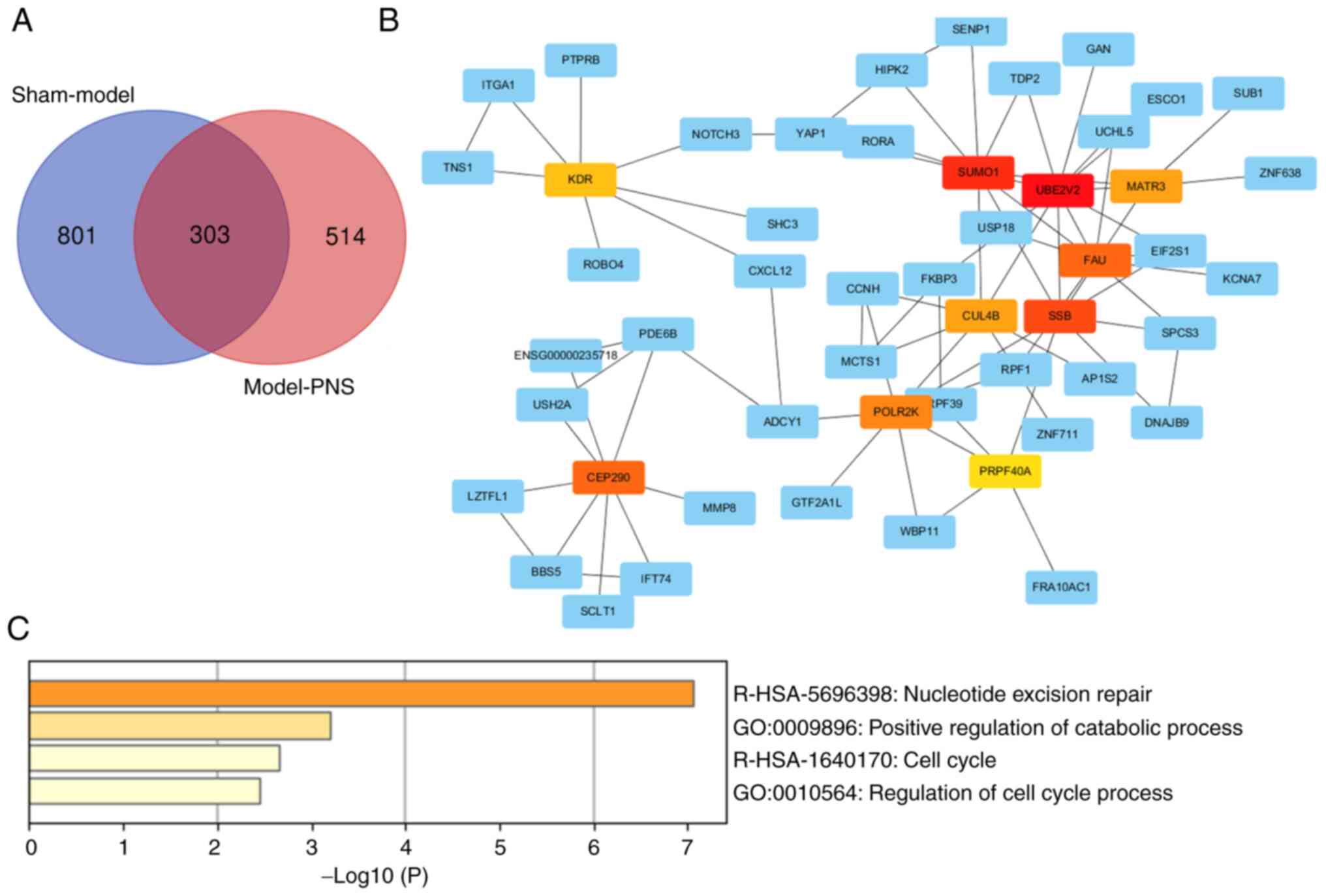
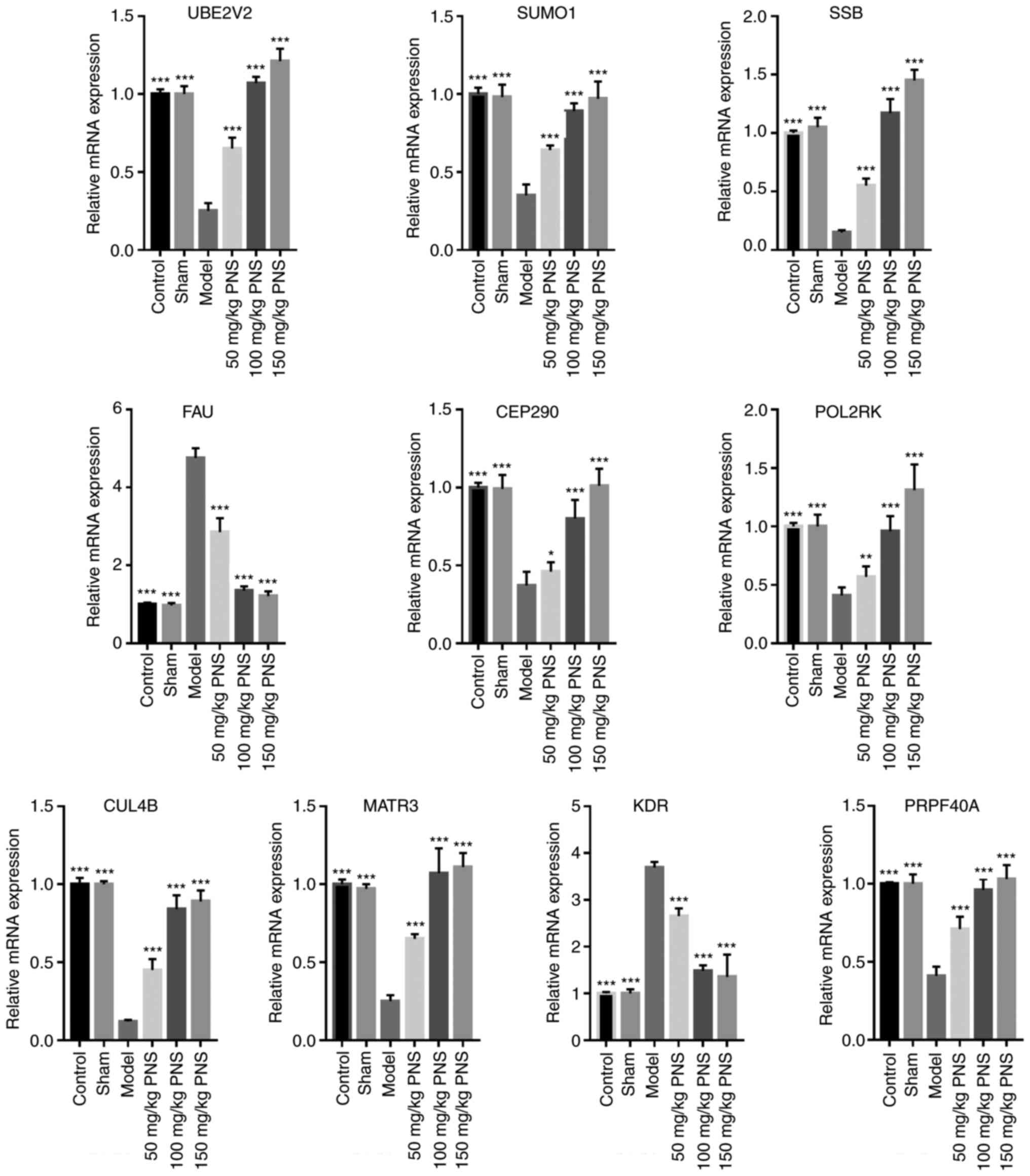
|
1
|
Fuentes B and Tejedor ED: Stroke: The
worldwide burden of stroke-a blurred photograph. Nat Rev Neurol.
10:127–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moskowitz MA, Lo EH and Iadecola C: The
science of stroke: mechanisms in search of treatments. Neuron.
67:181–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duan J, Cui J, Yang Z, Guo C, Cao J, Xi M,
Weng Y, Yin Y, Wang Y, Wei G, et al: Neuroprotective effect of
Apelin 13 on ischemic stroke by activating AMPK/GSK-3β/Nrf2
signaling. J Neuroinflammation. 16:242019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bi BL, Wang HJ, Bian H and Tian ZT:
Identification of therapeutic targets of ischemic stroke with DNA
microarray. Eur Rev Med Pharmacol Sci. 19:4012–4019.
2015.PubMed/NCBI
|
|
5
|
Wang J, Cao B, Han D, Sun M and Feng J:
Long Non-coding RNA H19 induces cerebral ischemia reperfusion
injury via activation of autophagy. Aging Dis. 8:71–84. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu N, Shan D, Li Y, Chen H, Gao Y and
Huang Y: Panax notoginseng saponins attenuate phenotype switching
of vascular smooth muscle cells induced by notch3 silencing. Evid
Based Complement Alternat Med. 2015:1621452015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li F, Zhao H, Han Z, Wang R, Tao Z, Fan Z,
Zhang S, Li G, Chen Z and Luo Y: Xuesaitong may protect against
ischemic stroke by modulating microglial phenotypes and inhibiting
neuronal cell apoptosis via the STAT3 Signaling pathway. CNS Neurol
Disord Drug Targets. 18:115–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Zhu L, Zou Y, Liu W, Zhang X, Wei
X, Hu B and Chen J: Panax notoginseng saponins promotes stroke
recovery by influencing expression of Nogo-A, NgR and p75NGF, in
vitro and in vivo. Biol Pharm Bull. 37:560–568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang PF, Song XY and Chen NH: Advances in
pharmacological studies of Panax notoginseng saponins on brain
ischemia-reperfusion injury. Yao Xue Xue Bao. 51:1039–1046.
