Introduction
There has been a consensus that bone marrow-derived
mesenchymal stem cells (BM-MSCs) are an ideal cell source for
cell-based therapy of various bone-related diseases (1,2)
because they can differentiate into osteoblasts, which are the most
important cells responsible for bone development and remodeling
(3). However, their clinical
application remains not widely accepted and treatment outcomes
still require additional confirmation with larger sample sizes and
control experiments (4). These
highlight the need to further understand the molecular mechanisms
of osteogenic differentiation in order to improve the
differentiation and therapeutic efficiency of BM-MSCs.
microRNAs (miRNAs) are a class of small, non-coding
RNAs of approximately 22 nt in length that exert roles by
negatively regulating the expression of target mRNAs via specific
binding to their 3′-untranslated region (3′-UTR). Recently, several
miRNAs have been found to be associated with the differentiation of
BM-MSCs. For example, Zhang et al (5) revealed that miR-664a-5p was
upregulated in osteoblast-differentiated human BM-MSCs compared
with control and this upregulation was positively correlated with
the expression of osteogenic genes (RUNX2, ALP and
OCN). Overexpression of miR-664a-5p stimulated the
osteogenic differentiation of BM-MSCs, whereas an opposite effect
was observed when it was knocked down. Luciferase reporting assay
revealed that HMGA2 mRNA was a direct target of miR-664a-5p.
Overexpression of HMGA2 rescued the effects of miR-664a-5p
on osteogenic differentiation (5).
Ge et al (6) reported that
overexpression of miR-374b significantly induced the osteogenic
differentiation of mouse BM-MSCs, showing increased ALP
activity and the expression of RUNX2, OPN and OCN.
Further analysis of the mechanism revealed that miR-374b may bind
to PTEN to mediate its functions (6). Huang et al (7) demonstrated that overexpression of
miR-320a downregulated HOXA10 and then significantly
inhibited osteogenesis of BM-MSCs, as determined by the
downregulation of the osteogenic markers RUNX2, ALP and
OCN and the inhibition of ALP activity and matrix
mineralization; while ectopic expression of HOXA10
attenuated miR-320a-induced inhibitory effects on osteogenic
differentiation of BM-MSCs. miR-125a-3p and miR-125b have been
revealed to specifically regulate the expression of GIT ArfGAP
1 (GIT1) and BMPR1b to inhibit the osteoblastic
proliferation and differentiation from BM-MSCs, respectively
(8,9). miR-223 was identified to be gradually
downregulated during osteogenic differentiation of BM-MSCs,
accompanied by the upregulated expression of its target gene
DHRS3 (10). These findings
revealed the importance of investigating the miRNAs and their
target genes to explain the osteogenic differentiation mechanisms
of BM-MSCs.
In our previous study, it was demonstrated that
miR-128 was downregulated by 58.8% in osteogenic treatment for
human BM-MSCs and that miR-128 may inhibit the osteogenic
differentiation through suppression of the vascular endothelial
growth factor pathway (11).
However, studies on miR-128 remain rare and its downstream
mechanisms for osteogenic differentiation are still not well
understood. The present study aimed to further elucidate the
molecular mechanisms of miR-128 during the osteogenic
differentiation of BM-MSCs by bioinformatic analyses of miRNA
(12) and mRNA sequencing datasets
which were collected from a public database. The predicted target
genes were also validated by in vitro experiments in which
miR-128 mimics were introduced. The present study may provide new
insights for elucidating the mechanisms of miR-128 and identifying
potential therapeutic targets to promote bone anabolism.
Materials and methods
High-throughput datasets
The high-throughput datasets of miRNAs and mRNAs in
BM-MSCs before or after osteoblast differentiation were available
from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession
number GSE107279 (platform: GPL11154, Illumina HiSeq 2000)
(12) and GSE112318 (platform:
GPL20795, HiSeq X Ten) (13),
respectively. GSE107279 (12)
included 3 undifferentiated human BM-MSC samples and 21
differentiated samples which were collected after induction for 12,
24 h, 3, 10 and 13 days in osteogenic medium. GSE112318 (13) contained one undifferentiated human
BM-MSC sample and one differentiated sample which was induced for 2
weeks in osteogenic medium. To identify the link between miRNAs and
mRNAs, the present study only used the 13-day data of GSE107279
dataset (12). Another reason for
selecting this time-point was that osteoblast differentiation was
induced in 13 days in the in vitro experiments of the
present study.
Differential analysis for miRNAs and
mRNAs
The reads data of GSE107279 (12) and GSE112318 (13) were downloaded from the GEO
repository and mean-normalized. The differentially expressed miRNAs
(DEMs) and genes (DEGs) between differentiated and undifferentiated
samples were identified using DESeq2 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html.).
package (14) in R (15). |log2fold change (FC)| >0.5 and
false discovery rate (FDR)-adjusted P<0.05 represented the
statistical threshold for DEMs and DEGs. A heat map was created
using the ‘pheatmap’ package (version: 1.0.8; http://cran.r-project.org/web/packages/pheatmap)
based on the Euclidean distance.
Target gene prediction for
miR-128-3p
The BLAST server (blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify
the nucleotide sequence similarity of human and rat miR-128-3p. The
target genes of miR-128-3p were predicted using the miRwalk
database (version 2.0; zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2)
(16). The miRwalk database
included 12 algorithms and the genes that were predicted in at
least 5 databases were collected. A Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was
drawn to screen the common genes predicted using the human and rat
as the species parameters (since rat BM-MSCs were used in
validation experiments) in miRwalk database and the shared genes
between predicted genes and DEGs, which were considered to be
possibly crucial targets of miR-128-3p. The miRNA-mRNA regulatory
network was visualized using the Cytoscape software (version 3.4;
www.cytoscape.org/) (17).
Protein-protein interaction (PPI)
network
The interactions between two target genes of
miR-128-3p were predicted using the STRING (Search Tool for the
Retrieval of Interacting Genes; version 10.0; http://stringdb.org/) database (18), with the relationship pairs having a
combined score >0.4 being used to construct the PPI network.
Several topological features of the nodes (proteins) in the PPI
network were calculated using the CytoNCA plugin in the Cytoscape
software (http://apps.cytoscape.org/apps/cytonca) (19) to screen hub genes, including degree
centrality (DC), eigenvector centrality (EC) and closeness
centrality (CC). In addition, several function-related modules were
also extracted from the PPI network using the Molecular Complex
Detection (MCODE; version, 1.4.2; http://apps.cytoscape.org/apps/mcode) plugin of the
Cytoscape software.
Function enrichment analysis for
target genes of miR-128-3p
Kyoto Encyclopedia of Genes and Genomes (KEGG;
version Release 92.0, October 1, 2019; genome.jp/kegg/) pathway
enrichment analysis was performed for target genes of DEMs using
the enricher tool (https://amp.pharm.mssm.edu/Enrichr/) (20). FDR<0.05 was considered to be
statistically significant.
Function validation of miR-128-3p for
the osteoblast differentiation from BM-MSCs
BM-MSCs of Wistar rats were purchased from the
Chinese Academy of Science Cell Bank and cultured in Alpha Minimum
Essential Medium (α-MEM; Gibco, Thermo Fisher Scientific, Inc.)
supplemented with GlutaMAX™-I, 10% fetal bovine serum and 1%
penicillin/streptomycin (all Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in a humidified atmosphere of 5% CO2 in air.
Transfection
miR-128-3p mimics and miR-normal control (miR-NC)
were obtained from Shanghai GenePharma Co., Ltd. On the day before
the transient transfections, the cells were seeded into 6-well
plates and incubated (37°C) to reach 65% confluency. Then,
miR-128-3p mimics (15, 30, 40 and 50 pmol) and the scrambled
control were transfected into BM-MSCs of Wistar rats using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
After 48 h of transfection, the cells were collected
to detect proliferation ability and the expression of miR-128-3p,
in order to screen the optimal treatment concentration of
miR-128-3p mimics. Subsequently, the cells were treated with this
concentration of miR-128-3p mimics for 13 days to observe its
functions on the osteoblast differentiation and the expression of
target genes.
Cell proliferation
Cell proliferation ability of BM-MSCs was determined
using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.). Briefly, cells were seeded into a 96-well plate at a density
of 2×103/well and cultured at 37°C, 5% CO2.
