Introduction
Acute promyelocytic leukemia (APL) accounts for
10–12% of all cases of acute myeloid leukemia worldwide, with a
characteristic chromosomal abnormality t(15;17)(q22;q23) and a
specific promyelocytic leukemia protein (PML)/retinoic acid
receptor α (RARα) fusion protein (1,2). APL
results from a blockade of granulocyte differentiation at the
promyelocytic stage and is associated with a high incidence of
coagulopathy, including disseminated intravascular coagulation,
fibrinolysis and proteolysis (3).
In APL, there is a tendency to bleed, disproportionate to
thrombocytopenia (4). APL blasts
in the bone marrow can lead to megakaryocyte inhibition. Besides,
the promyelocytes of APL are able to activate the coagulation
cascade and increase procoagulant activity in endothelial cells
(3). The coagulopathy would also
induce thrombocytopenia. Currently, combined all-trans retinoic
acid (ATRA) and arsenic trioxide (As2O3)
treatment is recommended as the first-line treatment for low-risk
APL worldwide (5), with a complete
remission rate of 94–100%, a 2-year overall survival rate of 97%
and a 4-year survival rate of 93% (6,7). It
has been reported that arsenic agent can bind to the PML portion of
PML/RARα protein and activate the ubiquitin-proteasome system,
leading to the degradation of PML/RARα protein (8) and the apoptosis of APL cells. When
ATRA combines with RARs or retinoid X receptors of PML/RARα
protein, RARα can further induce APL cell differentiation, and
promote myeloid and granulocyte maturation (9). However, the key pathways and proteins
involved in the degradation of PML/RARα protein and apoptosis
remain unclear; and certain patients relapse with resistance to
ATRA or As2O3. Therefore, there have been
numerous studies on the APL apoptosis pathway and the mechanism of
resistance to ATRA and As2O3. Several studies
have found mutations in the PML gene that contribute to resistance
to arsenic. Goto et al (10) reported A216V and L218P mutations of
the PML protein in two arsenic-resistant cases. In 2014, a previous
study found a PML gene mutation through gene sequencing and
proposed a ‘mutation hotspot’ (c202-s220) of PML in the case of
arsenic resistance (11). Patients
with PML mutations have a high fatality rate when arsenic
resistance occurs. A French study successfully established a mouse
model of the A216V mutation, which presented resistance to arsenic
treatment (12). ATRA resistance
has a more complex mechanism. Prolonged oral administration of ATRA
can lead to increased cytochrome P450 oxidase activity, which
results in decreased blood ATRA concentration (13). Elevation of cellular retinoic
acid-binding protein contributes to a decrease in free ATRA, and a
decreased ATRA concentration in the nucleus also results in reduced
efficacy (14,15). In the present study, using
proteomics research methods, a total of 21 differentially expressed
proteins between retinoic-resistant cell lines and non-resistant
cell lines were screened and identified, among which the expression
of cofilin-1 was upregulated.
Cofilin is a 21-kD actin-binding protein that is
universally present in eukaryotes and is crucially involved in
regulating the reorganization of the cytoskeleton and muscle
development (16). In humans,
there are two cofilin gene subtypes, cofilin-1 and cofilin-2, which
encode different proteins. The former is expressed in a variety of
tissues except for muscle, whereas the latter is mainly expressed
in muscle. Cofilin can bind to F-actin, accelerate the dissociation
of actin monomers from the filament, and lead to the
depolymerization of F-actin (17).
Besides the basic function of regulating the actin cytoskeleton,
cofilin plays a role in the metastasis, infiltration and apoptosis
of tumor cells (18,19). It was previously reported that at
the early stage of apoptosis, cofilin transferred from the
cytoplasm to the mitochondria and altered the permeability of
mitochondrial membranes, leading to the release of cytochrome C and
triggering cell apoptosis via the mitochondrial apoptosis pathway
(20).
In the present study, a significant increase of
cofilin-1 expression was found in retinoic acid-resistant NB4-R1
cells. Following which, the effects of cofilin-1 in
As2O3-induced apoptosis in NB4-R1 cells were
evaluated, and the possible underlying mechanisms were explored.
The present study could provide novel strategies for APL
treatment.
Materials and methods
Reagents and antibodies
As2O3 was purchased from
Sigma-Aldrich (Merck KGaA)_and dissolved at a concentration of 5 µM
in DMSO. Primary antibodies against poly (ADP-ribose) polymerase
(PARP; cat. no. 9532), cleaved PARP (cat. no. 5625), caspase
12/cleaved caspase 12 (cat. no. 2202), cofilin (cat. no. 5175),
cytochrome C (cat. no. 11940), cytochrome c oxidase subunit 4
isoform 1 mitochondrial (COX IV; cat. no. 4850) and GAPDH (cat. no.
5174) were purchased from Cell Signaling Technology, Inc. The
annexin V-FITC Apoptosis kit was purchased from BD Biosciences.
Cell culture
Human APL cell lines, NB4-R1 and NB4 cells were
obtained from the Institute of Hematology, Shanghai Ruijin Hospital
(Shanghai, China). The NB4 cell line was derived from a patient
with APL who underwent relapse and was established by Dr Lanotte
(Saint Louis Hospital, France) in 1991, with characteristic
chromosome translocation t (15;17) and positive PML-RARα (L type)
gene. The NB4-R1 cell line is an APL subclone resistant to ATRA and
is derived from NB4 cells. The NB4-R1 and NB4 cells were maintained
in RPMI-1640 medium (Cytiva) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin in a humidified atmosphere of 5%
CO2 at 37°C. The culture medium was replaced every 3
days.
Two-dimensional gel electrophoresis
(2-DE) and image analysis
Cells were collected and solubilized in lysis buffer
containing 7 mol/l urea, 2 mol/l thiourea, 4% (w/v)
3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate
(CHAPS), 1% (w/v) dithiothreitol (DTT), 1% protease inhibitor
cocktail (v/v), and 2% (v/v) IPG buffer at 11,000 IU/min on ice.
The agents used were purchased from Promega Corporation.
Suspensions were then placed at 4°C for 1 h followed by
centrifugation at 10,000 × g for 30 min at 4°C. Supernatants were
obtained and protein concentrations were measured using the
Bradford method. Each protein sample (90 µg) was loaded on 24-cm
immobilized pH gradient (IPG) strips (pH 3–10; non-linear; Cytiva).
Following the rehydration of the IPG-strips and isoelectric
focusing, the strips were equilibrated. Subsequently, 2-DE was
performed in an Ettan-Dalt twelve electrophoresis system (Cytiva).
Following silver-staining, as previously described (21), all the gels were scanned using
ImageScanner™ (Cytiva) and the resulting images were used for
detection, quantification, matching and analysis using ImageMaster™
2D Platinum software (version 5.0; Cytiva). Each sample was run
three times to minimize the variation and an average gel was made
to represent the medium protein expression level of each group. The
protein spots that changed in all three gels were compared between
each conditioned group. The differentially expressed proteins were
further identified by matrix-assisted laser desorption ionization
(MALDI) time-of-flight (TOF) mass spectrometry (MS).
