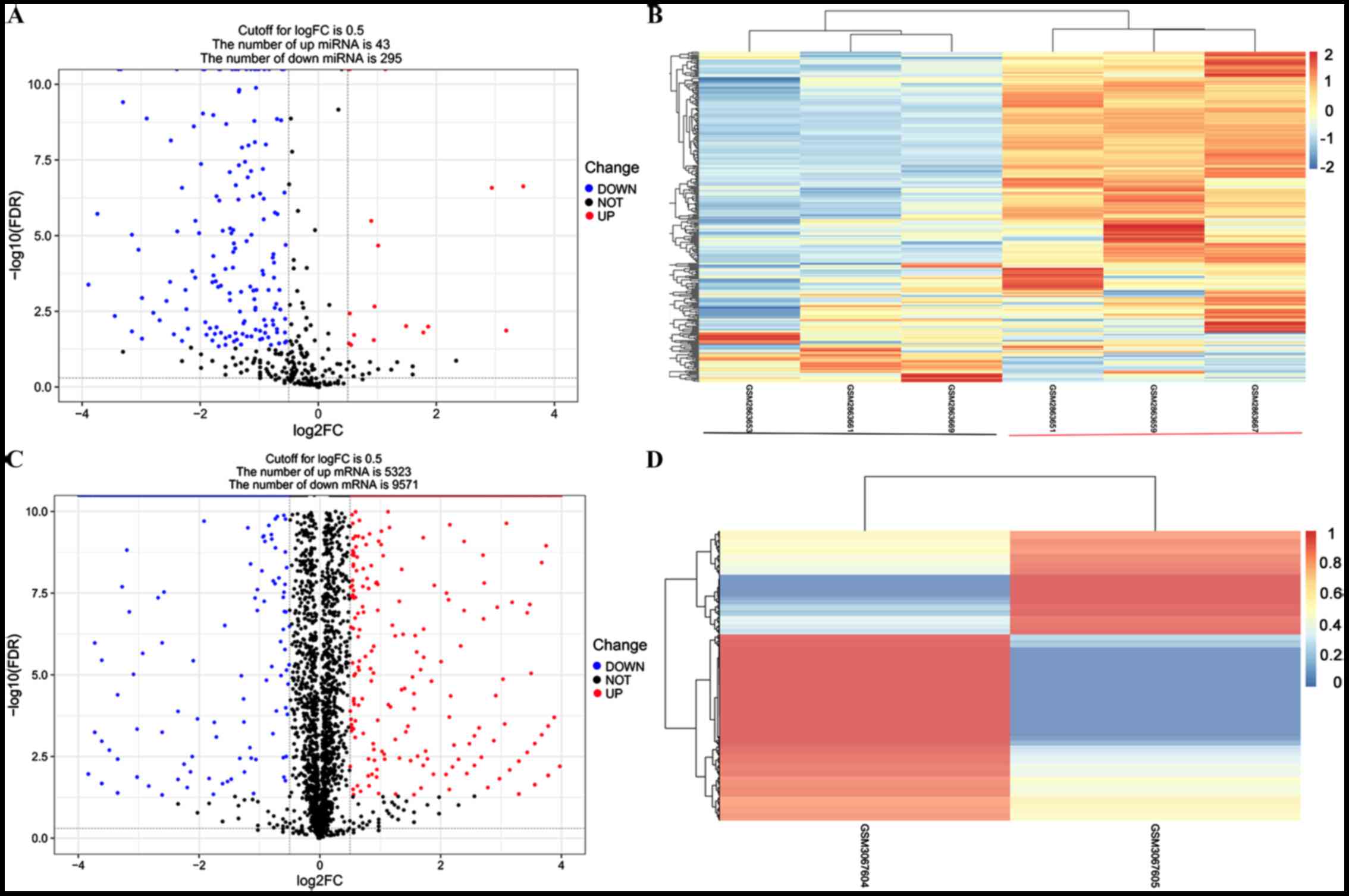
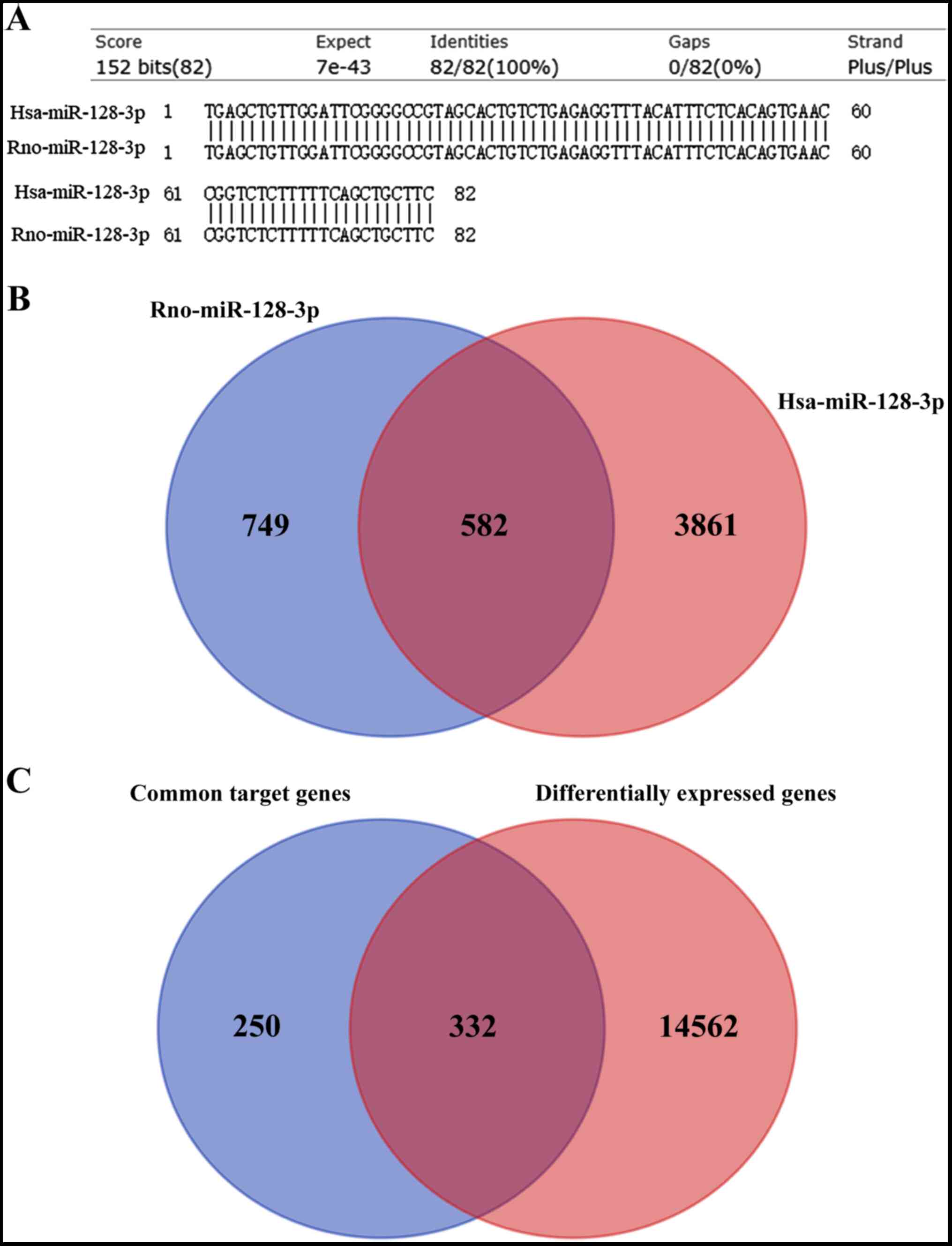
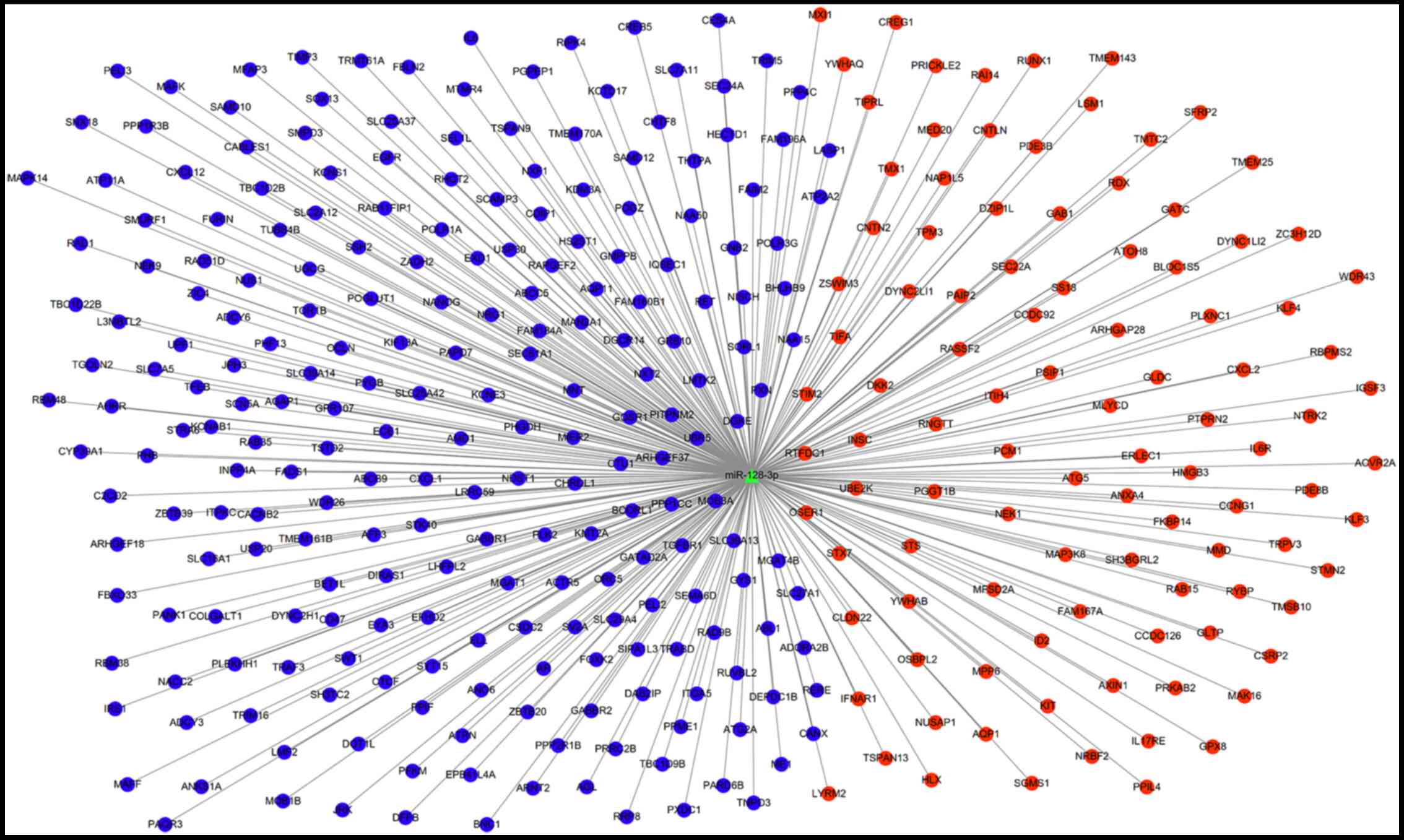
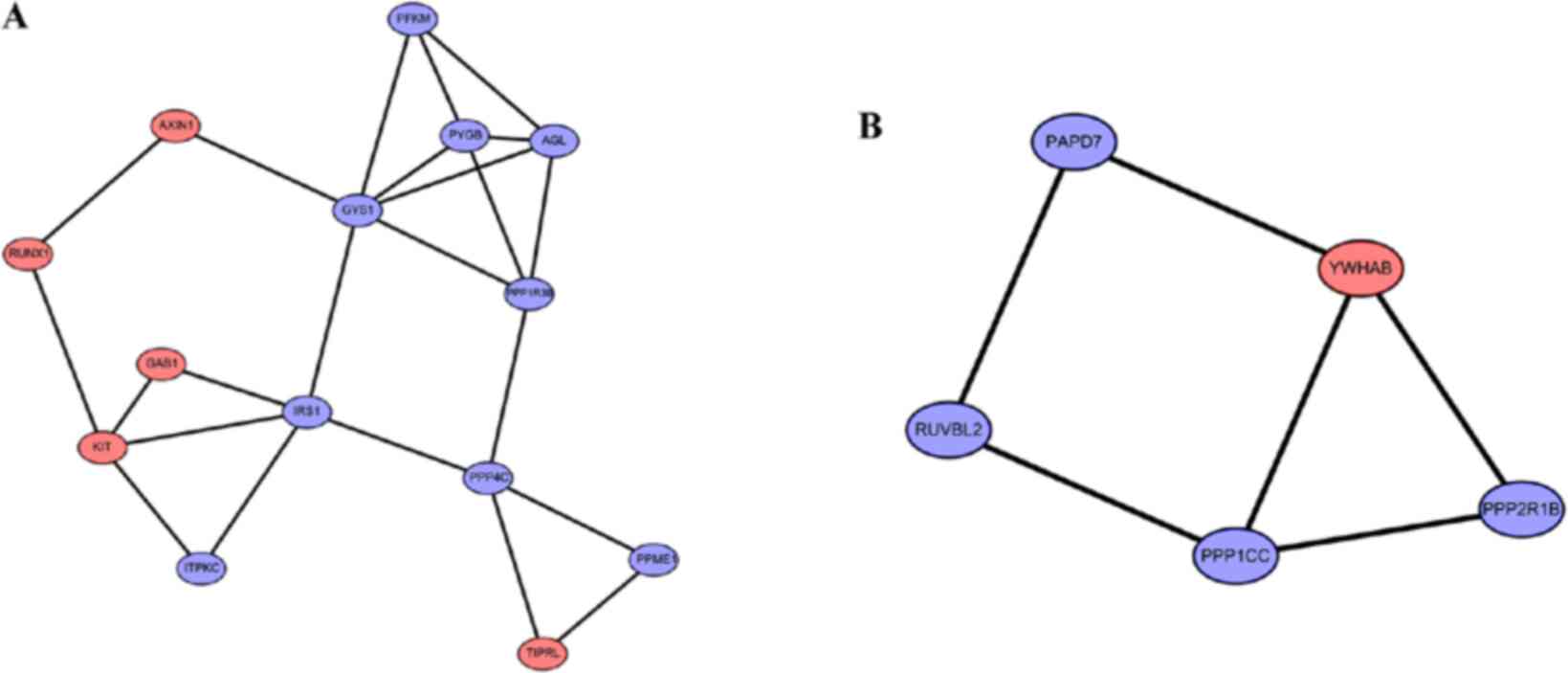
|
1
|
Confalonieri D, Schwab A, Walles H and
Ehlicke F: Advanced therapy medicinal products: A guide for bone
marrow-derived MSC application in bone and cartilage tissue
engineering. Tissue Eng Part B Rev. 24:155–169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gopal K, Amirhamed HA and Kamarul T:
Advances of human bone marrow-derived mesenchymal stem cells in the