Then, 10 µl of CCK-8 solution was added to assay the cell growth
status. At 2 h following CCK-8 treatment, the absorbance of each
well at 450 nm wavelengths was measured by a microplate reader.
Reverse transcription-quantitative
(RT-q) PCR
RT-qPCR was used for the analysis of gene expression
according to the manufacturer's protocols. Briefly, total RNA was
extracted from 2×103 cells using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). The high-purity and
intact RNA (OD260/OD280>1.9) was converted into cDNA by the
GoScript Reverse Transcription System (Promega Corporation).
RT-qPCR was then performed on a CFX96 Real-Time System/C1000
Thermal Cycler (Bio-Rad Laboratories, Inc.) using AceQ®
RT-qPCR SYBR-Green Master Mix (Vazyme Biotech Co., Ltd.). The
reaction conditions for RT-qPCR were pre-incubation at 95°C for 10
min, followed by amplification with 39 cycles of 10 sec
denaturation at 95°C, 20 sec annealing at 65°C and 10 sec
elongation at 72°C. U6 and actin were used as the internal
reference for miRNA and mRNA detection, respectively. Relative
expression levels were analyzed using the 2−ΔΔCq method
(21). The primers used are listed
in Table I.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Gene | Primer sequence
(5′-3′) |
|---|
| ACTIN | F:
GAGACCTTCAACACCCCAGC |
|
| R:
ATGTCACGCACGATTTCCC |
|
RUNX1-rat | F:
GGTTTCGCAGCGTGGTAAAA |
|
| R:
GCACTGTGGGTACGAAGGAA |
| KIT-rat | F:
TACTTGGAGCCTGCACCATT |
|
| R:
TATCGCTGCAGGAAGACTCC |
|
AXIN1-rat | F:
GTGCCCCTACCTCACATTCC |
|
| R:
CTCACCTTCCTCCTCCATGC |
|
YWHAB-rat | F:
GTCTCCAGAGATTTGGGCCG |
|
| R:
CCATTGTCATTCCCTGACTCCA |
|
GAB1-rat | F:
GAGAGCTAGGTTCTCGCCAC |
|
| R:
TCCTCTTCCATGCATAACGCT |
|
NTRK2-rat | F:
TTACGGTTTGTCACCCGACC |
|
| R:
GTGCTTGGTTCAGCTCTTGC |
| RUNX2-rat | F:
GCCAATCCCTGAGTGTGACT |
|
| R:
CCTGGTGGTGTCACTGAATG |
|
BGLAP-rat | F:
CACCGTTTAGGGCATGTGTT |
|
| R:
TCCTGGAGAGTAGCCAAAGC |
| miR-128-3p-rat | F:
CGCGTCACAGTGAACCGGT |
|
| R:
AGTGCAGGGTCCGAGGTATT |
| U6 |
AACGCTTCACGAATTTGCGT |
Osteogenic differentiation
Osteogenic differentiation of BM-MSCs was induced
using standard osteogenic medium consisting of phenol red-free
α-MEM, 10% FBS, ascorbic acid (50 µg/ml), dexamethasone
(10−8 M) and β-glycerophosphate (10−2 M). The
medium was changed twice every week. The osteoblastic
differentiation potency of BM-MSCs was determined by Alizarin red
staining: After induction culture for 2 weeks, the cells were
washed with PBS and fixed with 4% paraformaldehyde for 10 min and
then stained using 2% Alizarin red for 20 min at room temperature
to evaluate calcium nodule formation.
Statistical analysis
All experiments were performed for at least three
biological repeats. Statistical analysis was performed using SPSS
18.0 (SPSS, Inc.) and graphs were prepared in GraphPad Prism 5.0
(GraphPad Software, Inc.). All data were expressed as the mean ±
standard deviation. Statistical differences between two groups were
determined using two-tailed paired Student's t-test; while
statistical comparisons among more than two groups were performed
using one-way ANOVA followed by post hoc Tukey's test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of miR-128 as a DEM in
osteoblast-differentiated BM-MSCs
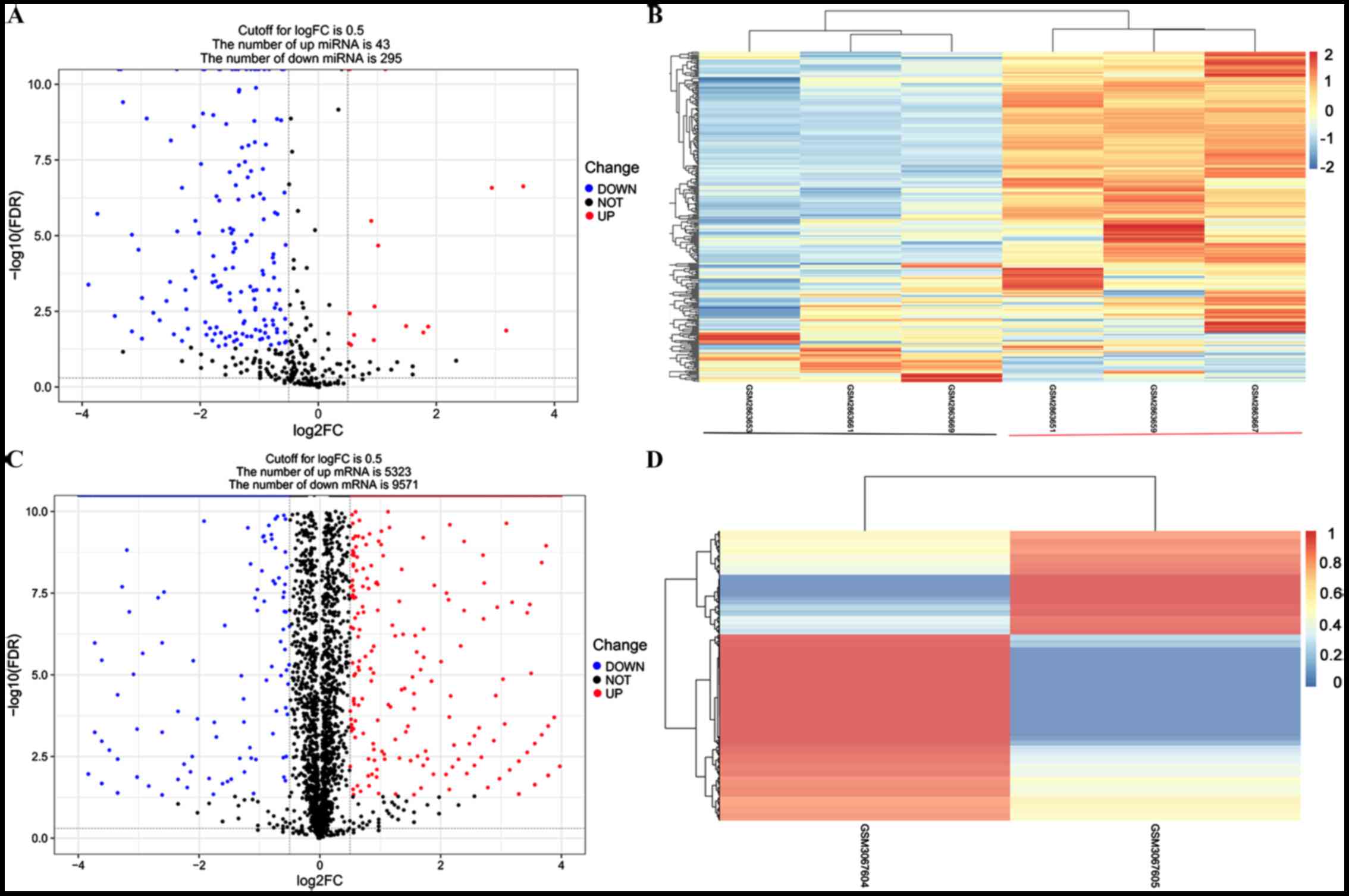
Among the 654 miRNAs included in the GSE107279
dataset, 338 of them were found to be differentially expressed
between differentiated and undifferentiated BM-MSCs under the
stated threshold, including 295 downregulated and 43 upregulated
DEMs (Fig. 1A and B). miR-128 was
one of the significantly downregulated DEMs in differentiated
BM-MSCs, with a log2FC of −0.77 and an FDR-adjusted P-value of
1.93×10−12.