2016.(In Chinese).
|
|
10
|
Xie W, Meng X, Zhai Y, Zhou P, Ye T, Wang
Z, Sun G and Sun X: Panax notoginseng saponins: A review of its
mechanisms of antidepressant or anxiolytic effects and network
analysis on phytochemistry and pharmacology. Molecules. 23:9402018.
View Article : Google Scholar
|
|
11
|
Xu C, Wang W, Wang B, et al: Analytical
methods and biological activities of Panax notoginseng
saponins: Recent trends. J Ethnopharmacol. 236:443–465. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi YH, Li Y, Wang Y, Xu Z, Fu H and Zheng
GQ: Ginsenoside-Rb1 for ischemic stroke: A systematic review and
meta-analysis of preclinical evidence and possible mechanisms.
Front Pharmacol. 11:2852020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nabavi SF, Sureda A, Habtemariam S and
Nabavi SM: Ginsenoside Rd and ischemic stroke; a short review of
literatures. J Ginseng Res. 39:299–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie CL, Wang WW, Xue XD, Zhang SF, Gan J
and Liu ZG: A systematic review and meta-analysis of
Ginsenoside-Rg1 (G-Rg1) in experimental ischemic stroke. Sci Rep.
5:77902015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng L, Lin J, Huang Q, Liang P, Huang J,
Jian C, Lin C and Li X: Panax notoginseng saponins attenuate
oxygen-glucose deprivation/reoxygenation-induced injury in human
SH-SY5Y Cells by regulating the expression of inflammatory factors
through miR-155. Biol Pharm Bull. 42:462–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi X, Yu W, Yang T, Liu W, Zhao Y, Sun Y,
Chai L, Gao Y, Dong B and Zhu L: Panax notoginseng saponins provide
neuroprotection by regulating NgR1/RhoA/ROCK2 pathway expression,
in vitro and in vivo. J Ethnopharmacol. 190:301–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi X, Yu W, Liu L, Liu W, Zhang X, Yang
T, Chai L, Lou L, Gao Y and Zhu L: Panax notoginseng saponins
administration modulates pro-/anti-inflammatory factor expression
and improves neurologic outcome following permanent MCAO in rats.
Metab Brain Dis. 32:221–233. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Longa EZ, Weinstein PR, Carlson S and
Cummins R: Reversible middle cerebral artery occlusion without
craniectomy in rats. Stroke. 20:84–91. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bederson JB, Pitts LH, Tsuji M, Nishimura
MC, Davis RL and Bartkowski H: Rat middle cerebral artery
occlusion: Evaluation of the model and development of a neurologic
examination. Stroke. 17:472–476. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
The Gene Ontology Resource: 20 years and
still GOing strong. Nucleic Acids Res. 47:D330–D338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang L, Wang C, Zhao S, Ge R, Guan S and
Wang JH: PKC and CaMK-II inhibitions coordinately rescue
ischemia-induced GABAergic neuron dysfunction. Oncotarget.
8:39309–39322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen BH, Park JH, Lee YL, Kang IJ, Kim DW,
Hwang IK, Lee CH, Yan BC, Kim YM, Lee TK, et al: Melatonin improves
vascular cognitive impairment induced by ischemic stroke by
remyelination via activation of ERK1/2 signaling and restoration of
glutamatergic synapses in the gerbil hippocampus. Biomed
Pharmacother. 108:687–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Secondo A, Bagetta G and Amantea D: On the
role of store-operated calcium entry in acute and chronic
neurodegenerative diseases. Front Mol Neurosci. 11:872018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hattori M and Minato N: Rap1 GTPase:
Functions, regulation, and malignancy. J Biochem. 134:479–484.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kooistra MR, Dube N and Bos JL: Rap1: A
key regulator in cell-cell junction formation. J Cell Sci.