treatment of cartilage defects: A systematic review. Exp Biol Med
(Maywood). 239:663–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guntur AR and Rosen CJ: The skeleton: A
multi-functional complex organ. New insights into osteoblasts and
their role in bone formation: The central role of PI3Kinase. J
Endocrinol. 211:123–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shin YS, Yoon JR, Kim HS and Lee SH:
Intra-articular injection of bone marrow-derived mesenchymal stem
cells leading to better clinical outcomes without difference in MRI
outcomes from baseline in patients with knee osteoarthritis. Knee
Surg Relat Res. 30:206–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Liu Y, Wu M, Wang H, Wu L, Xu B,
Zhou W, Fan X, Shao J and Yang T: MicroRNA-664a-5p promotes
osteogenic differentiation of human bone marrow-derived mesenchymal
stem cells by directly downregulating HMGA2. Biochem Biophys Res
Commun. 521:9–14. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ge JB, Lin JT, Hong HY, Sun YJ, Li Y and
Zhang CM: MiR-374b promotes osteogenic differentiation of MSCs by
degrading PTEN and promoting fracture healing. Eur Rev Med
Pharmacol Sci. 22:3303–3310. 2018.PubMed/NCBI
|
|
7
|
Huang J, Meng Y, Liu Y, Chen Y, Yang H,
Chen D, Shi J and Guo Y: MicroRNA-320a regulates the osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting HOXA10. Cell Physiol Biochem. 38:40–48. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu XM, Gu YL and Ren GQ: miR-125a-3p
targetedly regulates GIT1 expression to inhibit osteoblastic
proliferation and differentiation. Exp Ther Med 2016. 12:4099–4106.
