Introduction
Cutaneous squamous cell carcinoma (CSCC) is one of
the most common cancer types, accounting for 25–35% of cutaneous
malignancies (1). CSCC develops
from epidermal keratinocytes or adnexal squamous epithelial cells
(2). Currently, immunosuppression
factors that are sensitive to sunlight or UV radiation and genetic
factors are thought to be closely associated with CSCC (3). Treatments for cutaneous squamous cell
carcinoma include excision, radiotherapy, photodynamic therapy and
topical drug treatment (4).
DEAD-box (DDX) RNA helicases are a large
ATP-dependent RNA helicases family, which originates from a wide
number of organisms ranging from prokaryotes, including viruses, to
eukaryotes such as yeast, plants and animals (5). DDX is involved in almost every
cellular process of RNA metabolism, from RNA synthesis to RNA
degradation and plays important roles during cell development and
growth in almost all organisms (6). DDX46 is a member of the DDX RNA
helicase family located on chromosome 5q31.1 and plays a critical
role in transcript splicing and ribosome assembly (7). Previous studies (7,8) have
shown that DDX46 is abnormally expressed and plays an oncogenic
role in various tumors, and is also involved in tumorigenesis and
cancer progression. However, the role of DDX46 in CSCC is still not
fully understood.
Therefore, in the present study, three pairs of CSCC
tissues and corresponding adjacent tissues were collected, and the
expression level of DDX46 was assessed. Furthermore, the effect of
DDX46 silencing on CSCC cell proliferation, apoptosis and autophagy
was analyzed. To our best knowledge, the present study is the first
to identify the possible role of DDX46 in CSCC.
Materials and methods
Clinical specimens and cell lines
Human CSCC A431 and SCL-1 cells, and human
keratinocyte HaCaT cells were purchased from Shanghai Enzyme
Biological Technology Co., Ltd. In addition, three pairs of CSCC
tissues and corresponding adjacent tissues were collected from the
Department of Plastic Surgery, The First Hospital of Jilin
University from September 2015 to November 2018. All the specimens
were confirmed for CSCC by clinical and pathological diagnosis.
Consent was obtained from the patients and their families. The
present study was informed and approved by the Ethics Committee of
The First Hospital of Jilin University.
Reagents and instruments
DMEM and FBS were purchased from Santa Co., Ltd.
DMSO, crystal violet and tryptase were purchased from Sigma-Aldrich
(Merck KGaA). Cell Counting Kit-8 (CCK-8) kits, Annexin V/PI cell
apoptosis detection kits and TaqMan™ reverse transcription reagents
(cat. no. 4304134) were obtained from Santa Cruz Biotechnology,
Inc. TRIzol® kit, the transfection reagent Lipofectamine
3000® and 3,3′-diaminobenzidine (DAB) reagent kit were
provided by Thermo Fisher Scientific, Inc. Rabbit anti-DDX46
(1:500; cat. no. ab72083) antibody was purchased from Abcam. The
forward (F) and reverse (R) primers sequences for RT-qPCR of DDX46
were designed and synthesized by Arraystar, Inc., and were as
follows: DDX46 F, 5′-AAAATGGCGAGAAGAGCAACG-3′ and R,
5′-CATCATCGTCCTCTAAACTCCAC-3′; GAPDH F, 5′-TGACTTCAACAGCGACACCCA-3′
and R, CACCCTGTTGCTGTAGCCAAA-3′. The preparation and packaging of
DDX46 gene RNAi target sequence and shDX46 lentivirus (shDDX46)
were designed and synthesized by Arraystar, Inc.
RT-qPCR
Total RNAs of CSCC tissues and cells were extracted
with TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Total RNA was reverse transcribed into cDNA at
room temperature using TaqMan™ reverse transcription reagents (cat.
no. 4304134; Thermo Fisher Scientific, Inc.). qPCR was subsequently
performed using the iScript™ cDNA Synthesis kit (cat. no. 1708890;
Bio-Rad Laboratories, Inc.). The PCR conditions were as follows:
95°C for 3 min, followed by 40 cycles of 95°C for 10 sec, 60°C for
30 sec and 72°C for 30 sec, according to the manufacturer's
protocol of the PCR kit. The relative levels were assessed by
2−ΔΔCq method (9);
ΔCq=Cqtarget-CqGAPDH.
Western blotting
Total protein of CSCC tissues and cells were
collected with RIPA buffer (Sigma-Aldrich; Merck KGaA). Protein
concentrations were determined with a BCA assay (Bio-Rad
Laboratories, Inc.). Then, a total of 20 µg protein per lane was
transferred to the nitrocellulose membrane after 10% SDS-PAGE and
blocked by 5% TBST (0.1% Tween-20) for 1 h at room temperature. The
membranes were incubated with rabbit anti-DDX46 (1:1,000; cat. no.
ab72083; Abcam), rabbit anti-Bcl-2 antibody (1:1500; cat. no.
ab59348; Abcam), rabbit anti-Survivin antibody (1:1,000; cat. no.
ab469; Abcam), rabbit anti-Bax antibody (1:1,000; cat. no. ab53154;
Abcam), rabbit anti-Cleaved Caspase-3 antibody (1:1,000; cat. no.
ab2302; Abcam), rabbit anti-Beclin 1 antibody (ab62557) (1:1,000;
cat. no. ab62557; Abcam), rabbit anti-microtubule-associated
protein 1A/1B-light chain 3 (LC3) antibody (1:1,000; cat. no.
ab128025; Abcam) and anti-β-actin (1:2,000; cat. no. ab8227; Abcam)
at 4°C overnight. The PVDF membrane was washed three times with
TBST and incubated at room temperature for 2 h with horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat. no.
ab97051; Abcam). Blot bands were visualized by ECL systems (Pierce;
Thermo Fisher Scientific, Inc.). Densitometry was performed by
using ImageJ v1.47 software (National Institutes of Health)
(10).
Cell transfection
The preparation and packaging of DDX46 gene RNAi
target sequence (shDDX46) and negative control sequence (shNC) were
designed and synthesized by Arraystar, Inc. short hairpin (sh)RNA
sequence of DDX46 was 5′-AGAAATCACCAGGCTCATA-3′, and the negative
control shRNA (shNC) sequence was 5′-TTCTCCGAACGTGTCACGT-3′. The
quantity of siRNA transfected was 50 nM. Cells
(1×105/well) were transfected using Lipofectamine 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Then, 48 h
after transfection, the transfection efficiency was evaluated by
labeling vectors with green fluorescence protein (GFP) to ensure
that cells were successfully transfected, and evaluated by RT-qPCR
using the same steps as mentioned above).
CCK-8 assay
Transfected cells in the logarithmic growth stage
were collected and inoculated into 96-well plates with density of
2×103 cells per pore. Each group was assessed with three
multiple pores. After adherence, the cells were replaced with 100
ml RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.).containing 10 ml CCK-8 reagent and incubated for 2 h at 37°C
according to the manufacturer's instructions. The absorbance
(D) of each pore were measured at 450 nm by enzyme label.
