Introduction
The immune system is the body's natural defense
mechanism against pathogens such as viruses, bacteria, and other
intermediates (1). Inflammation
occurs when a variety of harmful substances, ranging from
infectious microorganisms (such as bacteria, viruses or fungi) to
transgenic cells (such as cancer cells), reside in a specific
tissue of the host or circulate in the blood (2–6). In
response to reactions from these immune cells participating in
inflammation are the inflammatory mediators, such as IL-1β and
TNF-α, which are synthesized and secreted during an inflammatory
response. Therefore, understanding the role of these inflammatory
intermediaries and their associated mechanisms is one of the major
goals for controlling the inflammatory response (5,6).
Regulating the inflammatory response is critical in the treatment
of inflammatory-related diseases such as systemic inflammatory
response syndrome (SIRS), severe tissue damage, and septic shock.
These inflammatory regulating agents include steroids, non-steroids
and biological agents. However, development of new drugs are
required to overcome certain drawbacks of the existing
anti-inflammatory drugs, such as adverse side effects and high
prices (7). Recently, natural
substances such as medicinal plants have drawn attention as
resources for acquiring anti-inflammatory drugs that are easy to
procure in large quantities at low cost, and are expected to have
fewer side effects (8,9).
Morus alba, commonly known as mulberry in
Korea, grows very well in a variety of climatic conditions, ranging
from temperate areas to tropical areas, and is found in northern
China, India, the Middle East, Southern Europe, and more recently
in North America (10). Mulberry
leaves are generally used to feed silkworms, and they contain
numerous active compounds that possess various pharmacological
activities (11). Many studies
have reported the anti-bacterial, anti-oxidant, anti-hypoglycemic,
anti-cancer, anti-obesity, anti-ER stress, and anti-inflammatory
activities of the roots and shells (9,12–16).
Especially, extracts of mulberry leaves fermented with C.
militaris (EMfC) treatment is reported to inhibit fat
accumulation in the HFD-induced obese C57BL/6 mice through
regulation of lipogenesis and lipolysis (17). These effects of EMfC are tightly
correlated with the suppression of ER stress and ER stress-induced
apoptosis (18).
C. militaris, an insect pathogen belonging to
Ascomycota, is traditionally used for elevating the immune
response, and as an anti-cancer and anti-aging agent in Chinese and
East Asian medicine (19). Several
studies report that the cordycepin isolated from C.
militaris has pharmacological activity as an anti-cancer
(20), anti-bacterial (21), and anti-oxidant (22) agent. Similar effects have been
reported for mixtures of C. militaris and secondary
metabolite products secreted from mushrooms (23). We therefore hypothesized that
certain products of mulberry leaves fermented with C.
militaris would exert protection against an lipopolysaccharide
(LPS)-induced inflammatory response and the autophagy pathway.
In the current study, we examined the basic
mechanisms involved in the anti-inflammatory and anti-autophagic
activity of EMfC in LPS-stimulated RAW264.7 macrophage cells. Our
results provide new data indicating that EMfC is associated with
the prevention of inflammation and autophagy-related diseases via
the regulation of iNOS-mediated COX-2 induced pathway, MAPK
signaling pathways, and PI3K/mTOR pathway.
Materials and methods
Cell culture
RAW264.7 cells (ATCC) were cultured in Dulbecco
Modified Eagle's Medium (DMEM; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Welgene),
L-glutamine, penicillin, and streptomycin (Thermo Fisher
Scientific, Inc.), in a humidified incubator at 37°C under 5%
CO2 and 95% air.
Preparation of EMfCs
EMfC samples were prepared in accordance with a
previous papers (17). The samples
of mulberry leaves were collected in October 2015 from plantations
in the Sangju district of Korea, and characterized by Professor
Young Whan Choi at the Department of Horticultural Bioscience,
Pusan National University. Voucher specimens of mulberry leaves
were deposited in the herbarium (accession no. Mul-PDRL-1) of the
Pusan National University (Miryang, Korea). The C. militaris
used for fermentation was kindly provided by Professor Sang Mong
Lee of the Department of Life Science and Environmental
Biochemistry, Pusan National University. The Jeongeup Agriculture
Cooperative Federations for Silkworm Farming (Jeongeup, Korea)
supplied the silkworm pupae powder.
Briefly, fresh mulberry leaves were first completely
dried in a hot-air drying machine (JSR) for 24 h at 60°C, and
subsequently powdered using an electric blender. The powdered dried
mulberry leaves were sterilized by autoclaving at 121°C for 60 min,
and mixed with 50% silkworm powder (SWP). This mixture was
inoculated with 10% C. militaris (v/w) and incubated in a
shaking incubator (#SI-600R; Lab Companion) maintained at 150 rpm
and 25°C, and fermented for 4 weeks. Subsequently, the pellet of
fermented mixture was harvested by centrifuging the flask at 3,000
rpm for 10 min. To prepare the extracts of fermented mulberry
powder, the harvested pellet was mixed with the solvent (95% EtOH)
in a fixed liquid ratio (mulberry powder:solvent, ratio 1:10), and
sonicated for 1 h using a JAC ultrasonic device (KODO). This
sonicated pellet was collected using centrifugation at 3,000 rpm
for 10 min. The resultant pellet was resuspended in 9 ml of the
solvent, and further sonicated using the same conditions. This
procedure was repeated once more, and the resultant supernatant was
collected, filtered through a 0.4 µm filter (#HAWP04700, Millex-LH;
Merck Millipore), and evaporated using a vacuum evaporator (#R-300;
BUCHI Corporation). Finally, lyophilization of the EMfC was
achieved using a circulating extraction equipment (IKA
Labortechnik). The extracts were dissolved in DMSO (#D2660;
Sigma-Aldrich; Merck KGaA) at a concentration of 50 mg/ml before
use.
Scavenging activity of free
radical
The scavenging capability of the
2,2-diphenyl-1-picrylhydrazyl (DPPH) radical was measured at eight
different concentrations of EMfC (0-1,000 µg/ml), according to the
method described in a previous study (24). Briefly, each sample of EMfC (100
µl) was mixed with 100 µl of 0.1 mM DPPH (Sigma-Aldrich; Merck
KGaA) prepared in 50% DMSO solution. After 30 min of incubation at
room temperature, absorbance of the reaction mixture was recorded
using a Versa-max plate reader (Molecular Devices) at a wavelength
of 517 nm. The percent drop in the absorbance, relative to that of
the control, determined the DPPH radical scavenging activity of the
EMfC. The concentration of EMfC resulting in a 50% loss in DPPH
activity was determined to be the IC50.
Cell viability assay
Cell viability was determined using the tetrazolium
compound 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium
bromide (MTT) (Sigma-Aldrich; Merck KGaA) assay. Briefly, RAW264.7
cells were seeded at a density of 2×104 cells/0.2 ml
medium per well, and incubated for 24 h in a humidified
CO2 incubator at 37°C. On attaining 70–80% confluency,
each well was assigned to one of the five experimental groups:
Untreated group, Vehicle+LPS treated group, EMfCLo+LPS (low
concentration of EMfC, 100 µg/ml) treated group, EMfCMid+LPS
(medium concentration of EMfC, 200 µg/ml) treated group, and
EMfCHi+LPS (high concentration of EMfC, 400 µg/ml) treated group.
The cells were exposed to either DMSO (Vehicle+LPS treated group)
or the assigned concentration of EMfC for 2 h, and subsequently
stimulated with 1 µg/ml LPS (Sigma-Aldrich; Merck KGaA) for 24 h in
a 37°C incubator. The supernatants were subsequently discarded, and
0.2 ml of fresh DMEM was added to each well, followed by addition
of 50 µl of MTT solution (2 mg/ml in 1X PBS). The cells were
incubated at 37°C for 4 h, after which the MTT was removed and
cells were lysed by adding 150 µl DMSO/well. The optical density
was measured at 570 nm using a microplate reader (Molecular Devices
VERSA max Plate reader). The morphological features of RAW264.7
cells in each treated group were also observed using a light
microscope (Leica Microsystems).
Analysis of intracellular reactive
oxygen species (ROS) level
ROS levels were measured by staining with
2′,7′-dichlorofluorescein diacetate (DCFH-DA) (Sigma-Aldrich; Merck
KGaA). Briefly, RAW264.7 cells were seeded at a density of
1×105 cells/ml in 24-well plates for 24 h, and exposed
to three different concentrations of EMfC (100, 200 or 400 µg/ml)
or DMSO for 2 h in a 37°C incubator. The cells were subsequently
stimulated with 1 µg/ml LPS for further 24 h. Cells were then
incubated with 25 µM DCFH-DA for 30 min at 37°C, washed twice with
PBS, and observed for green fluorescence at ×200 magnification
using a fluorescent microscope (Eclipse TX100; Nikon).
