Introduction
Gastric cancer (GC) is one of the most common
malignancies and the second leading cause of cancer-related
mortality worldwide (1).
Reportedly, ~380,000 new cases of GC are recorded every year in
China alone (2). Despite therapy
for GC improving in recent years, the overall 5-year survival rate
remains unsatisfactory (3).
Therefore, it is critically important to explore target genes and
to understand their underlying molecular mechanisms for the
prognosis and treatment of GC.
Long non-coding RNAs (lncRNAs) are a group of
transcripts >200 nucleotides long that lack a protein-coding
function (4). An increasing number
of studies have indicated that lncRNAs exert important functions in
the progression of cancer by regulating the proliferation,
apoptosis, cell cycle, migration and invasion of tumor cells
(5–7). A number of studies have demonstrated
that lncRNAs serve a regulatory role in GC as oncogenes or tumor
suppressor genes (8,9). For example, lncRNA human ovarian
cancer specific transcript 2 has been reported to accelerate cell
proliferation and metastasis in GC (10). In addition, Pan et al
(11) observed that lncRNA
differentiation antagonizing non-protein coding RNA promoted GC
cell proliferation and invasion by activating spalt-like
transcription factor 4. In recent years, lncRNA VIM antisense RNA 1
(VIM-AS1) has been reported to promote cell migration and invasion
through regulating the epithelial-mesenchymal transition (EMT) in
colorectal cancer and preeclampsia (12,13).
However, to the best of our knowledge, the effect of VIM-AS1 on GC
remains to be determined.
The Wnt/β-catenin pathway has been reported to be
involved in the occurrence and development of GC (14). Numerous lncRNAs may regulate the
proliferation and metastasis of several types of tumor cells
through modulating the Wnt/β-catenin pathway (15). For example, growth-arrest
associated lncRNA 1 has been reported to inhibit the growth of GC
via inactivation of the Wnt/β-catenin pathway (16). Another study reported that the
downregulation of lncRNA TP73 antisense RNA 1 (TP73-AS1) inhibited
the proliferation and invasion of GC cells by regulating the
Wnt/β-catenin pathway (17).
However, to the best of our knowledge, it remains unknown as to
whether VIM-AS1 affects GC progression through modulating the
Wnt/β-catenin pathway.
In the present study, the role of VIM-AS1 in GC and
its underlying molecular mechanisms were explored. The results of
the present study demonstrated that VIM-AS1, which is considered to
be an oncogene, promoted cell proliferation, migration, invasion
and EMT by regulating frizzled 1 (FZD1) and activating the
Wnt/β-catenin pathway in GC. Thus, this study may provide a
strategy and facilitate the development of lncRNA-directed
diagnostic and therapeutic strategies against GC.
Materials and methods
Patients and clinical tissue
samples
A total of 50 GC tissues and 30 adjacent non-tumor
tissues (located >5 cm from the tumor) were collected from 50
patients (24 men and 26 women; age range, 26–70 years; mean age,
47.7±6.3 years) who underwent surgical resection at Shouguang
People's Hospital (Shouguang, China) between April 2014 and March
2019. None of the patients received radiotherapy or chemotherapy
before the operation. Diagnosis was confirmed in all patients
through pathological examination. GC tissues and adjacent non-tumor
tissues were stored in liquid nitrogen until use. This study was
approved by the Ethics Committee of Shouguang People's Hospital and
all patients provided written informed consent. The clinical
characteristics of the patients are presented in Table I.
 | Table I.Association between the expression of
VIM-AS1 and the clinicopathological factors of 50 patients with
gastric cancer. |
Table I.
Association between the expression of
VIM-AS1 and the clinicopathological factors of 50 patients with
gastric cancer.
|
Characteristics | Number of
cases | VIM-AS1
expression | P-value |
|---|
| Age, years |
|
|
|
|
<50 | 23 | 5.155±1.422 | 0.481 |
|
≥50 | 27 | 5.049±1.736 |
|
| Sex |
|
|
|
|
Female | 26 | 5.101±1.486 | 0.490 |
|
Male | 24 | 5.095±1.716 |
|
| TNM stage |
|
|
|
|
I/II | 17 | 4.471±0.682 |
<0.001a |
|
III/IV | 33 | 5.960±1.166 |
|
| Tumor size, cm |
|
|
|
|
>5 | 19 | 5.340±1.594 | 0.128 |
|
<5 | 31 | 4.950±1.586 |
|
| Lymph node
metastasis |
|
|
|
|
Yes | 35 | 5.998±1.177 |
<0.001a |
| No | 15 | 4.031±0.861 |
|
| Distant
metastasis |
|
|
|
|
Yes | 13 | 6.683±1.016 |
<0.001a |
| No | 37 | 4.597±1.188 |
|
| Histological
differentiation |
|
|
|
|
High/middle | 24 | 4.761±1.374 | 0.106 |
|
Low | 26 | 5.409±1.724 |
|
Cell culture and transfection
The human GC cell lines AGS and HGC-27, and the
normal gastric mucosa epithelial cell line GES-1, were purchased
from the Cell Bank of Chinese Academy of Sciences. AGS and HGC-27
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.), whereas GES-1 cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA) and 100 U/ml penicillin sodium
(Sigma-Aldrich; Merck KGaA). All cells were cultured in a
humidified 5% CO2 incubator at 37°C. AGS and HGC-27
cells were harvested at 70–80% confluence and were then transfected
with 40 nM VIM-AS1 small interfering (si)RNA (si-VIM-AS1), VIM-AS1
negative control siRNA (si-NC), miR-8052 mimics, miR-8052 mimics
negative control (mimics NC), miR-8052 inhibitor and miR-8052
inhibitor negative control (inhibitor NC) using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
cells were transfected for 24 h at 37°C and stored for subsequent
experiments. Untransfected AGS and HGC-27 cells served as the
control cells. si-VIM-AS1, si-NC, miR-8052 mimics, mimics NC,
miR-8052 inhibitor and inhibitor NC were designed and synthesized
by Sangon Biotech Co., Ltd. The sequences used were as follows:
si-VIM-AS1, 5′-GCTTGCAGAATCTTTGCTTTT-3′; si-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-8052 mimics,
5′-CGGGACUGUAGAGGGCAUGAGC-3′; mimics NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-8052 inhibitor,
5′-GCUCAUGCCCTCTACAGUCCCG-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′.
Cell Counting Kit-8 (CCK-8) assay
The effects of VIM-AS1 on GC cell proliferation were
assessed using the CCK-8 kit (Beyotime Institute of Biotechnology),
according to the manufacturer's instructions. Briefly, the
transfected AGS and HGC-27 cells were seeded into a 96-well plate
at a density of 1×104 cells/well. Subsequently, 10 µl
CCK-8 solution was added to each well at different time points,
followed by incubation at 37°C for 4 h. Finally, the optical
density of each well at a wavelength of 450 nm was detected using a
microplate reader.
Colony formation assay
After 24 h of transfection, AGS and HGC-27 cells
(1×103) were plated into a 12-well plate and incubated
in a humidified 5% CO2 incubator at 37°C. After 2 weeks
of incubation, the cells were fixed in 4% paraformaldehyde at 37°C
for 20 min and stained with 0.1% crystal violet at 37°C for 10 min.
Subsequently, the numbers of cell colonies were calculated under a
light microscope.
Flow cytometric analysis
The apoptotic ability of AGS and HGC-27 cells was
evaluated by flow cytometry. Briefly, the transfected AGS and
HGC-27 cells (1×105) were digested with trypsin and then
resuspended in 1X binding buffer. Subsequently, 5 µl Annexin
V-fluorescein isothiocyanate (FITC) and 5 µl propidium iodide from
the Annexin V-FITC apoptosis kit (BD Biosciences) were added to the
cell suspension and maintained in the dark for 15 min at room
temperature. Early and late apoptosis were then detected by a
FACSCalibur instrument (BD Biosciences) within 1 h of staining. The
results were analyzed by FlowJo software (version 7.6.3; FlowJo
LLC).