Protein identification by MS
The selected protein spots were excised from
silver-stained gels, cut into small pieces, and dehydrated in 50 µl
acetonitrile (ACN) for 5 min at room temperature. The gel pieces
were dried following removal of the acetonitrile and incubated in
50 µl 10 mM DTT at 56°C for 1 h, followed by an alkylating
incubation in 50 µl 55 mM iodoacetamide in the dark. Subsequently,
the spots were dehydrated with 50 µl ACN, rehydrated in 5 µl
trypsin for 30 min, and then 10 µl 25 mM ammonium bicarbonate was
added. Proteolysis continued overnight at 37°C and was then stopped
by adding 10 µl 2% formic acid and desalted using C18 ZipTips (EMD
Millipore). The resulting peptides were concentrated, mixed with
α-cyano-4-hydroxycinnamic acid (α-HCCA), deposited on a 384-well
MALDI target and air-dried. All samples were analyzed in the
positive-ion, reflectron mode on a TOF Ultraflex II mass
spectrometer (Bruker Corporation). The accelerating potential was
20 kV with eight shots per second. Trypsin autodigestion peaks were
used for internal calibration.
Database searching using MS/MS
data
All peptide mass fingerprinting (PMF) and MS/MS data
were used for protein identification using the MASCOT search
program (http://www.matrixscience.com) based
on the Uniprot protein database (http://www.uniprot.org). Up to one missed trypsin
cleavage per peptide was allowed, although most matches did not
contain any missed cleavages. A mass tolerance of 100 ppm was the
window of error allowed for matching the peptide mass values.
Proteins with a score >56 identified by MASCOT were considered
significant and were verified manually for spectral quality.
Lentiviral vector infection
Three short hairpin RNAs (shRNAs) targeting the
human cofilin-1 gene (GenBank no. NM_005507) for RNA interference
were designed using siRNA Wizard™ software version 3.1 (https://www.invivogen.com/sirnawizard/design.php).
The efficacy of the sequence for cofilin-1 knockdown was evaluated
using western blotting (data not shown) and the most effective one
was screened as follows: 5′-GACAGGGATCAAGCATGAA-3′. The scrambled
sequence (5′-AAUCGCAUAGCGUAUGCCGUU-3′) was used as a negative
control. Following heating and annealing, the sequence was ligated
into the AgeI and EcoRI sites of pGCSIL-GFP
(containing human U6 promoter; Shanghai GeneChem Co., Ltd.) to
generate a pGCSIL-GFP-cofilin vector, which was then transformed
into E. coli. Positive recombinant clones were selected by
DNA sequencing. The new recombinant viral vector was generated by
co-transfecting 293T cells (American Type Culture Collection) with
the lentivirus expression plasmid and packaging plasmids (pHelper
1.0 and pHelper 2.0) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. A total of 48 h after transfection, the
recombinant lentiviral vector was harvested for the subsequent
viral infection.
NB4-R1 cells were divided into two groups: i) Sc,
infected with negative control lentiviral vector; and ii)
shCofilin, infected with the pGCSIL-GFP-cofilin lentiviral vector.
Cells were cultured at a density of 6×105/well in 6-well
plates with addition of 8 µg/ml polybrene (Sigma-Aldrich; Merck
KGaA) and infected with specific or negative control lentiviral
vectors (viral titer range, 2×108−2×109
TU/ml). Following incubation for 48 h, cells were observed under a
fluorescence microscope (magnification, ×200 or ×400). The
knockdown efficiency of transfection was analyzed by reverse
transcription-quantitative PCR (RT-qPCR) and western blotting. For
the subsequent experiments, 48 h after infection, cells were
divided to four groups, sc, sc+As2O3,
shCofilin and shCofilin+As2O3. Cells is in
the sc+As2O3 or
shCofilin+As2O3 groups were then treated with
2.5 µM of As2O3, while the other two groups
were treated with an equal volume of culture medium. One day later,
cells were collected for the subsequent experiments.
Overexpression plasmid
Cofilin-1 overexpression plasmid (pcDNA3.1-cofilin)
and a control vector were designed and constructed by Shanghai
GeneChem Co., Ltd. For transfection, NB4-R1 cells at a density of
5×105/ml in 6-well plates were transfected with 3 µg
expression plasmid in each group using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. After 48 h of transfection, cells were
divided into three groups, control, As2O3 and
As2O3 + oeCofilin groups. The cells were then
treated with 2.5 µM of As2O3 (or an equal
volume of culture medium for the control). One day later, cells
were collected for subsequent experiments. The present study aimed
to investigate the effects of As2O3 on
drug-resistant cell NB4-R1, so NB4 cells were not used for the
overexpression experiments.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell viability was determined using the MTT method.
The NB4-R1 cells were inoculated in 96-well plates at
5×103 cells/well and then treated with
As2O3 of different concentrations (0, 0.625,
1.25, 2.50, 5.0, 7.5 and 10 mmol/l). After 24 h, 20 µl MTT reagent
(cat. no. ab211091; Abcam) was added into each well and incubated
for 4 h, then 150 µl DMSO was added and the plates were oscillated
until the crystalline matter was fully dissolved. The absorbance at
490 nm was measured using a spectrophotometric plate reader.
Isolation of mitochondria
The cytoplasm and mitochondria extraction was
conducted using a Mitochondria Isolation kit (cat. no. MITOISO2;
Sigma-Aldrich; Merck KGaA) in accordance with the manufacturer's
protocols. Briefly, cells from each group were collected, washed
with ice-cold PBS, and subsequently were subjected to a 5 min 600 ×
g centrifugation step at 4°C. The supernatant was discarded and
cells were incubated and homogenized with Extraction Buffer A on
ice for 10 min. Then, the homogenate was pelleted for 10 min at 700
× g at 4°C. The post-nuclear supernatant was subjected to a 25 min,
10,000 × g centrifugation step at 4°C to pellet the mitochondrial
fraction. The post-mitochondrial supernatant was collected and
regarded as the remaining cellular cytoplasm for further analysis.
The pellet was suspended with Fractionation Buffer Mix as the
mitochondrial fraction. To confirm the protein origin, GAPDH was
applied as the internal reference for cytoplasm, while COX IV, a
membrane protein in the inner mitochondrial membrane, was the
internal reference for mitochondria.
Western blot analysis
NB4-R1 cells were cultured in 6-well plates and
following the different treatments, proteins were collected and
lysed with RIPA buffer (Thermo Fisher Scientific, Inc.) containing
a protease and phosphatase inhibitor cocktail (Sigma-Aldrich; Merck
KGaA) for western blotting. Protein concentration was determined
using the BCA method (Pierce; Thermo Fisher Scientific, Inc.) and
lysates (20–40 µg/sample) were subjected to 12% SDS-PAGE and
transferred onto PVDF membranes. Membranes were blocked at room
temperature for 1 h in 5% non-fat dry milk in Tris-HCl buffer
followed by incubation with primary antibodies overnight at 4°C.