Nitrogen oxide (NO) concentration
analysis
NO concentration in the culture supernatant was
measured using Griess reagent (1% sulfanilamide, 5% phosphoric
acid, 0.1% N-(1-naphthyl) ethylenediamine dihydrochloride;
Sigma-Aldrich; Merck KGaA), as described previously (25,26).
Briefly, RAW264.7 cells were treated with vehicle or EMfC (100, 200
or 400 µg/ml) for 2 h, followed by LPS stimulation (1 µg/ml) and
incubation for 24 h. After harvesting the culture supernatant, each
sample (100 µl) was mixed with the same volume of Griess reagent,
and incubated at room temperature for 10 min. Absorbance was read
at 540 nm using a VersaMax microplate reader (Molecular Devices).
The concentration of NO in the cell culture fluid was calculated by
comparing with a standard curve of sodium nitrite
(NaNO2).
Reverse transcription-quantitative
(RT-q)PCR analysis for cytokine gene expression
Total RNA was isolated by cell lysis using RNAzol
CS104 (Tel-Test Inc.), followed by cell reverse transcription and
PCR. Next, 10 pmol of the sense and antisense primers were added,
and the reaction mixture was subjected to 28–32 cycles of
amplification, conducted in a Perkin-Elmer Thermal Cycler, using
the following cycle: 30 sec at 94°C, 30 sec at 62°C, and 45 sec at
72°C. The primer sequences used for target gene expression
identification were as follows: iNOS, sense,
5′-CACTTGGAGTTCACCCAGT-3′ and antisense primer,
5′-ACCACTCGTACTTGGGATGC-3′; COX-2, sense, 5′-GAGGTGTATC-3′,
antisense primer, 5′-CCAGGAGGATGGAGTTGTTGTAGAG-3′; TNF-α, sense,
5′-CCTGTAGCCCACGTCGTAGC-3′ and antisense primer,
5′-TTGACCTCAGCGCTGACTTG-3′; IL-1β, sense,
5′-GCACATCAACAAGAGCTTCAGGCAG-3′ and antisense primer,
5′-GCTGCTTGTGAGGTGCTGATGTAC-3′; IL-6, sense,
5′-TTGGGACTGATGTTGTTGACA-3′ and antisense primer,
5′-TCATCGCTGTTGATACAATCAGA-3′. The experiment was repeated three
times, and all samples were analyzed in triplicate. The final PCR
products were separated on 1–2% agarose gel and visualized by
ethidium bromide staining. The density of specific bands was
quantified using a Kodak Electrophoresis Documentation and Analysis
System 120 (Eastman Kodak).
RT-qPCR analysis for cytokine gene
expression
RAW264.7 cells were homogenized with Polytron PT-MR
3100 D Homogenizer (Kinematica AG) in TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.), as per the manufacture's protocol. After
ethanol precipitation, total RNAs were harvested by centrifugation
at 10,000 × g for 15 min, and their concentration was subsequently
determined using the Nano-300 Micro-Spectrophotometer (Allsheng
Instruments Co., Ltd.). Total complementary DNA (cDNA) against mRNA
was synthesized using 200 units of SuperScript II reverse
transcriptase (Thermo Fisher Scientific, Inc.). RT-qPCR was
conducted with the cDNA template (1 µl), 2X Power SYBR Green (6 µl;
Toyobo Life Science), and specific primers used in the RT-PCR
analysis in appropriate buffer solution. The cycle quantification
value (Cq) was determined as described in the Livak and
Schmittgen's method (27).
Enzyme-linked immunosorbent assay
(ELISA) for IL-6 cytokine
Cells were pretreated with either Vehicle (DMSO) or
different concentrations of EMfC (100, 200 or 400 µg/ml) for 1 h,
prior to stimulation with 1 µg/ml LPS for 24 h. The culture
supernatant of RAW264.7 cells was assayed for IL-6 using an IL-6
ELISA kit (Biolegend), according to the manufacturer's
instructions.
Western blot analysis
RAW264.7 cells were treated with Vehicle (DMSO) or
EMfC (100, 200 or 400 µg/ml) for 2 h, followed by 1 µg/ml LPS
stimulation for 15 min. The treated cells were lysed using the
Pro-Prep Protein Extraction Solution (iNtRON Biotechnology). The
cell lysate was centrifuged at 13,000 rpm for 5 min, and
concentration of total protein was quantified using the SMARTTM BCA
Protein Assay Kit (Thermo Fisher Scientific, Inc.). The proteins
were separated by 4–20% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) for 2 h. The gels were transferred to
nitrocellulose membranes for 2 h at 40 V. The membranes were
subsequently blocked by incubating with 3% skim milk for 1 h at
room temperature. Each membrane was then incubated separately,
overnight at 4°C, with the following primary antibodies:
Anti-SAPK/JNK (Cell Signaling Technology), anti-p-SAPK/JNK
(Thr183/Tyr185) (Cell Signaling Technology), anti-ERK (K-23) (Santa
Cruz Biotechnology), anti-p-ERK (E-4) (Santa Cruz Biotechnology),
anti-p38 MAPK (Cell Signaling Technology), anti-p-p38 MAP Kinase
(Thr180/Tyr182) (Cell Signaling Technology), anti-PI3K (Cell
Signaling Technology), anti-p-PI3K (Cell Signaling Technology),
anti-mTOR (Cell Signaling Technology), anti-p-mTOR (Cell Signaling
Technology), anti-LC3 (Cell Signaling Technology), anti-Beclin
(Cell Signaling Technology), and anti-β actin (Sigma-Aldrich; Merck
KGaA). The probed membranes were then washed with washing buffer
(137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and
0.05% Tween-20) and subsequently incubated with 1:1,000 diluted
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
secondary antibody (Invitrogen; Thermo Fisher Scientific, Inc.), at
room temperature for 1 h. Finally, membrane blots were developed
using the Amersham ECL Select Western Blotting detection reagent
(GE Healthcare). The chemiluminescence signals that originated from
the specific bands were detected using FluorChemi®FC2
(Alpha Innotech Co.).
Autophagic vacuole analyses
For analyzing autophagic vacuoles, the cells were
treated similar to previous experiments, and subsequently washed
and stained using the Autophagy LC3-antibody-based kit (Millipore),
according to the manufacturer's instructions. The cells were
incubated with Autophagy Reagent A in Earle's balanced salt
solution (EBSS) for 5 h at 37°C, followed by washing with ice cold
HBSS. The cells were then stained with anti-LC3 Alexa
Fluor® 555 in 1X Autophagy Reagent B on ice for 30 min
in the dark, and washed with ice cold 1X Assay Buffer. The stained
samples were quantified by flow cytometry using a Muse Cell
Analyzer (Millipore) in duplicates. Based on manufacture's
instruction, at least 1,000 events for each sample should be
acquired to assure statistically significant determination of
stained cells number. These quantified results were presented as
autophagy induction ratio (test sample fluorescence relative to
control) after analysis using the MuseSoft 1.4.0.0. software.
Statistical analysis
One-way ANOVA (SPSS for Windows, Release 10.10;
Standard Version; SPSS Inc.) followed by Tukey's post hoc test, was
used to identify significant differences between the Vehicle and
the EMfC treated groups. All values are reported as the means ± SD,
and P<0.05 is considered to indicate a statistically significant
difference.
Results
Antioxidant ability of EMfC
To evaluate the antioxidant ability of EMfC, we
measured the DPPH scavenging activity and ability to suppress ROS
production at varying concentrations of EMfC and LPS-stimulated
RAW264.7 cells. As shown in Fig.
1A, the inhibitory activity against DPPH radicals gradually
increases dose-dependently from 1–1,000 µg/ml EMfC, with an
IC50 value of 579.6703 µg/ml. Also, treatment of EMfC
shows strong antioxidant activity at serial concentrations that
reduce the level of ROS production during the inflammatory response
of macrophages induced by LPS (Fig.
1B). These results indicate that subsequent to LPS stimulation,
the therapeutic function of EMfC in RAW264.7 cells is probably
associated with antioxidant activity.