Wound-healing assay
The GC cell motility capacity was assessed by a
wound-healing assay. The transfected AGS and HGC-27 cells
(3×105 cells/well) were plated into a 6-well plate and
incubated for 24 h at room temperature to form a 90% confluent
monolayer. The cell layer was then scratched with a sterile plastic
tip and washed with PBS. Thereafter, serum-free medium was added to
each well, and the scratched monolayer was cultured for 48 h. The
images were captured at 0 and 48 h after scratching under a light
microscope, after which the wound-healing rate was calculated and
recorded. The formula used to calculate the wound-healing rate was
as follows: [wound width (0 h)-wound width (48 h)]/wound width (0
h) ×100%.
Transwell assay
The migratory and invasive capabilities of AGS and
HGC-27 cells were determined using Transwell chambers (Corning,
Inc.), according to the manufacturer's protocol. For the migration
assay, the transfected AGS and HGC-27 cells (1×105
cells/ml) in serum-free medium were seeded into the upper chamber.
For the invasion assay, the transfected AGS and HGC-27 cells
(1×105 cells/ml) were plated into the upper chamber,
which was pre-coated with Matrigel at room temperature (BD
Biosciences). Subsequently, 500 µl RPMI-1640 medium containing 10%
FBS was added to the lower chamber and cells were cultured for 24 h
in an incubator at 37°C. The chambers were then removed, and the
cells were fixed with 4% paraformaldehyde for 20 min at room
temperature and stained with crystal violet for 10 min at room
temperature. The numbers of migrating or invading cells were
counted under a light microscope in five representative fields.
Dual luciferase reporter gene
assay
TargetScan 7.2 (http://www.targetscan.org/vert_72/) and DIANA tools
(http://carolina.imis.athena-innovation.gr/) were used
to predict the targeted relationship among VIM-AS1, miR-8052 and
FZD1. For the dual luciferase reporter gene assay, AGS and HGC-27
cells (6×104 cells/well) were co-transfected with
PsiCHECK-2 luciferase plasmids (Promega Corporation) carrying
VIM-AS1-wild-type (Wt) or VIM-AS1-mutant (Mut) 3′untranslated
regions (UTRs) and miR-8052 mimics or mimics NC at 37°C using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). In addition, FZD1-Wt or FZD1-Mut 3′UTRs and
miR-8052 mimics or mimics NC were transfected into AGS and HGC-27
cells at 37°C using Lipofectamine 3000. After 48 h of transfection,
the luciferase activity was detected using a dual luciferase kit
(Promega Corporation), according to the manufacturer's protocols.
The firefly luciferase activity was normalized using renilla
luciferase activity.
Reverse transcription-quantitative
(RT-q)PCR
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from GC
tissues and cells. RNA was reverse transcribed to cDNA using
Reverse EasyScript One-Step gDNA Removal and cDNA Synthesis
SuperMix (Beijing Transgen Biotech Co., Ltd.). The conditions for
reverse transcription were as follows: 25°C for 5 min, 55°C for 20
min and 85°C for 5 min. Subsequently, qPCR was performed with a ABI
7500 Real-time PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using TransStart TipTop Green qPCR SuperMix
(Beijing Transgen Biotech Co., Ltd.) with the following primer
sequences: VIM-AS1, forward 5′-ACTGTAATGGACTCGTGGTG-3′ and reverse
5′-CGTCGTGTTGTCCTGATG-3′; miR-8052, forward
5′-CGGGACTGTAGAGGGCATGAGC-3′ and reverse 5′-AACAATTGGAGGGCTGCGG-3′;
U6, forward 5′-GCTTCGGCACATATACTAAAAT-3′, and reverse
5′-CGCTTCACGAATTTGCGTGTCAT-3′; β-catenin, forward
5′-TGCAGTTCGCCTTCACTATG-3′ and reverse 5′-ACTAGTCGTGGAATGGCACC-3′;
C-myc, forward 5′-CACAGCAAACCTCCTCACAG-3′ and reverse
5′-GGATAGTCCTTCCGAGTGGA-3′; cyclin D1, forward
5′-GAGACCATCCCCCTGACGGC-3′ and reverse 5′-TCTTCCTCCTCCTCGGCGGC-3′;
and GAPDH, forward 5′-GCTCTCTGCTCCTCCTGTTC-3′ and reverse
5′-ACGACCAAATCCGTTGACTC-3. The results were analyzed using the
2−ΔΔCq method (18).
mRNA and lncRNA expression levels were normalized to the levels of
GAPDH, whereas miRNA expression levels were normalized to U6. The
RT-qPCR reaction conditions were as follows: Initial denaturation
at 94°C for 120 sec, followed by 40 cycles at 95°C for 30 sec, 60°C
for 30 sec, 72°C for 90 sec, and a final extension step at 72°C for
5 min.
Western blot analysis
Total protein was extracted from AGS and HGC-27
cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology). A BCA protein assay kit (Beyotime Institute of
Biotechnology) was used to measure the protein concentrations.
Protein samples (50 µg) were separated by 10% SDS-PAGE and were
then transferred onto polyvinylidene difluoride (PVDF) membranes.
Membranes were blocked with 5% non-fat milk for 2 h at room
temperature, and incubated with the primary antibodies overnight at
4°C. Subsequently, the PVDF membranes were incubated with the
horseradish peroxidase-conjugated anti-rabbit IgG secondary
antibody (1:5,000; cat. no. A3687; Sigma-Aldrich; Merck KGaA) and
the anti-mouse IgG secondary antibody (1:5,000; cat. no. 2216; Cell
Signaling Technology, Inc.) at room temperature for 1 h. Finally,
the protein bands were visualized with an enhanced
chemiluminescence kit (Beyotime Institute of Biotechnology) and
analyzed with a Molecular Imager® ChemiDoc™ XRS system
(Bio-Rad Laboratories, Inc.). The primary antibodies used in this
study were as follows: Cleaved caspase-3 (1:1,000; cat. no. 9664),
caspase-3 (1:1,000; cat. no. 9662), Bax (1:1,000; cat. no. 2772),
E-cadherin (1:1,000; cat. no. 14472), N-cadherin (1:1,000; cat. no.
13116), vimentin (1:1,000; cat. no. 5741), β-catenin (1:1,000; cat.
no. 8480), cyclin D1 (1:500; cat. no. 55506), C-myc (1:1,000; cat.
no. 18583) and GAPDH (1:1,000; cat. no. 5174) (all from Cell
Signaling Technology, Inc.); Bcl-2 (1:1,000; cat. no. ab32124),
matrix metalloprotease (MMP)-2 (1:1,000; cat. no. ab97779), MMP-9
(1:1,000; cat. no. ab76003), and FZD1 (1:500; cat. no. ab83044)
(all from Abcam).
Tumor formation assay in a nude mouse
model
A total of 12 athymic BALB/c female nude mice (age,
4 weeks; weight, 18–22 g) were supplied by the Laboratory Animal
Centre of Chinese Academy of Sciences. The procedures employed for
the animal studies were approved by the Animal Use Committee of
Shouguang People's Hospital. All mice were housed in a
temperature-controlled environment (23±2°C), with 50±5% humidity,
under a 12-h light/dark cycle and were provided with free access to
food and water. The mice were randomly assigned to two groups (n=6
mice/group). Briefly, HGC-27 cells (1×107 cells/mouse)
transfected with si-VIM-AS1 or si-NC were subcutaneously injected
into the right dorsal flank of BABL/c nude mice. The tumor volume
was determined every 5 days using the following formula:
Volume=length × width2 x1/2. After 25 days, the mice
were euthanized with 100 mg/kg (1.5%) sodium pentobarbital by tail
vein injection and xenograft tumor weights were recorded. In
addition, the humane endpoint was that mice were sacrificed when
tumor size reached 2,000 mm3. None of the mice were
sacrificed during the 25 days.