Primary antibodies (1:1,000 dilution) against COX IV and GAPDH
served as loading controls, GAPDH for cytoplasm (22) and COX IV for mitochondria (23). Membranes were then incubated with
HRP-conjugated anti-rabbit IgG secondary antibody (1:1,000; cat.
no. 7074; Cell Signaling Technology, Inc.) for 1 h at room
temperature. Following a thorough rinse, immunoreactive bands were
detected using an enhanced chemiluminescent substrate (Pierce;
Thermo Fisher Scientific, Inc.) and were visualized by the
charge-coupled device imaging system (Bio-Rad Laboratories, Inc.).
The protein expression was quantified using ImageJ 3.5 software
(National Institutes of Health). The values were normalized to the
GAPDH band.
RT-qPCR
NB4-R1 cells were cultured in 6-well plates and
treated with different concentrations (0.625, 1.25, 2.5, 5, 7.5 and
10 µmol/l) of As2O3 for 24 h. Total RNA was
extracted from cultured cells using TRIzol® reagent
(Thermo Fisher Scientific, Inc.). RT was performed using the
RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.) and the reaction was carried out in a
thermocycler at 25°C for 5 min, 42°C for 60 min, and 70°C for 5
min. The relative abundance of mRNA in each sample was determined
by qPCR using the SYBR Premix Ex Taq™ II kit (Takara Biotechnology
Co., Ltd.) on an iQ™ Multicolor Real-time PCR Detection System
(Bio-Rad Laboratories, Inc.). The corresponding primers were
designed and synthesized by Takara Biotechnology Co., Ltd., as
follows: Cofilin-1 forward, 5′-CACCTTTGTCAAGATGCT-3′ and reverse,
5′-GGAGCTGGCATAAATCAT-3′; GAPDH forward, 5′-GTCATCCCAGAGCTGAAC-3′
and reverse, 5′-TCAGTGTAGCCCAAGATG-3′. The PCR cycles were
performed under the following conditions: 95°C for 30 sec, 40
cycles at 95°C for 5 sec and 60°C for 30 sec. Data were analyzed
using the 2−ΔΔCq method (24) and GAPDH served as the internal
control. The results are presented as the mean ± standard deviation
(SD) of triplicate reactions from three separate experiments.
Flow cytometry analysis
NB4-R1 cells with or without cofilin-1 shRNA
treatment were routinely cultured in the 6-well plates, and cell
density was adjusted to 4.0×105/well. Subsequently,
cells were treated with As2O3 or an equal
amount of solvent for 24 h according to the grouping. The cells
were then collected, washed with cold PBS, stained with Annexin
V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) using
an Annexin V-FITC Apoptosis Detection kit (Merck KGaA) according to
the manufacturer's instructions, and then analyzed by a FACSCalibur
(BD Biosciences) accompanied with CellQuest™ software (version 5.1;
BD Biosciences). Each experiment was performed in triplicate wells
and repeated three times.
Fluorescence
NB4-R1 cells were cultured at a density of
1×105/well in 6-well plates, following the different
treatments. After fixation in 4% formaldehyde in PBS (pH 7.2) at
room temperature for 15 min, cells were stained with 20 µM DAPI in
antifade solution (Qbiogene, Inc.) at room temperature for 5 min.
The stained cells were visualized using a BX51 fluorescent
microscope (magnification, ×200 or ×400) equipped with a DP70
digital camera (Olympus Corporation).
Statistical analysis
All the data are reported as the mean ± SD from at
least three independent in vitro experiments. Shapiro-Wilk
test was used to test normality of data. Differences among groups
were analyzed using one-way analysis of variance, followed by
Tukey's post hoc test. All statistical analyses were performed
using the SPSS statistical software (version 19.0; IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
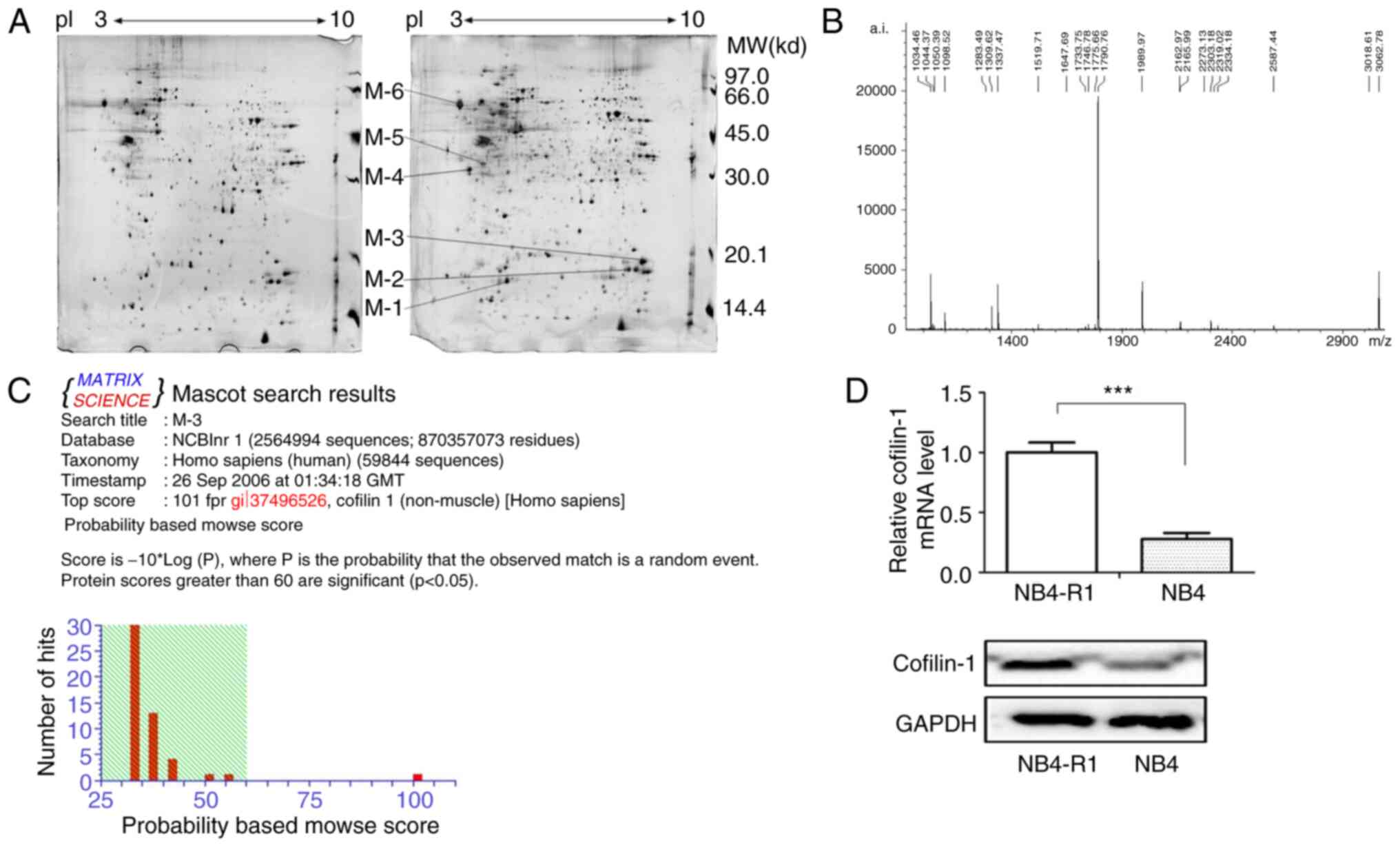
Protein expression profiles of NB4-R1
and NB4 cells
In the present study, 2-DE and silver staining were
used to analyze the proteomic profile of NB4-R1 and NB4 cells from
three independent biological replicates. The representative gel
images are shown in Fig. 1, which
indicates the total differential spots. In total, >1,000 protein
spots per gel (1,068±33 for NB4-R1 cells and 1,160±51 for NB4
cells) were detected using ImageMaster™ 2D Platinum software. One
2-DE gel of the NB4 cells was randomly selected as the reference
gel, and other gels were matched with it for analysis. The results
showed that the matching rate of protein spots was 81% in NB4 cells
and 78% in NB4-R1 cells. The matching rate between NB4-R1 and NB4
cells was 67%. Moreover, there were 21 protein spots found to be
significantly up- or downregulated, ≥2 or ≤0.5-fold, respectively,
in NB4-R1 cells (data not shown). Among these, six proteins were
upregulated, M1-M6 shown in Fig.