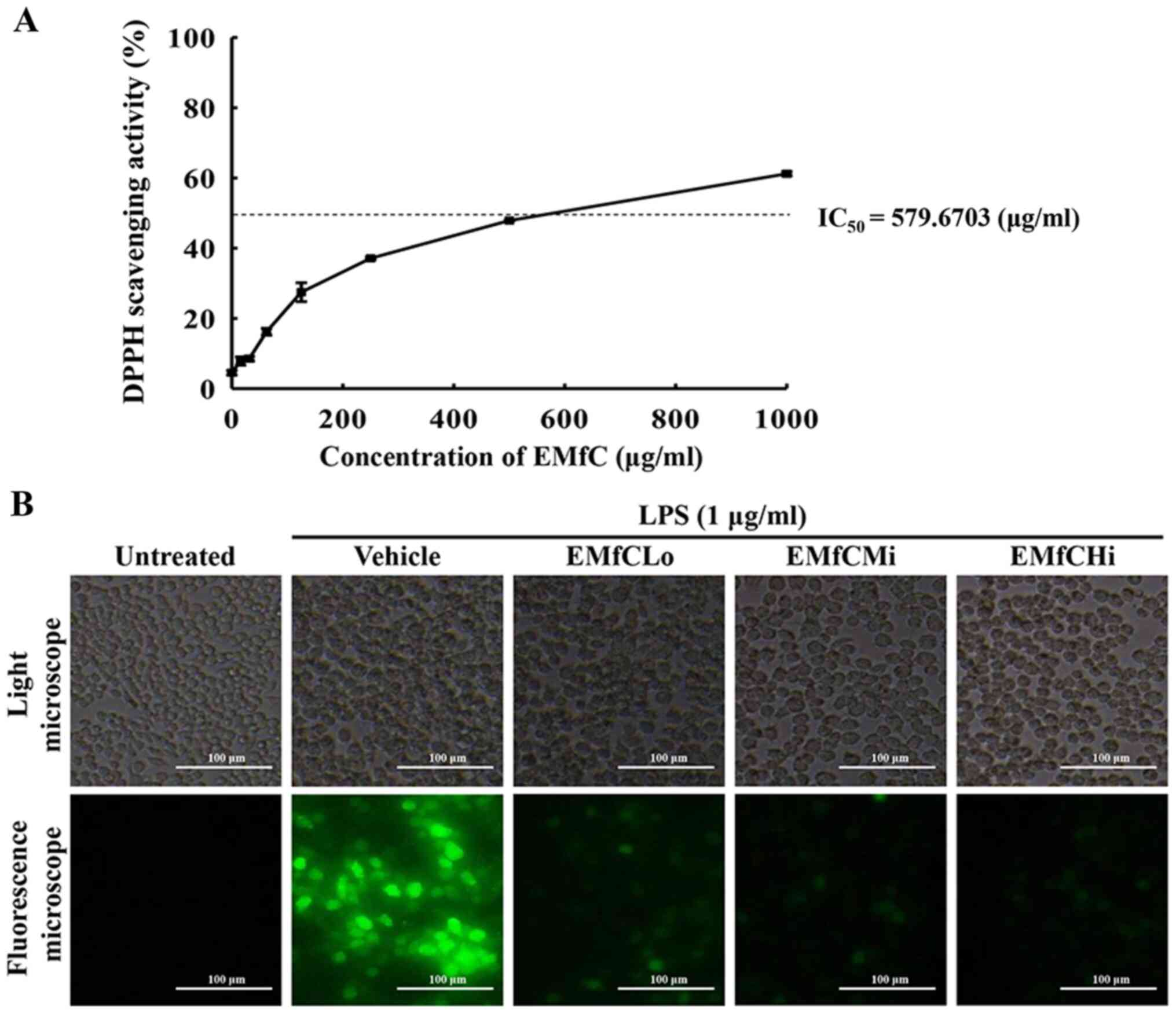 | Figure 1.Determination of DPPH radical
scavenging activity and intracellular ROS production. (A) DPPH
radical scavenging activity of EMfC was assayed in a mixture
containing 0.1 mM DPPH and varying concentrations of EMfC (0-1,000
µg/ml). For each dose, two to three wells were evaluated, and the
optical density was measured in duplicate for each sample. (B)
Determination of intracellular ROS production. Following DCFH-DA
treatment of EMfC+LPS-treated RAW264.7 cells, green fluorescence
was examined in the subset groups of cells using a fluorescence
microscope. Magnification, ×200. DPPH,
2,2-diphenyl-1-picrylhydrazyl; EMfC, ethanol extracts of mulberry
leaves fermented with C. militaris; LPS, lipopolysaccharide;
ROS, reactive oxygen species; Lo, low; Mi, medium; Hi, high. |
Inhibitory effects of EMfC on the
LPS-induced inflammatory response in RAW264.7 cells
To investigate whether exposure to EMfC prevents the
LPS-induced inflammatory response, alterations in the NO
concentration, the mRNA expressions of iNOS/COX-2 and inflammatory
cytokines (TNF-α, IL-6 and IL-1β), phosphorylation of MAPK pathway
members, and cell numbers at each stage of the cell cycle, were
measured in RAW264.7 cells pretreated with three different doses of
EMfCs and subsequently stimulated with LPS. Until now, it is well
known that the NO produced by iNOS (especially the active oxygen
species produced during inflammatory responses in LPS-stimulated
macrophages) plays an important role as a mediator in the
inflammatory response. As shown in Fig. 2B, exposure to EMfC dramatically
reduces the level of NO in the inflammatory response of
LPS-stimulated macrophages, without any significant cell toxicity
(Fig. 2A). Also, treatment with
EMfC significantly increases the concentration of NO in LPS-induced
macrophages in the Vehicle+LPS group, as compared to that of the
Untreated group. However, as compared to the Vehicle+LPS group, the
NO concentration of cells pretreated with EMfCMi and EMfCHi show
significantly reduced NO levels, whereas the EMfCLo treated group
shows similar NO levels (Fig. 2B).
Furthermore, RT-PCR and RT-qPCR were applied to examine alterations
in the iNOS-mediated COX-2 induced pathway in EMfC+LPS treated
cells. Enhanced levels of iNOS and COX-2 mRNA were detected in the
Vehicle+LPS group as compared to the Untreated groups. Especially,
these increased expression levels of iNOS and COX-2 mRNA were
strongly reduced after exposure to EMfC, as compared to the
Vehicle+LPS group (Fig. 2C and
D).
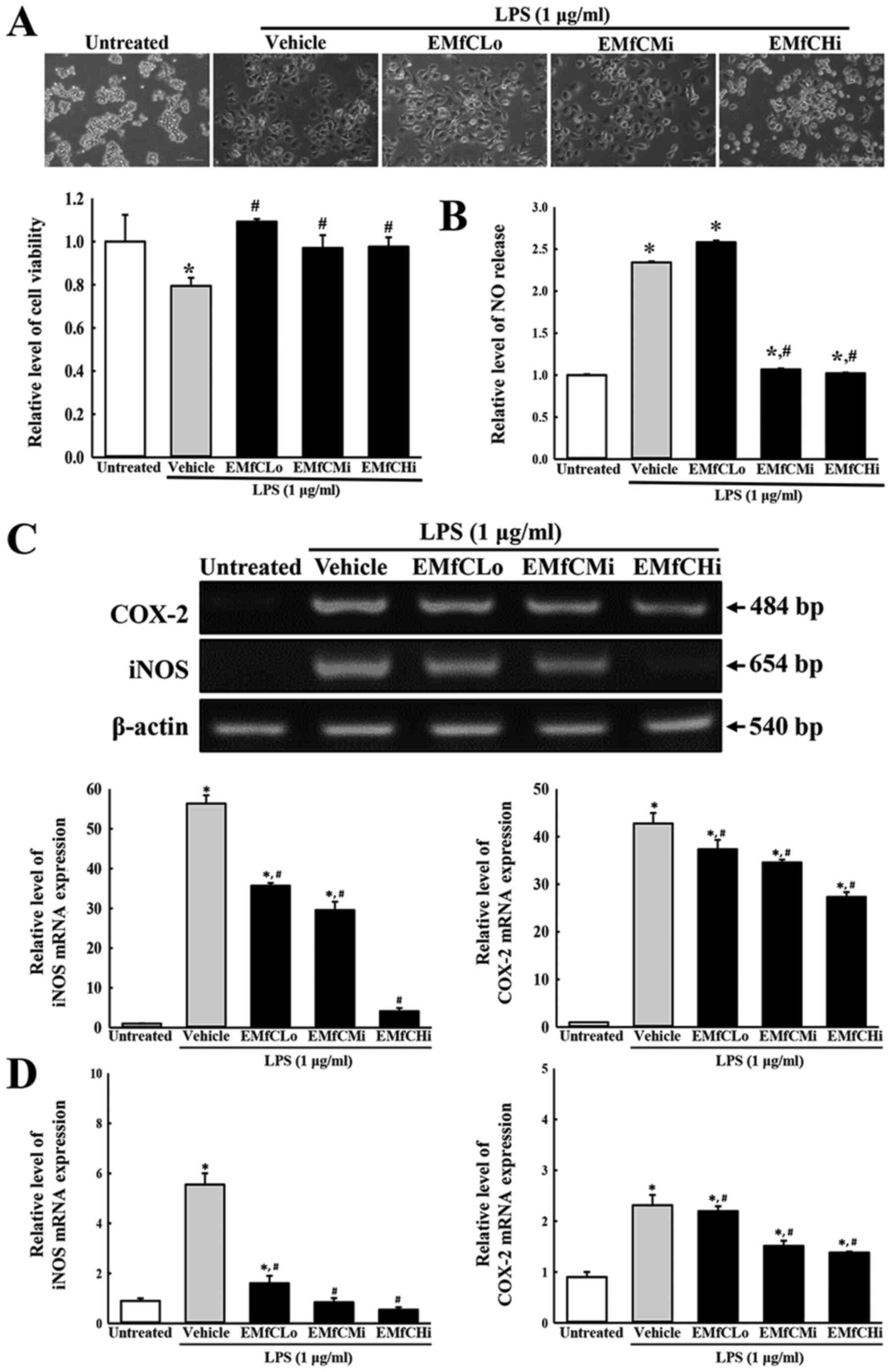 | Figure 2.Determination of cytotoxicity, NO
production and iNOS/COX-2 transcription. (A) Cytotoxicity of EMfC.