The D values of the cells in each group were measured at 24,
48, 72 and 96 h. The growth curves of each group cells were drawn
according to the measured D value.
Colony formation assay
Transfected cells in the logarithmic growth stage
were digested with 0.5% trypsin (Gibco; Thermo Fisher Scientific,
Inc.) at room temperature for 5 sec and seeded in 6-well plates
with 1×105/well. After cultured for 2 weeks, 6-well
plates were washed twice with PBS and cells were fixed with 2%
paraformaldehyde (PFA) at 37°C for 30 min. Then, cells were stained
with 0.5% crystal violet for 15 min at 37°C. Cell colony forming
units were imaged under a confocal microscope (Nikon Corporation;
magnification, ×400) and analyzed using ImageJ v1.47 software
(National Institutes of Health) (10).
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Transfected cells in the logarithmic growth stage
were digested with 0.5% trypsin (Gibco; Thermo Fisher Scientific,
Inc.) at room temperature for 5 sec and seeded in 24-well plates
with 1×104/well. After cultured for 24 h, 5 µl EdU
reagent (Sigma-Aldrich; Merck KGaA)was added to each dish and
incubated for 2 h at 37°C. Then, plates were washed twice with PBS
and cell were fixed with 2% PFA for 30 min at 37°C. EdU positive
cells were visualized and photographed under fluorescence
microscope (BD Biosciences; magnification, ×400), and analyzed
using ImageJ v1.47 software (National Institute of Health).
Flow cytometry
Transfected cells in the logarithmic growth stage
were digested with 0.5% trypsin (Gibco; Thermo Fisher Scientific,
Inc.) at room temperature for 5 sec and seeded in 6-well plates
