Introduction
Myocardial ischemia (MI) is characterized by damage
of blood vessels (1). According to
the World Health Organization, ischemic heart disease has become
the leading cause of human mortality worldwide (2), resulting in over 8 million deaths in
2013 (3). Although timely blood
reperfusion is the most effective therapy for MI, this can lead to
myocardial injury, known as myocardial ischemia/reperfusion injury
(MI/RI) (4). MI/RI is associated
with myocardial tissue injury, inflammation and heart failure
(5–7), and is the leading cause of mortality
following cardiac surgery (8).
There are few effective drugs for MI/RI (9), primarily due to the poor permeability
of the cytomembrane (10).
Therefore, identification of novel therapeutic targets to improve
the prognosis of patients with MI/RI is necessary.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNA transcripts that are involved in the progression of
cardiac disease. For instance, silencing of lncRNA metastasis
associated lung adenocarcinoma transcript 1 (MALAT1) has been
demonstrated to ameliorate acute myocardial infarction via sponging
microRNA (miR)-320 (11). In
addition, lncRNA 2810403D21Rik/Mirf enhances ischemia/reperfusion
(I/R) injury via directly targeting miR-26a (12). Moreover, overexpression of lncRNA
taurine-upregulated gene 1 promoted hypoxia-induced injury in
cardiomyocytes via the miR-145-5p-Binp3 axis (13). Research has demonstrated that
lncRNAs, including lncRNA urothelial carcinoma associated 1
(14), MALAT 1 (15) and hypoxia-inducible factor
1α-antisense RNA 1, are key regulators of MI/RI (16). Hypoxia/reoxygenation (H/R)
injury-related factor in myocytes (HRIM; E230034O05Rik gene) is an
lncRNA 1,470 bp in length located on chromosome 20p12 (including
three exons) (17). Due to the
upregulation of E230034O05Rik in H/R or I/R, this gene was selected
for functional analysis in the present study (18). Hypoxia/reoxygenation injury-related
factor in myocytes (HRIM) is co-expressed with autophagy- and
apoptosis-associated proteins, including zinc finger DHHC-type
palmitoyltransferase 7 (19),
prostaglandin I2 synthase (20)
and keratin, type I cytoskeletal 23 (21). Knockdown of HRIM has been
demonstrated to increase cell viability via mediating autophagy to
protect against myocardial H/R injury (22). Consistently, silencing of HRIM has
also been shown to alleviate MI/RI-induced damage (16).
NF-κB is an essential protein transcription factor
involved in cell proliferation, differentiation and apoptosis
(23). The NF-κB signaling pathway
has been demonstrated to be associated with a number of
pathological processes in the heart, including inflammation, injury
and apoptosis (24). The canonical
NF-κB pathway proceeds via activating the IκB kinase complex (IKK).
IKK phosphorylates IκB proteins, typically IkBα, leading to
dissociation from the NF-κBp65/p50 complex (25,26).
The NF-κB p65/p50 complex then translocates to the nucleus, where
it activates gene transcription (25,27).
p65 is a major subunit of NF-κB (28). Upregulation of NF-κB p65 can
enhance activation of the NF-κB signaling pathway (29), and inhibition of NF-kB p65 activity
has been shown to protect against MI/RI in rats (30). Moreover, downregulation of NF-κB
attenuated MI/RI in rats (31).
However, whether the effects of HRIM are mediated via the NF-κB
signaling pathway in MI/RI remains unclear.
The present study aimed to determine the biological
function of HRIM in MI/RI rats and H9C2 cells as well as its
underlying mechanism of action. These results may provide a novel
target for the treatment of MI/RI.
Materials and methods
Animals
Male Sprague-Dawley rats (n=44; weight, 220–260 g;
age, 7 weeks) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd. Rats were fed standard chow and water
and raised under specific pathogen-free conditions at 22°C and 50%
relative humidity with an artificial 12-h light/dark cycle. The
present study was approved by the Animal Ethics Committee of
Qingdao Municipal Hospital (approval no. 2020-L-054).
MI/RI model in rats
After one week of adjustment, rats were divided into
Control (n=10), Sham (n=10) and MI/RI (n=24) groups. Rats without
any treatment were defined as the Control group. The Sham group was
subjected to all procedures except ligation. The MI/RI model was
induced via ligation of the left anterior descending coronary
artery (LAD) as previously described, with some modifications
(30). Briefly, rats were
anesthetized with pentobarbital sodium (50 mg/kg) via
intraperitoneal injection. Subsequently, the LAD was ligated with a
6.0 thread and the ligation line was released after 45 min,
followed by reperfusion for 24 h. Thus, an MI/RI model was
established. In order to determine the effect of HRIM on MI/RI
rats, MI/RI rats were divided into the following groups: MI/RI
(MI/RI rats without treatment), MI/RI + small interfering RNA
(siRNA) negative control (si-NC; MI/RI rats injected with 200
µg/kg/d si-NC via the tail vein following surgery) and MI/RI + HRIM
siRNA (si-HRIM; MI/RI rats injected with 200 µg/kg/d si-HRIM via
the tail vein following surgery) (n=8/group). Both si-NC (forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) and si-HRIM (forward,
5′-GCGCCAUUGCUGCAAAUUATT-3′ and reverse,
5′-UAAUUUGCAGCAAUGGCGCTT-3′) were purchased from Shanghai
GenePharma Co., Ltd.
Transthoracic echocardiography
(TTE)
TTE was performed using a Vevo770 scanner
(VisualSonics, Inc.). The left ventricular ejection fraction
(LVEF), left ventricular fractional shortening (LVFS), left
ventricular end-diastolic diameter (LVEDD), ventricular
end-systolic diameter (LVESD), left ventricular systolic pressure
(LVSP), left ventricular end-diastolic pressure (LVEDP), maximum
velocity of LV contraction (+dp/dtmax) and maximum
velocity of LV diastolic function (-dp/dtmax) were
measured via TTE in rats.
2,3,5-Triphenyltetrazolium chloride
(TTC) staining
At one week after establishment of the MI/RI model,
rats were anesthetized via intraperitoneal injection of
pentobarbital sodium (50 mg/kg) and sacrificed by neck dislocation.
Myocardial tissue was isolated, sectioned (thickness, 3 mm),
stained with 1% TTC (cat. no. A610558; Sangon Biotech Co., Ltd.)
for 30 min at 37°C, and fixed in 10% formalin for 10 min at 37°C.
After washing with water, infarct size was quantified using the
Motic Med 6.0 Digital Medical Image Analysis system (Motic
Instruments). Infarcted myocardium was stained white, whereas
surviving myocardium was stained brick red. The infarct size was
calculated as follows: Infarct size (%)=(weight of white
sections/weight of whole cardiac) ×100%.
Hematoxylin-eosin (HE) staining
Myocardial tissue was collected, fixed in 4%
paraformaldehyde at 37°C for 24 h, dehydrated with an ethanol
gradient (70% ethanol for 2 h, 80% ethanol overnight, 90% ethanol
for 2 h, 100% I ethanol for 1 h and 100% II ethanol for 1 h)
vitrified with xylene, waxed, embedded in paraffin and cut into
sections (thickness, 4 µm). The sections were then stained with
0.5% hematoxylin for 5 min at 37°C, followed by 0.5% eosin for 1
min at 37°C. The ratio of inflammatory cells/field (5 random
fields) in myocardial tissues was calculated by light microscopy
(magnification, ×400). Stained cells were analyzed using
Image-ProPlus (version 6.0; Media Cybernetics, Inc.)