LPS-treated RAW264.7 cells were incubated in the absence or
presence of EMfC (100, 200 and 400 µg/ml) for 24 h. Cell morphology
was observed under a microscope. Magnification, ×200. Cell
viability was estimated using the MTT assay. For each group, two to
three wells were used in the MTT assay, and optical density was
measured in duplicate for each sample. (B) NO production. RAW264.7
cells (5×105 cells/ml) were treated with the Vehicle or
the indicated concentrations of EMfC, in the absence or presence of
LPS (1 µg/ml) for 24 h. The supernatants were subsequently isolated
and analyzed for nitrite. NO production was determined by measuring
nitrite accumulation in the culture medium using the Griess
reaction. For each group, two to three wells were used in the
preparation of culture supernatant, and the Griess reaction was
performed in duplicate for each sample. (C) Transcription levels of
iNOS/COX-2. Alterations in the levels of COX-2 and iNOS were
measured by RT-PCR. After the intensity of each band was determined
using an imaging densitometer, the relative levels of COX-2 and
iNOS mRNA were calculated based on the intensity of actin. (D)
Quantification of iNOS/COX-2 transcripts using RT-qPCR. For each
group, two to three wells were used in the preparation of total
RNA, and RT-PCR and RT-qPCR analysis was performed in duplicate for
each sample. Data are presented as the mean ± SD. *P<0.05 vs.
Untreated group; #P<0.05 vs. Vehicle+LPS-treated
group. NO, nitrogen oxide; COX-2, cyclooxygenase-2; iNOS, nitric
oxide synthase; EMfC, ethanol extracts of mulberry leaves fermented
with C. militaris; LPS, lipopolysaccharide; RT-qPCR, reverse
transcription-quantitative PCR; Lo, low; Mi, medium; Hi, high. |
Moreover, RT-PCR analysis for inflammatory cytokine
mRNA revealed that LPS stimulation induces a significant increase
in the generation of TNF-α, IL-6 and IL-1β mRNA in RAW264.7 cells
treated with Vehicle+LPS, as compared to controls (Untreated
group). However, compared to the Vehicle+LPS treated group, the
mRNA levels of these cytokines in the EMfC+LPS treated groups were
significantly reduced in a dose-dependent manner (Fig. 3A). A dose-dependent reduction of
IL-6 was also observed in protein production in the IL-6 ELISA
assay (Fig. 3B). Similar results
of inflammatory cytokine expressions were obtained by RT-qPCR
analysis (Fig. 3C).
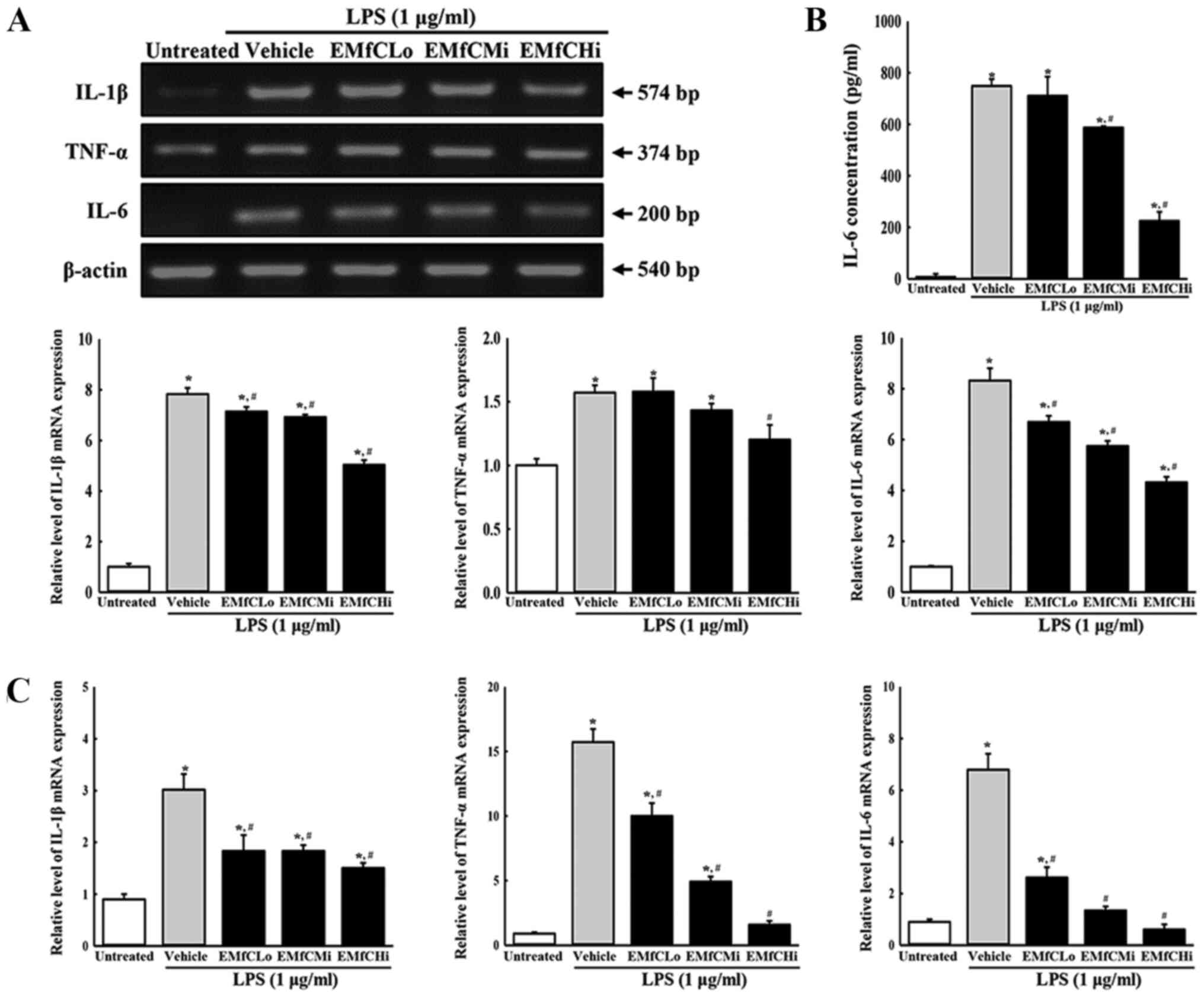 | Figure 3.Determination of inflammatory
cytokine levels. (A) Transcription levels of inflammatory
cytokines. LPS-stimulated RAW264.7 cells were treated with either
Vehicle or varying concentrations of EMfC for 3 h, and the mRNA
expression levels of TNF-α, IL-1β and IL-6 were determined by
RT-PCR. After the intensity of each band was determined using an
imaging densitometer, the relative mRNA expression levels of TNF-α,
IL-1β and IL-6 were calculated based on the intensity of actin. (B)
Secreted protein levels of IL-6. Alterations in the levels of IL-6
protein in culture supernatants of differently treated RAW264.7
cells were measured by ELISA. (C) Quantification of IL-1β, TNF-α
and IL-6 transcripts using RT-qPCR. For each group, two to three
wells were used in the preparation of total RNA and culture
supernatant, and RT-PCR, ELISA and RT-qPCR analysis was performed
in duplicate for each sample. Data are presented as the mean ± SD.
*P<0.05 vs. Untreated group; #P<0.05 vs.
Vehicle+LPS-treated group. EMfC, ethanol extracts of mulberry
leaves fermented with C. militaris; LPS, lipopolysaccharide;
RT-qPCR, reverse transcription-quantitative PCR; Lo, low; Mi,
medium; Hi, high. |
A typical signal transduction involved in
inflammation is the MAPK pathway. This pathway is reportedly
involved in inflammatory cytokine expression, control of cell
response to stress, cell growth, and differentiation. In an
inflammatory response, the MAPK signaling pathway plays a crucial
role in regulating the inflammatory cytokines. To investigate the
relationship between EMfC function and MAPK pathways, the
phosphorylation levels of ERK, JNK and p38 were evaluated in
macrophages exposed to EMfC and stimulated with LPS. As presented
in Fig. 4, phosphorylation levels
of ERK, JNK and p38 increase significantly after LPS treatment in
the Vehicle+LPS group, as compared to the Untreated group. However,
administration of EMfC to LPS-stimulated RAW264.7 cells
significantly inhibits the phosphorylation of ERK, JNK and p38, as
compared to the Vehicle+LPS group.