1A.
The M3 spot was further studied using PMF and
MALDI-TOF-MS analysis (Fig. 1B)
and the data were analyzed using the MASCOT search program
(http://www.matrixscience.com) (25,26).
The results revealed that M3 corresponded to cofilin-1 (Fig. 1C). The RT-qPCR and western blotting
results further confirmed the significantly increased expression of
cofilin-1 at the mRNA and protein levels in NB4-R1 cells, compared
with that in the NB4 cells (Fig.
1D).
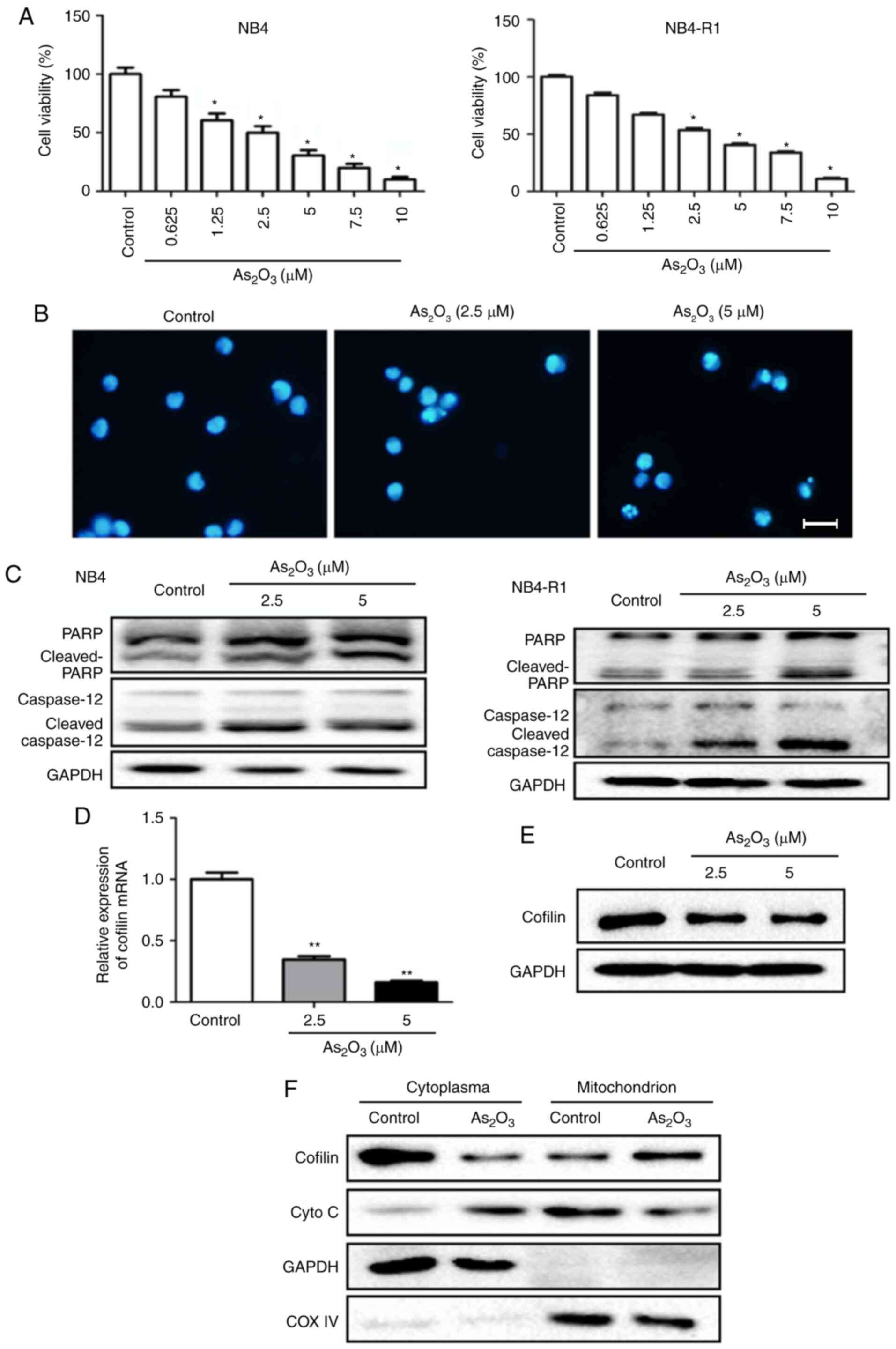
As2O3 suppresses
cell viability and promotes apoptosis in NB4-R1 or NB4 cells
NB4 and NB4-R1 cells were treated with different
concentrations (0.625, 1.25, 2.5, 5, 7.5 and 10 µmol/l) of
As2O3 for 24 h, and the results showed that
cell viability was significantly inhibited by
As2O3 at a concentration ≥2.5 µmol/l in both
cell types (Fig. 2A).