with 1×104/well. After cultured for 24 h, the cells were
collected and washed twice with pre-cooled PBS. Then, cells were
re-suspended with 1X combined buffer (Sigma-Aldrich; Merck KGaA)
and the cell density was adjusted to ~1×106/ml. Then,
100 ml cell fluid was added to the flow tube, 5 µl Annexin V-FITC
and 5 µl PI solution were added into flow tube to stain cells for
20 min in the dark at 4°C. Then, 400 ml combined buffer was added
and the apoptotic rate was detected by a FACSCalibur flow cytometer
(Becton-Dickinson and Company) for cell-cycle distribution analysis
using Cell Quest Software (v5.1; Becton-Dickinson and Company).
Statistical analysis
SPSS v17.0 (SPSS, Inc.) software were used for
statistical analyses. Data are presented as the mean ± SD of three
experimental repeats. Student's t-test and χ2 test were
performed among two groups, one-way ANOVA followed by Tukey's
multiple comparison tests were performed among three groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
DDX46 is overexpressed in CSCC
tissues
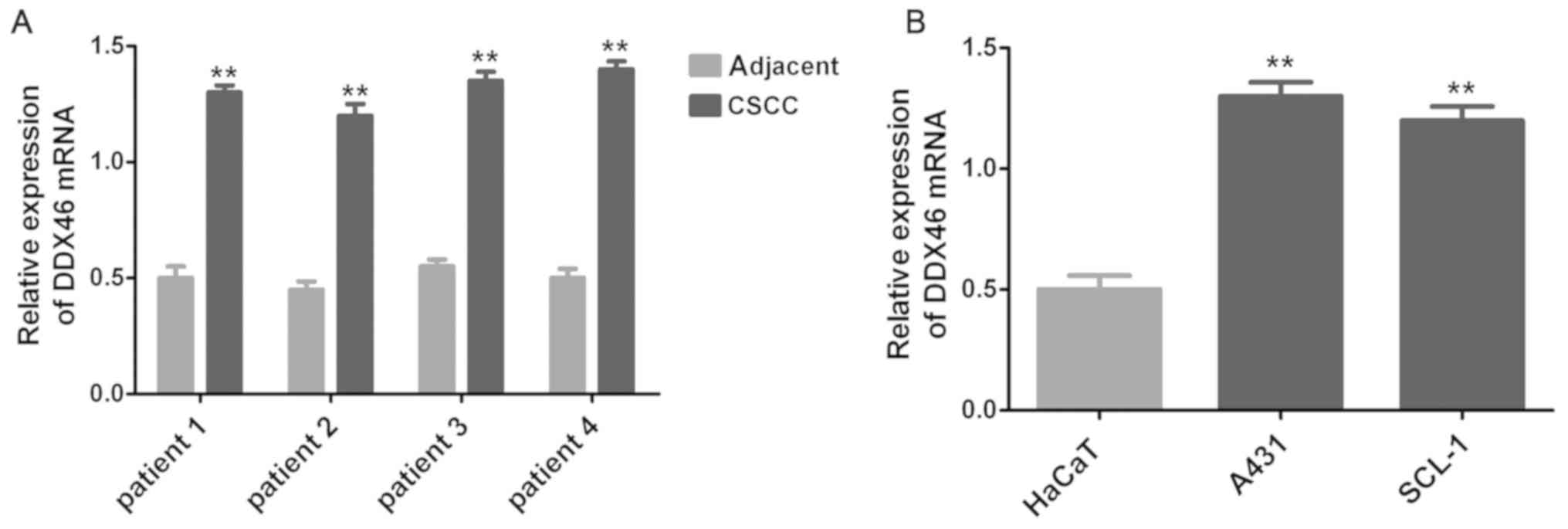
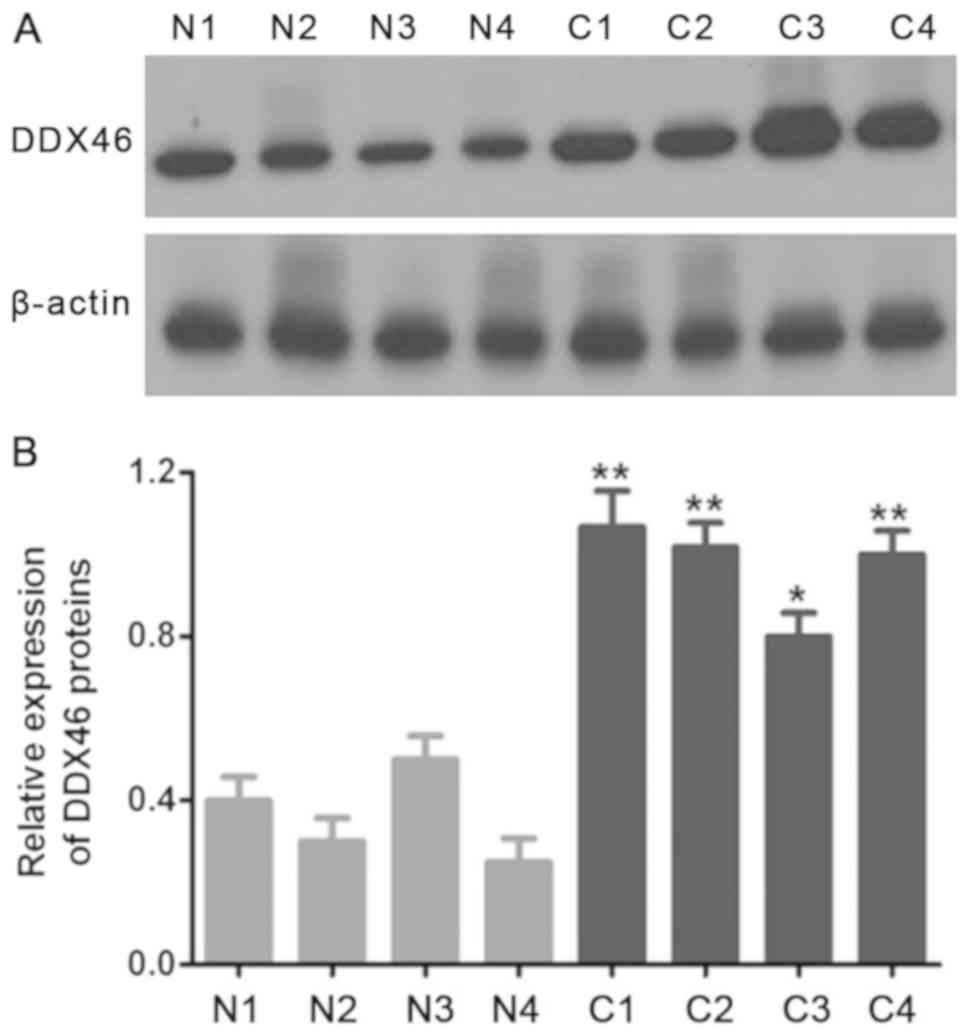
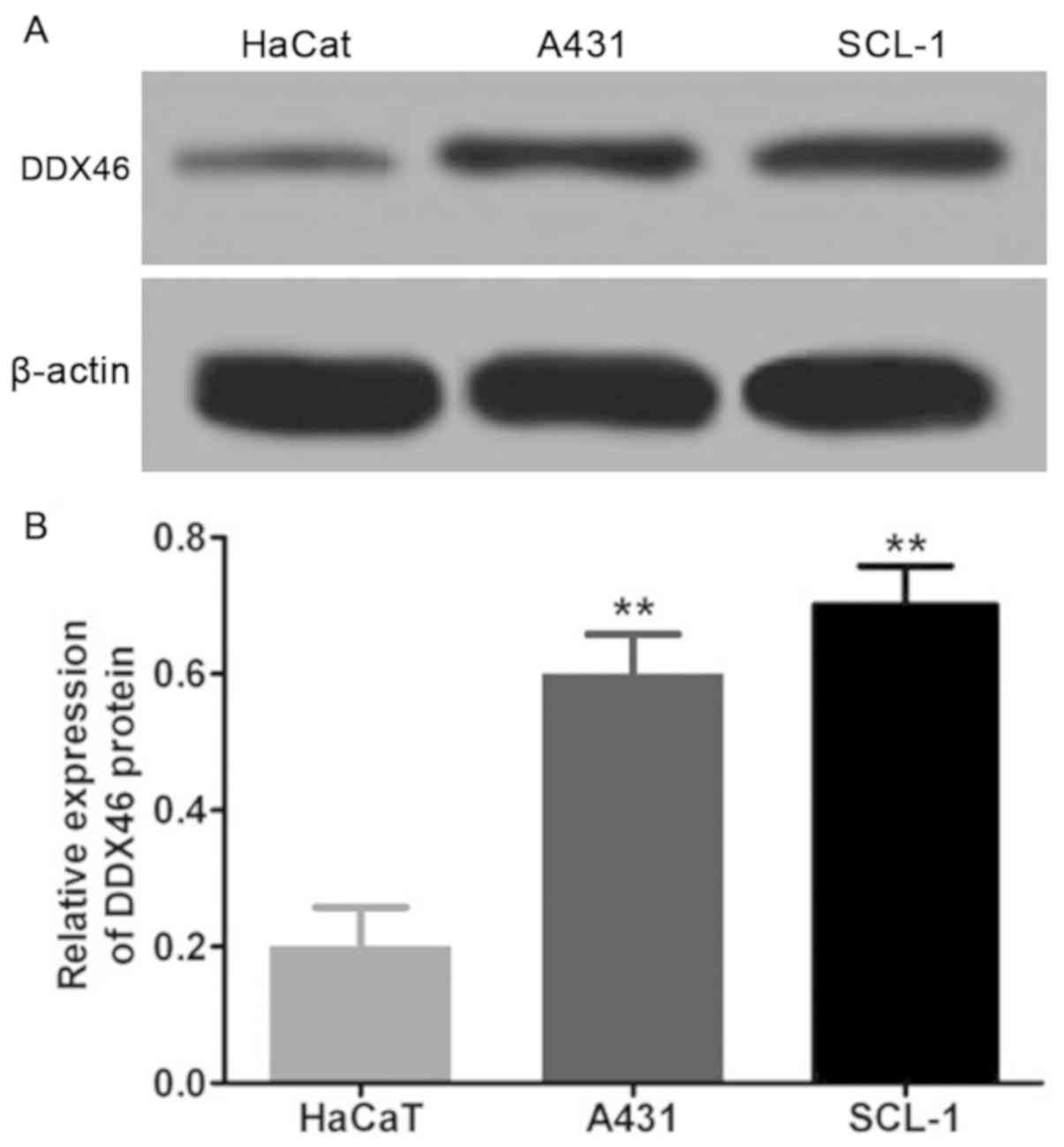
In the present study, the expression level of DDX46
in three pairs of CSCC tissues and corresponding adjacent tissues
was assessed using RT-qPCR and Western blot assay. The RT-qPCR and
Western blot results demonstrated that DDX46 mRNA (P<0.01;
Fig. 1) and protein (P<0.05;
Figs. 2 and 3) expression levels were significantly
overexpressed in CSCC cells, and also in CSCC tissues (n=4)
compared with corresponding adjacent tissues.