120:17–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chrzanowska-Wodnicka M: Rap1 in
endothelial biology. Curr Opin Hematol. 24:248–255. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chrzanowska-Wodnicka M: Distinct functions
for Rap1 signaling in vascular morphogenesis and dysfunction. Exp
Cell Res. 319:2350–2359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Y, Wang X, Zhou X, Cheng B, Li G and
Bai B: Temporal expression of apelin/apelin receptor in ischemic
stroke and its therapeutic potential. Front Mol Neurosci. 10:12017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu S, Cao Q, Xu P, Ji W, Wang G and Zhang
Y: Rolipram stimulates angiogenesis and attenuates neuronal
apoptosis through the cAMP/cAMP-responsive element binding protein
pathway following ischemic stroke in rats. Exp Ther Med.
11:1005–1010. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bai H, Zhao L, Liu H, Guo H, Guo W, Zheng
L, Liu X, Wu X, Luo J, Li X, et al: Adiponectin confers
neuroprotection against cerebral ischemia-reperfusion injury
through activating the cAMP/PKA-CREB-BDNF signaling. Brain Res
Bull. 143:145–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim YS, Yoo A, Son JW, Kim HY, Lee YJ,
Hwang S, Lee KY, Lee YJ, Ayata C, Kim HH and Koh SH: Early
activation of phosphatidylinositol 3-kinase after ischemic stroke
reduces infarct volume and improves long-term behavior. Mol
Neurobiol. 54:5375–5384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Yu Y, Yang J, Zhao X and Li Z:
Dissecting Xuesaitong's mechanisms on preventing stroke based on
the microarray and connectivity map. Mol Biosyst. 11:3033–3039.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao Y, Long MJC, Wang Y, Zhang S and Aye
Y: Ube2V2 Is a rosetta stone bridging redox and ubiquitin codes,
coordinating DNA damage responses. ACS Cent Sci. 4:246–259. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li P, Stetler RA, Leak RK, Shi Y, Li Y, Yu
W, Bennett MVL and Chen J: Oxidative stress and DNA damage after
cerebral ischemia: Potential therapeutic targets to repair the
genome and improve stroke recovery. Neuropharmacology. 134:208–217.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Wang Y, Zhu A, Huang D, Deng S,
Cheng J, Zhu MX and Li Y: SUMO-specific protease 1 protects neurons
from apoptotic death during transient brain ischemia/reperfusion.
Cell Death Dis. 7:e24842016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Perina D, Korolija M, Hadzija MP, Grbeša
I, Belužić R, Imešek M, Morrow C, Marjanović MP, Bakran-Petricioli
T, Mikoč A and Ćetković H: Functional and structural
characterization of FAU gene/protein from marine sponge suberites
domuncula. Mar Drugs. 13:4179–4196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rachel RA, Yamamoto EA, Dewanjee MK,
May-Simera HL, Sergeev YV, Hackett AN, Pohida K, Munasinghe J,
Gotoh N, Wickstead B, et al: CEP290 alleles in mice disrupt
tissue-specific cilia biogenesis and recapitulate features of
syndromic ciliopathies. Hum Mol Genet. 24:3775–3791. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He YM, Xiao YS, Wei L, Zhang JQ and Peng
CH: CUL4B promotes metastasis and proliferation in pancreatic
cancer cells by inducing epithelial-mesenchymal transition via the
Wnt/β-catenin signaling pathway. J Cell Biochem. 119:5308–5323.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Zhou X, Zhang Y, Yang J, Xu Y, Zhao
Y and Wang X: CUL4B regulates autophagy via JNK signaling in
diffuse large B-cell lymphoma. Cell Cycle. 18:379–394. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jean LeBlanc N, Menet R, Picard K, Parent
G, Tremblay ME and ElAli A: Canonical Wnt pathway maintains
blood-brain barrier integrity upon ischemic stroke and its
activation ameliorates tissue plasminogen activator therapy. Mol
Neurobiol. 56:6521–6538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang P, Shao BZ, Deng Z, Chen S, Yue Z and
Miao CY: Autophagy in ischemic stroke. Prog Neurobiol.
163-164:98–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng J, Dai Q, Han K, Hong W, Jia D, Mo
Y, Lv Y, Tang H, Fu H and Geng W: JNK-IN-8, a c-Jun N-terminal
kinase inhibitor, improves functional recovery through suppressing
neuroinflammation in ischemic stroke. J Cell Physiol.
235:2792–2799. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng Y, Wang J, He G, Qu F and Zheng M:
Mobilization of endothelial progenitor cell in patients with acute
ischemic stroke. Neurol Sci. 39:437–443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Marti-Fabregas J, Delgado-Mederos R,
Crespo J, Peña E, Marín R, Jiménez-Xarrié E, Fernández-Arcos A,
Pérez-Pérez J, Martínez-Domeño A, Camps-Renom P, et al: Circulating
endothelial progenitor cells and the risk of vascular events after
ischemic stroke. PLoS One. 10:e01248952015. View Article : Google Scholar : PubMed/NCBI
|