Oxygen-glucose
deprivation/reoxygenation (OGD/R)-induced in vitro model
MI/RI was mimicked in vitro by establishing
an OGD/R model using myocardial cells (H9C2 cells). H9C2 cells were
purchased from the Cell Bank of the Chinese Academy of Sciences and
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS at 37°C and 5% CO2. In order
to establish an OGD/R model, serum/glucose-free DMEM was used. H9C2
cells were placed in an anaerobic chamber (95% N2 and 5%
CO2) at 37°C for 3 h. The medium was then replaced with
normal culture medium and incubated for 0, 4 and 8 h to reoxygenate
the H9C2 cells. The OGD3h/R0h, OGD3h/R4h and OGD3h/R8h groups
received reoxygenation at 3 h post-OGD for 0, 4 and 8 h,
respectively. In addition, H9C2 cells without treatment were
regarded as the Normal group.
Cell transfection
For transfection experiments, H9C2 cells at >80%
confluency were harvested and divided into three groups: OGD/R
(H9C2 cells with reoxygenation at 3 h post-OGD for 8 h), OGD/R +
si-NC (cells transfected with 50 nmol si-NC for 48 h) and OGD/R +
si-HRIM (cells transfected with 50 nmol si-HRIM for 48 h). Asatone
(an activator of the NF-κB pathway; cat. no. HY-N6826; MedChem
Express) (32) was used to further
investigate the association between HRIM and the NF-κB pathway.
Thus, the OGD/R + si-HRIM + Asatone group comprised cells
transfected with 50 nmol si-HRIM and treated with 20 µmol/l asatone
at 37°C for 48 h. Cell transfection was performed using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 25°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from myocardial tissue or
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and subsequently reverse-transcribed into cDNA
using the PrimeScript RT reagent kit (Takara Bio, Inc.) according
to the manufacturer's instructions. Subsequently, qPCR was
performed using the MiScript SYBR Green PCR kit (Qiagen, Inc.) and
a ABI 7500HT Fast Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) under the following conditions: 95°C for 3
min and 40 cycles of 95°C for 15 sec and 60°C for 30 sec, and a
final extension step at 72°C for 10 min. Relative expression levels
were calculated via the 2−ΔΔCq method (33). HRIM expression levels were
normalized to those of GAPDH. The primer sequences are presented in
Table I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Name | Sequence
(5′→3′) |
|---|
| HRIM | F:
AGGGGAGGGGGAAAGTAGAA |
|
| R:
CACATGGAAGCCAGTGGTCA |
| GAPDH | F:
GACGGCCGCATCTTCTTGT |
|
| R:
CACACCGACCTTCACCATTTT |
Western blotting
A Nuclear and Cytoplasmic Protein Extraction kit
(Beyotime Institute of Biotechnology) was used to extract nuclear
and cytoplasmic proteins according to the manufacturer's protocol.
Myocardial tissue from Sprague-Dawley rats or H9C2 cells were lysed
using lysis buffer on ice to extract the total protein. Protein
concentration was determined using a bicinchoninic acid protein
concentration assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Proteins (20 µg) were separated via 10–12% SDS-PAGE and transferred
onto polyvinylidene fluoride membranes. The membranes were blocked
with 5% non-fat milk in Tris-buffered saline, containing 0.01%
Tween-20 at 4°C overnight. The membranes were then incubated with
the following primary antibodies (all 1:1,000; all Cell Signaling
Technology, Inc.) overnight at 4°C: Anti-NF-κB p65 (cat. no. 8242),
anti-phosphorylated (p)-NF-κB p65 (cat. no. 3033), anti-IκBα (cat.
no. 4812) and anti-p-IκBα (cat. no. 4812), followed by incubation
with horseradish peroxidase-labeled goat anti-rabbit IgG secondary
antibody (1:5,000; cat. no. ab205718; Abcam) for 1 h at 37°C. The
protein bands were visualized using enhanced chemiluminescence
exposure solution (Pierce; Thermo Fisher Scientific, Inc.) and
quantified with Quantity One 1-D analysis software (version 4.62;
Bio-Rad Laboratories, Inc.). β-actin (1:1,000; cat. no. sc-517582;
Cell Signaling Technology, Inc.) was used as the internal
reference.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) reagent (BD Biosciences)
was used to evaluate the proliferation of H9C2 cells according to
the manufacturer's instructions. H9C2 cells (5×103
cells/well) were seeded onto a 96-well plate and subjected to
OGD/R. CCK-8 solution (10 µl/well) was then added and incubated for
2 h at 37°C. The optical density at 450 nm (OD450) was
measured using a microplate reader (BioTek Instruments Inc.). Wells
containing only culture medium and CCK-8 reagent were used as the
blank group.
Flow cytometry
After 48 h transfection in 24-well plates
(3×105 cells/well), cells were collected in 10 ml
centrifuge tubes and centrifuged twice at 1,000 × g for 5 min at
4°C. Subsequently, the supernatant was collected and resuspended in
binding buffer (140 mM NaCl, 2.5 mM CaCl2, 10 mM
HEPES/NaOH, pH 7.4) to adjust the cell density to
1–5×106 cells/ml. Next, 100 µl cell suspension was
cultured with 5 µl V-FITC and 10 µl PI for 15 min in the dark at
25°C. An additional 400 µl 1X binding buffer was added to each
tube. The apoptotic rate was then detected using a FACSArray BD
Bioanalyzer flow cytometer (BD Biosciences), and data were analyzed
via CytoDiff CXP software (version 2.0; Beckman Coulter, Inc.).
ELISA
Following cell transfection, cells from each group
were centrifuged at 3,000 × g at 4°C for 10 min and the resulting
supernatant was collected. The supernatant was collected and levels
of TNF-α, IL-1β, IL-6, lactate dehydrogenase (LDH) and creatine
kinase (CK) were measured via ELISA kits (TNF-α, cat. no. KRC3011;
IL-1β, cat. no. BMS627; IL-6, cat. no. BMS625; all Thermo Fisher
Scientific, Inc.; LDH, cat. no. ab183367; CK, cat. no. ab264617;
both Abcam). The absorbance of each pore was measured at a
wavelength of 450 nm via an enzyme mark sensing instrument
(Molecular Devices LLC).
Statistical analysis
Statistical analysis was performed via SPSS software
23.0 (IBM Corp.). Data are presented as the mean ± SD of three
independent repeats. Comparisons between groups were performed via
ANOVA followed by post hoc Tukey's multiple comparisons test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
MI/RI affects myocardial function and
upregulates HRIM expression levels in rats
Once the MI/RI model was established, hemodynamic
indices were evaluated via TTE. Compared with the Control and Sham
groups, LVEF, LVFS, LVSP, +dp/dtmax, and
-dp/dtmax significantly decreased while LVEDD, LVESD,
and LVEDP were significantly increased in the MI/RI group
(P<0.01 and P<0.01; Fig.
1A). TTC staining results demonstrated that myocardial infarct
size was significantly higher in the MI/RI group compared with the
Sham group (P<0.001; Fig. 1B).
HE staining revealed that the ratio of inflammatory cells/field in
myocardial tissue was significantly increased in the MI/RI group
compared with the Sham group (P<0.001; Fig. 1C). Additionally, HRIM expression
levels were significantly upregulated in the MI/RI group compared
with the Sham group (P<0.001; Fig.
1D).
