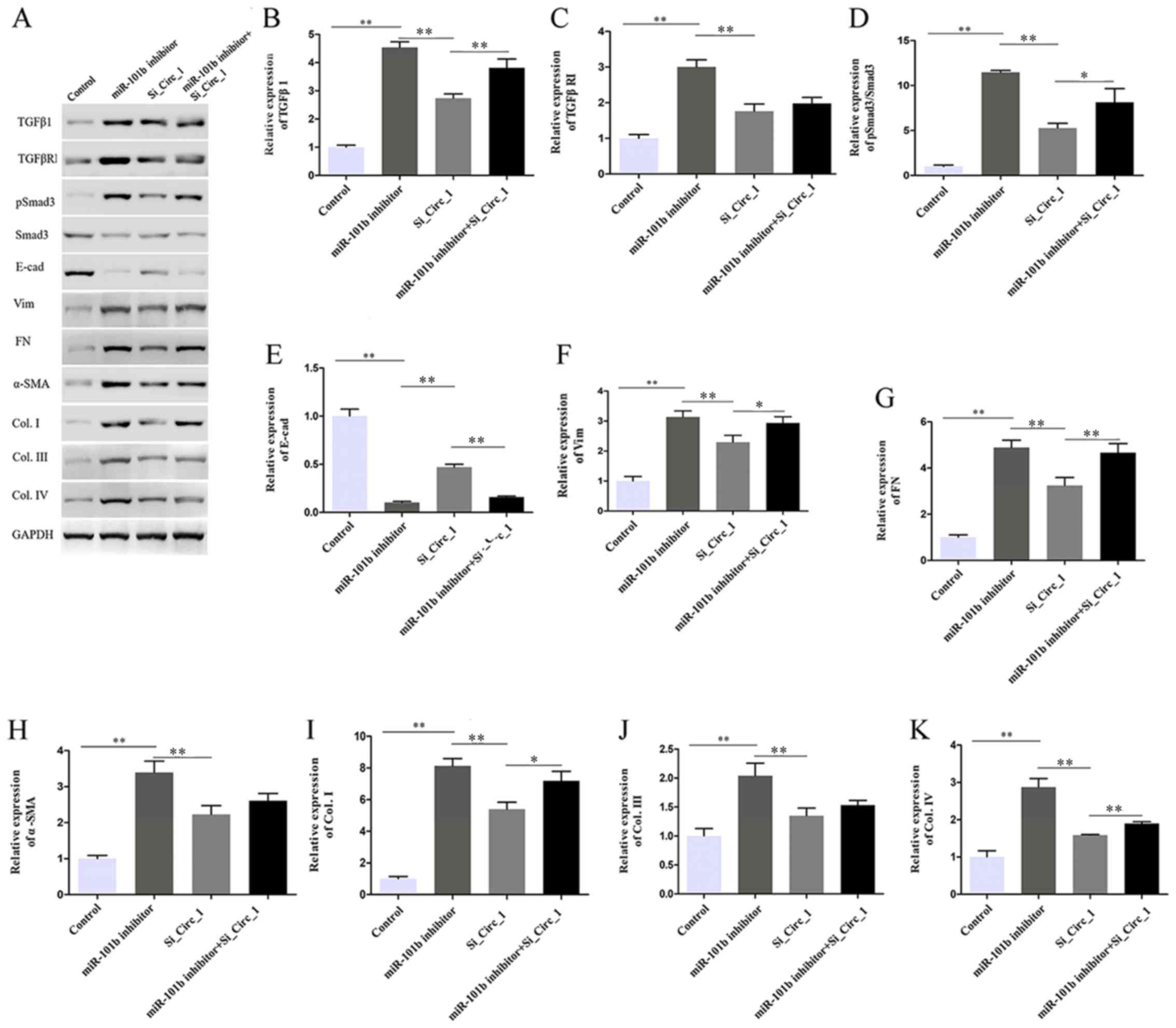 | Figure 6.Effect of circ_0000491 and miRNA-101b
inhibition on extracellular matrix and fibrosis-associated protein
expression via targeting TGFβRI. (A) Western blotting showed that
miR-101b inhibitor reversed the effect of si_circRNA_0000491 on the
protein expression levels of (B) TGFβ1, (C) TGFβRI, (D)
pSmad3/Smad3, (E) E-cad, (F) Vim, (G) FN, (H) α-SMA, (I) Col. I,
(J) Col. III and (K) Col. IV. *P<0.05, **P<0.01. Col.,
collagen; circRNA, circular RNA; E-cad, E-cadherin; FN,
fibronectin; miR/miRNA, microRNA; p, phosphorylated; si, small
interfering; SMA, smooth muscle actin; TGFβR1, TGFβ receptor 1;
Vim, vimentin. |
Discussion
DN is one of the major devastating complications of
diabetes mellitus (DM). Notably, 2 in every 10 patients with either
type 1 or type 2 DM, will develop DN after 10–20 years (21). The number of patients with DN was
382 million in 2013 and is estimated to reach 592 million by 2035
(22). It is evident that abundant
circRNAs exist in the eukaryotic transcriptomes and accumulating
evidence suggests that they may have a vital role in regulating a
series of human diseases and cellular functions (23). circRNAs are closed RNA transcripts
that are conserved among different biological systems. However, to
the best of our knowledge, the role of circRNA in DN and the
mechanisms of action by which circRNAs regulate the accumulation of
the ECM and fibrosis has not been fully described. The present
study aimed to investigate the circRNA microarray in the DN mice
model and then clarify the underlying biological characteristics of
circRNAs on the DN physiological and pathological processes.
In the present study, a subset of circRNAs were
identified through the circRNA microarray analysis. Heat maps and
volcano plots showed a total of 40 circRNAs were differentially
expressed, including 18 upregulated circRNAs and 22 downregulated
circRNAs. Among these abnormal circRNAs, the present study selected
an upregulated circRNA, circRNA_0000491, and investigated the
pathological phenotype associated with EMT, one of the known
underlying mechanisms of DN. The present study initially assessed
the potential effect of circRNA_0000491 on ECM and
fibrosis-associated protein expression, then performed
loss-of-function experiments through siRNA transfection. The
loss-of-function experiments demonstrated that compromised