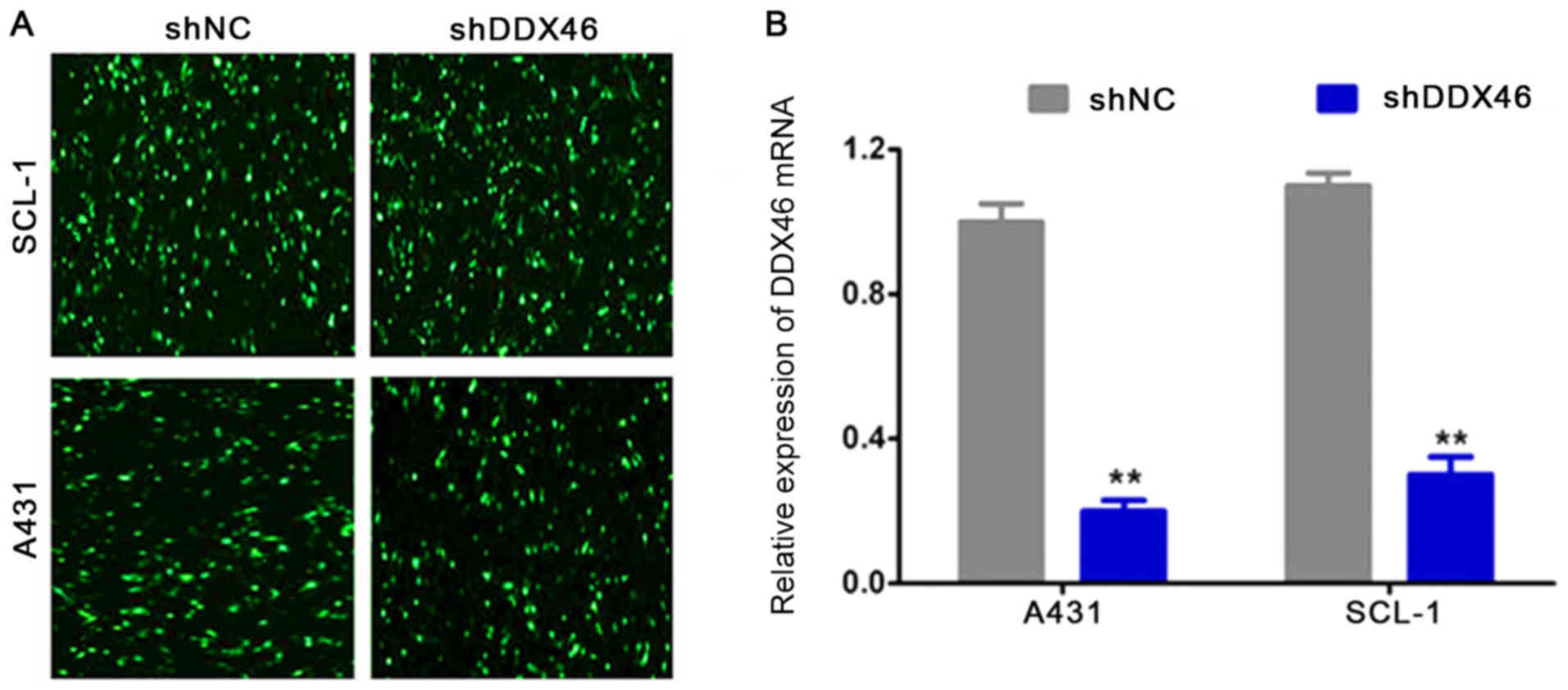
shRNA is successfully transfected into
CSCC cells
shDDX46 and shNC were transfected into A431 and
SCL-1 cells. Then, 48 h after transfection using a fluorescence
microscope GFP was observed in A431 and SCL-1 cells (Fig. 4A). RT-qPCR results indicated that
DDX46 mRNA expression levels were significantly decreased in the
shDDX46 group compared with the shNC group (P<0.05; Fig. 4B).
DDX46 silencing inhibits cell
proliferation
CCK-8, colony formation and EdU assay were used to
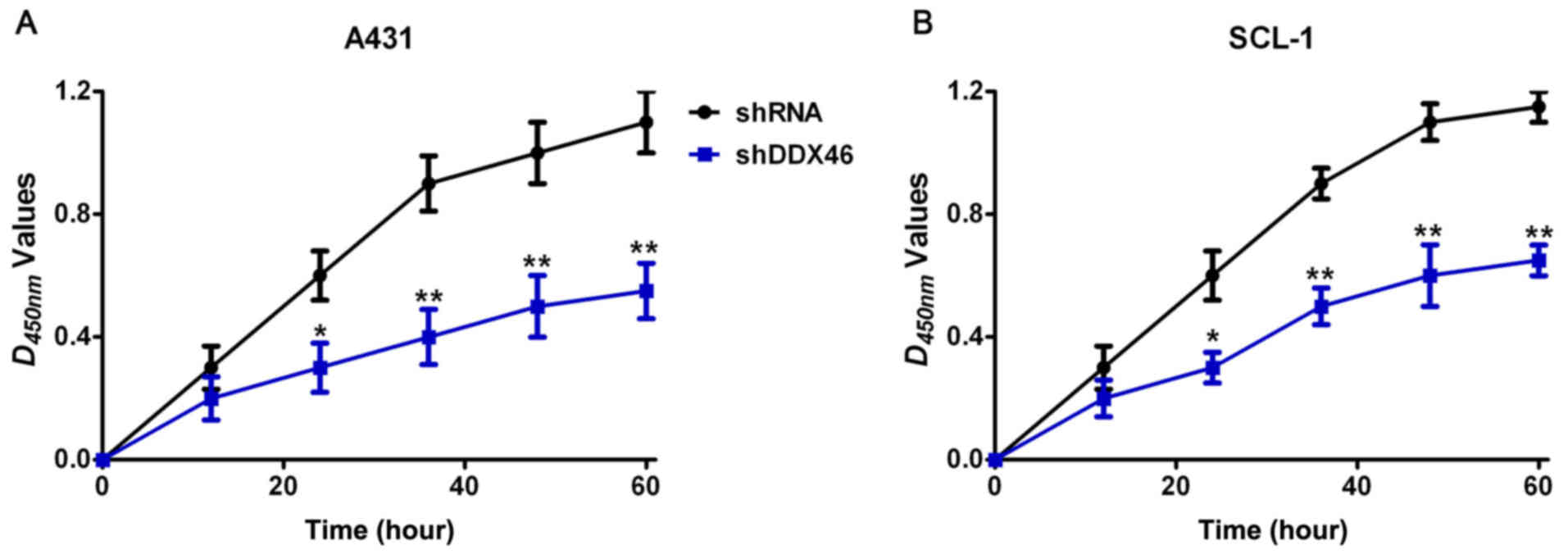
analyze the effect of DDX46 on cell proliferation. CCK-8 assay
results demonstrated that the proliferative ability of cells was
significantly decreased in the shDDX46 group compared with the
control group (P<0.05; Fig. 5).