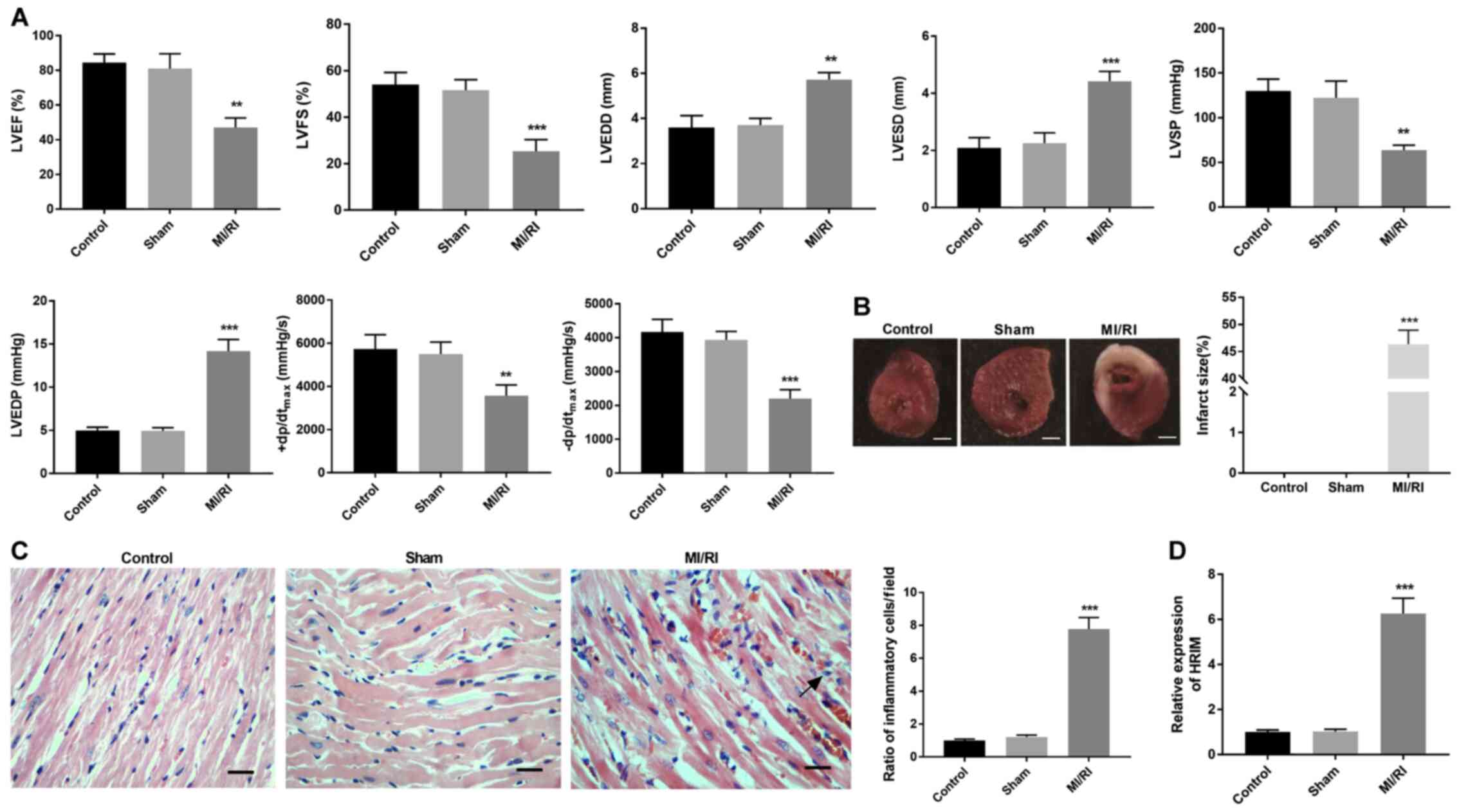 | Figure 1.MI/RI affects myocardial function in
rats. (A) Hemodynamics indices were evaluated via transthoracic
echocardiography. (B) Myocardial infarct size was identified via
2,3,5-triphenyltetrazolium chloride staining. Scalebar, 2 mm. (C)
Ratio of inflammatory cells/field in myocardial tissue was observed
via hematoxylin-eosin staining. Scale bar, 100 µm. (D) Expression
levels of HRIM in myocardial tissue were detected via reverse
transcription-quantitative PCR. n=3. **P<0.01 and ***P<0.001
vs. Sham. MI/RI, myocardial ischemia/reperfusion injury; HRIM,
hypoxia/reoxygenation injury-related factor in myocytes; LVEF, left
ventricular ejection fraction; LVFS, left ventricular fractional
shortening; LVEDD, left ventricular end-diastolic diameter; LVESD,
ventricular end-systolic diameter; LVSP, left ventricular systolic
pressure; LVEDP, left ventricular end-diastolic pressure;
(+dp/dtmax, maximum velocity of LV contraction;
-dp/dtmax, maximum velocity of LV diastolic
function. |
OGD/R induces myocardial injury and
upregulates HRIM expression levels in myocardial cells
H9C2 cells were subjected to OGD/R to mimic MI/RI
conditions in vitro. Following reoxygenation at 3 h post-OGD
for 0, 4 and 8 h, the survival rate of H9C2 cells significantly
decreased compared with normal cells (P<0.01 and P<0.001;
Fig. 2A). RT-qPCR results
demonstrated that the expression levels of HRIM in H9C2 cells
significantly increased in the OGD3h/R0h, OGD3h/R4h and OGD3h/R8h
groups compared with the Normal group (P<0.001; Fig. 2B). The OGD3h/R8h group was selected
for subsequent experiments due to the relative high HRIM
expression.
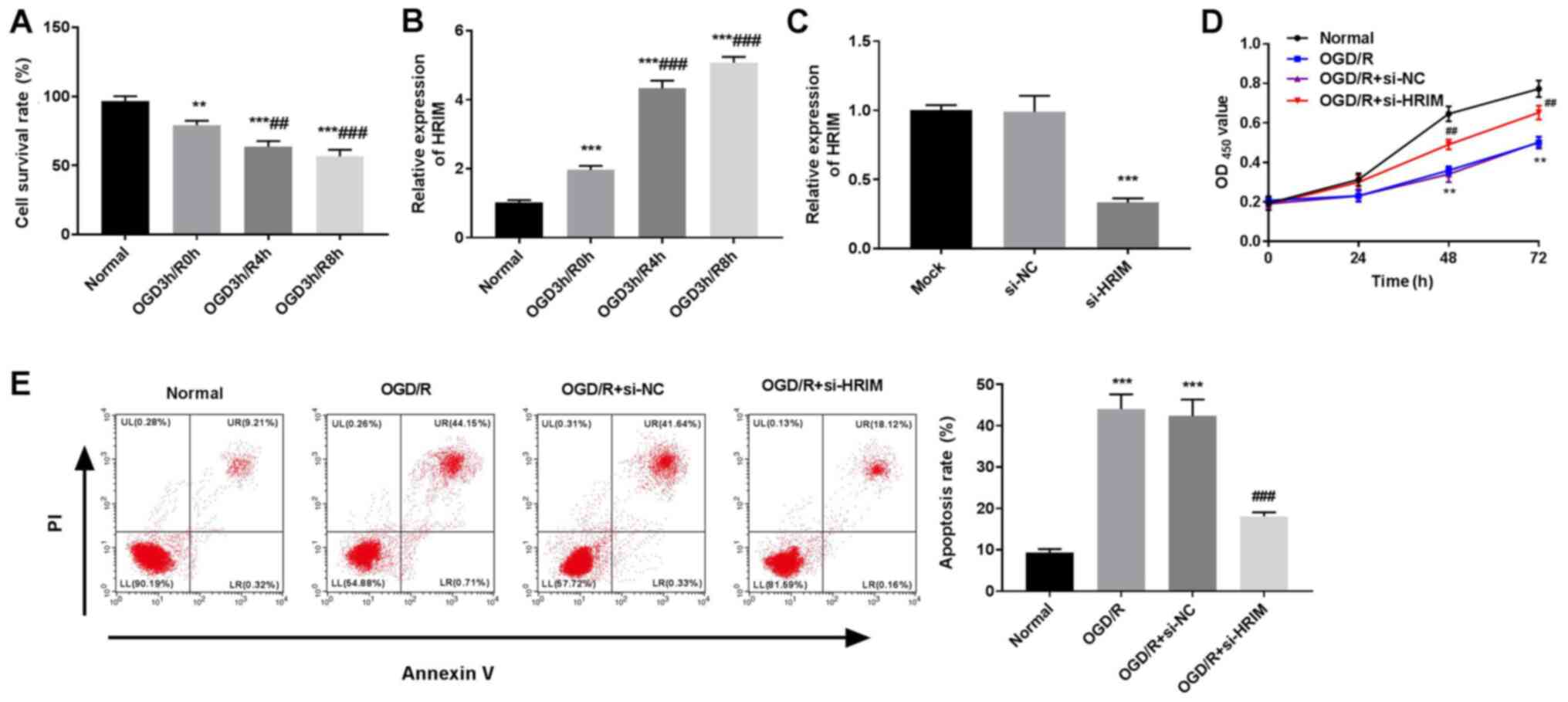 | Figure 2.Silencing of HRIM enhances
proliferation and attenuates apoptosis of OGD/R cells. Normal group
comprised untreated H9C2 cells; OGD3h/R0h, OGD3h/R4h and OGD3h/R8h
groups received reoxygenation at 3 h post-OGD for 0, 4 and 8 h,
respectively. (A) Cell survival rate was assessed via CCK-8 assay.