Identification of the crucial target
genes of miR-128 for osteoblast-differentiated BM-MSCs
Among the 19,652 genes included in the GSE112318
dataset, 14,894 of them were revealed to be differentially
expressed between differentiated and undifferentiated BM-MSCs under
the stated threshold, including 9,571 downregulated and 5,323
upregulated DEGs (Fig. 1C and
D).
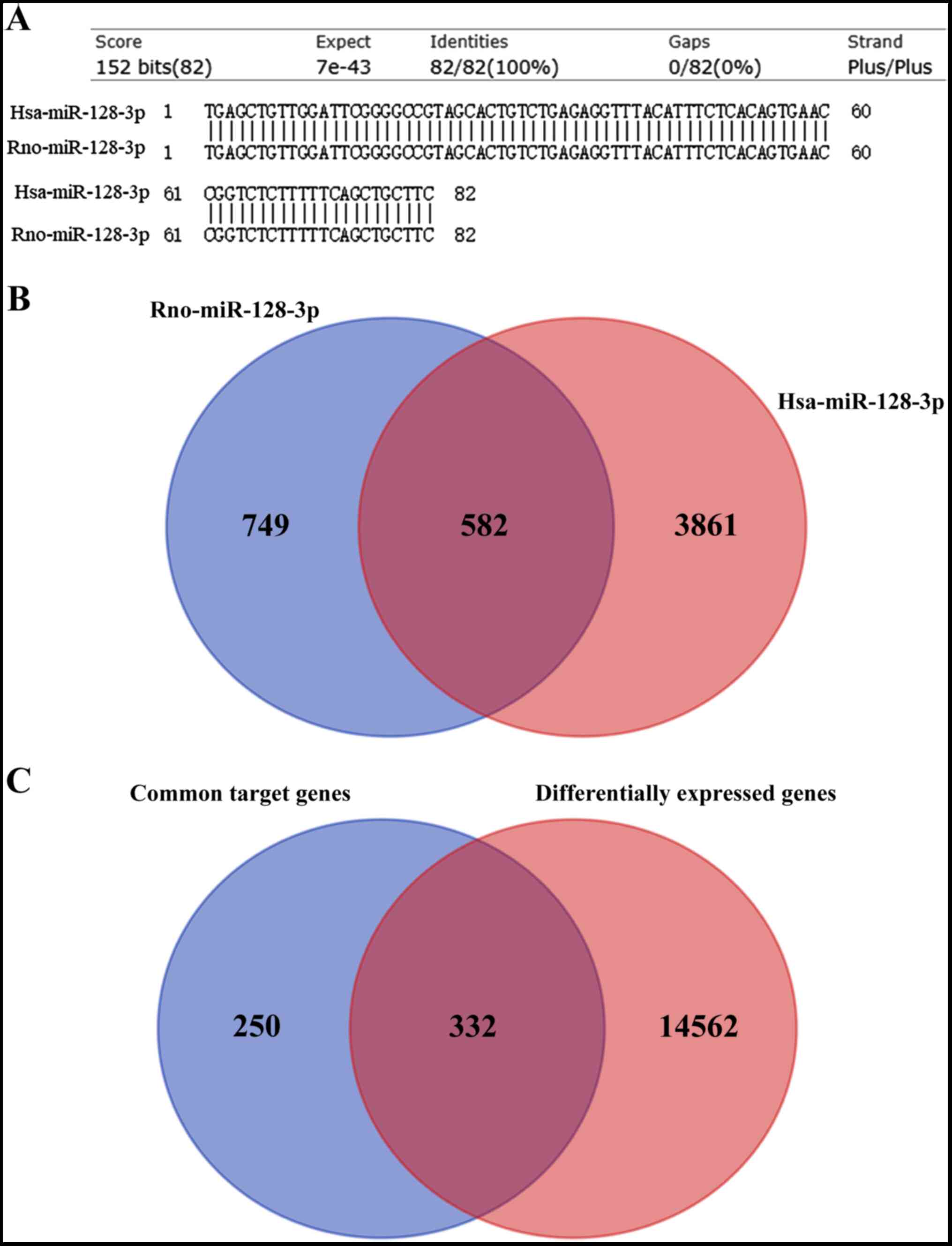
The sequence of has-miR-128-3p and Rno-miR-128-3p
was 100% identical (Fig. 2A).
Thus, the targets of miR-128-3p were predicted using human and rat
as species, respectively. Using the miRwalk 2.0 database, 4,443
target genes were predicted to interact with human miR-128-3p in
the 3′UTR, while 1,331 were predicted to be regulated by rat
miR-128-3p via the 3′UTR. Among them, 582 were shared between the
two species (Fig. 2B).
Furthermore, 332 of 582 genes were overlapped with the DEGs
(Fig. 2C), including 103
upregulated and 229 downregulated (which were used for establishing
the miRNA-mRNA network, as shown in Fig. 3), suggesting these 103 upregulated
DEGs may be underlying target genes of downregulated
miR-128-3p.
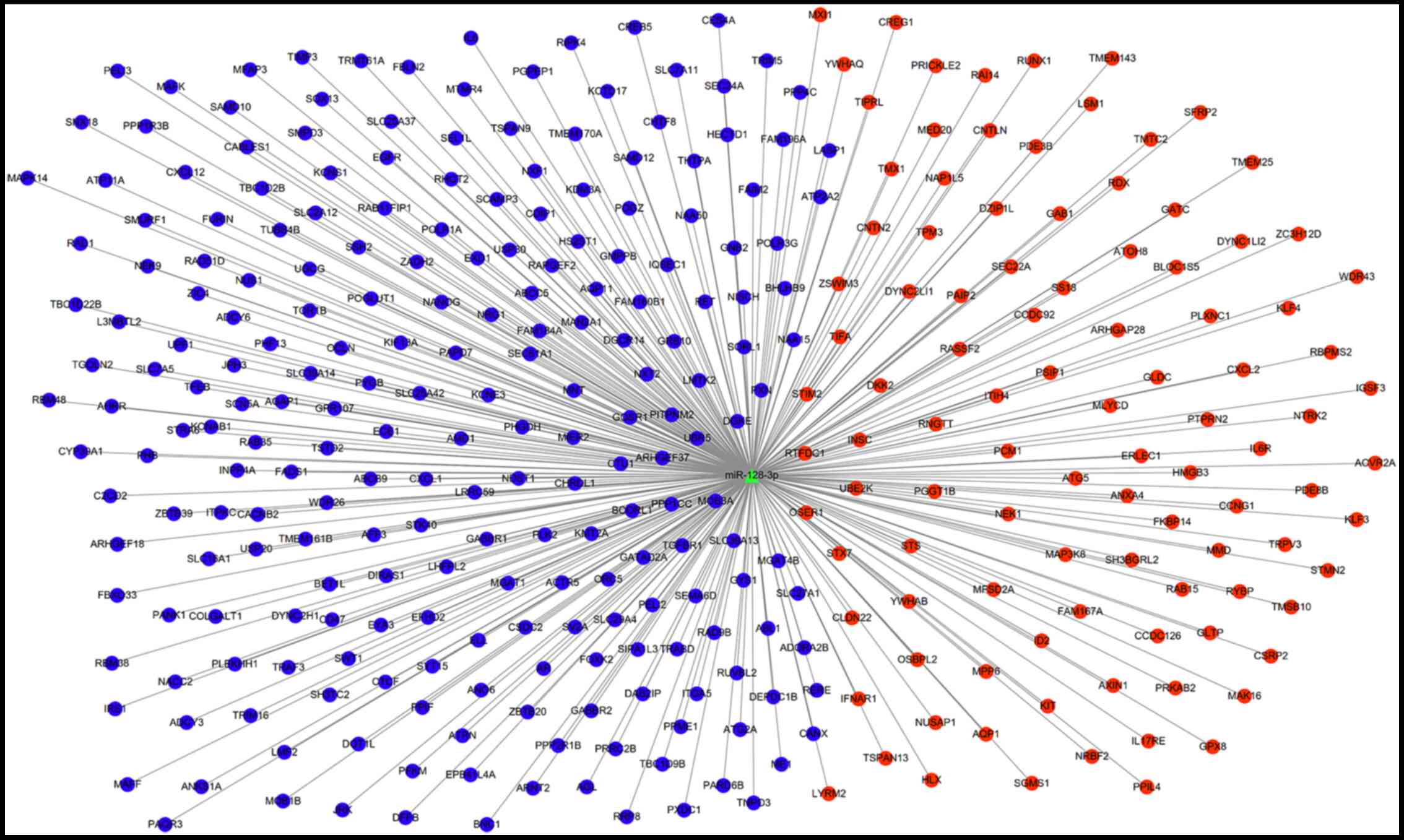
In order to further screen crucial targets for
miR-128-3p, the PPI network and function modules were analyzed. A
total of 264 DEGs were identified to interact with other genes to
form 539 interaction pairs (such as NTRK2-GAB1, GAB1-IRS1,
YWHAB-PPP1CC, AXIN1-GYS1 and GYS1-IRS1, which was used
for constructing the PPI network. After calculating the topological
properties, 26 DEGs (including 6 upregulated: KIT, NTRK2, YWHAB,
GAB1, AXIN1 and RUNX1) were found to be shared in the
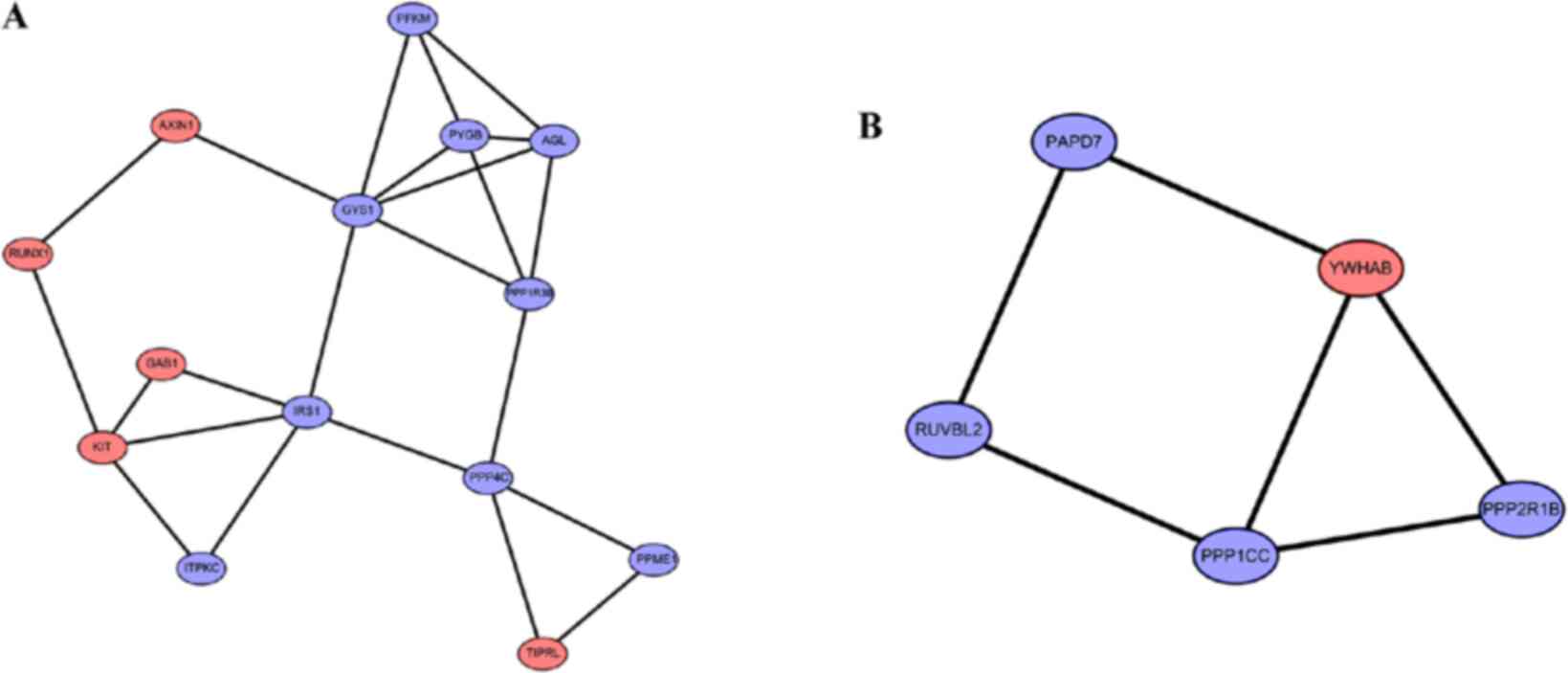
top 40 genes ranking for DC, CC and EC (Table II). In addition, 11 modules were
extracted from the PPI network. Among them, only module 4
(RUNX1, KIT, GAB1 and AXIN1; in which RUNX1
may be especially key since it could interact with KIT and
AXIN1) and module 7 (YWHAB) included the shared
upregulated DEGs identified by topological ranking (Fig. 4; Table III). These findings indicated
that these 5 upregulated genes may be particularly crucial targets
of miR-128-3p.
 | Table II.Topological properties of genes in
the PPI network. |
Table II.
Topological properties of genes in
the PPI network.
|
|
|
|
|
|
|
| Expression |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Gene | DC | Gene | CC | Gene | EC | Common | logFC | FDR |
|---|
| EGFR | 39 | EGFR | 0.068059 | EGFR | 0.39794025 | TGOLN2 | −1.08 | 0 |
| IL6 | 32 | IL6 | 0.067039 | IL6 | 0.34596485 | KIT | 1.88 |
2.43×10−209 |
| AR | 22 | AR | 0.066937 | AR | 0.23498377 | PPP1CC | −0.64 | 0 |