Statistical analysis
All experimental data from triplicate experiments
were expressed as the mean ± standard error of the mean. The
significance differences between two groups were analyzed using a
paired Student's t-test or one-way ANOVA followed by Tukey's
multiple comparison post hoc test for multiple groups. Survival
rates were assessed by the Kaplan-Meier method, and survival curves
were compared by log-rank tests. All statistical analyses were
performed using SPSS 23.0 Statistical Software (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
VIM-AS1 expression is upregulated in
GC tissues and associated with the prognosis of GC
The association between VIM-AS1 expression and
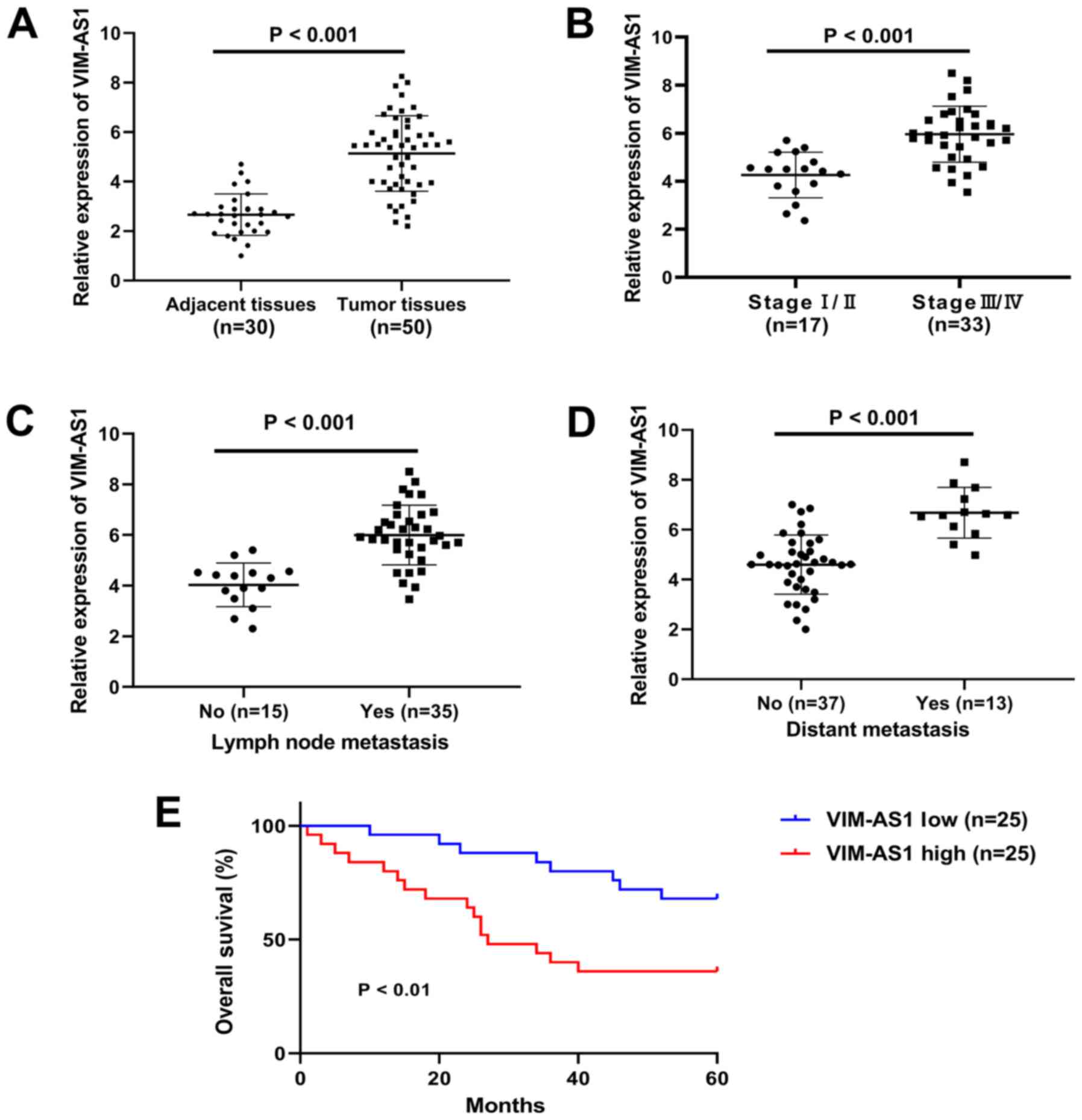
clinicopathological features was reported in Table I. The data indicated that the
expression of VIM-AS1 in patients with GC was significantly
associated with TNM stages (P<0.001), lymph node metastasis
(P<0.001) and distant metastasis (P<0.001; Table I). However, no significant
association was observed between the expression of VIM-AS1 and
other clinical characteristics, such as age, sex, tumor size and
histological differentiation (Table
I). To further evaluate the association between VIM-AS1 and GC,
RT-qPCR was performed to explore the expression of VIM-AS1 in GC
tissues. RT-qPCR results demonstrated that the expression levels of
VIM-AS1 in GC tissues were significantly higher compared with those
in cancer-adjacent tissues (P<0.001; Fig. 1A). The expression of VIM-AS1 was
also significantly increased in stage III/IV cancer tissues
compared with in stage I/II cancer tissues (P<0.001; Fig. 1B). In addition, the expression of
VIM-AS1 was significantly upregulated in tissues from patients with
lymph node metastasis and distant metastasis compared with in those
from patients without metastasis (P<0.001; Fig. 1C and D). In addition, the patients
with low VIM-AS1 expression had significantly longer overall
survival compared with patients with high VIM-AS1 expression
(P<0.01; Fig. 1E). These
results suggested that VIM-AS1 upregulation may be associated with
the progression of GC.
Silencing VIM-AS1 inhibits
proliferation of AGS and HGC-27 cells
To further investigate the effect of VIM-AS1 on GC
in vitro, the expression levels of VIM-AS1 were measured in
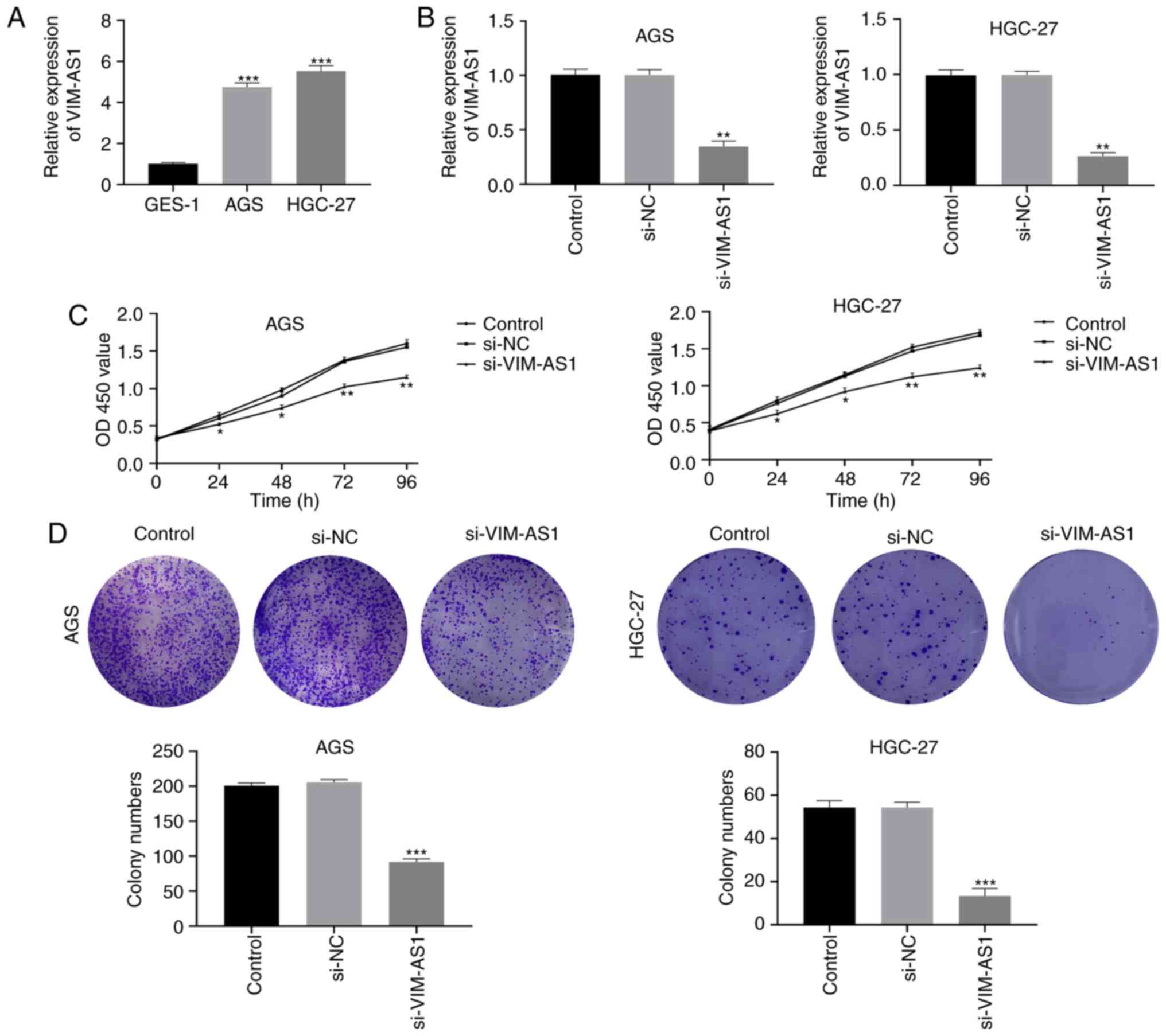
GC cell lines. The results demonstrated that the expression of
VIM-AS1 was significantly elevated in AGS and HGC-27 cells compared
with that in GES-1 cells (P<0.001; Fig. 2A). Thus, si-VIM-AS1 and si-NC were
transfected into AGS and HGC-27 cells (Fig. 2B). The results demonstrated that