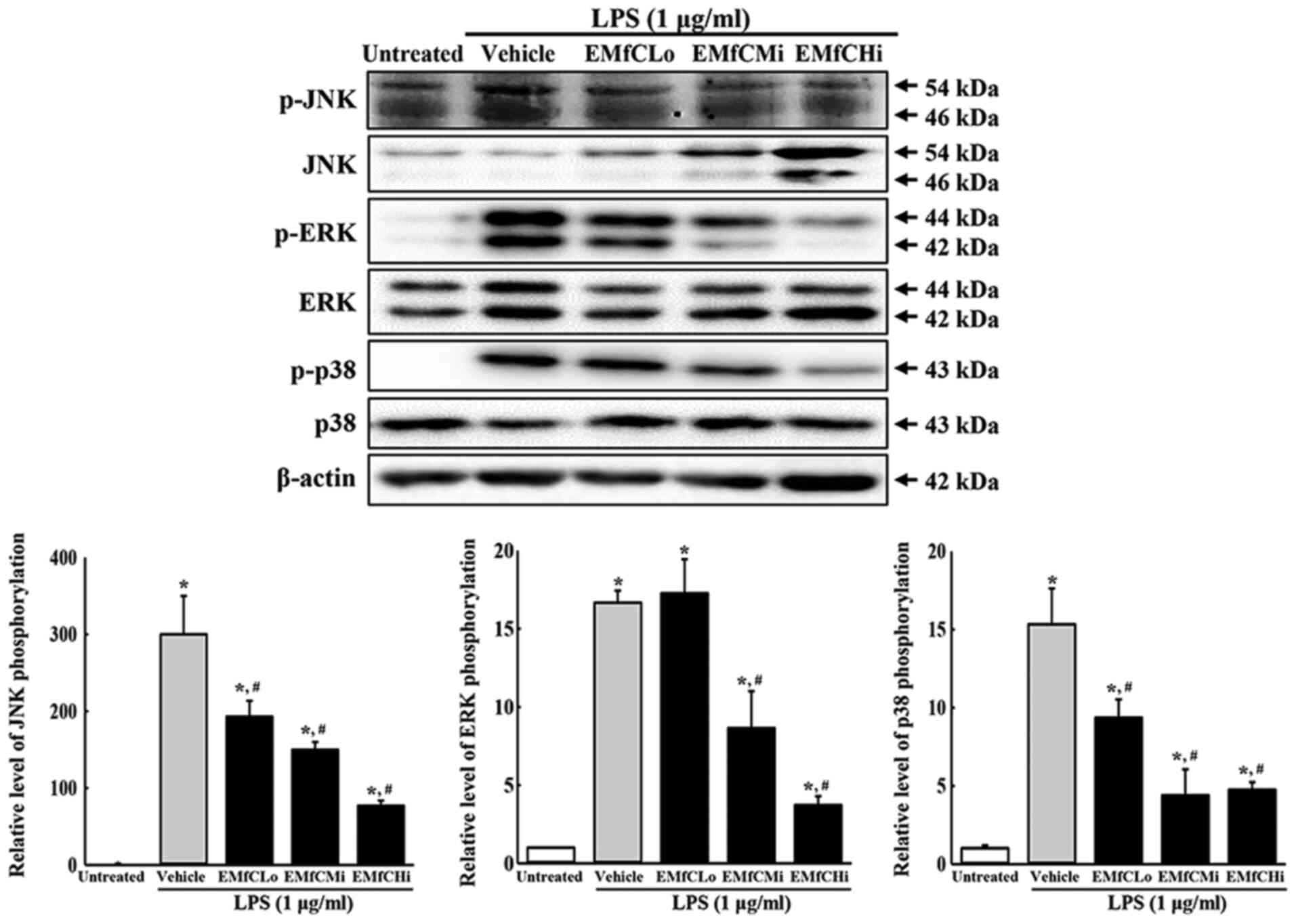 | Figure 4.Detection of MAPK pathway. Alteration
in the levels of p-PI3K/PI3K, p-mTOR/mTOR and β-actin proteins were
measured by western blotting. After the intensity of each band was
determined using an imaging densitometer, the relative levels of
the six proteins were calculated, based on the intensity of actin
protein. For each group, two to three dishes were used in the
preparation of the cell homogenates, and western blot analysis was
performed in duplicate for each sample. Data are presented as the
mean ± SD. *P<0.05 vs. Untreated group; #P<0.05
vs. Vehicle+LPS-treated group. p-, phosphorylated; EMfC, ethanol
extracts of mulberry leaves fermented with C. militaris;
LPS, lipopolysaccharide; Lo, low; Mi, medium; Hi, high. |
We further examined whether the suppression effect
of EMfC reflects on the cell cycle arrest in RAW264.7 cells,
pretreated at each concentration followed by induction of an
inflammatory response with LPS. As shown in Fig. 5, the S phase of the LPS-treated
inflammation in the Vehicle+LPS group decreases from 16.8 to 7.2%,
the G1 phase increases from 46.1 to 70.3%, and the G2/M phase
decreases from 37.1 to 22.5%. These results suggest that a G1
arrest is caused by the LPS-stimulated inflammatory reaction.
Contrarily, the G1 cycles in cells exposed from low to high EMfC
concentrations were 59.6, 53.6 and 51.7% respectively, compared to
the Vehicle+LPS group, and show minimal changes in the S phase.
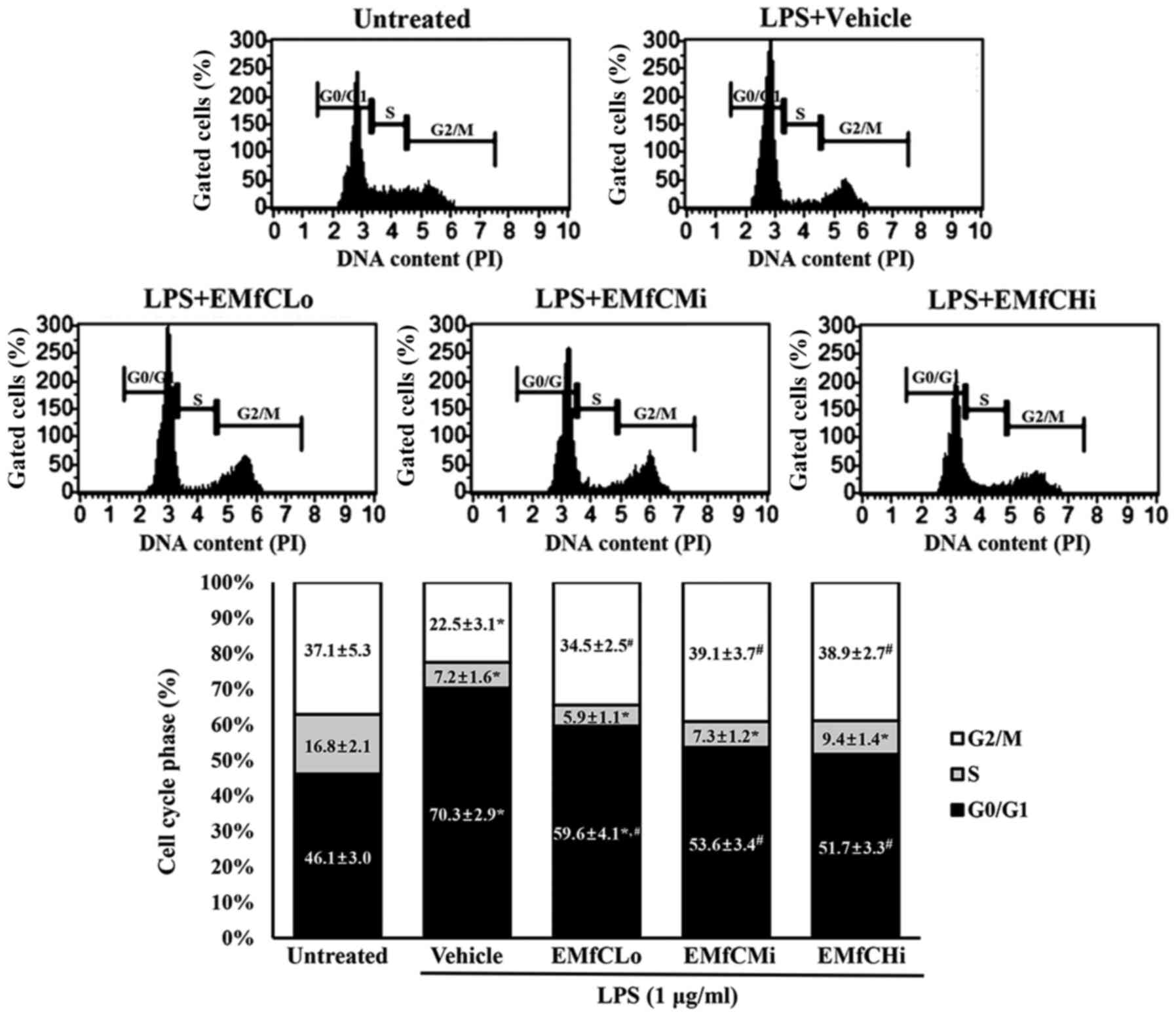 | Figure 5.Detection of cell cycle arrest. PI
staining was performed to determine the cell cycle distribution, by
flow cytometric analysis of the DNA content of nuclei of
EMfC+LPS-treated RAW264.7 cells. Following exposure to EMfC, the
numbers of cells in the G0/G1, S and
G2/M stage were determined. For each group, two to three
wells were used for PI staining, and the cell number at each phase
was counted in duplicates for each sample. Data are presented as
the mean ± SD. *P<0.05 vs. Untreated group;
#P<0.05 vs. Vehicle+LPS treated group. PI, propidium
iodide; EMfC, ethanol extracts of mulberry leaves fermented with
C. militaris; LPS, lipopolysaccharide; Lo, low; Mi, medium;
Hi, high. |
Taken together, our results suggest that EMfC
prevents the LPS-induced inflammatory response through regulation
of the iNOS-mediated COX-2 induced pathway, inflammatory cytokine
transcription, MAPK signaling pathway, and cell cycle arrest.