| ABL1 | 18 | IRS1 | 0.066785 | IRS1 | 0.2286748 | FURIN | −2.45 | 0 |
| PPP1CC | 18 | ABL1 | 0.066667 | ABL1 | 0.20841703 | PPP4C | −1.00 | 0 |
| IRS1 | 17 | MAPK14 | 0.066348 | CXCL12 | 0.19866262 | CANX | −0.55 | 0 |
| CXCL12 | 16 | YWHAB | 0.066265 | KIT | 0.18829939 | EGFR | −1.68 | 0 |
| KIT | 14 | TGFBR1 | 0.066033 | MAPK14 | 0.1772947 | NTRK2 | 4.77 | 0 |
| PXN | 14 | PPP1CC | 0.065885 | PXN | 0.16721927 | CXCL1 | −3.06 | 0 |
| CANX | 14 | PPP2R1B | 0.065852 | GAB1 | 0.1594914 | TGFBR1 | −0.79 | 0 |
| MAPK14 | 12 | PXN | 0.06577 | NANOG | 0.14420108 | IRS1 | −0.96 | 0 |
| ADCY3 | 12 | PPP4C | 0.065753 | CXCL1 | 0.14116184 | PPP2R1B | −2.08 | 0 |
| ADCY6 | 12 | NANOG | 0.065737 | RET | 0.1360971 | MAPK14 | −0.69 | 0 |
| TGFBR1 | 11 | CXCL12 | 0.065721 | TGFBR1 | 0.12943284 | ATP2A2 | −2.00 | 0 |
| YWHAB | 11 | KIT | 0.065606 | CANX | 0.12310072 | ABL1 | −1.99 | 0 |
| GNB2 | 11 | CANX | 0.065557 | KLF4 | 0.12178243 | YWHAB | 0.51 | 0 |
| CXCL1 | 10 | AXIN1 | 0.065557 | NTRK2 | 0.116883084 | GAB1 | 0.53 |
6.04×10−81 |
| NTRK2 | 10 | YWHAQ | 0.06546 | YWHAB | 0.112461776 | NANOG | −2.66 |
2.03×10−51 |
| GABBR2 | 10 | OCLN | 0.065379 | IL6R | 0.11212979 | CXCL12 | −9.14 | 0 |
| PPP2R1B | 10 | SMURF1 | 0.06525 | OCLN | 0.11207541 | AXIN1 | 0.78 |
4.50×10−27 |
| AXIN1 | 10 | NTRK2 | 0.065234 | GRB10 | 0.10549404 | AR | −1.01 | 0 |
| SMURF1 | 10 | KLF4 | 0.065169 | PPP1CC | 0.09967908 | RET | −10.3 |
6.15×10−99 |
| GAB1 | 9 | CXCL1 | 0.065137 | CXCL2 | 0.098889045 | IL6 | −1.15 | 0 |
| NANOG | 9 | ATP2A2 | 0.065105 | TGOLN2 | 0.09819438 | RUNX1 | 0.71 | 0 |
| OCLN | 9 | TGOLN2 | 0.065057 | ATG5 | 0.0979064 | OCLN | −10.2 |
1.76×10−94 |
| PPP4C | 9 | GAB1 | 0.065009 | GNB2 | 0.093786895 | PXN | −0.97 | 0 |
| KMT2A | 9 |
DYNC1LI2 | 0.064993 | TIMP3 | 0.09199475 |
|
|
|
| GYS1 | 9 | RUNX1 | 0.064977 | ITGA5 | 0.09169616 |
|
|
|
| RUVBL2 | 9 | ATG5 | 0.064929 | ITPKC | 0.08899418 |
|
|
|
| RET | 8 | GYS1 | 0.064897 | GABBR2 | 0.08424522 |
|
|
|
| CXCL2 | 8 | RDX | 0.064865 | RUNX1 | 0.08272818 |
|
|
|
| TGOLN2 | 8 | RET | 0.064865 | PPP2R1B | 0.08233988 |
|
|
|
| GABBR1 | 8 | FURIN | 0.064833 | ADCY3 | 0.08222689 |
|
|
|
| FURIN | 8 | ITPKC | 0.064769 | ADCY6 | 0.08222689 |
|
|
|
| ATP2A2 | 8 | KMT2A | 0.064753 | YWHAQ | 0.082175 |
|
|
|
|
DYNC1LI2 | 8 | GRB10 | 0.064753 | PPP4C | 0.08180887 |
|
|
|
| PYGB | 8 | TIMP3 | 0.064722 | AXIN1 | 0.080903396 |
|
|
|
| BET1L | 8 | SEC61A1 | 0.06469 | FURIN | 0.07912381 |
|
|
|
| RUNX1 | 7 | PHB | 0.064674 | ATP2A2 | 0.07898864 |
|
|
|
| CACNB2 | 7 | IL6R | 0.064643 | ITIH4 | 0.07645497 |
|
|
|
 | Table III.Modules extracted from the PPI
network. |
Table III.