VIM-AS1 expression in the si-VIM-AS1 group was significantly higher
compared with that in the si-NC group (P<0.01; Fig. 2B), which indicated that the
transfection was successful. Subsequently, the effects of VIM-AS1
on GC cell proliferation were detected by CCK-8 and colony
formation assays. The results of the CCK-8 assay demonstrated that
silencing VIM-AS1 significantly suppressed the proliferation of AGS
and HGC-27 cells 24 (P<0.05), 48 (P<0.05), 72 (P<0.01) and
96 h (P<0.01) post-transfection (Fig. 2C). In addition, colony formation
assay demonstrated that the numbers of AGS and HGC-27 cell colonies
were significantly decreased in the si-VIM-AS1 group compared with
those in the si-NC group (P<0.001; Fig. 2D). These results suggested that
silencing VIM-AS1 suppressed the proliferative ability of AGS and
HGC-27 cells.
Silencing VIM-AS1 promotes apoptosis
of AGS and HGC-27 cells
To investigate the effect of VIM-AS1 on GC cell
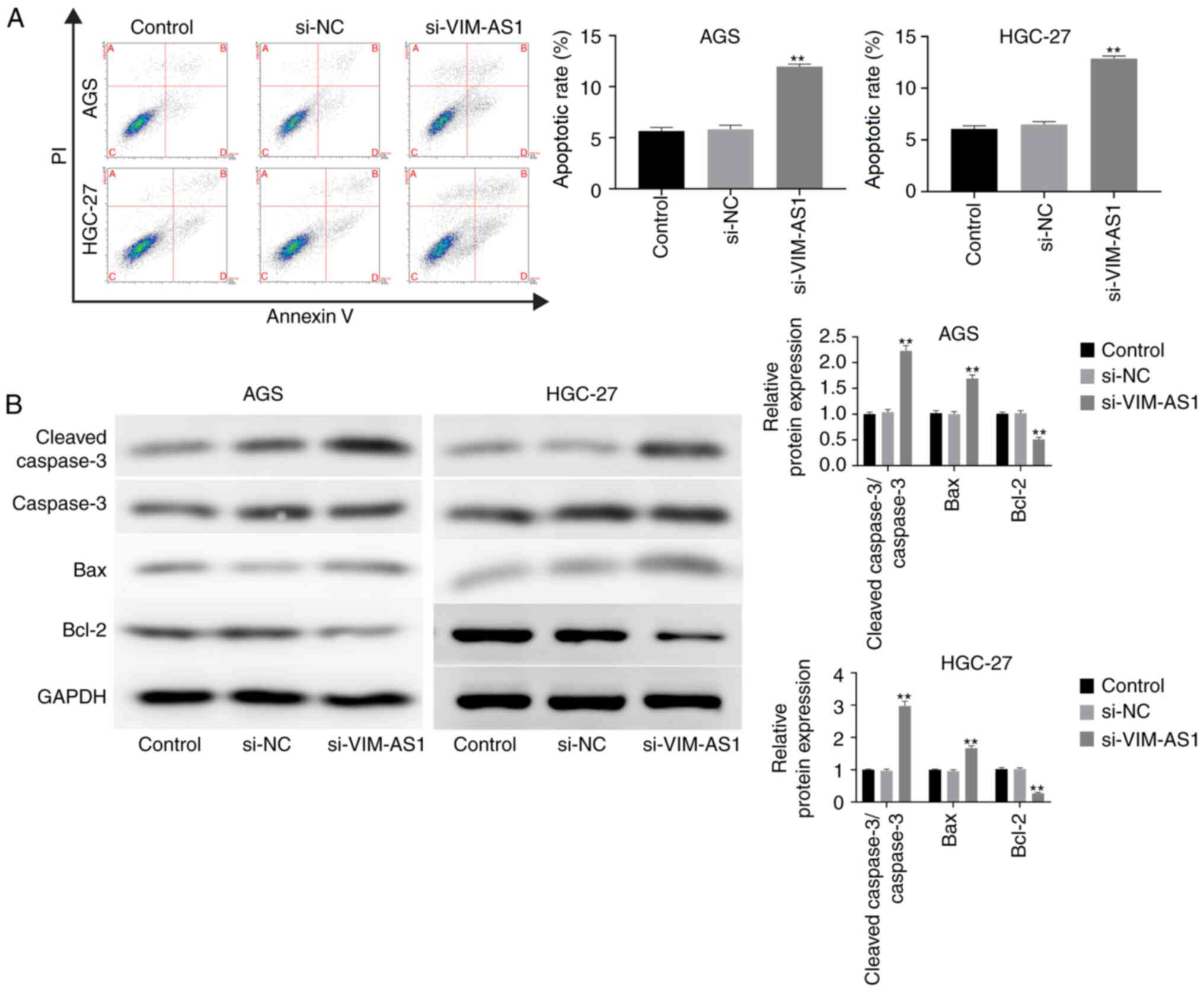
apoptosis, flow cytometry was performed. The results demonstrated
that the apoptotic rate of AGS and HGC-27 cells was significantly
enhanced in the si-VIM-AS1 group compared with that in the si-NC
group (P<0.01; Fig. 3A). In
addition, the results of western blotting demonstrated that
silencing VIM-AS1 significantly increased the expression levels of
cleaved caspase-3/caspase-3 and Bax, but decreased Bcl-2 expression
levels in AGC and HGC-27 cells compared with those in the si-NC
group (P<0.01; Fig. 3B). These
results suggested that silencing VIM-AS1 facilitated the apoptotic
ability of AGC and HGC-27 cells.
Silencing VIM-AS1 inhibits migration,
invasion and EMT in AGS and HGC-27 cells
The results of the wound-healing assay demonstrated
that cell migration was significantly inhibited in
si-VIM-AS1-transfected AGC and HGC-27 cells compared with that in
the si-NC-transfected cells (P<0.01; Fig. 4A). The Transwell assay revealed
that silencing VIM-AS1 significantly reduced the extent of
migration and invasion of AGC and HGC-27 cells compared with in the
si-NC group (P<0.001; Fig. 4B and
C). It has been reported that EMT serves an important role in
the migration and invasion of GC cells (19). Therefore, the effect of VIM-AS1 on
EMT in AGC and HGC-27 cells was investigated. The results of
western blotting demonstrated that silencing VIM-AS1 significantly
elevated E-cadherin expression (P<0.01), but reduced the
expression of N-cadherin (P<0.01), vimentin (P<0.01), MMP-2
(P<0.01) and MMP-9 (P<0.05) in AGC and HGC-27 cells compared
with those in the si-NC group (Fig.