Suppression effects of EMfC on the
LPS-induced autophagy pathway in RAW264.7 cells
Several studies have reported the involvement of
autophagy in LPS-induced inflammation. Therefore, to determine
whether EMfC affects the autophagy pathway, we measured the surface
and total cellular expression of LC3 using the anti-LC3 antibody,
in LPS-stimulated RAW264.7 macrophages exposed to different
concentrations of EMfC. As shown in Fig. 6A, the LC3 level on the cell surface
is remarkably enhanced by LPS stimulation in the Vehicle+LPS group,
as compared to the Untreated group. However, a dose-dependent
inhibition was observed after exposure to EMfC. Also, Western blot
analysis revealed similar inhibition patterns for the total
cellular expression of LC3 (Fig.
6B). These results indicate that EMfC inhibits the LPS-induced
autophagy.
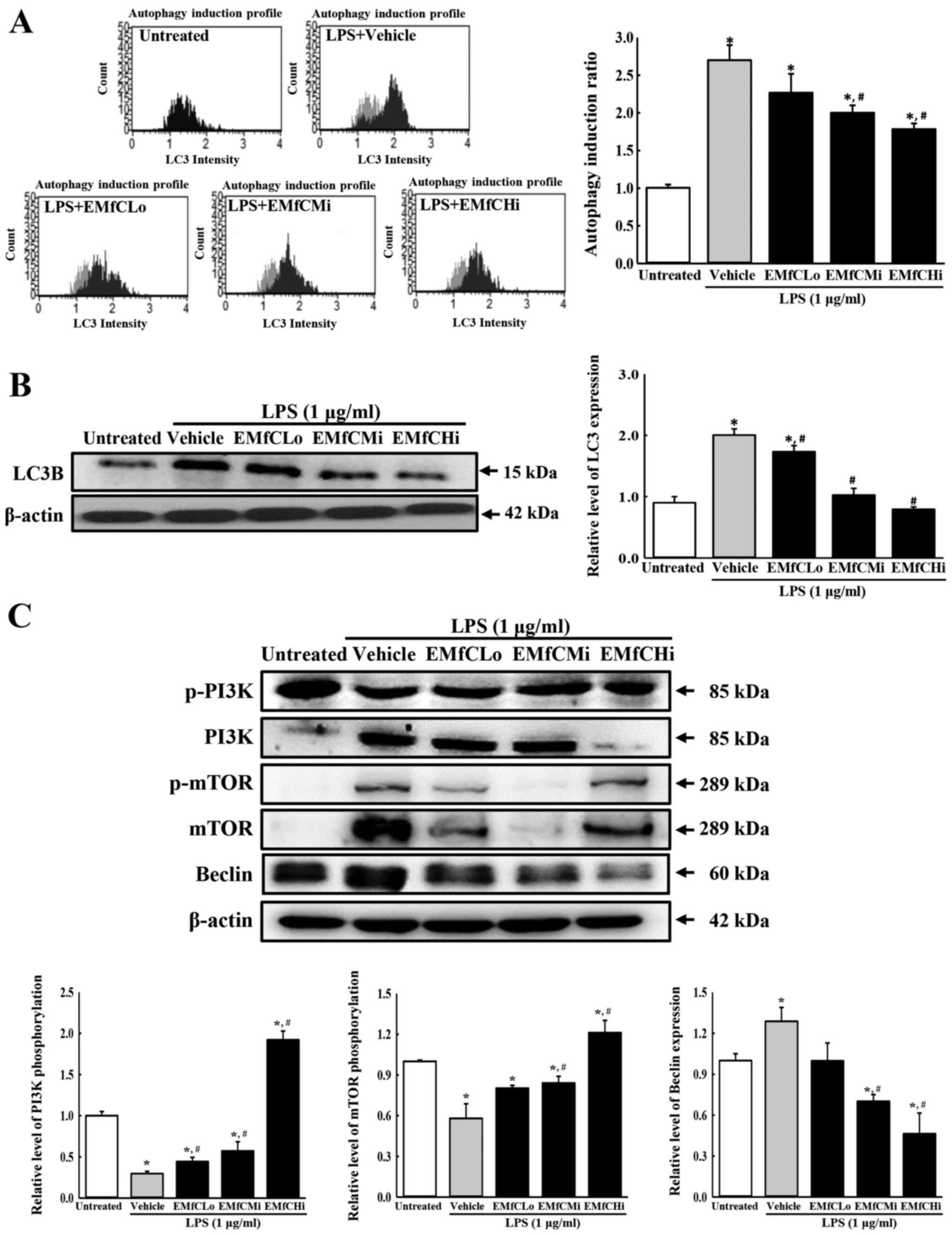 | Figure 6.Detection of autophagic vacuole and
PI3K/mTOR signaling pathway. (A) Autophagy induction profile and
ratio. RAW264.7 cells were treated with EMfC+LPS for 24 h, washed
and stained using the Autophagy LC3-antibody-based kit for FACS
analysis. For each group, two to three dishes were used in the
preparation of the LC3-stained cells, and FACS analysis was
performed in duplicate for each sample. The autophagy induction
ratio was presented as test sample fluorescence relative to
control. (B) Expression levels of LC3B. Alterations in the levels
of LC3 and β-actin proteins were measured by western blotting. (C)
Expression levels of PI3K/mTOR signaling pathway members.
Alterations in the levels of p-PI3K/PI3K, p-mTOR/mTOR, Beclin and
β-actin proteins were measured by western blotting. After the
intensity of each band was determined using an imaging
densitometer, relative levels of the five proteins were calculated
based on the intensity of actin protein. For each group, two to
three dishes were used in the preparation of cell homogenates, and
western blot analysis was performed in duplicate for each sample.
Data are presented as the mean ± SD. *P<0.05 vs. Untreated
group; #P<0.05 vs. Vehicle+LPS-treated group. EMfC,
ethanol extracts of mulberry leaves fermented with C.
militaris; LPS, lipopolysaccharide; p-, phosphorylated; FACS,
fluorescence-activated cell sorting; Lo, low; Mi, medium; Hi,
high. |
To determine the signal transduction mechanism that
inhibits the autophagy generation in LPS-stimulated macrophages by
EMfC, we measured the phosphorylation of PI3K and mTOR, and
production of Beclin protein using Western blot analysis. As seen
in Fig. 6C, the phosphorylation
levels of PI3K and mTOR in LPS-stimulated RAW264.7 cells are
significantly reduced in the Vehicle+LPS group, compared to
controls (Untreated group). However, the phosphorylation of PI3K
and mTOR are strongly upregulated in a dose-dependent manner after
EMfC treatment. The expression of Beclin, controlled by PI3K
activation, is strongly increased in the Vehicle+LPS group, where
phosphorylation of PI3K and mTOR is reduced after LPS exposure; as
expected, Beclin levels decrease with increasing phosphorylation of
PI3K and mTOR after EMfC exposure. These results indicate that EMfC
inhibits the LPS-induced autophagy via the regulation of PI3K/mTOR
pathway in RAW264.7 cells.