2016. View Article : Google Scholar
|
|
9
|
Wang H, Xie Z, Hou T, Li Z, Huang K, Gong
J, Zhou W, Tang K, Xu J and Dong S: MiR-125b regulates the
osteogenic differentiation of human mesenchymal stem cells by
targeting BMPR1b. Cell Physiol Biochem. 41:530–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang S, Liu Y, Zheng Z, Zeng X, Liu D,
Wang C and Ting K: MicroRNA-223 suppresses osteoblast
differentiation by inhibiting DHRS3. Cell Physiol Biochem.
47:667–679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Yao C, Wei Z and Dong Q: miR-128
promoted adipogenic differentiation and inhibited osteogenic
differentiation of human mesenchymal stem cells by suppression of
VEGF pathway. J Recept Signal Transduct Res. 37:217–223. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang CC, Venø MT, Chen L, Ditzel N, Le D,
Dillschneider P, Kassem M and Kjems J: Global microrna profiling in
human bone marrow skeletal-stromal or mesenchymal-stem cells
identified candidates for bone regeneration. Mol Ther. 26:593–605.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Cai M, Zhong J, Yang L, Xiao J, Jin
F, Xue H, Liu X, Liu H, Zhang Y, et al: The long noncoding RNA
lnc-ob1 facilitates bone formation by upregulating Osterix in
osteoblasts. Nat Metab. 1:485–496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
R Core Team. R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2012, simplehttp://www.R-project.org/
|
|
16
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:697. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou W, Greenblatt MB, Brady N, Lotinun S,
Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al: The
microtubule-associated protein DCAMKL1 regulates osteoblast
function via repression of Runx2. J Exp Med. 210:1793–1806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matta C, Szűcs-Somogyi C, Kon E, Robinson
D, Neufeld T, Altschuler N, Berta A, Hangody L, Veréb Z and Zákány
R: Osteogenic differentiation of human bone marrow-derived
mesenchymal stem cells is enhanced by an aragonite scaffold.
Differentiation. 107:24–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Motohashi N, Alexander MS, Casar JC and
Kunkel LM: Identification of a novel microRNA that regulates the
proliferation and differentiation in muscle side population cells.
Stem Cells Dev. 21:3031–3043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo Y, Zhang Y, Miao G, Zhang Y, Liu Y and
Huang Y: Runx1 regulates osteogenic differentiation of BMSCs by
inhibiting adipogenesis through Wnt/β-catenin pathway. Arch Oral
Biol. 97:176–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji C, Xiaohua L, Li X, Tingting Y, Chaoqun
D and Jinyong L: RUNX1 plays an important role in mediating
BMP9-Induced osteogenic differentiation of mesenchymal stem cells
line C3H10T1/2, murine multi-lineage cells lines C2C12 and MEFs.
Int J Mol Sci. 18:13482017. View Article : Google Scholar
|
|
27
|
Saito T, Ohba S, Yano F, Seto I, Yonehara
Y, Takato T and Ogasawara T: Runx1 and Runx3 are downstream
effectors of nanog in promoting osteogenic differentiation of the
mouse mesenchymal cell line C3H10T1/2. Cell Reprogram. 17:227–234.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakamura T, Hamada F, Ishidate T, Anai K,
Kawahara K, Toyoshima K and Akiyama T: Axin, an inhibitor of the
Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and
APC and reduces the beta-catenin level. Genes Cells. 3:395–403.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Figeac N and Zammit PS: Coordinated action
of Axin1 and Axin2 suppresses β-catenin to regulate muscle stem
cell function. Cell Signal. 27:1652–1665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gwak J, Hwang SG, Park HS, Choi SR, Park
SH, Kim H, Ha NC, Bae SJ, Han JK, Kim DE, et al: Small
molecule-based disruption of the Axin/β-catenin protein complex
regulates mesenchymal stem cell differentiation. Cell Res.