4D). These results suggested that silencing VIM-AS1 inhibited
migration, invasion and EMT in AGS and HGC-27 cells.
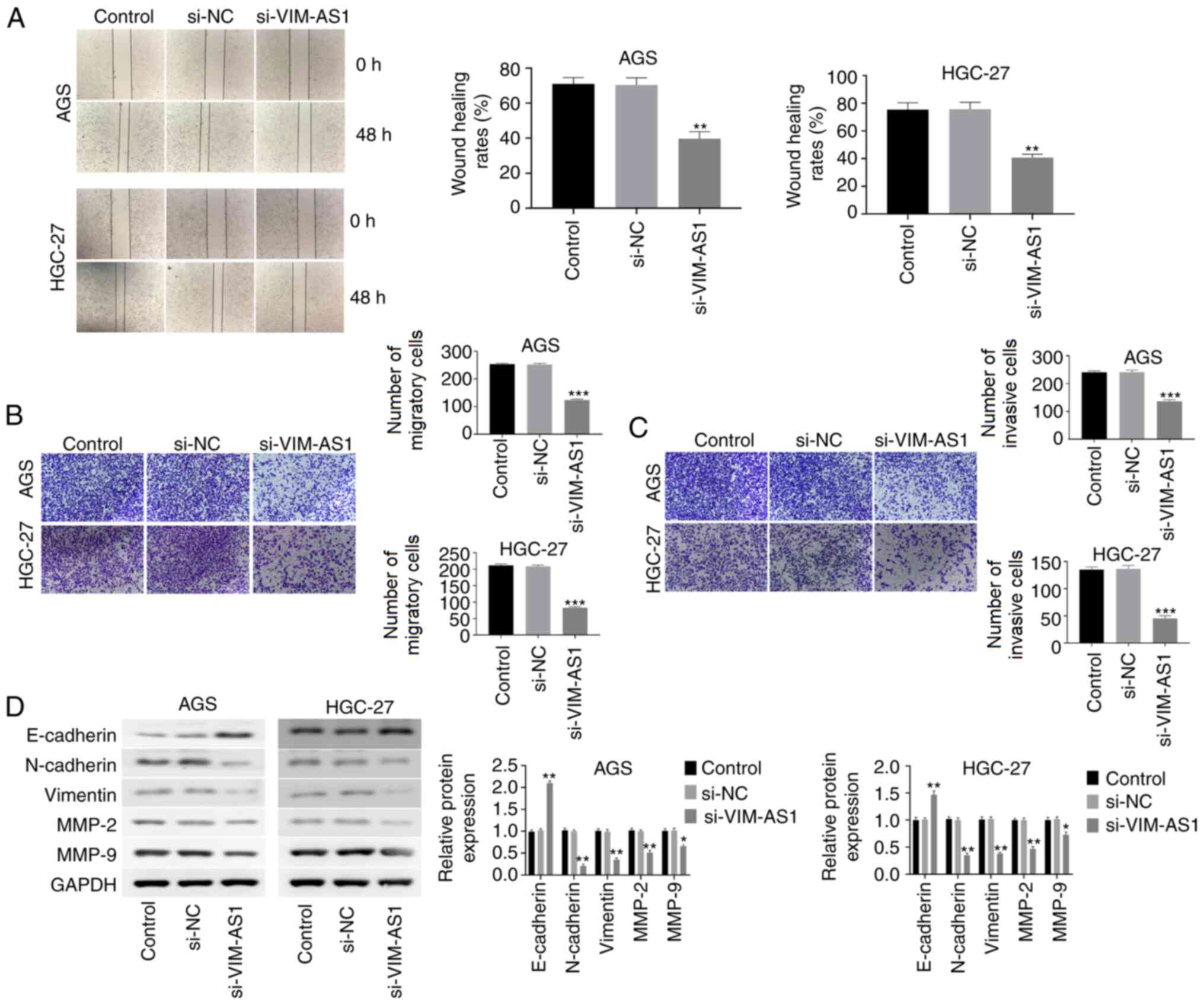 | Figure 4.Silencing VIM-AS1 inhibits the
migration, invasion and EMT of AGS and HGC-27 cells. (A)
Wound-healing assay was used to investigate the migratory ability
of AGS and HGC-27 cells (magnification, ×40). (B) Transwell assay
was performed to detect the migratory ability of AGS and HGC-27
cells (magnification, ×100). (C) Transwell assay was performed to
detect the invasive ability of AGS and HGC-27 cells (magnification,
×100). (D) Protein expression levels of E-cadherin, N-cadherin,
vimentin, MMP-2 and MMP-9 were measured by western blotting.
*P<0.05, **P<0.01, ***P<0.001 vs. si-NC group. VIM-AS1,
VIM antisense RNA 1; si, small interfering RNA; NC, negative
control; MMP, matrix metalloproteinase. |
Silencing VIM-AS1 inhibits the
expression of FZD1 in AGS and HGC-27 cells
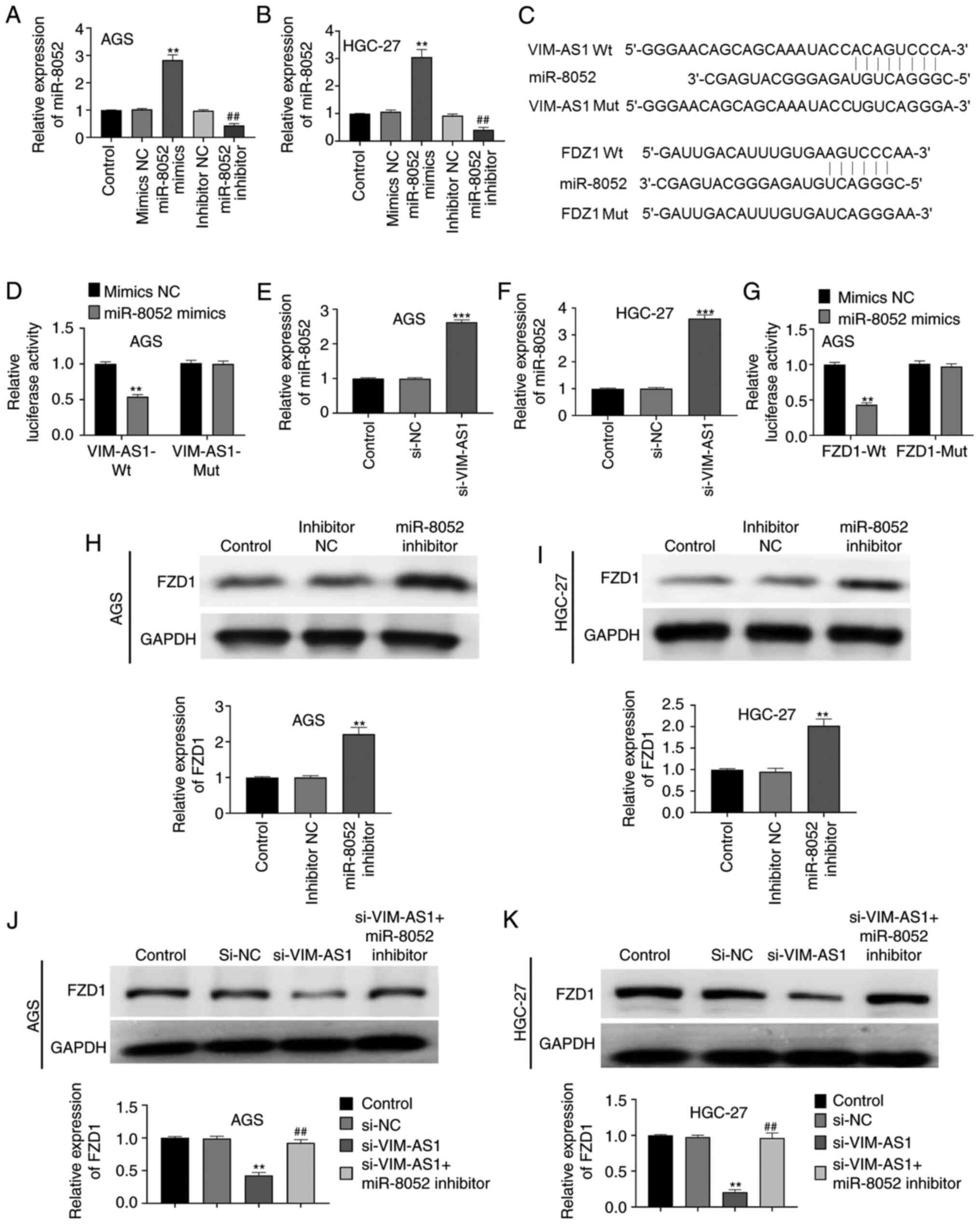
Compared with in the mimics NC or inhibitor NC
group, transfection with miR-8052 mimics significantly increased
miR-8052 expression in AGS and HGC-27 cells, whereas transfection
with the miR-8052 inhibitor significantly decreased miR-8052
expression, thus indicating that miR-8052 mimics and miR-8052
inhibitor were successfully transfected into AGS and HGC-27 cells
(P<0.01; Fig. 5A and B). The
binding sites between VIM-AS1, miR-8052 and FZD1 were predicted
using TargetScan 7.2 and DIANA tools (Fig. 5C). In addition, dual luciferase
reporter gene assay demonstrated that compared with in the mimics
NC group, miR-8052 mimics significantly reduced luciferase activity
in the VIM-AS1-Wt (P<0.01) and FZD1-Wt groups (P<0.01), but
had no significant effect on the luciferase activity in the
VIM-AS1-Mut and FZD1-Mut groups (P>0.05; Fig. 5D and G). Additionally, silencing