Immunofluorescence staining further indicated the inhibitory effect
of As2O3 on NB4-R1 cell growth (Fig. 2B). A similar result that
As2O3 suppressed the proliferation was also
seen in NB4 cells (data not shown). Besides, western blotting
demonstrated that both cleaved-PARP and cleaved-caspase 12 were
notably increased following As2O3 treatment
at a concentration of 2.5 and 5 µmol/l in NB4-R1 cells. Similar
results were found in NB4 cells (Fig.
2C).
Effects of As2O3
on cofilin-1 expression and transfer in NB4-R1 cells
Cytoplasmic and mitochondrial cofilin levels in
NB4-R1 cells were detected using RT-qPCR and western blotting, with
or without As2O3 treatment. The results
showed that following As2O3 treatment at 5
µmol/l for 24 h, both the mRNA level and cytoplasmic protein
content of cofilin-1 were significantly decreased, whereas the
protein level of cofilin-1 in mitochondria was markedly increased
(Fig. 2D-F). This indicated that
As2O3 could decrease the production of
cofilin-1, as well as induce cofilin-1 transfer to the mitochondria
from the cytoplasm in NB4-R1 cells. Besides, experiments were
conducted to study the effects of As2O3 on
cofilin-1 translocation in NB4 cells. The results showed that
As2O3 treatment could also induce cofilin-1
transfer to the mitochondria from the cytoplasm in NB4 cells (data
not shown).
Effects of As2O3
on the production and transfer of cytochrome C in NB4-R1 cells
In NB4-R1 cells, mitochondrial expression of
cytochrome C was notably decreased, whereas the cytoplasmic level
was markedly increased after As2O3 treatment,
compared with the control group (Fig.
2F). Combined with the result of increased cleaved-caspase 12
following As2O3 treatment (Fig. 2C), the data suggested that
cytochrome C was released to the cytoplasm, which activated the
mitochondrial apoptosis pathway at the early stage of cell
apoptosis.
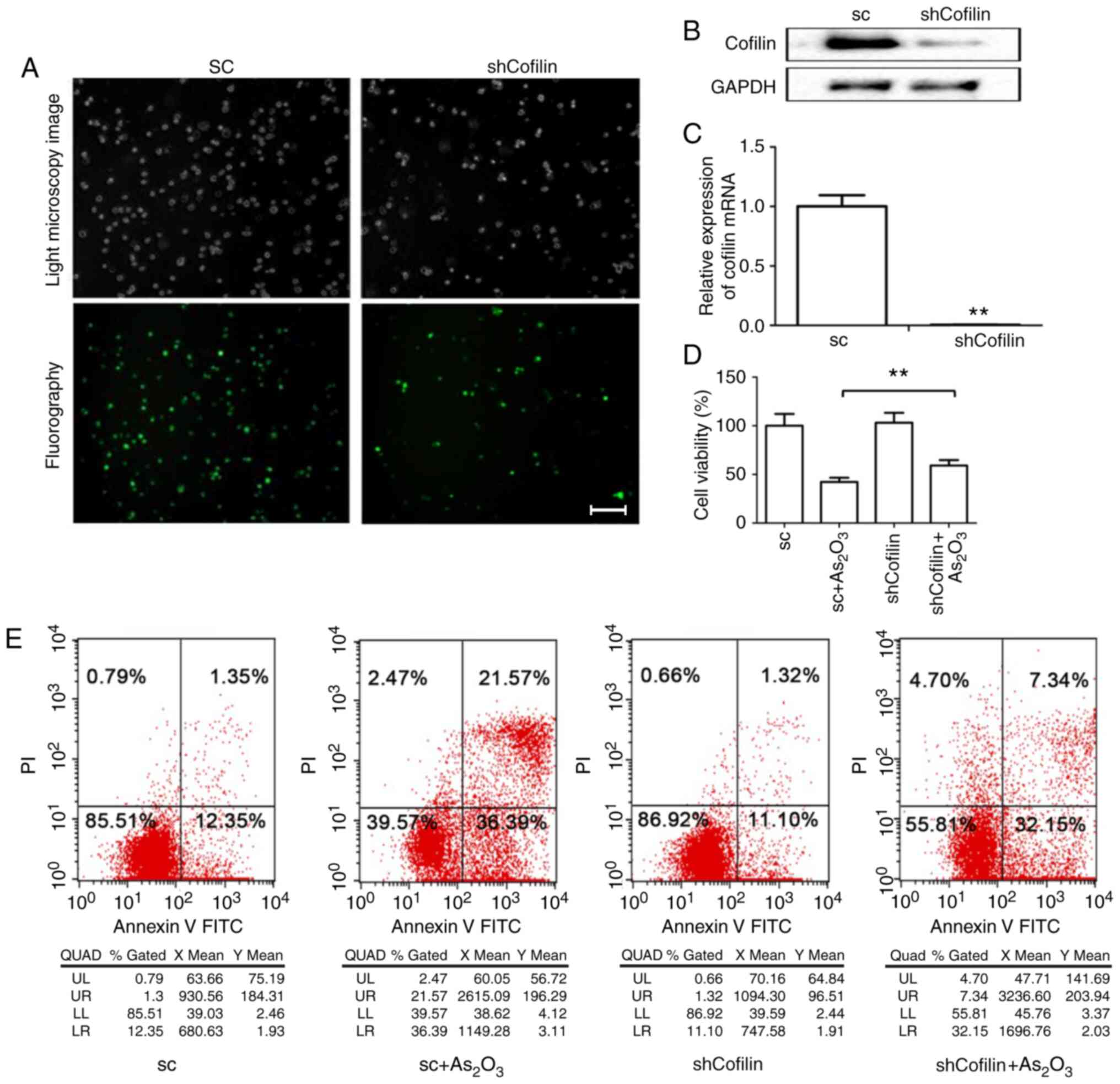
Role of cofilin-1 in
As2O3-induced apoptosis of NB4-R1 cells
A stable shCofilin cell line was obtained with a
downregulated expression of the cofilin gene (target gene sequence:
GACAGGGATCAAGCATGAA) by transfection of NB4-R1 cells with shRNA.
Immunofluorescence staining demonstrated that the recombinant viral
vectors containing shRNA were successfully transfected into the
NB4-R1 cells, and RT-qPCR and western blotting indicated that
cofilin expression was significantly decreased by shRNA (Fig. 3A-C). No significant alterations in
viability and apoptosis were found between the shRNA control and
shCofilin groups. Compared with the sc +
As2O3 group, cell viability was increased
(Fig. 3D) and apoptosis was
significantly decreased in the shCofilin +
As2O3 group (Fig. 3E).