Modules extracted from the PPI
network.
| Cluster | Score (density ×
number of nodes) | Nodes | Edges | Node IDs |
|---|
| 1 | 8 | 8 | 28 | CXCL12, GABBR2,
GABBR1, ADCY3, CXCL1, CXCL2, GNB2, ADCY6 |
| 2 | 5.2 | 11 | 26 | PXN, AR, IL6R,
TGFBR1, IL6, EGFR, RET, ABL1, NANOG, OCLN, KLF4 |
| 3 | 4 | 4 | 6 | TRMT61A, WDR43,
RRP8, MAK16 |
| 4 | 3.538 | 14 | 23 | GAB1, KIT, PPME1,
RUNX1, PYGB, AGL, AXIN1, ITPKC, PPP4C, TIPRL, PPP1R3B, GYS1, IRS1,
PFKM |
| 5 | 3 | 3 | 3 | STMN2, FAIM2,
CNTN2 |
| 6 | 3 | 3 | 3 | BET1L, SEC22A,
STX7 |
| 7 | 3 | 5 | 6 | YWHAB, PAPD7,
PPP2R1B, RUVBL2, PPP1CC |
| 8 | 3 | 3 | 3 | FAM160B1, IGSF3,
POGLUT1 |
| 9 | 3 | 3 | 3 | RHOT2, ARHGAP28,
DEPDC1B |
| 10 | 3 | 3 | 3 | ANO6, ATP11A,
CD47 |
| 11 | 3 | 3 | 3 | SGMS1, SMPD3,
UGCG |
Function enrichment analysis of the genes in the PPI
network revealed that 30 KEGG pathways were enriched, including
Insulin resistance (PPP1CC and IRS1), Hippo signaling
pathway (PPP1CC, YWHAB and AXIN1), Pathways in cancer
(RUNX1, AXIN1 and KIT), PI3K-Akt signaling pathway
(YWHAB, KIT and NTRK2), Tight junction
(RUNX1), Proteoglycans in cancer (GAB1) and Rap1
signaling pathway (KIT) (Table
IV).
 | Table IV.KEGG pathways enriched for the genes
in the PPI network. |
Table IV.
KEGG pathways enriched for the genes
in the PPI network.
| Term | P-value | Genes |
|---|
| Insulin
resistance |
4.22×10−3 | PPP1CC, PYGB,
GYS1, SLC27A1, PRKAB2, IL6, IRS1, PPP1R3B, CREB5 |
|
Vasopressin-regulated water
reabsorption |
3.64×10−3 | DYNC2LI1,
DYNC2H1, DYNC1LI2, ADCY3, ADCY6, CREB5 |
| Adrenergic
signaling in cardiomyocytes |
2.48×10−3 | PPP1CC, CACNB2,
PPP2R1B, TPM3, ATP2A2, ADCY3, SCN5A, MAPK14, ADCY6, CREB5 |
| Hippo signaling
pathway |
4.32×10−3 | MOB1B, PPP1CC,
PARD6B, RASSF2, PPP2R1B, YWHAQ, YWHAB, ID2, AXIN1, TGFBR1 |
| TNF signaling
pathway |
6.73×10−3 | IL6, TRAF3,
DAB2IP, MAP3K8, CXCL1, MAPK14, CXCL2, CREB5 |
| Morphine
addiction |
1.07×10−2 | GABBR2, GABBR1,
GNB2, PDE3B, ADCY3, PDE8B, ADCY6 |
| Pathways in
cancer |
1.18×10−2 | RET, ARNT2,
TPM3, AXIN1, ADCY3, ADCY6, EGFR, TGFBR1, RUNX1, AR, IL6, CXCL12,
TRAF3, GNB2, KIT, ABL1, IL6R, IFNAR1 |
| PI3K-Akt signaling
pathway |
1.10×10−2 | NTRK2, IRS1,
YWHAB, EGFR, GYS1, IL6, PPP2R1B, YWHAQ, GNB2, KIT, ITGA5, IL6R,
IFNAR1, CREB5 |
| Tight junction |
1.55×10−2 | CLDN22, PRKAB2,
OCLN, PARD6B, PPP2R1B, RDX, ARHGEF18, RAPGEF2, RUNX1 |
| Human
cytomegalovirus infection |
2.69×10−2 | IL6, CXCL12,
GNB2, PXN, ADCY3, MAPK14, IL6R, ADCY6, EGFR, CREB5 |
| Hypertrophic
cardiomyopathy |
2.61×10−2 | CACNB2, PRKAB2,
IL6, TPM3, ATP2A2, ITGA5 |
| Salmonella
infection |
2.55×10−2 | DYNC2H1,
DYNC1LI2, IL6, CXCL1, MAPK14, CXCL2 |
| Hepatitis C |
2.61×10−2 | CLDN22, OCLN,
PPP2R1B, YWHAQ, YWHAB, TRAF3, EGFR, IFNAR1 |
| AMPK signaling
pathway |
2.43×10−2 | GYS1, PRKAB2,
PPP2R1B, IRS1, MLYCD, PFKM, CREB5 |
| Dilated
cardiomyopathy | 2.73×
10−2 | CACNB2, TPM3,
ATP2A2, ADCY3, ITGA5, ADCY6 |
| mRNA surveillance
pathway |
2.56×10−2 | PPP1CC, UPF1,
NXF1, RNGTT, PPP2R1B, NXT2 |
| Oocyte meiosis |
2.54×10−2 | PPP1CC, AR,
PPP2R1B, YWHAQ, YWHAB, ADCY3, ADCY6 |
| Proteoglycans in
cancer |
2.55×10−2 | PPP1CC, RDX,
PXN, GAB1, TIMP3, NANOG, ITGA5, MAPK14, EGFR |
| Hepatitis B |
2.46×10−2 | IL6, YWHAQ,
YWHAB, TRAF3, MAPK14, TGFBR1, IFNAR1, CREB5 |
| Central carbon
metabolism in cancer |
2.62×10−2 | RET, SLC7A5,
KIT, PFKM, EGFR |
| cGMP-PKG signaling
pathway |
2.50×10−2 | PPP1CC, IRS1,
PDE3B, PPIF, ATP2A2, ADCY3, ADCY6, CREB5 |
| Relaxin signaling
pathway |
2.46×10−2 | GNB2, ADCY3,
MAPK14, TGFBR1, EGFR, ADCY6, CREB5 |
| Rap1 signaling
pathway |
2.36×10−2 | PARD6B, ADORA2B,
KIT, RAPGEF2, ADCY3, MAPK14, SIPA1L3, ADCY6, EGFR |
| FoxO signaling
pathway |
2.46×10−2 | PRKAB2, IL6,
IRS1, PLK2, MAPK14, TGFBR1, EGFR |
| Insulin signaling
pathway |
2.91×10−2 | PPP1CC, PYGB,
GYS1, PRKAB2, IRS1, PPP1R3B, PDE3B |
| Longevity
regulating pathway |
2.82×10−2 | PRKAB2, IRS1,
ADCY3, ADCY6, ATG5, CREB5 |
| Glucagon signaling
pathway |
2.86×10−2 | PYGB, GYS1,
PRKAB2, PPP4C, PDE3B, CREB5 |
| Thyroid hormone
synthesis |
3.31×10−2 | CANX, GPX8,
ADCY3, ADCY6, CREB5 |
| Phagosome |
4.47×10−2 | DYNC2H1,
SEC61A1, DYNC1LI2, STX7, CANX, ITGA5, TUBB4B |
| Cushing
syndrome |
4.80×10−2 | KMT2A, AXIN1,
ADCY3, PDE8B, EGFR, ADCY6, CREB5 |
Validation of the function on the
osteoblast differentiation of miR-128-3p and the expression of
target genes
To confirm the differential characteristics of
miR-128-3p and their target genes during the osteoblast
differentiation, RT-qPCR was performed. As anticipated, the results
demonstrated that osteogenic induction can lead to a reduction in
the expression of miR-128-3p and an increase in the expression of
its potential target genes (KIT, NTRK2, YWHAB, GAB1, AXIN1
and RUNX1; Fig. 5A).