Expression levels of HRIM in (B) OGD/R and (C) transfected cells
were detected via reverse transcription-quantitative PCR.
**P<0.01 and ***P<0.001 vs. Normal or si-NC;
##P<0.01 and ###P<0.001 vs. OGD3h/R0h.
(D) OD450 value at 0, 24, 48 and 72 h were observed via
CCK-8 assay. (E) Apoptosis rate of H9C2 cells detected via flow
cytometry. n=3. **P<0.01 and ***P<0.001 vs. Normal;
##P<0.01 and ###P<0.001 vs. OGD/R +
si-NC. HRIM, hypoxia/reoxygenation injury-related factor in
myocytes; OGD/R, oxygen-glucose deprivation/reoxygenation; CCK-8,
Cell Counting Kit-8; OD450, optical density at 450 nm;
si, small interfering RNA; NC, negative control. |
In order to examine the role of HRIM, effective HRIM
knockdown was performed in H9C2 cells via si-HRIM transfection
(P<0.001; Fig. 2C). The CCK-8
assay results demonstrated that the OD450 value of the
OGD/R group was lower compared with the Normal group at 48 and 72 h
post-culturing (P<0.01), and that silencing of HRIM
significantly increased the OD450 value of H9C2 cells
compared with the OGD/R + si-NC group at 48 and 72 h post-culturing
(P<0.01; Fig. 2D).
Additionally, apoptosis of H9C2 cells significantly increased
following OGD/R compared with the Normal group (P<0.001),
whereas HRIM knockdown decreased H9C2 cell apoptosis compared with
the OGD/R + si-NC group (Fig.
2E).
Silencing of HRIM protects against
inflammation and injury in H9C2 cells
In order to investigate the effect of HRIM knockdown
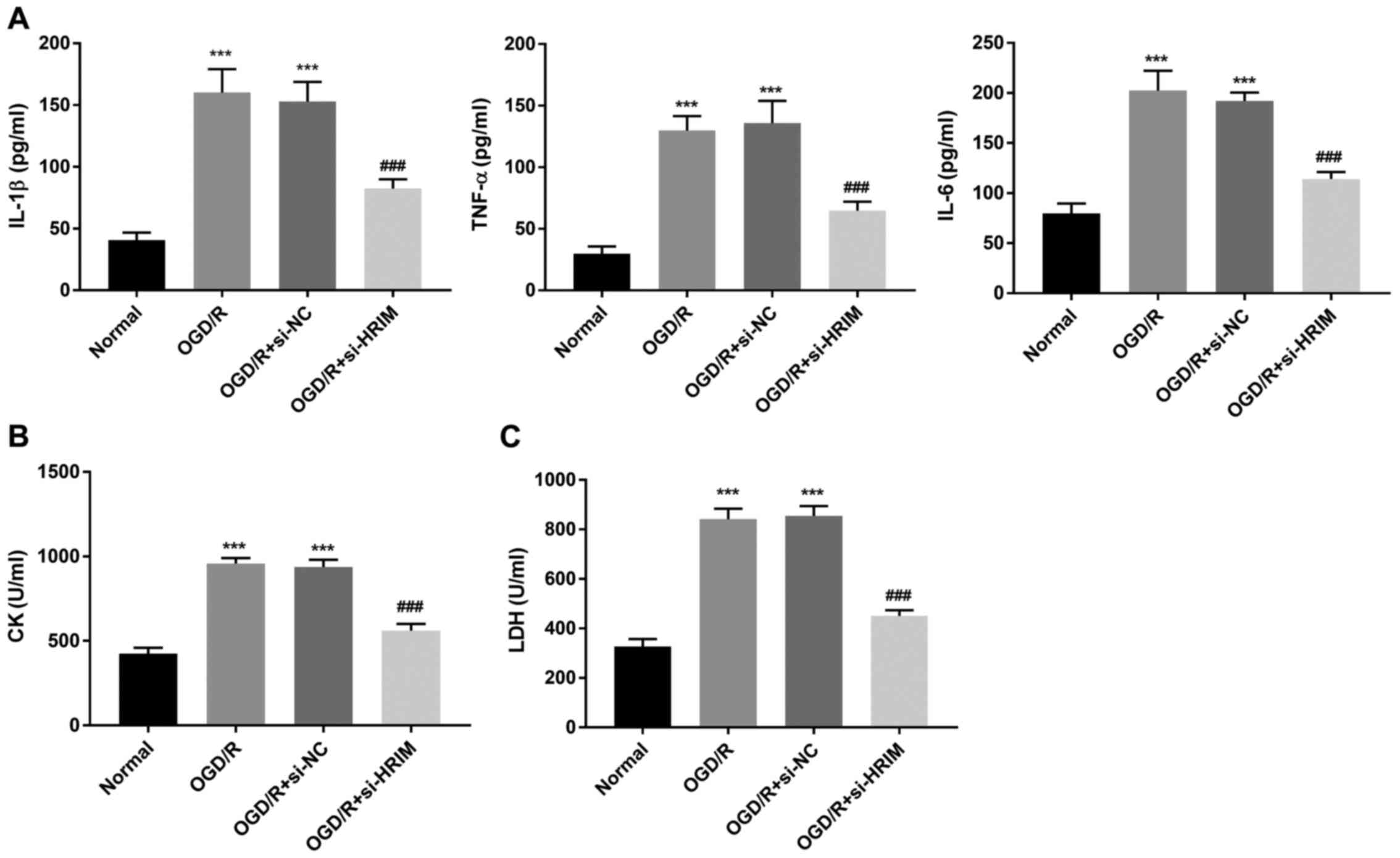
on inflammation in H9C2 cells, levels of IL-1β, TNF-α and IL-6 were
determined via ELISA. Compared with the Normal group, the levels of
IL-1β, TNF-α and IL-6 in H9C2 cells were significantly increased in
the OGD/R group (P<0.001); however, HRIM knockdown significantly
attenuated these effects (P<0.001; Fig. 3A). Moreover, compared with the
Normal group, the levels of LDH and CK in H9C2 cells were
significantly elevated in the OGD/R group (P<0.001), and HRIM
inhibition significantly decreased these levels in the OGD/R +
si-HRIM group (P<0.001; Fig. 3B and
C).