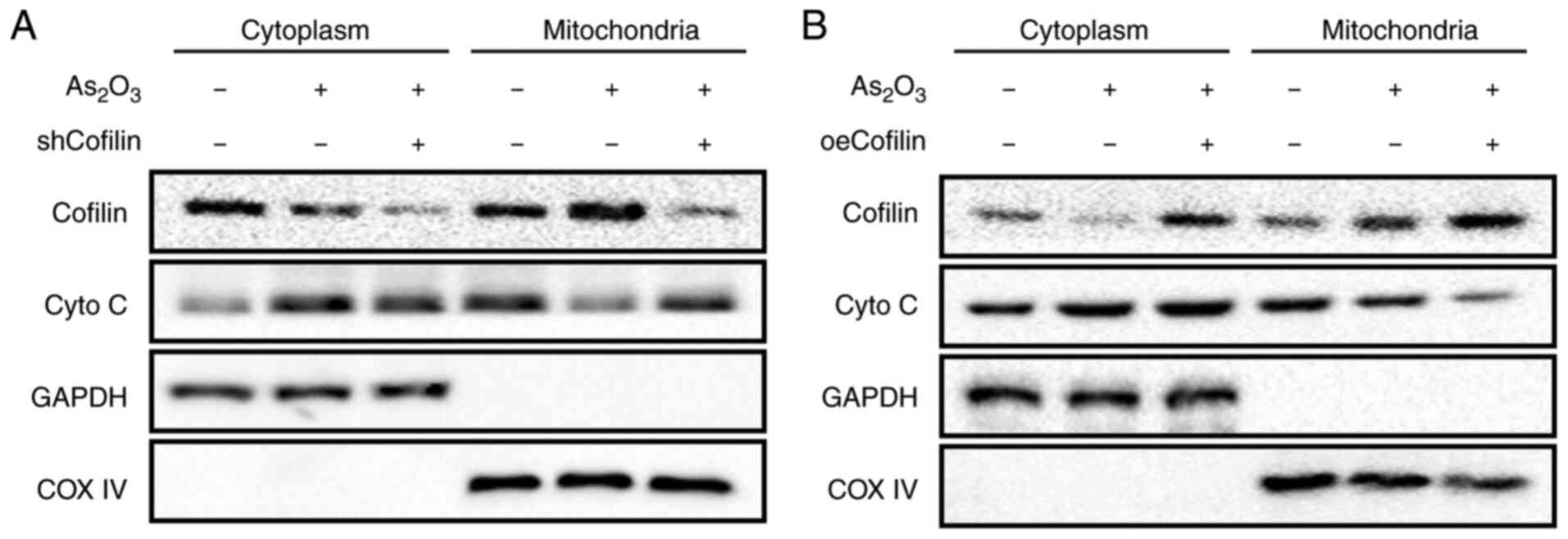
Association between cofilin-1
expression and mitochondrial cytochrome C release
The cytoplasmic and mitochondrial levels of
cytochrome C in the shCofilin and oeCofilin groups with or without
As2O3 treatment were detected using western
blotting. The results showed that in the absence of
As2O3 treatment, the cytoplasmic and
mitochondrial levels of cytochrome C were hardly affected by either
shCofilin or oeCofilin (data not shown).
As2O3 treatment markedly increased cofilin
expression and decreased cytochrome C expression in the
mitochondria, whereas cytoplasmic cytochrome C expression was
remarkably increased (Fig. 4A and
B). This result was consistent with the finding in Fig. 2F. However, in the shCofilin +
As2O3 group, mitochondrial cytochrome C
levels were notably increased while its cytoplasmic level was
markedly reduced, compared with the As2O3
group (Fig. 4A). Opposite results
were found in the oeCofilin + As2O3 group;
the mitochondrial level of cytochrome C was notably decreased and
the cytoplasmic level of cytochrome C was marginally increased,
compared with the As2O3 group (Fig. 4B). It revealed that alterations in
cofilin-1 levels had little influence on the release of cytochrome
C from the mitochondria, whereas
As2O3-induced cofilin-1 translocation was the
primary factor triggering cytochrome C release.
Discussion
Differential proteomics has been widely used in
hematological tumors. In the study, comparative proteomics were
applied to screen and identify the differentially expressed
proteins between retinoic acid-sensitive cell lines and
drug-resistant cell lines. Key proteins that may be related to
apoptosis and drug resistance were identified and the effects of
cofilin-1 on As2O3-induced apoptosis in
NB4-R1 cells were further investigated. The results of the primary
study indicated that cofilin-1 serves a role in the mitochondrial
apoptosis pathway in As2O3-treated NB4-R1
cells.
The combined treatment of ATRA and
As2O3 has achieved favorable clinical
efficacy in APL, which can be mainly attributed to the targeted
mechanism of inducing cell differentiation and apoptosis (27). However, these results are not seen
in patients with other types of acute leukemia as there are no
specific targeted drugs. Therefore, further studies are required to
illustrate the molecular mechanisms in APL and identify novel
factors that regulate differentiation and apoptosis. NB4-R1 cells
are naturally resistant to ATRA but sensitive to
As2O3, thus this cell line is a useful model
for the study of drug resistance and apoptosis (28,29).
Through protein spectrum analysis, the present study identified
that the expression of a number of proteins was significantly
increased in NB4-R1 cells, a retinoid-resistant cell line, compared
with that in NB4 cells, a retinoid-sensitive cell line.
Particularly, the expression of M3 protein was increased by
2.3-fold. Following the PMF and database retrieval, cofilin-1 was
identified for M3. Increased expression of cofilin-1 in non-small
cell lung cancer has been reported, suggesting cisplatin resistance
and poor prognosis (30). Liao
et al (31) found that
cofilin-1 expression was markedly increased in drug-resistant liver
cancer cell lines and an inhibitor of cofilin-1 could abolish drug
resistance and induce tumor cell apoptosis. Nevertheless, little is
known about the role of cofilin-1 in APL. Cofilin-1 may be involved
in the resistance of NB4-R1 cells to retinoic acid, but hardly
affect the pro-apoptotic potency of As2O3 in
NB4-R1. Thus, the present study further investigated the role of
cofilin-1 in the As2O3-induced apoptosis of
NB4-R1 cells.
The main pathways of apoptosis include extracellular
signal-triggered caspase activation and intracellular apoptotic
enzyme release from mitochondria, which activates caspase (32). At the early stage of apoptosis,
cytochrome C is released by mitochondria and binds to apoptotic
protease-activating factor 1, ATP/dATP and pro-caspase-9 in the
cytoplasm, forming an apoptotic complex. Then, the complex
activates caspase-9 and leads to apoptosis (20,33,34).
Increasing evidence has suggested that mitochondrial translocation
of cofilin seems to be necessary in regulating apoptosis. It was
reported that in the pathogenesis of Alzheimer's disease activated
cofilin could form a complex with p53 and promote its mitochondrial
localization, resulting in promotion of apoptosis (35,36).
Moreover, Li et al (37)
demonstrated that allyl isothiocyanate (AITC) could induce
dephosphorylation of cofilin, which then translocated to
mitochondria, leading to the release of cytochrome C and apoptosis.