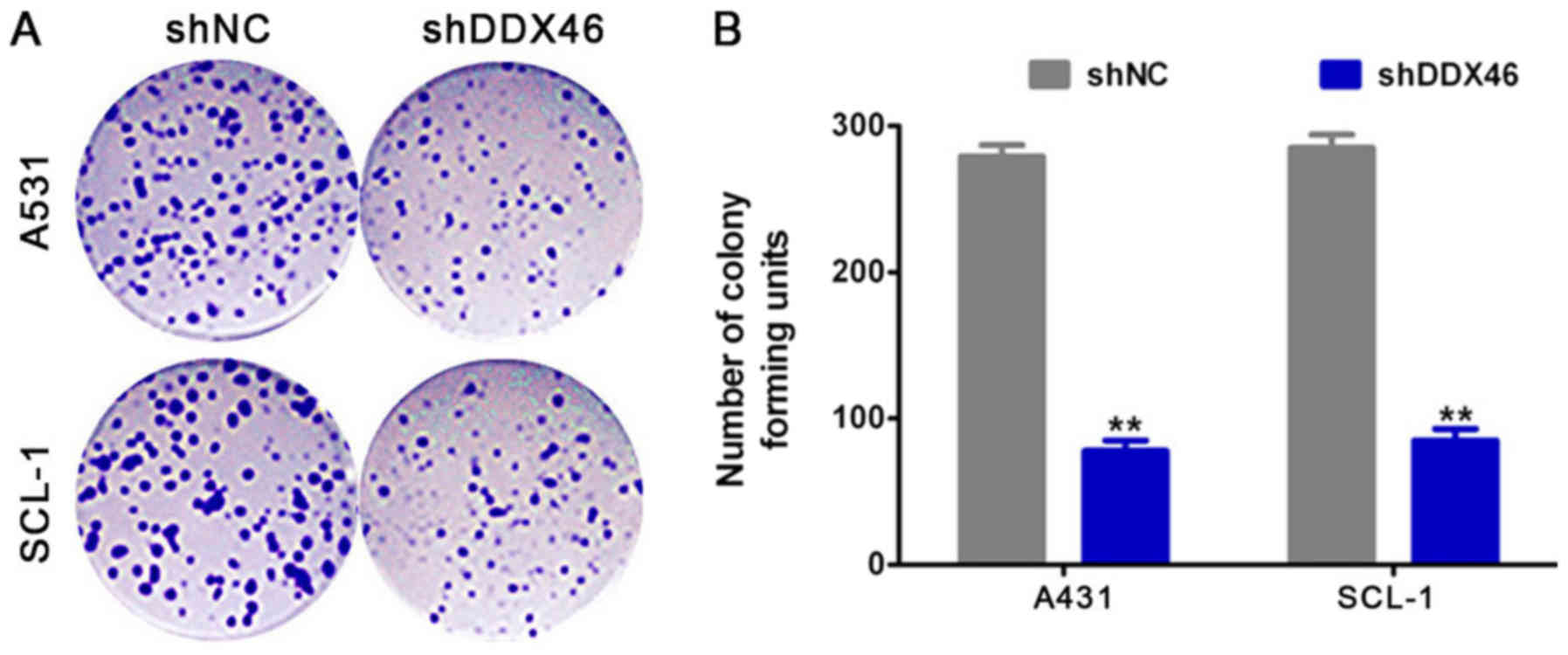
Furthermore, colony formation assay results suggested that the
number of colony-forming units were significantly reduced in the
shDDX46 group (P<0.05; Fig. 6)
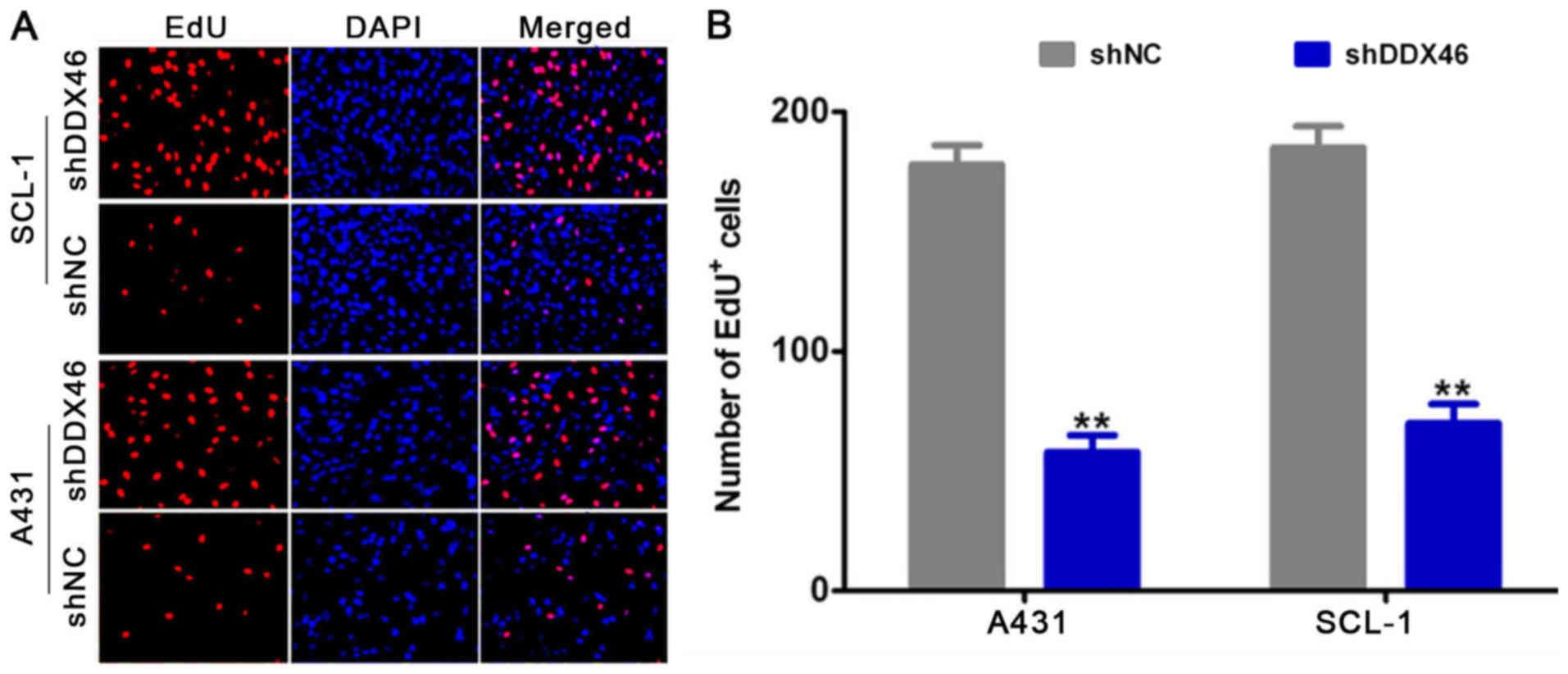
compared with the shNC group. In addition, it was found that the
number of EdU positive cells was significantly decreased in the
shDDX46 group compared with the shNC group (P<0.05; Fig. 7).