circRNA_0000491 expression significantly suppressed the protein
expression levels of Vim, FN and α-SMA, as well as Col. I, III and
IV, indicating that circRNA_0000491 participated in the ECM
accumulation of the EMT process. This suggested that silencing its
expression would be conducive to decrease the synthesis of
fibrosis-associated proteins.
Accumulating studies have demonstrated that circRNAs
are crucial regulators in the process of the transmission and
function of genetic information, usually acting as miRNA sponges.
Research on circRNAs in DM is a novel research field and further
research into specific circRNA expression on the ECM is still
required, despite recent advancements. For example, Wu et al
(24) investigated
hsa_circ_0005105 expression in osteoarthritis and reported that
hsa_circ_0005105 could promote ECM degradation through sponging
miR-26a targeting NAMPT. Zhao et al (25) indicated that hsa_circ_0054633 may
be a potential biomarker and have a diagnostic capability for
pre-diabetes and type 2 DM. In CFs, circRNA_000203 specifically
sponges miR-26b-5p and overexpression of circRNA_000203 may
eliminate the anti-fibrotic effect of miR-26b-5p in CFs,
accompanied by the suppression of collagen type III α 1 chain
(Col3a1) and α-SMA (26).
Furthermore, emerging evidence has demonstrated that circRNAs may
regulate cancer cell proliferation, migration and invasion as miRNA
sponge, not only in DN. For instance, Lili and Yue (27) demonstrated that significantly
upregulated expression levels of hsa_circ_0007534 are present in
breast cancer (BC) and knockdown of hsa_circ_0007534 inhibited BC
colony formation, cell proliferation and invasion, as well as
strengthening apoptosis in BC cells by acting as a miR-593 sponge
to raise mucin 19, oligomeric (MUC19) expression levels. In gastric
cancer, the knockdown of hsa_circ_0001368 results in accelerated
tumor growth in vivo and may act as a ceRNA to sponge
miR-6506-5p and play a tumor-suppressive role (28). Liu et al (29) suggested that circ_0080425 functions
as sponge, harboring miR-24-3p, which inhibits cell proliferation
and fibrosis in DN by targeting fibroblast growth factor 11.
Furthermore, Yao et al (30) demonstrated that circ_0000285
aggravates podocyte injury through sponging miR-654-3p and
activating MAPK6 in DN. Taken together, these results provide novel
insights into circRNAs and add to the growing amount of evidence
that circRNAs can sequester miRNAs.