22:237–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu ZQ, Brabletz T, Fearon E, Willis AL, Hu
CY, Li XY and Weiss SJ: Canonical Wnt suppressor, Axin2, promotes
colon carcinoma oncogenic activity. Proc Natl Acad Sci USA.
109:11312–11317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chimge NO, Little GH, Baniwal SK,
Adisetiyo H, Xie Y, Zhang T, O'Laughlin A, Liu ZY, Ulrich P, Martin
A, et al: RUNX1 prevents oestrogen-mediated AXIN1 suppression and
β-catenin activation in ER-positive breast cancer. Nat Commun.
7:107512016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Wang Y, Zhong T, Guo J, Li L,
Zhang H and Wang L: Transcriptional regulation of pig GYS1 gene by
glycogen synthase kinase 3β (GSK3β). Mol Cell Biochem. 424:203–208.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen RJ, Zhang G, Garfield SH, Shi YJ,
Chen KG, Robey PG and Leapman RD: Variations in glycogen synthesis
in human pluripotent stem cells with altered pluripotent states.
PLoS One. 10:e01425542015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Contaldo C, Myers TJ, Zucchini C, Manara
MC, Chiodoni C, Colombo MP, Nicoletti G, Lollini PL, Li T,
Longobardi L, et al: Expression levels of insulin receptor
substrate-1 modulate the osteoblastic differentiation of
mesenchymal stem cells and osteosarcoma cells. Growth Factors.
32:41–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ardawi MS: The maximal activity of
phosphate-dependent glutaminase and glutamine metabolism in the
colon and the small intestine of streptozotocin-diabetic rats.
Diabetologia. 30:109–114. 1987.PubMed/NCBI
|
|
37
|
Yu Y, Newman H, Shen L, Sharma D, Hu G,
Mirando AJ, Zhang H, Knudsen E, Zhang GF, Hilton MJ and Karner CM:
Glutamine metabolism regulates proliferation and lineage allocation
in skeletal stem cells. Cell Metab. 29:966–978.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matsumoto T, Ii M, Nishimura H, Shoji T,
Mifune Y, Kawamoto A, Kuroda R, Fukui T, Kawakami Y, Kuroda T, et
al: Lnk-dependent axis of SCF-cKit signal for osteogenesis in bone
fracture healing. J Exp Med. 207:2207–2223. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang YM, Zhang ZQ, Liu YY, Zhou X, Shi
XH, Jiang Q, Fan DL and Cao C: Requirement of Gαi1/3-Gab1 signaling
complex for keratinocyte growth factor-induced PI3K-Akt-mTORC1
activation. J Invest Dermatol. 135:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu L, Shao L, Li B, Zong C, Li J, Zheng
Q, Tong X, Gao C and Wang J: Extracellular signal-regulated
kinase1/2 activated by fluid shear stress promotes osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
through novel signaling pathways. Int J Biochem Cell Biol.
43:1591–1601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Q, Lei L, Yu T, Jiang T and Kang Y:
Effect of brain-derived neurotrophic factor on the neurogenesis and
osteogenesis in bone engineering. Tissue Eng Part A. 24:1283–1292.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hirohiko O, Kaya Y, Hiroyuki M, Jumpei T,
Kazuhiko O, Tatsuji H and Akihito Y: Role of protein phosphatase 2A
in osteoblast differentiation and function. J Clin Med. 6:232017.
View Article : Google Scholar
|
|
43
|
Stechschulte LA, Ge C, Hinds TD, Sanchez
ER and Lecka-Czernik B: Protein phosphatase PP5 controls bone mass
and the negative effects of rosiglitazone on bone through
reciprocal regulation of PPARγ (peroxisome proliferator-activated
receptor γ) and RUNX2 (runt-related transcription factor 2). J Biol
Chem. 291:24475–24486. 2016. View Article : Google Scholar : PubMed/NCBI
|