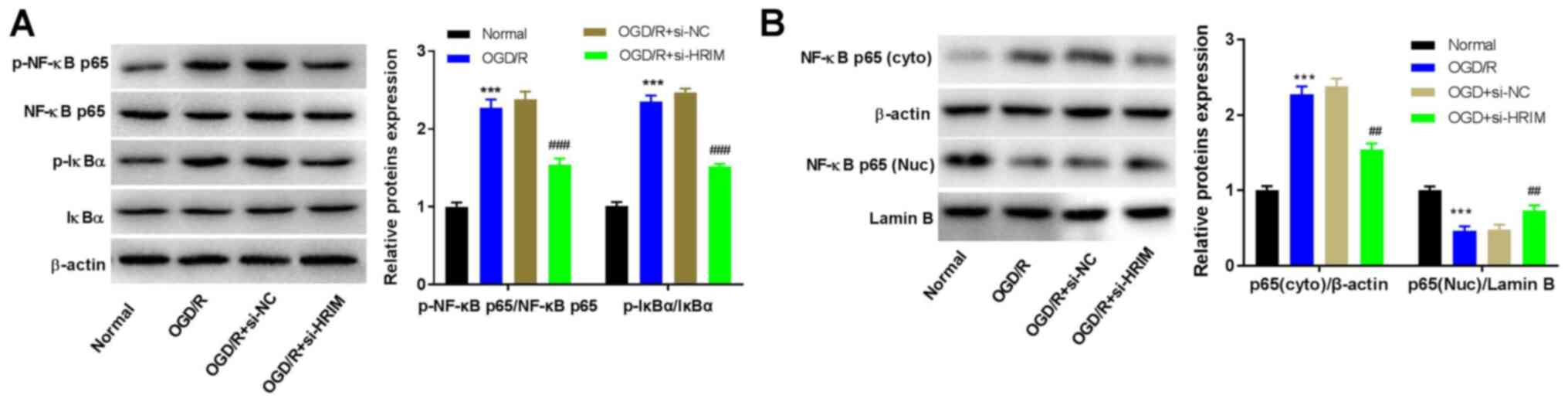
Silencing of HRIM inhibits the NF-κB
signaling pathway
In order to evaluate the effect of HRIM inhibition
on the NF-κB signaling pathway in H9C2 cells, the expression levels
of NF-κB-associated proteins were measured via western blotting.
The results demonstrated that the levels of p-NF-κB p65/NF-κB p65
and p-IkBα/IkBα were significantly increased in the OGD/R group
compared with the Normal group (P<0.001). However, compared with
the OGD/R + si-NC group, knockdown of HRIM significantly decreased
the levels of p-NF-κB p65 and p-IkBα (P<0.01; Fig. 4A). Furthermore, compared with the
Normal group, the levels of cytoplasmic NF-κB p65 significantly
increased (P<0.001), but those of nuclear NF-κB p65
significantly decreased in the OGD/R group (P<0.001); however,
HRIM knockdown attenuated these effects (P<0.01; Fig. 4B).
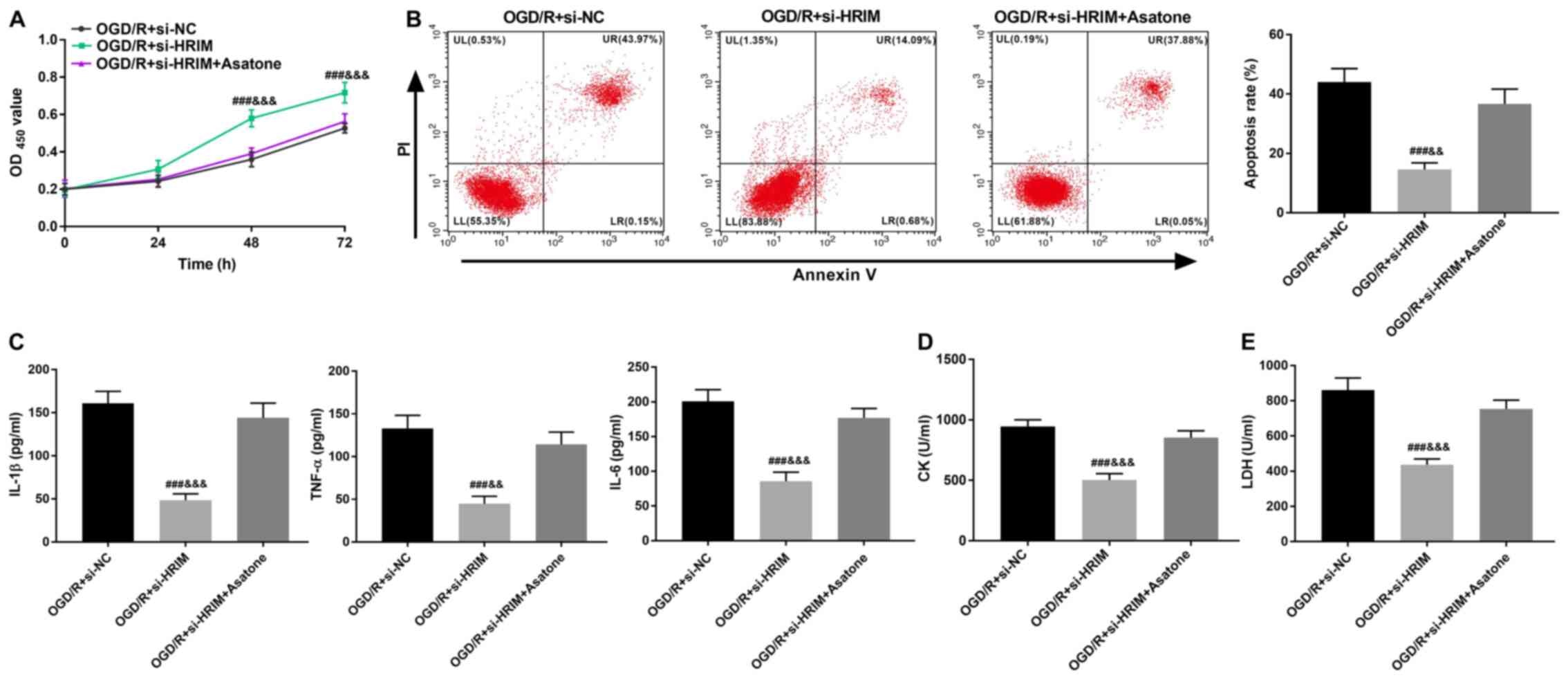
In order to investigate the association between HRIM
and NF-κB signaling, H9C2 cells were treated with the NF-κB
signaling pathway activator asatone. CCK-8 assay and flow cytometry
results demonstrated that HRIM knockdown significantly increased
the OD450 value at 48 and 72 h (P<0.001)
post-culturing and significantly decreased the apoptosis of H9C2
cells (P<0.001; Fig. 5A and B).
Moreover, HRIM knockdown significantly decreased the levels of
TNF-α, IL-1β, IL-6, CK and LDH in H9C2 cells (P<0.001; Fig. 5C-E). However, the inhibitory
effects of HRIM knockdown on H9C2 cells were reversed by treatment
with asatone (P<0.01).
Silencing of HRIM inhibits MI/RI
injury via inactivating the NF-κB signaling pathway
The biological function of HRIM in MI/RI rats was
investigated. TTE analysis demonstrated that silencing of HRIM
significantly increased the LVEF, LVFS, LVSP, +dp/dtmax
and -dp/dtmax (P<0.05), but significantly decreased
the LVEDD, LVESD and LVEDP in MI/RI rats compared with the MI/RI +
si-NC group (P<0.001; Fig. 6A).
Moreover, the ratio of inflammatory cells/field in myocardial
tissue was significantly decreased in the MI/RI + si-HRIM group
compared with that in the MI/RI + si-NC group (P<0.001; Fig. 6B). HRIM knockdown significantly
decreased the expression levels of HRIM in MI/RI rats (P<0.001)
compared with the MI/RI + si-NC group (Fig. 6C). Additionally, knockdown of HRIM
significantly decreased the levels of p-NF-κB p65/NF-κB p65,
p-IkBα/IkBα and cytoplasmic NF-κB p65 and significantly increased
nuclear NF-κB p65 levels in MI/RI rats compared with the MI/RI +
si-NC group (P<0.01; Fig. 6D and
E).