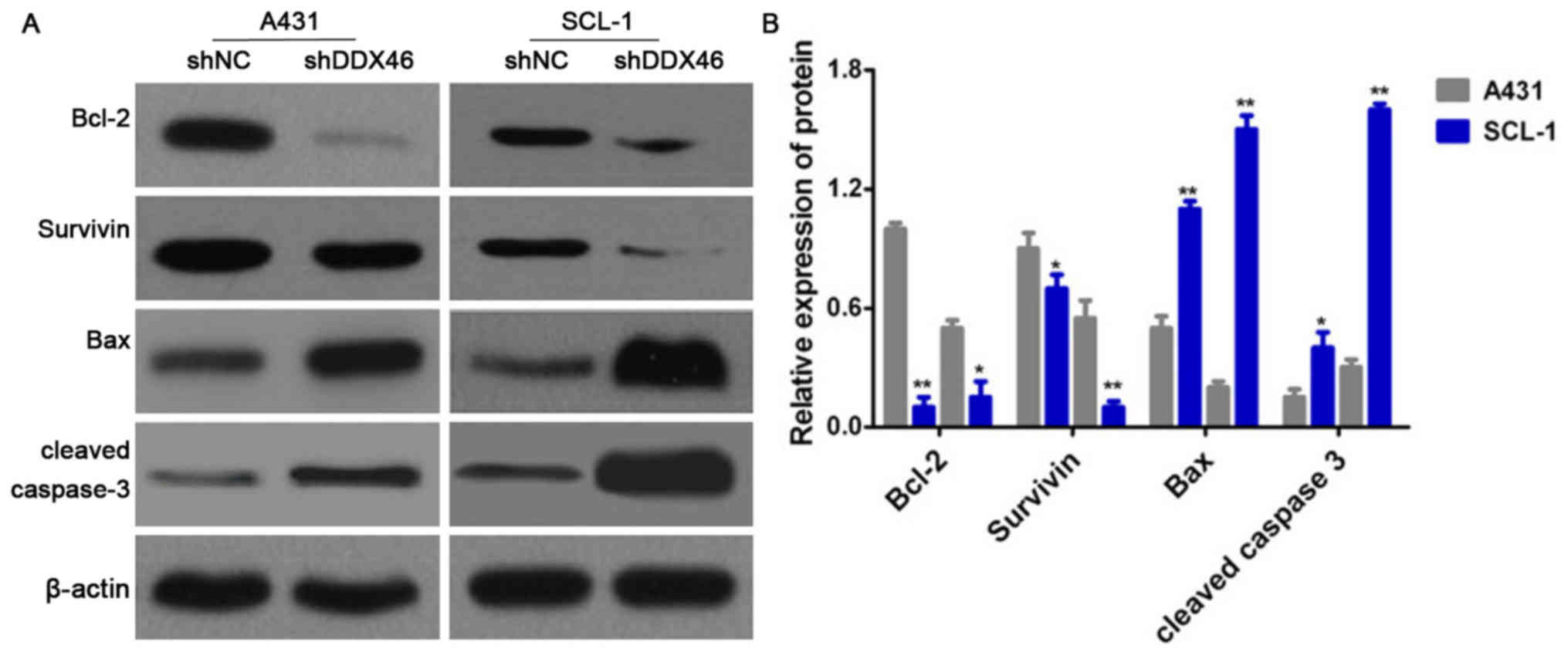
DDX46 silencing inhibits cell
apoptosis
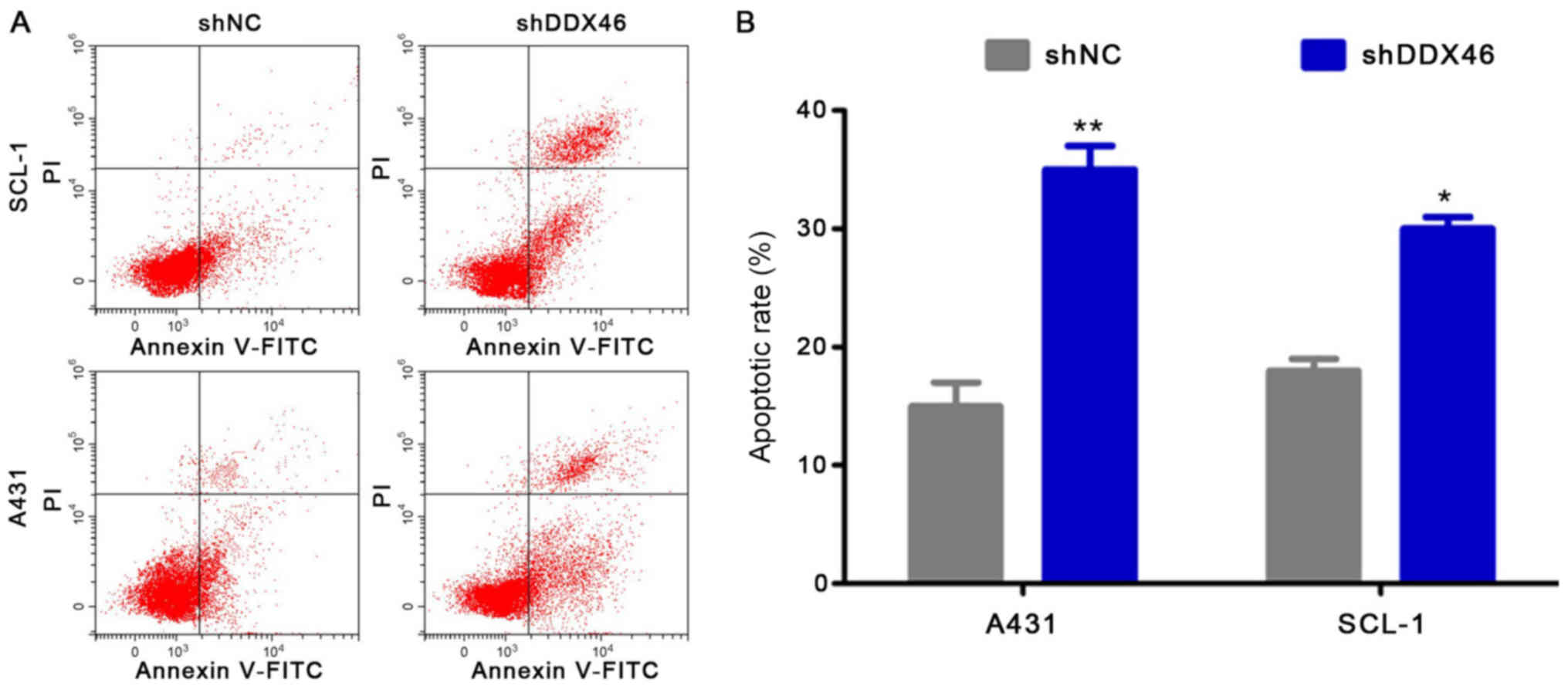
Flow cytometry was used to assess cell apoptosis and
Western blotting was used to measure the expression levels of the
apoptotic-related proteins Bcl-2, Survivin, Bax and cleaved
caspase-3. It was demonstrated that the apoptotic rate was
significantly increased in the shDDX46 group, compared with shNC
group (P<0.05; Fig. 8).