Furthermore, it was revealed that the underlying mechanisms of
cofilin activation by AITC might involve the ROCK1/PTEN/PI3K
signaling pathway (37). In the
present study, the results indicated that
As2O3 treatment significantly decreased the
expression of cytoplasmic cofilin-1 and increased the mitochondrial
expression of cofilin-1 in NB4-R1 cells. Downregulation of
cofilin-1 expression using specific shRNAs did not have much of an
effect on the proliferation and apoptosis of NB4-R1 cells. This
suggested that alterations in the location of cofilin-1 rather than
its expression level were the primary cause of
As2O3-induced apoptosis in NB4-R1 cells,
which was consistent with the aforementioned studies (35–37).
Besides, Xing et al (38)
demonstrated that isoalantolactone inhibits IKKβ kinase activity
and promotes apoptosis of glioblastoma cells by inducing
translocation of cofilin-1 to the mitochondria and the release of
mitochondrial cytochrome C to the cytoplasm, which activates
caspase-3/9. In human leukemia cells, cofilin-1 translocation to
the mitochondria could induce mitochondrial injury and cell
apoptosis (39). Xiao et al
(40) revealed that docetaxel
induced apoptosis of prostate cancer cells by inhibiting cofilin-1
and paxillin signaling pathways. These results were consistent with
the data of the present study. However, others found that genetic
ablation of cofilin does not affect apoptosis in mouse embryonic
fibroblasts (MEF), suggesting that cofilin activity is not
generally required for inducing apoptosis (17). The observation that cytochrome C
release from mitochondria proceeds normally in the absence of
cofilin in MEF suggests that cell-type-specific functions for
cofilin in apoptosis might exist. Further studies are needed to
explore the underlying mechanisms.
The current results confirmed for the first time
that the downregulation of cofilin-1 expression suppressed
As2O3-induced apoptosis in NB4-R1 cells. The
underlying mechanism may involve the reduction of cytochrome C
release from mitochondria following cofilin-1 translocation to the
mitochondria. These results suggested that the change of cofilin-1
location occurs earlier than the release of cytochrome C in the
mitochondrial apoptosis pathway. Cofilin-1 serves as a key
regulatory point in apoptosis, and it may be a novel potential drug
target for APL treatment. Both NB4 and NB4-R1 cells are sensitive
to As2O3 (41,42).
Dual induction therapy is effective in certain patients with APL
and arsenic-resistant APL. Besides, the effectiveness of
combination therapy is higher compared with that of single drug use
(43), which is consistent with
the real-world conclusions. Further studies are needed to
investigate mechanisms by which cofilin-1 regulates apoptosis in
APL cells.
In conclusion, the present study demonstrated that
during the process of As2O3-induced apoptosis
in NB4-R1 cells, cofilin-1 is transferred to mitochondria from the
cytoplasm, which promotes the release of cytochrome C from
mitochondria and further activates the mitochondrial apoptosis
pathway. Cofilin-1 may play a key role in the mitochondrial
apoptosis pathway and has the potential to be used as a drug target
in APL treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Research
and Development Project of Shaanxi Province (grant no.
2017sf-185).
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
HZ and PH were involved in developing the concept
and design of the study, and are guarantors of the integrity of the
study. HZ and XZ performed the experiments, and prepared and
revised the manuscript. HF and WW performed the experiments. ML and
JZ were responsible for the literature review and assisted with
data analysis. XW and HW performed the data analysis and revised
the manuscript. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park JH, Qiao B, Panageas KS, Schymura MJ,
Jurcic JG, Rosenblat TL, Altman JK, Douer D, Rowe JM and Tallman
MS: Early death rate in acute promyelocytic leukemia remains high
despite all-trans retinoic acid. Blood. 118:1248–1254. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grignani F, Ferrucci PF, Testa U, Talamo
G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C,
Nicoletti I, et al: The acute promyelocytic leukemia-specific
PML-RAR alpha fusion protein inhibits differentiation and promotes
survival of myeloid precursor cells. Cell. 74:423–431. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L, Chai W, Wang Y, Cao L, Xie M, Yang
M, Kang R and Yu Y: Reactive oxygen species regulate the
differentiation of acute promyelocytic leukemia cells through
HMGB1-mediated autophagy. Am J Cancer Res. 5:714–725.
2015.PubMed/NCBI
|
|
4
|
Bassi SC and Rego EM: Molecular basis for
the diagnosis and treatment of acute promyelocytic leukemia. Rev
Bras Hematol Hemoter. 34:134–139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanz MA, Fenaux P, Tallman MS, Estey EH,
Löwenberg B, Naoe T, Lengfelder E, Döhner H, Burnett AK, Chen SJ,
et al: Management of acute promyelocytic leukemia: Updated
recommendations from an expert panel of the European LeukemiaNet.
Blood. 133:1630–1643. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lo-Coco F, Avvisati G, Vignetti M, Thiede
C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona
E, et al: Retinoic acid and arsenic trioxide for acute
promyelocytic leukemia. N Engl J Med. 369:111–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Burnett AK, Russell NH, Hills RK, Bowen D,
Kell J, Knapper S, Morgan YG, Lok J, Grech A, Jones G, et al:
Arsenic trioxide and all-trans retinoic acid treatment for acute
promyelocytic leukaemia in all risk groups (AML17): Results of a
randomised, controlled, phase 3 trial. Lancet Oncol. 16:1295–1305.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY,
Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et
al: Arsenic trioxide controls the fate of the PML-RARalpha
oncoprotein by directly binding PML. Science. 328:240–243. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esnault C, Rahmé R, Rice KL, Berthier C,
Gaillard C, Quentin S, Maubert AL, Kogan S and de Thé H: FLT3-ITD
impedes retinoic acid, but not arsenic, responses in murine acute
promyelocytic leukemias. Blood. 133:1495–1506. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goto E, Tomita A, Hayakawa F, Atsumi A,
Kiyoi H and Naoe T: Missense mutations in PML-RARA are critical for
the lack of responsiveness to arsenic trioxide treatment. Blood.
118:1600–1609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu HH, Qin YZ and Huang XJ: Resistance to
arsenic therapy in acute promyelocytic leukemia. N Engl J Med.