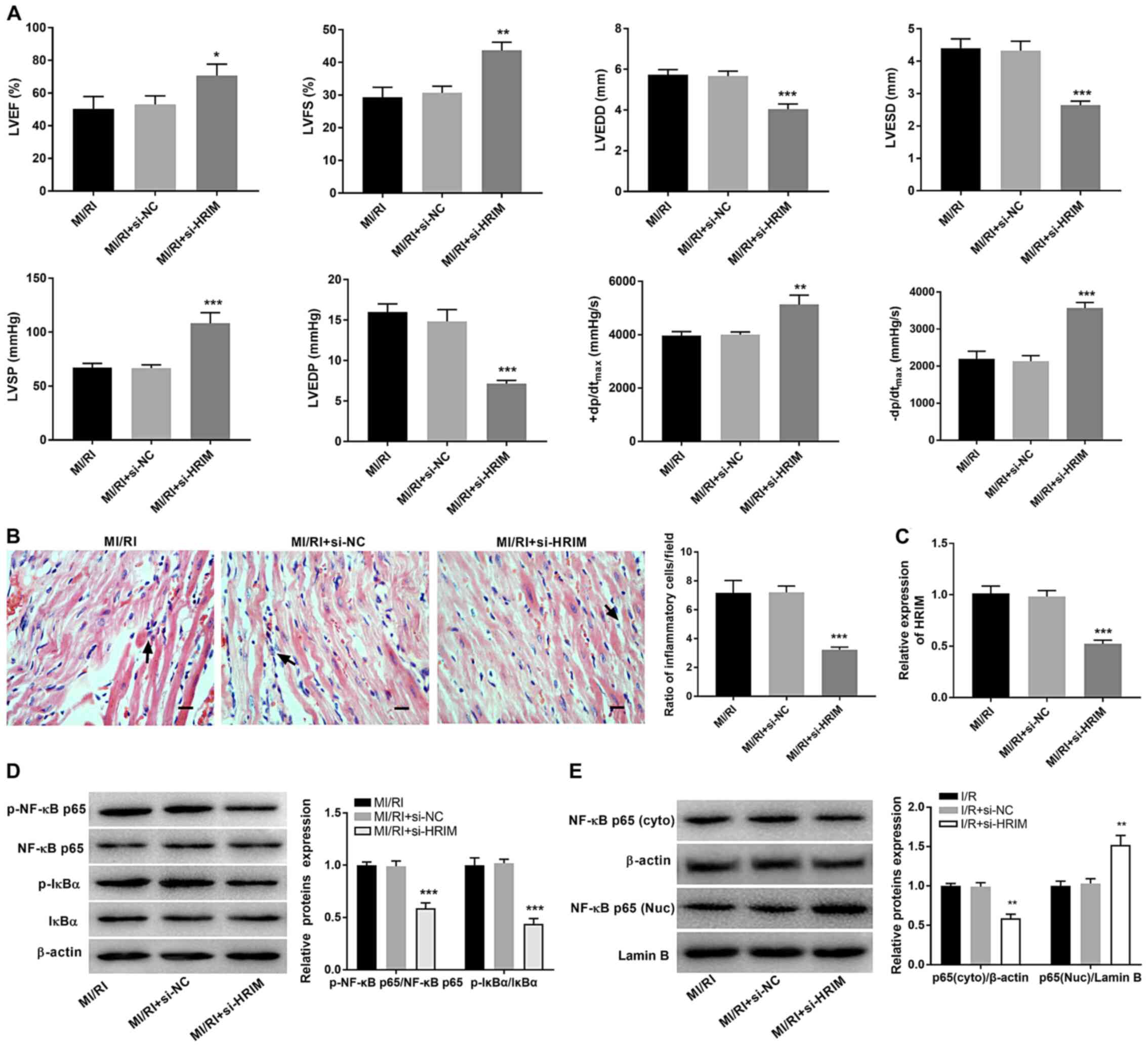 | Figure 6.Silencing of HRIM attenuates MI/RI
and inhibits the NF-κB signaling pathway in rats. (A) Hemodynamics
indices were evaluated via transthoracic echocardiography. (B) The
ratio of inflammatory cells/field in myocardial tissue was observed
via hematoxylin-eosin staining. Scale bar, 100 µm. (C) Expression
levels of HRIM in myocardial tissue were detected via reverse
transcription-quantitative PCR. Expression levels of (D) p-NF-κB
p65/NF-κB p65 and p-IkBα/IkBα and (E) NF-κB p65 (cyto) and (Nuc)
were detected via western blotting. n=3. *P<0.05, **P<0.01,
***P<0.001 vs. MI/RI + si-NC. HRIM, hypoxia/reoxygenation
injury-related factor in myocytes; MI/RI, myocardial
ischemia/reperfusion injury; cyto, cytoplasmic; Nuc, nuclear; si,
small interfering RNA; NC, negative control; LVEF, left ventricular
ejection fraction; LVFS, left ventricular fractional shortening;
LVEDD, left ventricular end-diastolic diameter; LVESD, ventricular
end-systolic diameter; LVSP, left ventricular systolic pressure;
LVEDP, left ventricular end-diastolic pressure;
(+dp/dtmax, maximum velocity of LV contraction;
-dp/dtmax, maximum velocity of LV diastolic function; p,
phosphorylated. |
Discussion
MI/RI is a major cause of cardiac dysfunction
(34). Numerous studies have
reported that abnormal expression levels of lncRNAs are associated
with elevation of MI/RI (14,35).
In the present study, the expression levels of HRIM in myocardial
tissue of MI/RI rats were significantly upregulated, which is
consistent with previously described lncRNAs (11,16).
The expression levels of the lncRNAs necrosis-related factor
(36), AK123487 (37) and lncRNA nuclear enriched abundant
transcript 1 (38) have previously
been shown to be increased in MI/RI cardiomyocytes. It was
hypothesized that HRIM may serve a key role in the treatment of
MI/RI. The present results demonstrated that knockdown of HRIM
significantly elevated the LVEF, LVFS, LVSP, +dp/dtmax
and -dp/dtmax, and decreased the LVEDD, LVESD, LVEDP and
ratio of inflammatory cells/field in myocardial tissue in MI/RI
rats. Hence, these findings indicated that silencing of HRIM may
improve cardiac function in MI/RI rats.
An OGD/R-induced model was established to mimic
MI/RI in myocardial cells in vitro. Following reoxygenation
at 3 h post-OGD for 0, 4 and 8 h, the survival rate of H9C2 cells
was decreased, whereas HRIM expression levels were increased. A
recent study has demonstrated the promoting effect of lncRNAs in
the pathogenesis of cardiac diseases, including secretion of
proinflammatory cytokines and myocardial cell apoptosis and damage
(39). Moreover, knockdown of HRIM
has been shown to increase the survival rate of H9C2 cells in H/R
via decreasing autophagy in myocytes (40). Downregulation of HRIM has also been
demonstrated to promote myocardial cell viability in MI/RI via
inducing autophagy (41).
Furthermore, inhibition of the lncRNA regulator of reprogramming
has been shown to increase cell viability and decrease apoptosis in
H9C2 cells (42). The present
results indicated that silencing of HRIM ameliorated MI/RI in rats
via increasing proliferation and decreasing apoptosis of myocardial
cells. Inflammation is a key feature of MI/RI (43). Evidence has demonstrated that
inhibition of proinflammatory cytokines can ameliorate MI/RI
(44,45). High levels of LDH and CK are key
biomarkers of myocardial damage (46). In the present study, knockdown of
HRIM significantly decreased the levels of TNF-α, IL-6, IL-1β, LDH
and CK in myocardial cells. Moreover, silencing of HRIM alleviated
MI/RI-induced inflammation and damage. These results indicated that
HRIM knockdown may protect myocardial cells against MI/RI.