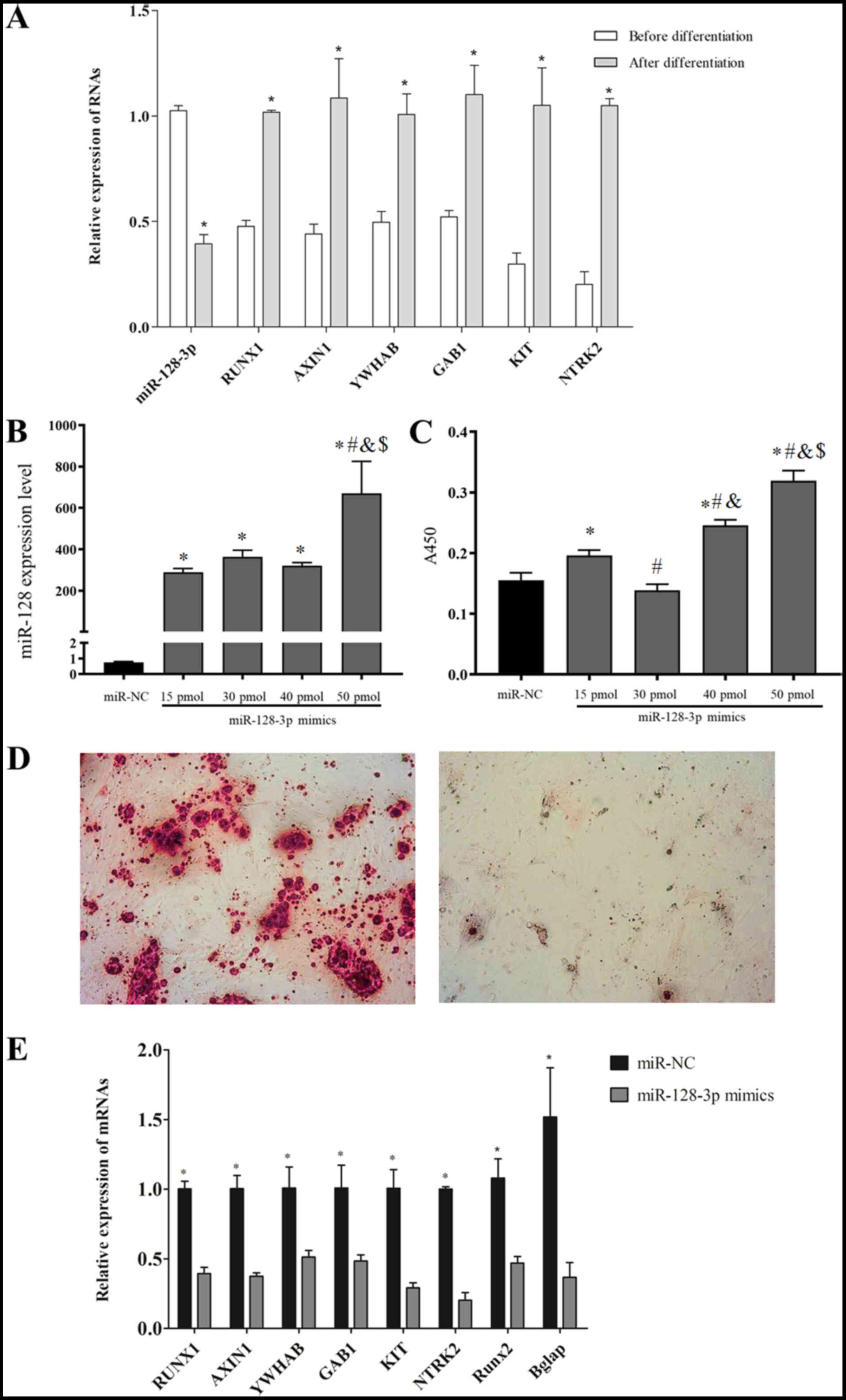 | Figure 5.Validation experiments. (A) The
expression of miR-128-3p and its target genes in BM MSCs before and
after osteoblast differentiation, as determined by RT-qPCR. (B)
After transfection of miR-128-3p into rat BM MSCs for 48 h, the
expression of miR-128-3p was detected. (C) The effects of
miR-128-3p overexpression on the viability of BM MSCs, as evaluated
by Cell Counting Kit-8 assay. (D) The effects of miR-128-3p
overexpression on osteoblast differentiation, as determined by
Alizarin red staining (magnification, ×100). (E) The effects of
miR-128-3p overexpression on the expression of its target genes, as
assessed via RT-qPCR. *P<0.05 vs. before differentiation or
miR-normal control (miR-NC). #P<0.05 vs. 15 pmol
miR-128-3p mimics. &P<0.05 vs. 30 pmol miR-128-3p
mimics. $P<0.05 vs. 40 pmol miR-128-3p mimics. miRNA,
microRNA; RT-qPCR, reverse transcription- quantitative PCR; BM
MSCs, bone marrow derived mesenchymal stem cells; miR NC, miR
normal control. |
To further validate the regulatory relationship
between miR-128-3p and its target genes, miR-128-3p mimic
transfection experiments were carried out. After transfection of
miR-128-3p into rat BM-MSCs for 48 h, the expression of miR-128-3p
(Fig. 5B) and the cell
proliferation (Fig. 5C) of BM-MSCs
were revealed to be the highest in the group with the treatment
concentration of 50 pmol, indicating the successful overexpression
of miR-128-3p. Thus, this concentration was used to investigate the
influence of miR-128-3p overexpression on the osteoblast
differentiation of BM-MSCs. In line with the sequencing data
analysis results and our previous study on human BM-MSCs (11), overexpression of miR-128-3p clearly
decreased calcium deposition (Fig.
5D) and significantly weakened the expression levels of
osteogenic specific markers Runx2 (22) and bone γ-carboxyglutamate protein
(BGLAP) (23) (Fig. 5E), further suggesting its
inhibitory effects on the osteoblast differentiation. Notably, the
expression levels of 6 target genes as aforementioned were
significantly decreased in the miR-128-3p-mimics group compared
with the miR-NC group (Fig.
5E).