Furthermore, Western blotting results indicated that the protein
expression levels of Bcl-2 and Survivin were significantly
decreased in the shDDX46 group, and that the protein expression
levels of Bax and cleaved caspase-3 protein were significantly
increased in shDDX46 group (P<0.05; Fig. 9).
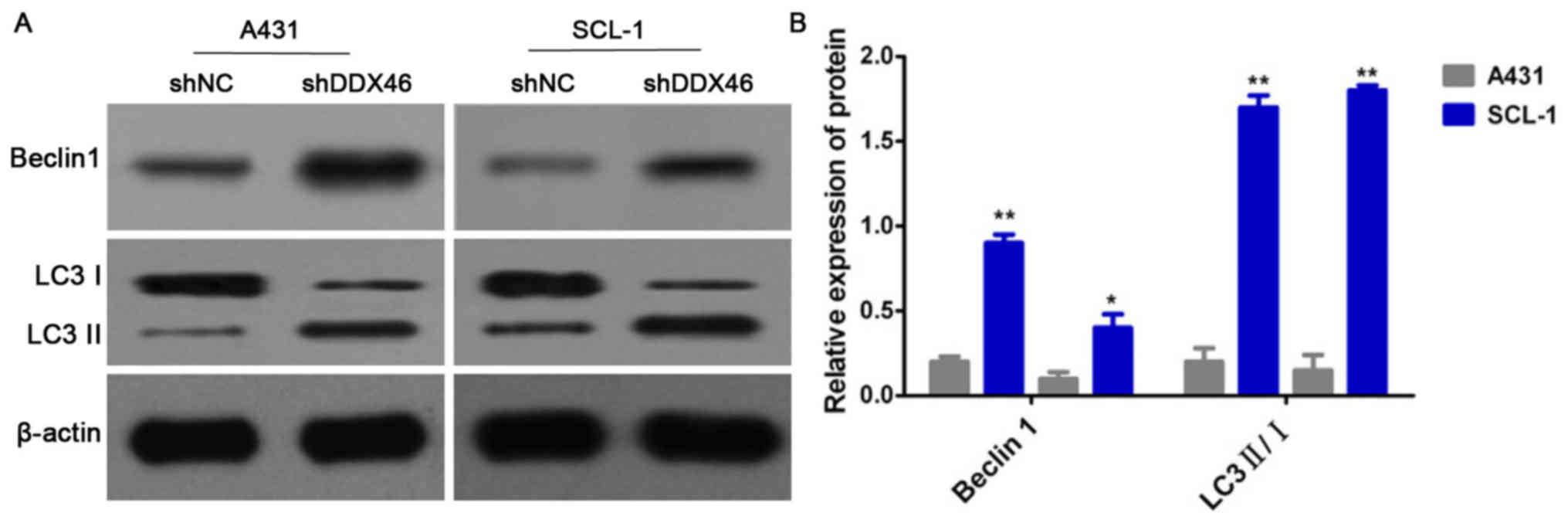
DDX46 silencing can activate cell
autophagy
Western blotting was used to detect the protein
expression levels of Beclin1 and LC3 II/I. It was demonstrated that
the protein expression levels of Beclin1 protein and the ratio of
LC3 II/I were significantly increased in the shDDX46 group compared
with the shNC group (P<0.05; Fig.
10).
Discussion
RNA helicase is essential for RNA metabolism,
including RNA splicing, transcription, transport, translation and
degradation (7). DDX is an
important ATP-dependent RNA helicase family, which exists in almost
all prokaryotes and eukaryotes (11). Furthermore, DDX plays an important
role in RNA metabolism, it is required for RNA functions, including
gene transcription, RNA precursor mothering, ribosome synthesis,
RNA output, translation initiation and RNA degradation (12,13).
Several DDX RNA helicase subtypes have been reported to be
abnormally expressed in various cancer types (14). Previous studies have shown that
DDX1 is overexpressed in retinoblastoma, transitional blastoma,
breast cancer and testicular cancer (15,16).
Moreover, DDX5 is upregulated in human melanoma and hepatocellular
carcinoma (17,18), and DDX6 is increased in
glioblastoma, rhabdomyosarcoma, lung cancer, colon cancer and liver
cancer (19). Thus, the DDX family
may be involved in the occurrence and progression cancer.
DDX46 is a member of the DDX RNA helicase family
with ATP-dependent RNA helicase, that is located on chromosome
5q31.1 and plays a critical role in transcript splicing and
ribosome assembly (20). Previous
studies (21–23) in zebrafish have shown that DDX46 is
required in the multilineage differentiation of hematopoietic stem
cells and in the development of digestive organs and the brain. Li
et al (24) reported that
DDX46 is significantly overexpressed in colorectal cancer and that
lentiviral DDX46 knockdown inhibits growth and induces apoptosis in
human colorectal cancer cells. Liu et al (25) showed that DDX46 is upregulated in
human bladder cancer 5637 and T24 cells. Li et al (26) found that DDX46 is significantly
upregulated in esophageal squamous cell carcinoma tissues and
cells, and that DDX46 knockdown decreases proliferation and
increased apoptosis in TE-1 and Eca-109 cells. Thus, DDX46 may be
involved in tumorigenesis and cancer progression. However, the role
of DDX46 in CSCC still remains unknown.
The present results suggested that DDX46 was
overexpressed in CSCC tissues, as indicated by results from IHC,
RT-qPCR and Western blotting. The present study used DDX46 RNAi
lentivirus to silence DDX46 expression in CSCC cells. It was found
that DDX46 silencing can inhibit cell proliferation. Furthermore,
DDX46 silencing can also induce cell apoptosis and activate
autophagy by regulating the protein expression levels of Bcl-2,
Survivin, Bax, cleaved caspase-3, Beclin1 and LC3 II/I. Thus, the
present results suggested that DDX46 was overexpressed in CSCC and
that DDX46 silencing can inhibit cell proliferation, the mechanism
of which may be associated with apoptosis and autophagy. Previous
studies have found that autophagy and apoptotic activity changes
occur in a variety of human tumors (26,27).
In some cases, autophagy inhibits cell apoptosis, which promotes
cell survival (28). In other
instances, autophagy can induce cell death, or function alongside
apoptosis to induce cell death in the case of apoptotic defect
(29,30). Thus, there is a close relationship
between autophagy and apoptosis during cell death, and these
pathways are interrelated and regulate each other. The present
results indicated that apoptosis and autophagy worked together to
inhibit CSCC cell proliferation.