NF-κB participates in inflammatory responses and
blockade of the NF-κB signaling pathway has been demonstrated to
ameliorate MI/RI (47). In
addition, total glucosides of peonies have been shown to alleviate
MI/RI in rats via decreasing NF-κB expression levels (48).
17-methoxyl-7-hydroxy-benzene-furanchalcone protects against MI/RI
by decreasing the activity of NF-κB p65 (46). In the present study, HRIM knockdown
decreased the expression levels of NF-κB-associated proteins,
including p-NF-κB p65/NF-κBp65 and p-IkBα/IkBα. p65 is a major
subunit of NF-κB (49). Under the
action of IKK, IkBα is phosphorylated and then dissociated from the
p65/p50 dimer, and p65 is dissociated in the cytoplasm (27). Phosphorylated p65 and p50 are then
translocated into the nucleus (50). The phosphorylation levels of IkBα
and p65 and translocation of p65 to the nucleus are key to
verifying NF-κB pathway activation (51). A previous study demonstrated that
silencing of the lncRNA C2dat1 inhibited focal cerebral I/R injury
via suppressing the NF-κB signaling pathway (48). The present study demonstrated that
silencing of HRIM suppressed NF-κB activity to alleviate MI/RI in
cells, and that treatment with asatone effectively reverses this
inhibitory effect. Hence, the present results indicate that
silencing of HRIM may protect against MI/RI via suppression of the
NF-κB signaling pathway in rats.
There are certain limitations in the present study.
Due to the lack of additional data, such as clinical samples with
follow-up information, it was difficult to determine the clinical
importance of HRIM. Furthermore, although the NF-κB signaling
pathway is regulated via HRIM, further investigation is required to
determine whether HRIM is directly involved in NF-κB signaling.
In summary, the present findings demonstrated that
HRIM expression levels were significantly increased in the
myocardial tissue of MI/RI rats and OGD/R-induced H9C2 cells.
Moreover, knockdown of HRIM decreased inflammation, apoptosis and
myocardial injury via inactivating the NF-κB signaling pathway.
Knockdown of HRIM also improved cardiac function in MI/RI rats.
Hence, HRIM may act as a potential therapeutic target for
MI/RI.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request
Authors' contributions
LN made substantial contributions to the conception
and design of the study. YZ and WP acquired the data. YZ and SL
interpreted the data. SL and WP performed the experiments. SL
performed the data analysis. LN and WP drafted the manuscript and
revised it critically for important intellectual content. YZ
revised the manuscript for critically important intellectual
content. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
This study was performed with the approval of the
Animal Ethics Committee of Qingdao Municipal Hospital (approval no.
2020-L-054).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fröhlich GM, Meier P, White SK, Yellon DM
and Hausenloy DJ: Myocardial reperfusion injury: Looking beyond
primary PCI. Eur Heart J. 34:1714–1722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roger VL, Go AS, Lloyd-Jones DM, Adams RJ,
Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, et
al: Heart disease and stroke statistics-2011 update: A report from
the American heart association. Circulation. 123:e18–e209. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
GBD 2013 Mortality and Causes of Death
Collaborators: Global, regional, and national age-sex specific
all-cause and cause-specific mortality for 240 causes of death,
1990–2013: A systematic analysis for the global burden of disease
study 2013. Lancet. 385:117–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu HJ, Wang DG, Yan J and Xu J:
Up-regulation of microRNA-135a protects against myocardial
ischemia/reperfusion injury by decreasing TXNIP expression in
diabetic mice. Am J Transl Res. 7:2661–2671. 2015.PubMed/NCBI
|
|
5
|
Liu NB, Wu M, Chen C, Fujino M, Huang JS,
Zhu P and Li XK: Novel molecular targets participating in
myocardial ischemia-reperfusion injury and cardioprotection.
Cardiol Res Pract. 2019:69351472019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boag SE, Andreano E and Spyridopoulos I:
Lymphocyte communication in myocardial ischemia/reperfusion injury.
Antioxid Redox Signal. 26:660–675. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamacher-Brady A, Brady NR and Gottlieb
RA: The interplay between pro-death and pro-survival signaling
pathways in myocardial ischemia/reperfusion injury: Apoptosis meets
autophagy. Cardiovasc Drugs Ther. 20:445–462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niemann B, Schwarzer M and Rohrbach S:
Heart and mitochondria: Pathophysiology and implications for
cardiac surgeons. Thorac Cardiovasc Surg. 66:11–19. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sanchez-Hernandez CD, Torres-Alarcon LA,
Gonzalez-Cortes A and Peon AN: Ischemia/reperfusion injury:
Pathophysiology, current clinical management, and potential
preventive approaches. Mediators Inflamm. 2020:84053702020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ong SB, Katwadi K, Kwek XY, Ismail NI,
Chinda K, Ong SG and Hausenloy DJ: Non-coding RNAs as therapeutic
targets for preventing myocardial ischemia-reperfusion injury.
Expert Opin Ther Targets. 22:247–261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu H, Wu J, Li D, Zhou J, Yu H and Ma L:
Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction
through miR-320-Pten axis. Biomed Pharmacother. 106:738–746. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang H, Su X, Wu Q, Shan H, Lv L, Yu T,
Zhao X, Sun J, Yang R, Zhang L, et al: LncRNA 2810403D21Rik/Mirf
promotes ischemic myocardial injury by regulating autophagy through
targeting Mir26a. Autophagy. 16:1077–1091. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Z, Zhao S, Li C and Liu C: LncRNA TUG1
serves an important role in hypoxia-induced myocardial cell injury
by regulating the miR1455pBinp3 axis. Mol Med Rep. 17:2422–2430.
2018.PubMed/NCBI
|
|
14
|
Yu SY, Dong B, Zhou SH and Tang L: LncRNA
UCA1 modulates cardiomyocyte apoptosis by targeting miR-143 in
myocardial ischemia-reperfusion injury. Int J Cardiol. 247:312017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao ZH, Hao W, Meng QT, Du XB, Lei SQ and
Xia ZY: Long non-coding RNA MALAT1 functions as a mediator in
cardioprotective effects of fentanyl in myocardial
ischemia-reperfusion injury. Cell Biol Int. 41:62–70. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue X and Luo L: LncRNA HIF1A-AS1
contributes to ventricular remodeling after myocardial
ischemia/reperfusion injury by adsorption of microRNA-204 to
regulating SOCS2 expression. Cell Cycle. 18:2465–2480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiong W, Qu Y, Chen H and Qian J: Insight
into long noncoding RNA-miRNA-mRNA axes in myocardial
ischemia-reperfusion injury: The implications for mechanism and
therapy. Epigenomics. 11:1733–1748. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang Z, Ye B, Wang Z, Han J, Lin L, Shan
P, Cai X and Huang W: Inhibition of LncRNA-HRIM increases cell
viability by regulating autophagy levels during
hypoxia/reoxygenation in myocytes. Cell Physiol Biochem.
46:1341–1351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu ST, Chang YL, Wang WM, Chung MH, Lin
WS, Chou WY and Huang SM: A non-covalent interaction between small
ubiquitin-like modifier-1 and Zac1 regulates Zac1 cellular
functions. Int J Biochem Cell Biol. 44:547–555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakayama T: Genetic polymorphisms of
prostacyclin synthase gene and cardiovascular disease. Int Angiol.