Discussion
The present study, using an integrated analysis of
miRNA and mRNA sequencing datasets and in vitro
verification, revealed that miR-128-3p may inhibit the osteoblast
differentiation of BM-MSCs by targeted downregulation of RUNX1,
NTRK2 and YWHAB. Subsequently, these target genes may
respectively regulate the expression of downstream AXIN1, KIT,
GAB1 and PPP1CC to mediate Insulin resistance, Hippo,
PI3K-Akt and Rap1 signaling pathways.
There is evidence that RUNX1 plays an important role
in mediating the osteogenic differentiation of stem cells and this
was associated with miR-128 (24).
For example, Luo et al (25) revealed that knockdown of RUNX1 in
murine BM-MSCs significantly inhibited osteogenesis (as evaluated
by lower ALP activity and reduced calcium nodule formation) and
decreased the expression levels of osteogenic-related genes
(RUNX2, OCN and OPN) compared with control cells. Ji
et al (26) also reported
that overexpression of RUNX1 enhanced bone morphogenetic
protein 9-induced osteogenic differentiation of murine mesenchymal
stem cell line (C3H10T1/2), whereas inhibitory effects were
observed after knockdown of RUNX1. Saito et al
(27) demonstrated that
RUNX1 was a downstream effector of Nanog to promote the
osteogenic differentiation of C3H10T1/2. In accordance with these
studies, the present study also screened RUNX1 as an
upregulated gene in differentiated BM-MSCs and its expression was
decreased after miR-128 mimic treatment. Further mechanism studies
indicated that RUNX1 may exert its promoting roles in
osteogenic differentiation by activating the canonical Wnt
signaling pathway (including upregulation of β-catenin, Lef1, Tcf1
and Wnt10b) (25). AXIN1
was previously considered as a negative regulator of the canonical
Wnt/β-catenin signaling pathway via degradation of β-catenin with
adenomatous polyposis coli (APC), glycogen synthase kinase-3β
(GSK-3β) and Casein kinase 1 (28). Thus, theoretically, upregulated
RUNX1 may lead to the downregulation of AXIN1 in the
osteogenic differentiation of BM-MSCs (29,30).
However, the results of the present study revealed that the
expression of AXIN1 was higher in differentiated compared
with undifferentiated BM-MSCs. These controversial findings may be
associated with the dual roles of AXIN1 similar to the
reports of its homologous gene AXIN2 in cancers (31). Furthermore, the study of Chimge
et al (32) reported that
silencing of RUNX1 resulted in the downregulation of
AXIN1, indicating their synergetic expression trend, which
appears to be in agreement with the results of the present study.
The present study predicted that RUNX1-AXIN1 may
downregulate GYS1 and then IRS1 to participate in the
osteogenic differentiation. This hypothesis can be indirectly
demonstrated by the fact that overexpression of GSK-3β (an
AXIN1 complex component) decreased the mRNA expression level
of GYS1 (33), while
pharmacological inhibition of GSK-3 induced the formation of
glycogen bodies (34). Also, an
increase in the IRS-1 protein level was revealed to inhibit
the osteoblastic differentiation of MSCs (35). In addition, insulin-deficiency may
trigger the lower expression of its downstream signaling molecule
insulin receptor substrate 1, which is reported to increase the
activity of glutaminase (36) and
then stimulate osteoblast differentiation from stem cells (37). Accordingly, the
miR-128-RUNX1-AXIN1-GYS1-IRS1 axis may be a credible
mechanism to explain the osteoblast differentiation processes. In
addition to AXIN1, RUNX1 was also observed to interact with
KIT/GAB1 to influence the osteogenic differentiation.
KIT is a type 3 transmembrane receptor for stem cell factor (SCF)
(38). Activation of the SCF-c-KIT
signaling pathway has been revealed to promote the recruitment of
stem cells and osteogenesis, which establishes a favorable
environment for bone healing and remodeling (38). As an adaptor protein of KIT, GAB1 A
complex of α-subunits of the G proteins (Gαi1/3) and Gab1 can
mediate the activation of PI3K-Akt signaling (39), which is suggested to be related
with enhanced osteogenesis of MSCs (40).
NTRK2, also known as TRKB, can bind
with the TrkB receptor to phosphorylate the downstream Erk1/2,
which promotes the expression of transcription factors, such as
Runx2 and Osterix that are associated with the osteoblast
differentiation of BM-MSCs (41).
In accordance with these findings, the present study also revealed
that NTRK2 was upregulated in differentiated BM-MSCs and
downregulated after miR-128-mimic transfection, indicating that
miR-128-NTRK2 may represent a crucial mechanism for the
osteoblast differentiation of BM-MSCs. In addition, the present
study also hypothesized that NTRK2 may interact with
GAB1 to become involved in osteogenesis of MSCs.
No studies, to the best of the authors' knowledge,
have investigated the roles of YWHAB in the osteogenic
differentiation and the regulatory relationship between
YWHAB and miR-128, implying that miR-128-YWHAB
interaction may be a novel mechanism for explaining the osteogenic
differentiation process of BM-MSCs. The present study predicted
PPP1CC may be one of the downstream targets of YWHAB.
PPP1CC is a member of the serine/threonine protein
phosphatase families. A previous study revealed that a reduction in
PP2A promoted the bone formation and the osteoblast differentiation
of MSCs through the regulation of bone-related transcription
factors such as Osterix (42).
Mice deficient in PP5 phosphatase were observed to have increased
osteoblast numbers and high bone formation, which is associated
with a substantial increase in osteoblast differentiation of MSCs
(43). Similar to other members,
downregulated PPP1CC by miR-128-YWHAB may also
contribute to the osteoblast differentiation.
However, there were some limitations to the present
study. First, the target genes of miR-128-3p and their possible
interactive downstream genes were predicted, however, the
connection between them and their associations with the
proliferation and osteoblast differentiation of BM-MSCs still
requires additional wet experiments (luciferase reporter assay,
co-immunoprecipitation, co-overexpression or co-knockdown) to
verify the conclusions. Second, the expression levels and functions
of target genes of miR-128-3p should be further tested in human
BM-MSCs. Third, in vivo studies should be performed to
investigate the differentiation and therapeutic efficiency of
BM-MSCs that were transfected with miR-128-3p inhibitors or target
genes of miR-128-3p. Fourth, the sample size of our included
sequencing datasets was small and thus, it may also be necessary to
launch a high-throughput analysis with a larger sample size in
order to identify more underlying targets for the osteoblast
differentiation of BM-MSCs.
The findings of the present study indicated that
miR-128-3p may inhibit the osteoblast differentiation of BM-MSCs by
targeted downregulation of RUNX1, YWHAB and
NTRK2.
Acknowledgements
Not applicable.
Funding
The present study was sponsored by the
Interdisciplinary Program of Shanghai Jiao Tong University
(Shanghai, China; grant no. YG2017QN21).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WZ, YZ and CC contributed to the conception and
design of this study. WZ and YZ performed the experiments. WZ, YZ,
JC and JW performed the statistical analyses. CY was involved in
the interpretation of the data. WZ and YZ drafted the manuscript.
CC edited and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Confalonieri D, Schwab A, Walles H and
Ehlicke F: Advanced therapy medicinal products: A guide for bone
marrow-derived MSC application in bone and cartilage tissue
engineering. Tissue Eng Part B Rev. 24:155–169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gopal K, Amirhamed HA and Kamarul T:
Advances of human bone marrow-derived mesenchymal stem cells in the
treatment of cartilage defects: A systematic review. Exp Biol Med
(Maywood). 239:663–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guntur AR and Rosen CJ: The skeleton: A
multi-functional complex organ. New insights into osteoblasts and
their role in bone formation: The central role of PI3Kinase. J
Endocrinol. 211:123–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shin YS, Yoon JR, Kim HS and Lee SH:
Intra-articular injection of bone marrow-derived mesenchymal stem
cells leading to better clinical outcomes without difference in MRI
outcomes from baseline in patients with knee osteoarthritis. Knee
Surg Relat Res. 30:206–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Liu Y, Wu M, Wang H, Wu L, Xu B,
Zhou W, Fan X, Shao J and Yang T: MicroRNA-664a-5p promotes
osteogenic differentiation of human bone marrow-derived mesenchymal
stem cells by directly downregulating HMGA2. Biochem Biophys Res
Commun. 521:9–14. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ge JB, Lin JT, Hong HY, Sun YJ, Li Y and
Zhang CM: MiR-374b promotes osteogenic differentiation of MSCs by
degrading PTEN and promoting fracture healing. Eur Rev Med
Pharmacol Sci. 22:3303–3310. 2018.PubMed/NCBI
|
|
7
|
Huang J, Meng Y, Liu Y, Chen Y, Yang H,
Chen D, Shi J and Guo Y: MicroRNA-320a regulates the osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting HOXA10. Cell Physiol Biochem. 38:40–48. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu XM, Gu YL and Ren GQ: miR-125a-3p
targetedly regulates GIT1 expression to inhibit osteoblastic
proliferation and differentiation. Exp Ther Med 2016. 12:4099–4106.