Mechanistically, in the present study, the
interaction between miR-101b and circRNA_0000491 was predicted
using bioinformatics analysis, and was confirmed by the luciferase
report assay. Thus, circRNA_0000491 may act as an endogenous
miR-101b sponge to promote ECM degradation. Additionally, the
target genes of miR-101b were predicted using bioinformatics
analysis. Subsequently, dual-luciferase reporter assays further
confirmed that TGFβRI serves as the target gene of miR-101b.
TGF-β1/Smad signaling is critical in the process of EMT.
Dysregulation of the signaling pathway can contribute to abnormal
ECM deposition, causing extensive kidney fibrosis (31,32).
In this pathway, TGF-β1 exerts biological effects through binding
to type II β-β receptor and subsequently recruits and activates
TGFβRI, then the activated TGFβR1 phosphorylates Smad2/3 (33,34).
Previous studies have demonstrated that inhibiting TGFβRI
significantly improves various disease, including pulmonary
fibrosis (35), hypertensive
nephropathy (36) and
tubulo-interstitial fibrosis (37), suggesting that inhibiting TGFβRI
may be a promising anti-fibrotic therapeutic strategy for DN. These
findings are consistent with the current study where it was found
that circRNA_0000491 knockdown inhibited TGFβ1, TGFβRI and pSmad3
protein expression levels (34,36).
In conclusion, the high expression levels of
circRNA_0000491 in db/db mice and high concentration
glucose-induced MCs was negatively correlated with miR-101b
expression. Furthermore, the results from the present study
indicated that the circRNA_0000491/miR-101b/TGFβRI axis may
regulate ECM and fibrosis-associated protein synthesis of DN.
Therefore, circRNA_0000491 may be considered as a DN-promoting gene
and may represent a novel insight for the treatment of DN.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81774217 and
81273623), Zhejiang Traditional Chinese Medicine Administration
(grant nos. 2017ZKL016 and 2019ZB096) and the Science and
Technology Commission of Hangzhou (grant no. 20150733Q42).
Availability of data and materials
The datasets analyzed/generated during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
XM, JWC, DYZ and YBH conceived and designed the
present study. XM, KL and LJC performed the experimental
procedures. KL, LJC and DZ analyzed the data. XM, JWC and YBH
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Welfare Committee of Zhejiang Chinese Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen B, Li Y, Liu Y and Xu Z: circLRP6
regulates high glucose-induced proliferation, oxidative stress, ECM
accumulation, and inflammation in mesangial cells. J Cell Physiol.
234:21249–21259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen C, Gong W, Li C, Xiong F, Wang S,
Huang J, Wang Y, Chen Z, Chen Q, Liu P, et al: Sphingosine kinase 1
mediates AGEs-induced fibronectin upregulation in diabetic
nephropathy. Oncotarget. 8:78660–78676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, He Z, Yang Q and Zhou G: XBP1
inhibits mesangial cell apoptosis in response to oxidative stress
via the PTEN/AKT pathway in diabetic nephropathy. FEBS Open Bio.
9:1249–1258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Z, Chen Q, Huang J, Gong W, Zou Y,
Zhang L, Liu P and Huang H: CK2α promotes advanced glycation end
products-induced expressions of fibronectin and intercellular
adhesion molecule-1 via activating MRTF-A in glomerular mesangial
cells. Biochem Pharmacol. 148:41–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu W, Han Q, Zhao L and Wang L: Circular
RNA circRNA_15698 aggravates the extracellular matrix of diabetic
nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol.
234:1469–1476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang P, Sun Y, Peng R, Chen W, Fu X,
Zhang L, Peng H and Zhang Z: Long non-coding RNA Rpph1 promotes
inflammation and proliferation of mesangial cells in diabetic
nephropathy via an interaction with Gal-3. Cell Death Dis.
10:5262019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zha F, Qu X, Tang B, Li J, Wang Y, Zheng
P, Ji T, Zhu C and Bai S: Long non-coding RNA MEG3 promotes
fibrosis and inflammatory response in diabetic nephropathy via
miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 11:3716–3730. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang T and Wang X, Du Q, Wu N, Liu X, Chen
Y and Wang X: The circRNA circP4HB promotes NSCLC aggressiveness
and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun.