29((2 Suppl)): S33–S42. 2010.
|
|
21
|
Birkenkamp-Demtroder K, Hahn SA, Mansilla
F, Thorsen K, Maghnouj A, Christensen R, Øster B and Ørntoft TF:
Keratin23 (KRT23) knockdown decreases proliferation and affects the
DNA damage response of colon cancer cells. PLoS One. 8:e735932013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turkieh A, Charrier H, Dubois-Deruy E,
Porouchani S, Bouvet M and Pinet F: Noncoding RNAs in cardiac
autophagy following myocardial infarction. Oxid Med Cell Longev.
2019:84386502019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hayden MS and Ghosh S: NF-κB, the first
quarter-century: Remarkable progress and outstanding questions.
Genes Dev. 26:203–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hall G, Hasday JD and Rogers TB:
Regulating the regulator: NF-kappaB signaling in heart. J Mol Cell
Cardiol. 41:580–591. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oeckinghaus A and Ghosh S: The NF-kappaB
family of transcription factors and its regulation. Cold Spring
Harb Perspect Boil. 1:a0000342009.
|
|
26
|
Thomas-Jardin SE, Dahl H, Nawas AF,
Bautista M and Delk NA: NF-κB signaling promotes
castration-resistant prostate cancer initiation and progression.
Pharmacol Ther. 211:1075382020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang M, Yan S and Zhou Y:
Trans-cinnamaldehyde reverses depressive-like behaviors in chronic
unpredictable mild stress rats by inhibiting NF-κB/NLRP3
inflammasome pathway. Evid Based Complement Alternat Med.
2020:45721852020.PubMed/NCBI
|
|
28
|
Li Q and Verma IM: NF-kappaB regulation in
the immune system. Nat Rev Immunol. 2:725–734. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abd El-Mohsen M, Bayele H, Kuhnle G,
Gibson G, Debnam E, Kaila Srai S, Rice-Evans C and Spencer JP:
Distribution of [3H]trans-resveratrol in rat tissues following oral
administration. Br J Nutr. 96:62–70. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang X, Huang J, Lin X, Qin F, Wen Q,
Chen C, Li Y, Ge W and Huang R: The effect of
17-methoxyl-7-hydroxy-benzene-furanchalcone on NF-κB and the
inflammatory response during myocardial ischemia reperfusion injury
in rats. J Cardiovasc Pharmacol. 63:68–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu L, Wei P, Cao Y, Zhang Q, Liu M, Liu
XD, Wang ZL and Zhang PY: Effect of total peony glucoside
pretreatment on NF-κB and ICAM-1 expression in myocardial tissue of
rat with myocardial ischemia-reperfusion injury. Genet Mol Res.
15:2016. View Article : Google Scholar
|
|
32
|
Chang HY, Chen YC, Lin JG, Lin IH, Huang
HF, Yeh CC, Chen JJ and Huang GJ: Asatone prevents acute lung
injury by reducing expressions of NF-[Formula: See text]B, MAPK and
inflammatory cytokines. Am J Chin Med. 46:651–671. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Duan W, Jin Z, Yi W, Yan J, Zhang
S, Wang N, Liang Z, Li Y, Chen W, et al: JAK2/STAT3 activation by
melatonin attenuates the mitochondrial oxidative damage induced by
myocardial ischemia/reperfusion injury. J Pineal Res. 55:275–286.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen J, Hu Q, Zhang BF, Liu XP, Yang S and
Jiang H: Long noncoding RNA UCA1 inhibits ischaemia/reperfusion
injury induced cardiomyocytes apoptosis via suppression of
endoplasmic reticulum stress. Genes Genomics. 41:803–810. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang K, Liu F, Liu CY, An T, Zhang J, Zhou
LY, Wang M, Dong YH, Li N, Gao JN, et al: The long noncoding RNA
NRF regulates programmed necrosis and myocardial injury during
ischemia and reperfusion by targeting miR-873. Cell Death Differ.
23:1394–1405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng C, Wu Z, Tian L, Li D, Wang X, He Y,
He Y, Jin W, Li M, Zhu Q, et al: Long noncoding RNA AK12348 is
involved in the regulation of myocardial ischaemia-reperfusion
injury by targeting PARP and caspase-3. Heart Lung Circ.
27:e51–e58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo M, Sun Q, Zhao H, Tao J and Yan D:
Long noncoding RNA NEAT1 sponges miR-495-3p to enhance myocardial
ischemia-reperfusion injury via MAPK6 activation. J Cell Physiol.
235:105–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang F, Qin Y, Wang Y, Li A, Lv J, Sun X,
Che H, Han T, Meng S, Bai Y and Wang L: LncRNA KCNQ1OT1 mediates
pyroptosis in diabetic cardiomyopathy. Cell Physiol Biochem.
50:1230–1244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie XJ, Fan DM, Xi K, Chen YW, Qi PW, Li
QH, Fang L and Ma LG: Suppression of microRNA-135b-5p protects
against myocardial ischemia/reperfusion injury by activating
JAK2/STAT3 signaling pathway in mice during sevoflurane anesthesia.
Biosci Rep. 37:BSR201701862017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu X, Dai S, Wu WJ, Tan W, Zhu X, Mu J,
Guo Y, Bolli R and Rokosh G: Stromal cell derived factor-1 alpha
confers protection against myocardial ischemia/reperfusion injury:
Role of the cardiac stromal cell derived factor-1 alpha CXCR4 axis.
Circulation. 116:654–663. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang W, Li Y and Wang P: Long non-coding
RNA-ROR aggravates myocardial ischemia/reperfusion injury. Braz J
Med Biol Res. 51:e65552018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hausenloy DJ and Yellon DM: Myocardial
ischemia-reperfusion injury: A neglected therapeutic target. J Clin
Invest. 123:92–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shin IW, Jang IS, Lee SM, Park KE, Ok SH,
Sohn JT, Lee HK and Chung YK: Myocardial protective effect by
ulinastatin via an anti-inflammatory response after regional
ischemia/reperfusion injury in an in vivo rat heart model. Korean J
Anesthesiol. 61:499–505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hadi NR, Alamran F, Yousif M and Zamil ST:
Antiapoptotic effect of simvastatin ameliorates myocardial
ischemia/reperfusion injury. ISRN Pharmacol. 2013:8150942013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu X, Cui B, Zhou X, Xu C, Lu Z and Jiang
H: Ethyl pyruvate reduces myocardial ischemia and reperfusion
injury by inhibiting high mobility group box 1 protein in rats. Mol
Biol Rep. 39:227–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He XH, Wang Y, Yan XT, Wang YL, Wang CY,
Zhang ZZ, Li H and Jiang HX: Transduction of PEP-1-heme oxygenase-1
fusion protein reduces myocardial ischemia/reperfusion injury in
rats. J Cardiovasc Pharmacol. 62:436–442. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S,
Zhao Z, Nepomuceno R, Bhuiyan MIH, Sun D, et al: Long non-coding
RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal
survival through the NF-κB signaling pathway following cerebral
ischemia. Cell Death Dis. 7:e21732016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu M, Gu J, Mei S, Xu D, Jing Y, Yao Q,
Chen M, Yang M, Chen S, Yang B, et al: Resveratrol delays
polycystic kidney disease progression through attenuation of
nuclear factor κB-induced inflammation. Nephrol Dial Transplant.
31:1826–1834. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tian F, Zang WD, Hou WH, Liu HT and Xue
LX: Nuclear factor-kB signaling pathway constitutively activated in
esophageal squamous cell carcinoma cell lines and inhibition of
growth of cells by small interfering RNA. Acta Biochim Biophys Sin
(Shanghai). 38:318–326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Krappmann D and Vincendeau M: Mechanisms
of NF-κB deregulation in lymphoid malignancies. Semin Cancer Biol.
39:3–14. 2016. View Article : Google Scholar : PubMed/NCBI
|