VIM-AS1 significantly elevated the expression levels of miR-8052 in
AGS and HGC-27 cells compared with those in the si-NC group
(P<0.001; Fig. 5E and F). To
further confirm the relationship among VIM-AS1, miR-8052 and FZD1,
western blotting was performed. The results demonstrated that the
knockdown of miR-8052 significantly increased the expression levels
of FDZ1 in AGS and HGC-27 cells compared with those in the
inhibitor NC group (P<0.01; Fig. 5H
and I). In addition, silencing VIM-AS1 significantly decreased
FZD1 expression in AGS and HGC-27 cells compared with those in the
si-NC group (P<0.01; Fig. 5J and
K). Notably, co-transfection of cells with si-VIM-AS1 and
miR-8052 inhibitor significantly reversed the effect of VIM-AS1
silencing on FZD1 expression in AGS and HGC-27 cells (P<0.01;
Fig. 5J and K). These results
suggested that silencing VIM-AS1 inhibited the expression of FZD1
in AGS and HGC-27 cells.
Silencing VIM-AS1 suppresses the
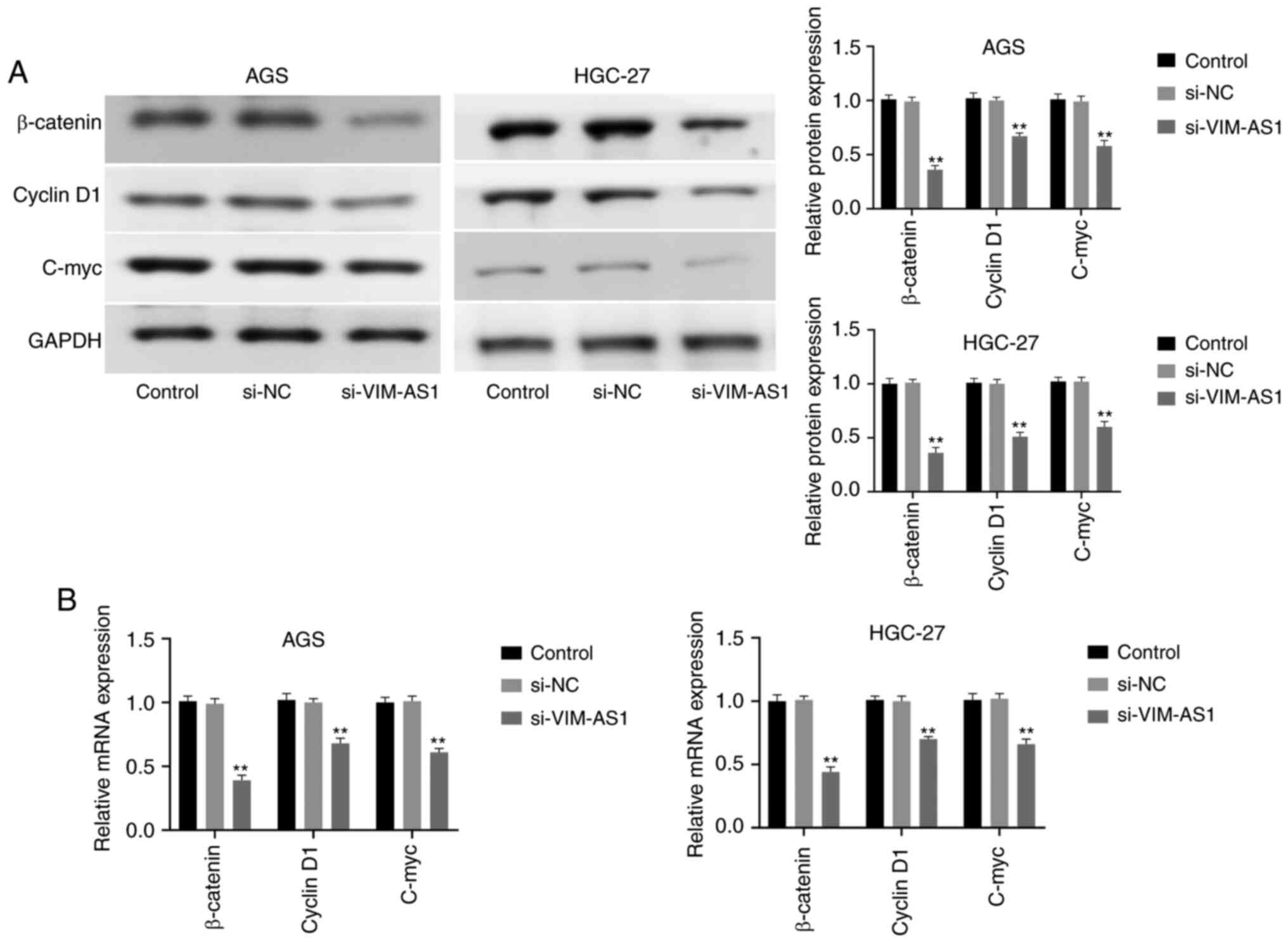
Wnt/β-catenin pathway in AGS and HGC-27 cells
To explore the effects of VIM-AS1 on the
Wnt/β-catenin pathway, the expression levels of β-catenin, cyclin
D1 and C-myc were measured by western blotting and RT-qPCR after
silencing VIM-AS1 in AGS and HGC-27 cells. The results demonstrated
that silencing VIM-AS1 significantly decreased the protein
expression levels of β-catenin (P<0.01), cyclin D1 (P<0.01)
and C-myc (P<0.01) in AGC and HGC-27 cells compared with those
in the si-NC group (Fig. 6A). In
addition, RT-qPCR confirmed that silencing VIM-AS1 significantly
decreased the mRNA expression levels of β-catenin (P<0.01),
cyclin D1 (P<0.01) and C-myc (P<0.01) compared with those in
the si-NC group (Fig. 6B). These
results suggested that silencing VIM-AS1 suppressed the
Wnt/β-catenin pathway in AGS and HGC-27 cells.