370:1864–1866. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lehmann-Che J, Bally C and de Thé H:
Resistance to therapy in acute promyelocytic leukemia. N Engl J
Med. 371:1170–1172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ozpolat B, Mehta K and Lopez-Berestein G:
Regulation of a highly specific retinoic acid-4-hydroxylase
(CYP26A1) enzyme and all-trans-retinoic acid metabolism in human
intestinal, liver, endothelial, and acute promyelocytic leukemia
cells. Leuk Lymphoma. 46:1497–1506. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de Thé H and Chen Z: Acute promyelocytic
leukaemia: Novel insights into the mechanisms of cure. Nat Rev
Cancer. 10:775–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang C, Ding M, Weng XQ, Sheng Y, Wu J,
Li ZY and Cai X: Combination of enzastaurin and ATRA exerts
dose-dependent dual effects on ATRA-resistant acute promyelocytic
leukemia cells. Am J Cancer Res. 9:906–926. 2019.PubMed/NCBI
|
|
16
|
Chua BT, Volbracht C, Tan KO, Li R, Yu VC
and Li P: Mitochondrial translocation of cofilin is an early step
in apoptosis induction. Nat Cell Biol. 5:1083–1089. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rehklau K, Gurniak CB, Conrad M, Friauf E,
Ott M and Rust MB: ADF/cofilin proteins translocate to mitochondria
during apoptosis but are not generally required for cell death
signaling. Cell Death Differ. 19:958–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Satoh M, Takano S, Sogawa K, Noda K,
Yoshitomi H, Ishibashi M, Mogushi K, Takizawa H, Otsuka M, Shimizu
H, et al: Immune-complex level of cofilin-1 in sera is associated
with cancer progression and poor prognosis in pancreatic cancer.
Cancer Sci. 108:795–803. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tahtamouni LH, Shaw AE, Hasan MH, Yasin SR
and Bamburg JR: Non-overlapping activities of ADF and cofilin-1
during the migration of metastatic breast tumor cells. BMC Cell
Biol. 14:452013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Zhou GL, Vedantam S, Li P and
Field J: Mitochondrial shuttling of CAP1 promotes actin- and
cofilin-dependent apoptosis. J Cell Sci. 121:2913–2920. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blough ER, Rennie ER, Zhang F and Reiser
PJ: Enhanced electrophoretic separation and resolution of myosin
heavy chains in mammalian and avian skeletal muscles. Anal Biochem.
233:31–35. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barber RD, Harmer DW, Coleman RA and Clark
BJ: GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression
in a panel of 72 human tissues. Physiol Genomics. 21:389–395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrientos A, Barros MH, Valnot I, Rötig
A, Rustin P and Tzagoloff A: Cytochrome oxidase in health and
disease. Gene. 286:53–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Villanueva J, Lawlor K, Toledo-Crow R and
Tempst P: Automated serum peptide profiling. Nat Protoc. 1:880–891.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Strupat K: Molecular weight determination
of peptides and proteins by ESI and MALDI. Methods Enzymol.
405:1–36. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McCulloch D, Brown C and Iland H: Retinoic
acid and arsenic trioxide in the treatment of acute promyelocytic
leukemia: Current perspectives. Onco Targets Ther. 10:1585–1601.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Xiao H, Zhang X, Liao W, Fu S and
Huang H: Restoration of CCAAT enhancer binding protein α P42
induces myeloid differentiation and overcomes all-trans retinoic
acid resistance in human acute promyelocytic leukemia NB4-R1 cells.
Int J Oncol. 47:1685–1695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bruel A, Benoit G, De Nay D, Brown S and
Lanotte M: Distinct apoptotic responses in maturation sensitive and
resistant t(15;17) acute promyelocytic leukemia NB4 cells. 9-cis
retinoic acid induces apoptosis independent of maturation and Bcl-2
expression. Leukemia. 9:1173–1184. 1995.PubMed/NCBI
|
|
30
|
Becker M, De Bastiani MA, Müller CB,
Markoski MM, Castro MA and Klamt F: High cofilin-1 levels correlate
with cisplatin resistance in lung adenocarcinomas. Tumour Biol.
35:1233–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao PH, Hsu HH, Chen TS, Chen MC, Day CH,
Tu CC, Lin YM, Tsai FJ, Kuo WW and Huang CY: Phosphorylation of
cofilin-1 by ERK confers HDAC inhibitor resistance in
hepatocellular carcinoma cells via decreased ROS-mediated
mitochondria injury. Oncogene. 36:1978–1990. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakagawa T, Zhu H, Morishima N, Li E, Xu
J, Yankner BA and Yuan J: Caspase-12 mediates
endoplasmic-reticulum-specific apoptosis and cytotoxicity by
amyloid-beta. Nature. 403:98–103. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Elena-Real CA, Díaz-Quintana A,
González-Arzola K, Velázquez-Campoy A, Orzáez M, López-Rivas A,
Gil-Caballero S, De la Rosa MÁ and Díaz-Moreno I: Cytochrome c
speeds up caspase cascade activation by blocking
143-3epsilon-dependent Apaf-1 inhibition. Cell Death Dis.
9:3652018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Babbitt SE, Sutherland MC, San Francisco
B, Mendez DL and Kranz RG: Mitochondrial cytochrome c biogenesis:
No longer an enigma. Trends Biochem Sci. 40:446–455. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu T, Wang F, LePochat P, Woo JA, Bukhari
MZ, Hong KW, Trotter C and Kang DE: Cofilin-mediated neuronal
apoptosis via p53 translocation and PLD1 regulation. Sci Rep.
7:115322017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang DE and Woo JA: Cofilin, a master node
regulating cytoskeletal pathogenesis in Alzheimer's disease. J
Alzheimers Dis. 72 (Suppl 1):S131–S144. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li GB, Cheng Q, Liu L, Zhou T, Shan CY, Hu
XY, Zhou J, Liu EH, Li P and Gao N: Mitochondrial translocation of
cofilin is required for allyl isothiocyanate-mediated cell death
via ROCK1/PTEN/PI3K signaling pathway. Cell Commun Signal.
11:502013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xing JS, Wang X, Lan YL, Lou JC, Ma B, Zhu
T, Zhang H, Wang D, Yu Z, Yuan Z, et al: Isoalantolactone inhibits
IKKβ kinase activity to interrupt the NF-κB/COX-2-mediated
signaling cascade and induces apoptosis regulated by the
mitochondrial translocation of cofilin in glioblastoma. Cancer Med.
8:1655–1670. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu Z, Jia X, Chen Y, Han X, Chen F, Tian
S, Su X, Li Z, Zhao J, Zhang X, et al: Identification and
characterization of key charged residues in the cofilin protein
involved in azole susceptibility, apoptosis, and virulence of
Aspergillus fumigatus. Antimicrob Agents Chemother. 62:e01659–17.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao P, Ma T, Zhou C, Xu Y, Liu Y and
Zhang H: Anticancer effect of docetaxel induces apoptosis of
prostate cancer via the cofilin-1 and paxillin signaling pathway.
Mol Med Rep. 13:4079–4084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong X, Ma N, Liu M and Liu Z: Effects of
As2O3 nanoparticles on cell growth and
apoptosis of NB4 cells. Exp Ther Med. 10:1271–1276. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chelbi-alix MK, Bobé P, Benoit G, Canova A
and Pine R: Arsenic enhances the activation of Stat1 by interferon
gamma leading to synergistic expression of IRF-1. Oncogene.
22:9121–9130. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
de Thé H, Pandolfi PP and Chen Z: Acute
promyelocytic leukemia: A paradigm for oncoprotein-targeted cure.
Cancer Cell. 32:552–560. 2017. View Article : Google Scholar : PubMed/NCBI
|