Discussion
Fermentation is the process wherein microbes produce
substances that are useful to humans (28). Traditionally, many foods have been
developed by fermentation, and numerous studies have proved the
beneficial effects of these fermented foods on health, by improving
the absorption rate of food and physiological activity of food
components. Recent studies have drawn attention of applying these
fermentation techniques to natural products, for obtaining more
useful and improved physiological activities. For example,
fermentation of red ginseng facilitates intraperitoneal absorption
by converting saponin, the main component of red ginseng, into
low-molecular weight material (29). In addition to increasing the
absorption rate, the study also found that fermentation enhances
the anti-inflammatory and antioxidant activity of red ginseng
(30). In another example,
fermented cultures using mushrooms such as Schizophyllum
commune, are reported to augment the antioxidant and
anti-inflammatory effects as compared to unfermented mushrooms
(31). A study on fermented C.
militaris reported the beneficial effects for hypertension
(32). Choi and Hwang (33) showed that the anti-inflammatory
activity of mulberry leaves is due to the reduction of NO and
alteration in the levels of cytokines. In the current study, we
evaluated the anti-inflammatory and anti-autophagy activities of
mulberry leaves fermented by C. militaris, and determined
the effects on NO production, iNOS-mediated COX-2 induced pathway,
inflammatory cytokines transcription, MAPK pathway, cell cycle
arrest, autophagic vacuole, and PI3K/mTOR pathway in LPS-stimulated
RAW264.7 cells exposed to the fermented product.
Oxidative stress is a condition in which the balance
of oxidative stimulators and inhibitors in the body is disturbed by
events such as inflammation. This ultimately causes oxidative
damage to cells and the human body. The active oxygen species
involved in oxidative damage is ROS, especially NO, which is known
to play a significant role in the inflammatory response (34). NO plays an important role in signal
delivery and killing bacteria under normal conditions, but
excessive NO production causes adverse side effects such as tissue
damage and genetic mutations (35). Thus, antioxidants that inhibit
oxidative stress by eliminating the active oxygen species
(including NO) are important candidates as anti-inflammatory drugs.
In this study, we observed a dose-dependent degradation of free
radicals and NO generation in LPS-stimulated RAW264.7 cells after
exposure to EMfC. No cell toxicity was observed at the EMfC
concentrations exerting the antioxidant and NO production
inhibitory effects. Moreover, EMfC also inhibited the expressions
of iNOS and COX-2 during activation of iNOS-mediated COX-2 induced
pathway.
Macrophages that engage in adaptive immunity as
antigen presenting cells and are involved in adaptive immunity by
inducing an inflammatory response by monitoring external
substances, are important cells in charge of the front line of the
immune system. In particular, during inflammatory reactions,
macrophages express toll-like receptors (TLRs) on the surface to
sensitize pathogens. TLR4 is responsible for the function of LPS
and inflammatory factors such as NO and prostaglandin E2 (PGE2),
and inflammatory cytokines such as TNF-α, IL-1β and IL-6 (26,36).
These inflammatory factors and inflammatory cytokines are the main
markers of inflammation (37). The
inflammatory factor PEG2 (active oxygen species induced by COX-2)
increases the permeability of blood vessels, resulting in fever,
abscess and pain during the inflammatory process. In addition,
inflammatory cytokines such as TNF-α, IL-1β and IL-6 also cause
fever, inflammation of endocardial cells, activation of
neutrophils, increased breakdown of muscles and fats, and, in
severe cases, septic shock (38).
We therefore evaluated the inhibitory effect of EMfC on the
production of inflammatory cytokines (TNF-α, IL-1β and IL-6) in
LPS-stimulated RAW264.7 cells. Our results indicate that EMfC
effectively and dose-dependently reduces the generation of
inflammatory factors and inflammatory cytokines induced by LPS.
The MAPK pathway corresponds to the upstream region
of the transcriptional factor NF-κB signal transduction, and is
involved in the LPS-induced inflammatory response involving TLR4.
It also regulates the growth and differentiation of cells, and
controls the cell response to cytokines and stresses. Therefore, we
investigated the MAPK pathway to determine the mechanism by which
EMfC regulates the LPS-induced inflammatory response. As presented
in Fig. 4, EMfC inhibits
phosphorylation of ERK1/2, p38 MAPK and JNK of the MAPK pathway.
Thus, although studies on NF-κB (the downstream signal transduction
of MAPK) were not conducted, EMfC was observed to play a role in
controlling MAPK, and it is assumed that EMFC also controls NF-κB.
Future research needs to examine the mechanism of EMFC that
controls NF-κB regulation. In addition, we examined LPS-induced
cell cycle changes, to determine whether regulation of the MAPK
pathway by EMfC is involved in cell growth and differentiation.
Interestingly, we observed that the G1 arrest caused after exposure
to LPS is restored by EMfC treatment.
Autophagy is the process that maintains cell
homeostasis, and is an important catabolism for disassembling
unnecessary or dysfunctional components of cells through the
lysosome (39). It is a
fundamental phenomenon in most tissues, and is also triggered by
lack of sugars and amino acids in the cell, as well as by hypoxia,
oxidative stress, and administration of chemotherapy agents
(40). It has recently been
reported that autophagy acts as an intermediary in diseases such as
cancer and diabetes, as well as in inflammatory responses (40,41).
In inflammation, autophagy helps to eliminate microbes in cells
through phagocytosis, and the phagocytosis antigens are mainly
involved in the presentation of major histocompatibility complex
class II molecules. Autophagy is also reported to be involved in
the inflammatory response, aiding the action of PAMP (cytosolic
pathogen-associated molecule pattern) and TLR7 (42,43).
In our study, we observed that EMfC exposure inhibits the
LPS-induced autophagy. In addition, EMfC also inhibits
phosphorylation of PI3K and mTOR, with a signal transduction that
regulates autophagy and ultimately inhibiting the LPS-induced
autophagy.
Our study has limitations, in that we did not
investigate the protective effects of EMfC with high or low
concentrations of LPS stimulation. In most previous studies,
RAW264.7 cells were stimulated by LPS at 1 mg/ml, although only a
few studies applied different concentrations (44–47).
Based on previous studies, our study also established 1 mg/ml as
the optimal concentration to evaluate the protective effects of
EMfC. Moreover, this LPS concentration successfully reduced the
viability of RAW264.7 cells by ~30% in our pilot study, where we
had screened the cell viability at various concentration of LPS
(data not shown). However, more studies are required to evaluate
the protective effects of EMfC in cell models with higher or lower
concentrations of LPS, although the protective effects of EMfC have
been proven at optimum concentrations of LPS.
The results of this study reveal that EMfC prevents
the inflammatory response and autophagy pathway in LPS-induced
macrophages. This anti-inflammatory mechanism of EMfC is associated
with the regulation of iNOS-mediated COX-2 induced pathway,
expression of inflammatory cytokines, MAPK signaling pathway, and
cell cycle progression. Furthermore, EMfC exposure inhibits the
LPS-induced autophagy through activation of the PI3K/mTOR pathway.
Taken together, our results indicate that EMfC has excellent
anti-inflammatory and anti-autophagy capabilities, and is a
competitive and potential drug candidate for inflammation-related
diseases.
Acknowledgements
The authors would like to thank Miss Jin Hyang Hwang
for directing the animal care and use at the Laboratory Animal
Resources Center, Pusan National University (Miryang, Republic of
Korea).
Funding
This study was supported by a grant from the Korea
Institute of Planning Evaluation for Technology of Food,
Agriculture, Forestry and Fisheries (grant no. 116027-032-HD030; to
DYW).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MRL, JEK, JJP, JYC, BRS, DSK and DYH participated in
designing the study, sample preparation, animal experiments and
data analyses. YWC and KMK helped with sample preparation and data
analysis. HKS majorly performed the first draft preparation and
data analysis. DYH was a major contributor in experimental design,
funding management and writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luo A, Leach ST, Barres R, Hesson LB,
Grimm MC and Simar D: The microbiota and epigenetic regulation of T
helper 17/regulatory T cells: In search of a balanced immune
system. Front Immunol. 8:4172017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hawiger J: Innate immunity and
inflammation: A transcriptional paradigm. Immunol Res. 23:99–109.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Artis D and Spits H: The biology of innate
lymphoid cells. Nature. 517:293–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Isailovic N, Daigo K, Mantovani A and
Selmi C: Interleukin-17 and innate immunity in infections and
chronic inflammation. J Autoimmun. 60:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rock KL, Lai JJ and Kono H: Innate and
adaptive immune response to cell death. Immunol Rev. 243:191–205.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Waisman A, Liblau RS and Becher B: Innate
and adaptive immune responses in the CNS. Lancet Neurol.
14:945–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beutler B, Krochin N, Milsark IW, Luedke C
and Cerami A: Control of cachectin (tumor necrosis factor)
synthesis: Mechanisms of endotoxin resistance. Science.
232:977–980. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou XL, Tong Q, Wang WQ, Shi CY, Xiong W,
Chen J, Liu X and Fang JG: Suppression of inflammatory responses by
dihydromyricetin, a flavonoid from ampelopsis grossedentata, via
inhibiting the activation of NF-κB and MAPK signaling pathways. J
Nat Prod. 78:1689–1696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HY, Hwang KW and Park SY: Extracts of
Actinidia arguta stems inhibited LPS-induced inflammatory
responses through nuclear factor-κB pathway in Raw 264.7 cells.