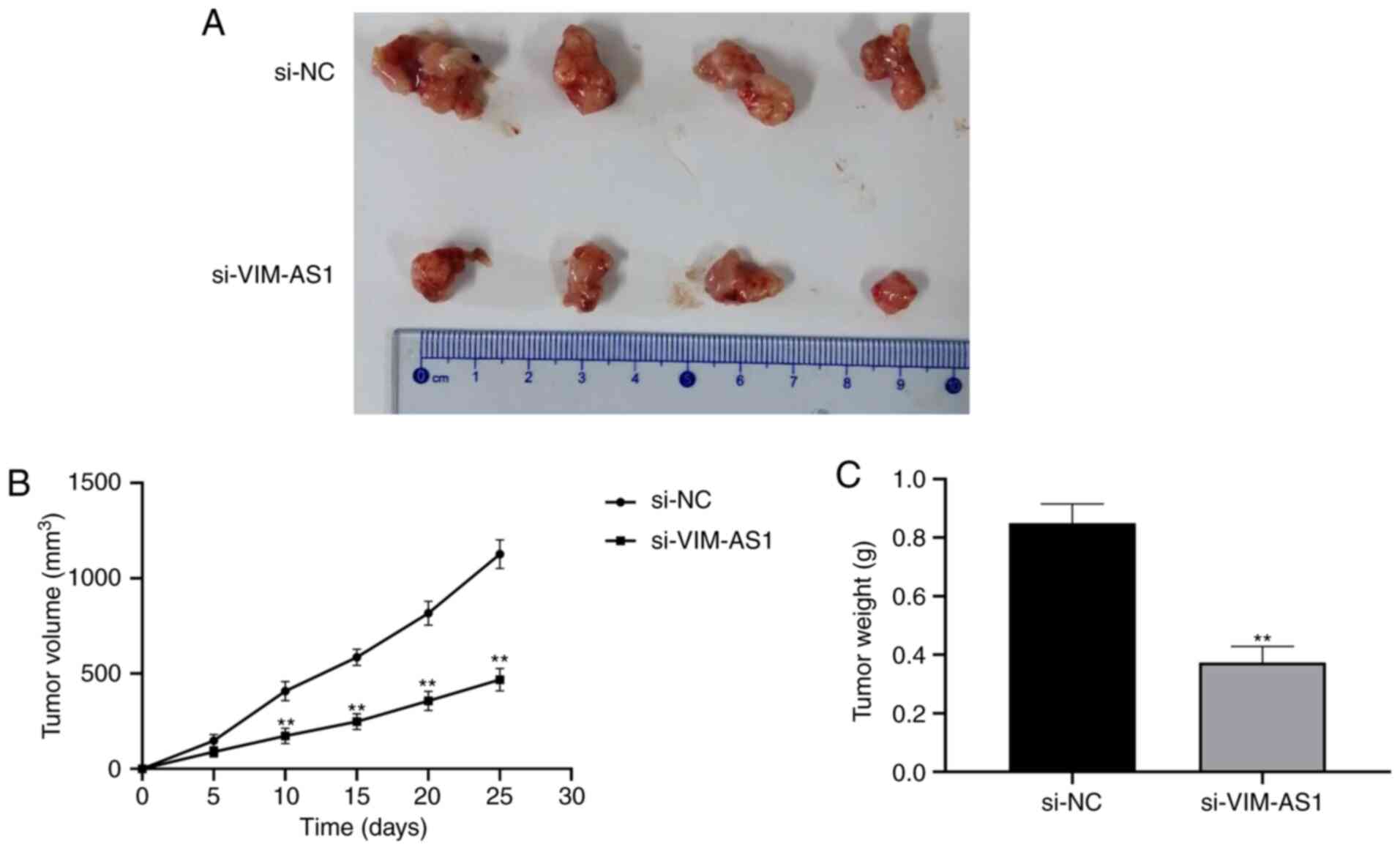
Silencing VIM-AS1 inhibits tumor
growth in nude mice
To explore whether VIM-AS1 affects tumorigenesis,
HGC-27 cells transfected with si-VIM-AS1 or si-NC were used in a
nude mice xenograft model (Fig.
7A). The results demonstrated that tumor volume was
significantly smaller and the weight of xenograft tumors was
significantly lighter in the si-VIM-AS1 group compared with in the
si-NC group (P<0.01; Fig. 7B and
C). These results suggested that silencing VIM-AS1 inhibited
tumor growth in nude mice.
Discussion
GC is one of the main causes of cancer-related death
in China (20). Patients with GC
that present with distant metastasis often have a poor prognosis,
and are unable to undergo optimal and timely surgical intervention.
Thus, it is important to search for innovative, novel and effective
targets to improve prognostic and therapeutic strategies for GC.
The results of the present study suggested that VIM-AS1 promoted
cell proliferation, migration, invasion and EMT by regulating FDZ1
and activating the Wnt/β-catenin pathway in GC.
Accumulating evidence has suggested that lncRNAs are
involved in diverse biological processes by modulating gene
expression (21). Recently,
lncRNAs have been reported to serve important roles in the
progression of numerous tumor types (22). Although VIM-AS1 is characterized as
an oncogene in colorectal cancer (12), to the best of our knowledge, its
relationship with GC has not been reported. In the present study,
VIM-AS1 was demonstrated to be highly expressed in GC tissues and
cell lines (AGS and HGC-27 cells). In addition, the expression of
VIM-AS1 was associated with the prognosis of GC and some clinical
parameters, including TNM stage, lymph node metastasis and distant
metastasis. The results of the present study suggested that VIM-AS1
may serve as an oncogene in GC. A previous study confirmed that
lncRNAs may be involved in regulating cell proliferation,
apoptosis, migration and invasion in GC (23). A recent study demonstrated that
hepatocellular carcinoma upregulated lncRNA promoted the
proliferation, migration and invasion of GC cells via regulation of
the miR-9-5p/myosin heavy chain 9 axis (24). Another study observed that the
lncRNA amine oxidase copper containing 4, pseudogene affected the
biological behavior of GC cells by modulating the mitogen-activated
protein kinase pathway (25). In
addition, Wang et al (26)
demonstrated that the lncRNA small nucleolar RNA host gene 7
promoted the proliferation and inhibited the apoptosis of GC cells
through regulation of p15 and p16 expression. The present study
investigated the role of VIM-AS1 expression in GC cells and
demonstrated that silencing VIM-AS1 significantly inhibited the
proliferation, migration and invasion, and enhanced the apoptosis
of AGS and HGC-27 cells. Additionally, western blotting results
demonstrated that silencing VIM-AS1 promoted the expression of
cleaved caspase-3 and Bax in AGS and HGC-27 cells, but suppressed
the expression of Bcl-2, which further suggested that silencing
VIM-AS1 may promote GC cells apoptosis.
EMT is a crucial step in the progression of cancer
cell metastasis, which is characterized by the loss of epithelial
features and the acquisition of mesenchymal features by cells
(27). Notably, the expression of
the epithelial marker E-cadherin is decreased in cancer cells
during the process of EMT, whereas the expression of mesenchymal
markers N-cadherin and vimentin is increased (28). In addition, a previous study
demonstrated that MMPs, including MMP-2 and MMP-9, which enhance
the invasion and metastasis of tumor cells, are highly expressed
during the process of EMT (29).
The results of the present study demonstrated that silencing
VIM-AS1 increased E-cadherin expression, and reduced the expression
levels of N-cadherin, vimentin, MMP-2 and MMP-9, which supported
the hypothesis that silencing VIM-AS1 suppressed the process of EMT
in GC cells. In addition, the results of the present study
demonstrated that silencing VIM-AS1 inhibited tumorigenesis in
vivo.
The Wnt/β-catenin pathway serves an important role
in a various types of cancer as it regulates numerous processes,
such as cell proliferation, apoptosis, migration and invasion
(30). The Wnt receptor FZD1 is
considered to be an essential component of the Wnt/β-catenin
pathway, and has been observed to be overexpressed in neuroblastoma
and breast cancer (30,31). Numerous studies have also
demonstrated that overactivation of the Wnt/β-catenin pathway is
attributable to the overexpression of FZD receptors in several
types of cancer (32,33). In addition, the Wnt/β-catenin
pathway regulates its downstream target genes, such as cyclin D1
and C-myc during the progression of cancer (34). β-catenin is reported to be a
critical molecule involved in the Wnt/β-catenin signaling pathway
in the extracellular ligands of tumor cells (35). Several studies have demonstrated
that lncRNAs may modulate the biological behaviors of GC cells by
regulating the Wnt/β-catenin pathway. For example, LINC00052 has
been reported to promote GC cell proliferation and metastasis
through activation of the Wnt/β-catenin pathway (36). Peng et al (16) suggested that growth
arrest-associated lncRNA 1 suppressed the growth of GC by
inhibiting activation of the Wnt/β-catenin pathway. Wang et
al (17) reported that
knockdown of lncRNA TP73-AS1 inhibited GC cell proliferation and
invasion through regulating the Wnt/β-catenin pathway. The results
of the present study demonstrated that silencing VIM-AS1
significantly inhibited FZD1 expression by targeting miR-8052. In
addition, silencing VIM-AS1 significantly reduced the protein and
mRNA expression levels of β-catenin, cyclin D1 and C-myc in AGS and
HGC-27 cells, which together suggested that silencing VIM-AS1 may
inhibit GC cell proliferation, migration, invasion and EMT in GC
cells by downregulating FZD1 and inactivating the Wnt/β-catenin
pathway. Notably, further studies are required to identify other
potential mechanisms of VIM-AS1 that are involved in GC progression
in vitro, as well as the role of VIM-AS1 and its underlying
molecular mechanisms in GC in vivo.