513:904–911. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang W, Feng J, Zhou H and Li Q:
Circ_0123996 promotes cell proliferation and fibrosisin mouse
mesangial cells through sponging miR-149-5p and inducing Bach1
expression. Gene. 761:1449712020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li S, Pei Y, Wang W, Liu F, Zheng K and
Zhang X: Circular RNA 0001785 regulates the pathogenesis of
osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2.
Cell Cycle. 18:1281–1291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y,
Shen G and Wang F: Inhibition of circHIPK3 prevents angiotensin
II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol.
292:188–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Zhang J and Zhao F: Circular RNA
identification based on multiple seed matching. Brief Bioinform.
19:803–810. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
20
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He M, Wang J, Yin Z, Zhao Y, Hou H, Fan J,
Li H, Wen Z, Tang J, Wang Y, et al: MiR-320a induces diabetic
nephropathy via inhibiting MafB. Aging (Albany NY). 11:3055–3079.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Leon BM and Maddox TM: Diabetes and
cardiovascular disease: Epidemiology, biological mechanisms,
treatment recommendations and future research. World J Diabetes.
6:1246–1258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W,
Chuang CY and Kuo HC: The circular RNA circBIRC6 participates in
the molecular circuitry controlling human pluripotency. Nat Commun.
8:11492017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Y, Zhang Y, Zhang Y and Wang JJ:
CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes
chondrocyte extracellular matrix degradation by sponging miR-26a.
Cell Biol Int. 41:1283–1289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Z, Li X, Jian D, Hao P, Rao L and Li
M: Hsa_circ_0054633 in peripheral blood can be used as a diagnostic
biomarker of pre-diabetes and type 2 diabetes mellitus. Acta
Diabetol. 54:237–245. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lili S and Yue X: Downregulation of
hsa_circ_0007534 suppresses breast cancer cell proliferation and
invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys
Res Commun. 503:2603–2610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin
JX, Chen QY, Cao LL, Huang CM and Zheng CH: Circular RNA
hsa_circ_0001368 suppresses the progression of gastric cancer by
regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun.
512:29–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu H, Wang X, Wang ZY and Li L:
Circ_0080425 inhibits cell proliferation and fibrosis in diabetic
nephropathy via sponging miR-24-3p and targeting fibroblast growth
factor 11. J Cell Physiol. 235:4520–4529. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao T, Zha D, Hu C and Wu X: Circ_0000285
promotes podocyte injury through sponging miR-654-3p and activating
MAPK6 in diabetic nephropathy. Gene. 747:1446612020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sutariya B, Jhonsa D and Saraf MN: TGF-β:
The connecting link between nephropathy and fibrosis.
Immunopharmacol Immunotoxicol. 38:39–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Liu L, Peng W, Liu H, Liang L,
Zhang X, Mao Y, Zhou X, Shi M, Xiao Y, et al: Ski-related novel
protein suppresses the development of diabetic nephropathy by
modulating transforming growth factor-β signaling and microRNA-21
expression. J Cell Physiol. 234:17925–17936. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tu X, Zhang H and Zhang J, Zhao S, Zheng
X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y and Zhang J: MicroRNA-101
suppresses liver fibrosis by targeting the TGFβ signalling pathway.
J Pathol. 234:46–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li L, Ma L, Wang D, Jia H, Yu M, Gu Y,
Shang H and Zou Z: Design and synthesis of matrine derivatives as
novel anti-pulmonary fibrotic agents via repression of the
TGFβ/smad pathway. Molecules. 24:11082019. View Article : Google Scholar
|
|
36
|
Ding H, Zhou Y and Huang H: MiR-101a
ameliorates AngII-mediated hypertensive nephropathy by blockade of
TGFβ/Smad3 and NF-κB signalling in a mouse model of hypertension.
Clin Exp Pharmacol Physiol. 46:246–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang M, Chen DQ, Wang MC, Chen H, Chen L,
Liu D, Zhao H and Zhao YY: Poricoic acid ZA, a novel RAS inhibitor,
attenuates tubulo-interstitial fibrosis and podocyte injury by
inhibiting TGF-β/Smad signaling pathway. Phytomedicine. 36:243–253.
2017. View Article : Google Scholar : PubMed/NCBI
|