2016. View Article : Google Scholar
|
|
9
|
Wang H, Xie Z, Hou T, Li Z, Huang K, Gong
J, Zhou W, Tang K, Xu J and Dong S: MiR-125b regulates the
osteogenic differentiation of human mesenchymal stem cells by
targeting BMPR1b. Cell Physiol Biochem. 41:530–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang S, Liu Y, Zheng Z, Zeng X, Liu D,
Wang C and Ting K: MicroRNA-223 suppresses osteoblast
differentiation by inhibiting DHRS3. Cell Physiol Biochem.
47:667–679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Yao C, Wei Z and Dong Q: miR-128
promoted adipogenic differentiation and inhibited osteogenic
differentiation of human mesenchymal stem cells by suppression of
VEGF pathway. J Recept Signal Transduct Res. 37:217–223. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang CC, Venø MT, Chen L, Ditzel N, Le D,
Dillschneider P, Kassem M and Kjems J: Global microrna profiling in
human bone marrow skeletal-stromal or mesenchymal-stem cells
identified candidates for bone regeneration. Mol Ther. 26:593–605.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Cai M, Zhong J, Yang L, Xiao J, Jin
F, Xue H, Liu X, Liu H, Zhang Y, et al: The long noncoding RNA
lnc-ob1 facilitates bone formation by upregulating Osterix in
osteoblasts. Nat Metab. 1:485–496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
R Core Team. R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2012, simplehttp://www.R-project.org/
|
|
16
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:697. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou W, Greenblatt MB, Brady N, Lotinun S,
Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al: The
microtubule-associated protein DCAMKL1 regulates osteoblast
function via repression of Runx2. J Exp Med. 210:1793–1806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matta C, Szűcs-Somogyi C, Kon E, Robinson
D, Neufeld T, Altschuler N, Berta A, Hangody L, Veréb Z and Zákány
R: Osteogenic differentiation of human bone marrow-derived
mesenchymal stem cells is enhanced by an aragonite scaffold.
Differentiation. 107:24–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Motohashi N, Alexander MS, Casar JC and
Kunkel LM: Identification of a novel microRNA that regulates the
proliferation and differentiation in muscle side population cells.
Stem Cells Dev. 21:3031–3043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo Y, Zhang Y, Miao G, Zhang Y, Liu Y and
Huang Y: Runx1 regulates osteogenic differentiation of BMSCs by
inhibiting adipogenesis through Wnt/β-catenin pathway. Arch Oral
Biol. 97:176–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji C, Xiaohua L, Li X, Tingting Y, Chaoqun
D and Jinyong L: RUNX1 plays an important role in mediating
BMP9-Induced osteogenic differentiation of mesenchymal stem cells
line C3H10T1/2, murine multi-lineage cells lines C2C12 and MEFs.
Int J Mol Sci. 18:13482017. View Article : Google Scholar
|
|
27
|
Saito T, Ohba S, Yano F, Seto I, Yonehara
Y, Takato T and Ogasawara T: Runx1 and Runx3 are downstream
effectors of nanog in promoting osteogenic differentiation of the
mouse mesenchymal cell line C3H10T1/2. Cell Reprogram. 17:227–234.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakamura T, Hamada F, Ishidate T, Anai K,
Kawahara K, Toyoshima K and Akiyama T: Axin, an inhibitor of the
Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and
APC and reduces the beta-catenin level. Genes Cells. 3:395–403.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Figeac N and Zammit PS: Coordinated action
of Axin1 and Axin2 suppresses β-catenin to regulate muscle stem
cell function. Cell Signal. 27:1652–1665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gwak J, Hwang SG, Park HS, Choi SR, Park
SH, Kim H, Ha NC, Bae SJ, Han JK, Kim DE, et al: Small
molecule-based disruption of the Axin/β-catenin protein complex
regulates mesenchymal stem cell differentiation. Cell Res.
22:237–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu ZQ, Brabletz T, Fearon E, Willis AL, Hu
CY, Li XY and Weiss SJ: Canonical Wnt suppressor, Axin2, promotes
colon carcinoma oncogenic activity. Proc Natl Acad Sci USA.
109:11312–11317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chimge NO, Little GH, Baniwal SK,
Adisetiyo H, Xie Y, Zhang T, O'Laughlin A, Liu ZY, Ulrich P, Martin
A, et al: RUNX1 prevents oestrogen-mediated AXIN1 suppression and
β-catenin activation in ER-positive breast cancer. Nat Commun.
7:107512016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Wang Y, Zhong T, Guo J, Li L,
Zhang H and Wang L: Transcriptional regulation of pig GYS1 gene by
glycogen synthase kinase 3β (GSK3β). Mol Cell Biochem. 424:203–208.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen RJ, Zhang G, Garfield SH, Shi YJ,
Chen KG, Robey PG and Leapman RD: Variations in glycogen synthesis
in human pluripotent stem cells with altered pluripotent states.
PLoS One. 10:e01425542015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Contaldo C, Myers TJ, Zucchini C, Manara
MC, Chiodoni C, Colombo MP, Nicoletti G, Lollini PL, Li T,
Longobardi L, et al: Expression levels of insulin receptor
substrate-1 modulate the osteoblastic differentiation of
mesenchymal stem cells and osteosarcoma cells. Growth Factors.
32:41–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ardawi MS: The maximal activity of
phosphate-dependent glutaminase and glutamine metabolism in the
colon and the small intestine of streptozotocin-diabetic rats.
Diabetologia. 30:109–114. 1987.PubMed/NCBI
|
|
37
|
Yu Y, Newman H, Shen L, Sharma D, Hu G,
Mirando AJ, Zhang H, Knudsen E, Zhang GF, Hilton MJ and Karner CM:
Glutamine metabolism regulates proliferation and lineage allocation
in skeletal stem cells. Cell Metab. 29:966–978.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matsumoto T, Ii M, Nishimura H, Shoji T,
Mifune Y, Kawamoto A, Kuroda R, Fukui T, Kawakami Y, Kuroda T, et
al: Lnk-dependent axis of SCF-cKit signal for osteogenesis in bone
fracture healing. J Exp Med. 207:2207–2223. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang YM, Zhang ZQ, Liu YY, Zhou X, Shi
XH, Jiang Q, Fan DL and Cao C: Requirement of Gαi1/3-Gab1 signaling
complex for keratinocyte growth factor-induced PI3K-Akt-mTORC1
activation. J Invest Dermatol. 135:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu L, Shao L, Li B, Zong C, Li J, Zheng
Q, Tong X, Gao C and Wang J: Extracellular signal-regulated
kinase1/2 activated by fluid shear stress promotes osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
through novel signaling pathways. Int J Biochem Cell Biol.
43:1591–1601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Q, Lei L, Yu T, Jiang T and Kang Y:
Effect of brain-derived neurotrophic factor on the neurogenesis and
osteogenesis in bone engineering. Tissue Eng Part A. 24:1283–1292.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hirohiko O, Kaya Y, Hiroyuki M, Jumpei T,
Kazuhiko O, Tatsuji H and Akihito Y: Role of protein phosphatase 2A
in osteoblast differentiation and function. J Clin Med. 6:232017.
View Article : Google Scholar
|
|
43
|
Stechschulte LA, Ge C, Hinds TD, Sanchez
ER and Lecka-Czernik B: Protein phosphatase PP5 controls bone mass
and the negative effects of rosiglitazone on bone through
reciprocal regulation of PPARγ (peroxisome proliferator-activated
receptor γ) and RUNX2 (runt-related transcription factor 2). J Biol
Chem. 291:24475–24486. 2016. View Article : Google Scholar : PubMed/NCBI
|