In conclusion, the results of the present study
demonstrated that VIM-AS1 was highly expressed in GC tissues and
cell lines, and that VIM-AS1 promoted cell proliferation,
migration, invasion and EMT possibly by regulating FDZ1 and
activating the Wnt/β-catenin pathway in GC. These findings support
the oncogenic role of VIM-AS1 in GC, which may encourage studies on
VIM-AS1 as a potentially diagnostic biomarker and therapeutic
target for GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XFL designed the study. JGS, XBL and RHY performed
the experiments and analyzed the data. JGS wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study involving human samples was approved by
the Ethics Committee of Shouguang People's Hospital (approval no.
201803). All patients provided written informed consent. The
procedures employed for the animal studies were approved by the
Animal Use Committee of Shouguang People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng S, Lv P, Su J, Miao K, Xu H and Li
M: Silencing of the long non-coding RNA RHPN1-AS1 suppresses the
epithelial-to-mesenchymal transition and inhibits breast cancer
progression. Am J Transl Res. 11:3505–3517. 2019.PubMed/NCBI
|
|
3
|
Dong Y, Wang ZG and Chi TS: Long noncoding
RNA Lnc01614 promotes the occurrence and development of gastric
cancer by activating EMT pathway. Eur Rev Med Pharmacol Sci.
22:1307–1314. 2018.PubMed/NCBI
|
|
4
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirano T, Yoshikawa R, Harada H, Harada Y,
Ishida A and Yamazaki T: Long noncoding RNA, CCDC26, controls
myeloid leukemia cell growth through regulation of KIT expression.
Mol Cancer. 14:902015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tordonato C, Di Fiore PP and Nicassio F:
The role of non-coding RNAs in the regulation of stem cells and
progenitors in the normal mammary gland and in breast tumors. Front
Genet. 6:722015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin XC, Zhu Y, Chen WB, Lin LW, Chen DH,
Huang JR, Pan K, Lin Y, Wu BT, Dai Y and Tu ZG: Integrated analysis
of long non-coding RNAs and mRNA expression profiles reveals the
potential role of lncRNAs in gastric cancer pathogenesis. Int J
Oncol. 45:619–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao WJ, Wu HL, He BS, Zhang YS and Zhang
ZY: Analysis of long non-coding RNA expression profiles in gastric
cancer. World J Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu D, Zhang MY, Chu Z and Zhang M: Long
non-coding RNA HOST2 enhances proliferation and metastasis in
gastric cancer. Neoplasma. 66:101–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rezanejad Bardaji H, Asadi MH and Yaghoobi
MM: Long noncoding RNA VIM-AS1 promotes colorectal cancer
progression and metastasis by inducing EMT. Eur J Cell Biol.
97:279–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao X, Jiang X, Liu Z, Zhou M, Zhang J,
Wang X and Li X: Long noncoding RNA VIM antisense RNA 1 (VIM-AS1)
plays an important role in development of preeclampsia by
regulation of epithelial mesenchymal transition. Med Sci Monit.
25:8306–8314. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng C, Li X, Yu Y and Chen J: LncRNA
GASL1 inhibits tumor growth in gastric carcinoma by inactivating
the Wnt/β-catenin signaling pathway. Exp Ther Med. 17:4039–4045.
2019.PubMed/NCBI
|
|
17
|
Wang Y, Xiao S, Wang B, Li Y and Chen Q:
Knockdown of lncRNA TP73-AS1 inhibits gastric cancer cell
proliferation and invasion via the WNT/β-catenin signaling pathway.
Oncol Lett. 16:3248–3254. 2018.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang L, Wu RL and Xu AM:
Epithelial-mesenchymal transition in gastric cancer. Am J Transl
Res. 7:2141–2158. 2015.PubMed/NCBI
|
|
20
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Renganathan A and Felley-Bosco E: Long
noncoding RNAs in cancer and therapeutic potential. Adv Exp Med
Biol. 1008:199–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He C, Yang W and Yang J, Ding J, Li S, Wu
H, Zhou F, Jiang Y, Teng L and Yang J: Long noncoding RNA MEG3
negatively regulates proliferation and angiogenesis in vascular
endothelial cells. DNA Cell Biol. 36:475–481. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu T, Liu Y, Wei C, Yang Z, Chang W and
Zhang X: LncRNA HULC promotes the progression of gastric cancer by
regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 121:1096072019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu CX, Shi XC, Bi H, Zhai LQ and Yang Q:
LncRNA AOC4P affects biological behavior of gastric cancer cells
through MAPK signaling pathway. Eur Rev Med Pharmacol Sci.
23:8852–8860. 2019.PubMed/NCBI
|
|
26
|
Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S
and Xia TS: LncRNA SNHG7 promotes the proliferation and inhibits
apoptosis of gastric cancer cells by repressing the P15 and P16
expression. Eur Rev Med Pharmacol Sci. 21:4613–4622.
2017.PubMed/NCBI
|
|
27
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Coussens LM, Fingleton B and Matrisian LM:
Matrix metalloproteinase inhibitors and cancer-trials and
tribulations. Science. 295:2387–2392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Flahaut M, Meier R, Coulon A, Nardou KA,
Niggli FK, Martinet D, Beckmann JS, Joseph JM, Mühlethaler-Mottet A
and Gross N: The Wnt receptor FZD1 mediates chemoresistance in
neuroblastoma through activation of the Wnt/beta-catenin pathway.
Oncogene. 28:2245–2256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Zhang X, Wu X, Li W, Su P, Cheng
H, Xiang L, Gao P and Zhou G: Interference of Frizzled 1 (FZD1)
reverses multidrug resistance in breast cancer cells through the
Wnt/β-catenin pathway. Cancer Lett. 323:106–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Milovanovic T, Planutis K, Nguyen A, Marsh
JL, Lin F, Hope C and Holcombe RF: Expression of Wnt genes and
frizzled 1 and 2 receptors in normal breast epithelium and
infiltrating breast carcinoma. Int J Oncol. 25:1337–1342.
2004.PubMed/NCBI
|
|
33
|
Merle P, Kim M, Herrmann M, Gupte A,
Lefrançois L, Califano S, Trépo C, Tanaka S, Vitvitski L, de la
Monte S and Wands JR: Oncogenic role of the frizzled-7/beta-catenin
pathway in hepatocellular carcinoma. J Hepatol. 43:854–862. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ara H, Takagishi M, Enomoto A, Asai M,
Ushida K, Asai N, Shimoyama Y, Kaibuchi K, Kodera Y and Takahashi
M: Role for Daple in non-canonical Wnt signaling during gastric
cancer invasion and metastasis. Cancer Sci. 107:133–139. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Karimaian A, Majidinia M, Bannazadeh Baghi
H and Yousefi B: The crosstalk between Wnt/β-catenin signaling
pathway with DNA damage response and oxidative stress: Implications
in cancer therapy. DNA Repair (Amst). 51:14–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shan Y, Ying R, Jia Z, Kong W, Wu Y, Zheng
S and Jin H: LINC00052 promotes gastric cancer cell proliferation
and metastasis via activating the Wnt/β-catenin signaling pathway.
Oncol Res. 25:1589–1599. 2017. View Article : Google Scholar : PubMed/NCBI
|