In conclusion, to the best of our knowledge, the
present study provides the first evidence that DDX46 was
significantly overexpressed in CSCC. Furthermore, it was found that
DDX46 silencing inhibited cell proliferation by inducing apoptosis
and activating autophagy. Thus, DDX46 may serve as a novel
potential therapeutic target for CSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG and QL designed the study and performed the
experiments. HJJ and DZ analyzed the data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Hospital of Jilin University (Jilin, China),
and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Que SKT, Zwald FO and Schmults CD:
Cutaneous squamous cell carcinoma: Incidence, risk factors,
diagnosis, and staging. J Am Acad Dermatol. 78:237–247. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ratushny V, Gober MD, Hick R, Ridky TW and
Seykora JT: From keratinocyte to cancer: The pathogenesis and
modeling of cutaneous squamous cell carcinoma. J Clin Invest.
122:464–472. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Umezono Y, Sato Y, Noto M, Yamada K,
Noguchi N, Hasunuma N, Osada SI and Manabe M: Incidence rate of
cutaneous squamous cell carcinoma is rapidly increasing in Akita
Prefecture: Urgent alert for super-aged society. J Dermatol.
46:259–262. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peters FS, Peeters AMA, Mandaviya PR, van
Meurs JB, Hofland LJ, van de Wetering J, Betjes MG, Baan CC and
Boer K: Differentially methylated regions in T cells identify
kidney transplant patients at risk for de novo skin cancer. Clin
Epigenetics. 10:812018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang F, Zhang D, Li G and Wang X:
Knockdown of DDX46 inhibits the invasion and tumorigenesis in
osteosarcoma cells. Oncol Res. 25:417–425. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nawaz G and Kang H: Rice OsRH58, a
chloroplast DEAD-box RNA helicase, improves salt or drought stress
tolerance in Arabidopsis by affecting chloroplast translation. BMC
Plant Biol. 19:172019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baek W, Lim CW and Lee SC: A DEAD-box RNA
helicase, RH8, is critical for regulation of ABA signaling and the
drought stress response via inhibition of PP2CA activity. Plant
Cell Environ. 41:1593–1604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng Q, Hou J, Zhou Y, Li Z and Cao X:
The RNA helicase DDX46 inhibits innate immunity by entrapping
m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol.
18:1094–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schindelin J, Rueden CT, Hiner MC and
Eliceiri KW: The ImageJ ecosystem: An open platform for biomedical
image analysis. Mol Reprod Dev. 82:518–529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pauszek R, Lamichhane R, Rajkarnikar Singh
A, van der Schans E and Millar D: The 5 nuclease domain of DNA
polymerase I mediates a novel DNA transfer pathway during
proofreading. Biophys J. 114:82a2018. View Article : Google Scholar
|
|
12
|
Sohn SO and Chay KO: The ATP-dependent RNA
helicase, DDX42 interacts with paxillin and regulates apoptosis and
polarization of Ba/F3 cells. Anim Cells Syst Seoul. 23:1–9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao J, Byrd AK, Zybailov BL, Marecki JC,
Guderyon MJ, Edwards AD, Chib S, West KL, Waldrip ZJ, Mackintosh
SG, et al: DEAD-box RNA helicases Dbp2, Ded1 and Mss116 bind to
G-quadruplex nucleic acids and destabilize G-quadruplex RNA. Chem
Commun (Camb). 55:4467–4470. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Curmi F and Cauchi RJ: The multiple lives
of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem Soc Trans.
46:329–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang J, Chen H, Wong CC, Liu D, Li T, Wang
X, Ji J, Sung JJ, Fang JY and Yu J: DEAD-box helicase 27 promotes
colorectal cancer growth and metastasis and predicts poor survival
in CRC patients. Oncogene. 37:3006–3021. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ribeiro de Almeida C, Dhir S, Dhir A,
Moghaddam AE, Sattentau Q, Meinhart A and Proudfoot NJ: RNA
helicase DDX1 converts RNA G-Quadruplex structures into R-Loops to
promote IgH class switch recombination. Mol Cell. 70:650–662.e8.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhong W, Li Z, Zhou M, Xu T and Wang Y:
DDX1 regulates alternative splicing and insulin secretion in
pancreatic β cells. Biochem Biophys Res Commun. 500:751–757. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu N, Jiang M, Han Y, Liu H, Chu Y, Liu H,
Cao J, Hou Q, Zhao Y, Xu B, et al: O-GlcNAcylation promotes
colorectal cancer progression by regulating protein stability and
potential catcinogenic function of DDX5. J Cell Mol Med.
23:1354–1362. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu
L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, et al: Retraction
note: DDX5 and its associated lncRNA Rmrp modulate TH17 cell
effector functions. Nature. 562:1502018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taniguchi K, Iwatsuki A, Sugito N,
Shinohara H, Kuranaga Y, Oshikawa Y, Tajirika T, Futamura M,
Yoshida K, Uchiyama K, et al: Oncogene RNA helicase DDX6 promotes
the process of c-Myc expression in gastric cancer cells. Mol
Carcinog. 57:579–589. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lumb JH, Li Q, Popov LM, Ding S, Keith MT,
Merrill BD, Greenberg HB, Li JB and Carette JE: DDX6 Represses
Aberrant Activation of Interferon-Stimulated Genes. Cell Rep.
20:819–831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirabayashi R, Hozumi S, Higashijima S and
Kikuchi Y: Ddx46 is required for multi-lineage differentiation of
hematopoietic stem cells in zebrafish. Stem Cells Dev.
22:2532–2542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hozumi S, Hirabayashi R, Yoshizawa A,
Ogata M, Ishitani T, Tsutsumi M, Kuroiwa A, Itoh M and Kikuchi Y:
DEAD-box protein Ddx46 is required for the development of the
digestive organs and brain in zebrafish. PLoS One. 7:e336752012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li M, Ma Y, Huang P, Du A, Yang X, Zhang
S, Xing C, Liu F and Cao J: Lentiviral DDX46 knockdown inhibits
growth and induces apoptosis in human colorectal cancer cells.
Gene. 560:237–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Zhang C, Huang D, et al:
Construction of DDX46 lentiviral vector and its expression
identification in human bladder cancer cells. Anhui Medical
Journal. 2018.(In Chinese).
|
|
26
|
Li B, Li YM, He WT, Chen H, Zhu HW, Liu T,
Zhang JH, Song TN and Zhou YL: Knockdown of DDX46 inhibits
proliferation and induces apoptosis in esophageal squamous cell
carcinoma cells. Oncol Rep. 36:223–230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mane SD and Kamatham AN: Ascorbyl stearate
stimulates cell death by oxidative stress-mediated apoptosis and
autophagy in HeLa cervical cancer cell line in vitro. 3 Biotech.
9:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Q, Kang J and Fu C: The independence
of and associations among apoptosis, autophagy, and necrosis.
Signal Transduct Target Ther. 3:18–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song S, Tan J, Miao Y, Li M and Zhang Q:
Crosstalk of autophagy and apoptosis: Involvement of the dual role
of autophagy under ER stress. J Cell Physiol. 232:2977–2984. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan X, Zhou R and Ma Z: Autophagy-cell
survival and death. Adv Exp Med Biol. 1206:667–696. 2019.
View Article : Google Scholar : PubMed/NCBI
|