Nutr Res. 34:1008–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Flaczyk E, Kobus-Cisowska J, Przeor M,
Korczak J, Remiszewski M, Korbas E and Buchowski M: Chemical
characterization and antioxidative properties of Polish variety of
Morus alba L. leaf aqueous extracts from the laboratory and
pilot-scale processes. Agric Sci. 4:141–147. 2013.
|
|
11
|
Thanchanit T, Surawej N and Pornanong A:
Mulberry leaves and their potential effects against cardiometabolic
risks: A review of chemical compositions, biological properties and
clinical efficacy. Pharm Biol. 56:109–118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gunjal S, Ankola AV and Bhat K: In vitro
antibacterial activity of ethanolic extract of Morus alba
leaf against pathogens. Indian J Dent Res. 26:533–536. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Raman ST, Ganeshan AK, Chen C, Jin C, Li
SH, Chen HJ and Gui Z: In vitro and in vivo
antioxidant activity of flavonoid extracted from mulberry fruit
(Morus alba L.). Pharmacogn Mag. 12:128–133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Xiang L, Wang C, Tang C and He X:
Antidiabetic and antioxidant effects and phytochemicals of mulberry
fruit (Morus alba L.) polyphenol enhanced extract. PLoS One.
8:e711442013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jo SP, Kim JK and Lim YH:
Antihyperlipidemic effects of stilbenoids isolated from Morus
alba in rats fed a high-cholesterol diet. Food Chem Toxicol.
65:213–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chan EW, Lye PY and Wong SK:
Phytochemistry, pharmacology, and clinical trials of Morus
alba. Chin J Nat Med. 14:17–30. 2016.PubMed/NCBI
|
|
17
|
Lee MR, Kim JE, Choi JY, Park JJ, Kim HR,
Song BR, Choi YW, Kim KM, Song H and Hwang DY: Anti-obesity effect
in high-fat-diet-induced obese C57BL/6 mice: Study of a novel
extract from mulberry (Morus alba) leaves fermented with
Cordyceps militaris. Exp Ther Med. 17:2185–2193.
2019.PubMed/NCBI
|
|
18
|
Lee MR, Bae SJ, Kim JE, Song BR, Choi JY,
Park JJ, Park JW, Kang MJ, Choi HJ, Choi YW, et al: Inhibition of
endoplasmic reticulum stress in high-fat-diet-induced obese C57BL/6
mice: Efficacy of a novel extract from mulberry (Morus alba)
leaves fermented with Cordyceps militaris. Lab Anim Res.
34:288–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Silva DD, Rapior S, Sudarman E, Stadler
M, Xu J, Alias SA and Hyde KD: Bioactive metabolites from
macrofungi: Ethnopharmacology, biological activities and chemistry.
Fungal Divers. 62:1–40. 2013. View Article : Google Scholar
|
|
20
|
Silva DD, Rapior S, Fons F, Bahkali AH and
Hyde KD: Medicinal mushrooms in supportive cancer therapies: An
approach to anti-cancer effects and putative mechanisms of action.
Fungal Divers. 55:1–35. 2012. View Article : Google Scholar
|
|
21
|
Kim JR, Yeon SH, Kim HS and Ahn YJ:
Larvicidal activity against plutella xylostella of
cordycepin form fruiting body of Cordyceps militaris. Pest
Manag Sci. 58:713–717. 2012. View
Article : Google Scholar
|
|
22
|
Ramesh T, Yoo SK, Kim SW, Hwang SY, Sohn
SH, Kim IW and Kim SK: Cordycepin (3′-deoxyadenosine) attenuates
age-related oxidative stress and ameliorates antioxidant capacity
in rats. Exp Gerontol. 47:979–987. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hung YP and Lee CL: Higher anti-liver
fibrosis effect of Cordyceps militaris-fermented product
cultured with deep ocean water via inhibiting proinflammatory
factors and fibrosis-related factors expressions. Mar Drugs.
15:1682017. View Article : Google Scholar
|
|
24
|
Oh H, Ko EK, Kim DH, Jang KK, Park SE, Lee
HS and Kim YC: Secoiridoid glucosides with free radical scavenging
activity from the leaves of Syringa dilatate. Phytother Res.
17:417–419. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun J, Zhang X, Broderick M and Fein H:
Measurement of nitric oxide production in biological systems by
using griess reaction assay. Sensors. 3:276–284. 2003. View Article : Google Scholar
|
|
26
|
Choi EA, Park HY, Yoo HS and Choi YH:
Anti-inflammatory effects of egg white combined with chalcanthite
in lipopolysaccharide-stimulated BV2 microglia through the
inhibition of NF-κB, MAPK and PI3K/Akt signaling pathways. Int J
Mol Med. 31:134–162. 2012.
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prescott LM, Harley JP and Klein DA:
Microbiology. 6th edition. McGraw-Hill; New York, NY: 2006
|
|
29
|
Trinh HT, Han SJ, Kim SW, Lee YC and Kim
DH: Bifidus fermentation increases hypolipidemic and hypoglycemic
effects of red ginseng. J Microbiol Biotechnol. 17:1127–1133.
2007.PubMed/NCBI
|
|
30
|
Kim YJ and Park WS: Anti-inflammatory
effect of quercetin on RAW 264.7 mouse macrophages induced with
polyinosinic-polycytidylic acid. Molecules. 21:4502016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song MH, Bae JT, Ko HJ, Jang TM, Lee JD,
Lee GS and Pyo HB: Anti-oxidant effect and anti-inflammatory of
fermented Citrus Unshiu peel extract by using
Schizophyllum commune. J Soc Cosmet Sci Korea. 37:351–356.
2011.
|
|
32
|
Liu RL, Ren SF and Wang YZ: Influence of
the fermentation Cordyceps sinensisfy powder on blood uric
acid and lipid in the cases of essential hypertension. World Clin
Drugs. 27:498–502. 2006.
|
|
33
|
Choi EM and Hwang JK: Effects of Morus
alba leaf extract on the production of nitric oxide,
prostaglandin E2 and cytokines in RAW264.7 macrophages.
Fitoterapia. 76:608–613. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
John JH: Antioxidant and prooxidant
mechanisms in the regulation of redox(y)-sensitive transcription
factors. Cell Signal. 14:879–897. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohshima H and Bartsch H: Chronic
infections and inflammatory processes as cancer risk factors:
Possible role of nitric oxide in carcinogenesis. Mutat Res.
305:253–264. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Muniandy K, Gothai S, Badran KMH, Suresh
Kumar S, Esa NM and Arulselvan P: Suppression of proinflammatory
cytokines and mediators in LPS-induced RAW 264.7 macrophages by
stem extract of alternanthera sessilis via the inhibition of
the NF-κB pathway. J Immunol Res. 2018:34306842018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin HI, Chu SJ, Wang D and Feng NH:
Pharmacological modulation of TNF production in macrophages. J
Microbiol Immunol Infect. 37:8–15. 2004.PubMed/NCBI
|
|
38
|
Abbas A, Lichtman A and Pillai S: Cellular
and Molecular Immunology. (6th edition). Seoul. 271–296. 2008.
|
|
39
|
Cuervo AM: Autophagy: In sickness and in
health. Trends Cell Biol. 14:70–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sridhar S, Botbol Y, Macian F and Cuervo
AM: Autophagy and disease: Always two sides to a problem. J Pathol.
226:255–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Deretic V: Multiple regulatory and
effector roles of autophagy in immunity. Curr Opin Immunol.
21:53–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Orvedahl A and Levine B: Eating the enemy
within: Autophagy in infectious diseases. Cell Death Differ.
16:57–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu J, Zhao Y and Aisa HA:
Anti-inflammatory effect of pomegranate flower in
lipopolysaccharide (LPS)-stimulated RAW264.7 macrophages. Pharm
Biol. 55:2095–2101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sugiyama Y, Hiraiwa Y, Hagiya Y, Nakajima
M, Tanaka T and Ogura SI: 5-Aminolevulinic acid regulates the
immune response in LPS-stimulated RAW 264.7 macrophages. BMC
Immunol. 19:412018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dong J, Li J, Cui L, Wang Y, Lin J, Qu Y
and Wang H: Cortisol modulates inflammatory responses in
LPS-stimulated RAW264.7 cells via the NF-κB and MAPK pathways. BMC
Vet Res. 14:302018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee DS, Hwang IH, Im NK, Jeong GS and Na
MK: Anti-inflammatory effect of dactyloquinone B and
cyclospongiaquinone-1 mixture in RAW264.7 macrophage and ICR mice.
Nat Prod Sci. 21:268–272. 2015. View Article : Google Scholar
|




















