Introduction
Intervertebral disc (IVD) degeneration is a common
aging-related physiological change and is mostly a non-morbid
condition (1). However, lower back
pain caused by IVD degeneration brings long-term economic burdens
for societies and families (2).
The normal physiological functions of IVDs are dependent on the
central highly-hydrated nucleus pulposus (NP) containing
considerable amounts of proteoglycans and aggrecans, as well as the
outer annulus fibrosus (2). Once
IVD occurs with aging-related or pathological damage-induced
degeneration, a series of alterations to the IVD microenvironment
can be observed, such as imbalances in extracellular matrix (ECM)
metabolism, reduced hydration, increased inflammatory cytokine
expression levels, as well as accumulating cellular senescence
(3,4). These alterations are closely
associated, and interactions among them can aggravate the rate of
degeneration (4).
Cell senescence is an irreversible cell-cycle arrest
that occurs in response to various external stimuli (5). Previous studies have demonstrated
that the expression of senescence-associated β-galactosidase
(SA-β-gal), a marker of senescence in IVD cells, is positively
associated with IVD pathological grading (6,7). NP
cells isolated from degenerated IVDs exhibit slower proliferation
and increased cellular senescence compared with those from
non-degenerated IVD (8,9). In addition, these senescent IVD cells
with a low rate of proliferation are metabolically viable and
exhibit altered expression of various catabolic cytokines and
degradative enzymes, which further aggravates the imbalances in ECM
metabolism, as well as reduces the hydration status in IVD
(1). These observations emphasize
the significant role of cellular senescence in the development of
IVD degeneration. To date, pathways such as p53-p21-retinoblastoma
protein (RB) and p16INK4a-RB have been reported to contribute to
degeneration of the IVD (1,10);
however, the underlying molecular mechanisms remain unclear.
Long non-coding RNAs (lncRNAs), first identified in
1991 from cDNA, are RNA transcripts >200 nucleotides long that
lack evident open reading frames (11). LncRNAs are widely expressed in
tissues and cells, and their expression patterns exhibit some
specificity (12). Studies have
reported that lncRNAs regulate gene expression via their
interactions with other RNAs or proteins, in spite of lacking
protein-coding capacity; consequently, lncRNAs serve various roles
in cellular, physiological or pathological processes including
cellular senescence (13,14). Although a number of lncRNAs such as
lncPolE and FAM83H-AS1 have been verified as players in IVD
degeneration (15,16), the lncRNAs involved in IVD
degeneration via the regulation of cell senescence are largely
unknown.
Microarray technology is a highly-effective and
convenient method to identify differentially expressed (DE) genes
or lncRNAs, providing abundant potential choices for mechanisms and
candidate target studies (17).
Thus, the present study aimed to evaluate cellular senescence of NP
cells in patients with moderate IVD degeneration, identify and
analyze DE lncRNAs, and confirm the role of cellular
senescence-associated lncRNA in IVD degeneration, as well as to
identify novel mechanisms and potential therapeutic targets
involved in IVD degeneration.
Materials and methods
Sample collection
A total of 6 NP specimens were obtained from three
healthy controls whose IVD was mechanically damaged in car
accidents, and three patients with grade III IVD degeneration
between March 2017 and June 2018. In the control patients who had
undergone mechanical trauma, only those diagnosed with NP
herniation and the IVD degeneration confirmed to be grade I before
surgery were considered; the herniated NP tissue was resected
within 3 days after the accident. The degenerative grade was
evaluated by magnetic resonance imaging according to the Pfirrmann
grading system (18). Patients
with degenerative spinal stenosis, idiopathic scoliosis, tumors,
infections or previous lumbar disc surgery were excluded from the
study. Ages of the participants (males, 4; females, 2) ranged
between 28 and 33 years, and the mean age was 29.3 years. All NP
tissue samples were carefully collected under sterile conditions.
The present study was approved by the Ethics Review Board of
Liuzhou Traditional Chinese Medicine Hospital, and the study was
performed in accordance with the Declaration of Helsinki.
Cells and cell culture
NP cell isolation was performed as previously
described (19). Briefly, NP
tissues were washed with PBS three times and cut into small pieces
(1–2 mm3). F-12 complete medium (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% fetal bovine serum (FBS;
HyClone; Cytiva) and 1% penicillin/streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc.), was used to culture the tissue pieces at
37°C with 5% CO2. The sample fragments were removed when
NP cells migrated out of the tissues. The remaining NP cells were
cultured and amplified in F-12 complete medium. NP cells obtained
from specimens from healthy participants were used as a control
group, whereas those from patients served as the degeneration
group.
Reverse-transcription quantitative PCR
(RT-qPCR)
Total RNA was isolated from NP cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and 1 µg RNA and random primers were used for cDNA synthesis
according to the protocol of the PrimeScript RT reagent kit (TaKaRa
Biotechnology Co., Ltd.). SYBR® Premix Ex Taq™ (TaKaRa
Biotechnology Co, Ltd.) was used to determine the selected lncRNA
and mRNA expression levels using 7500 Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. The reaction was incubated at 95°C
for 5 min, followed by 42 cycles of 95°C for 10 sec and 60°C for 60
sec. The specific primers used were procured from Sangon Biotech
Co., Ltd., and their sequences are listed in Table I. GAPDH was used as a normalization
control for lncRNAs and mRNAs. Data were analyzed using the
2−ΔΔCq method (20).
 | Table I.Primer sequences for lncRNAs and
genes. |
Table I.
Primer sequences for lncRNAs and
genes.
| Target | Sequences
(5′→3′) |
|---|
|
lnc-ST8SIA5-1:2 | F:
GGAAACCTTTTGCCCTGGAG |
|
| R:
TGAGAGGAAAGCAAGGGAGG |
| lnc-HRK-2:1 | F:
AGGACACGGGAAGCTTTTCT |
|
| R:
CCAACAACGTCAGAACCCAG |
| CCL5 | F:
CCAGCAGTCGTCTTTGTCAC |
|
| R:
CTCTGGGTTGGCACACACTT |
| PNPT1 | F:
GCGAGCACTATGGAGTAGCG |
|
| R:
GCAGTGTCACCTGACTGTACTA |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
|
| R:
GGCTGTTGTCATACTTCTCATGG |
SA-β-gal staining assay
The number of senescent NP cells was determined
using a SA-β-gal staining kit (Beyotime Institute of Biotechnology)
according to the manufacturer's instructions. Briefly,
3×105 cells/well were seeded into 6-well plates. When
confluency reached ~80%, the cells were fixed with 1 ml fixative
solution for 15 min at room temperature, washed three times with
PBS and stained with 1 ml specific working solution overnight at
37°C. The next day, five random visual fields (magnification, ×400)
were captured by light microscopy and the the rate of
SA-β-gal-positive cells was calculated as the following equation:
SA-β-gal-positive cells rate=SA-β-gal-positive cell number/total
cell number ×100%.
Viability assay
A total of 4×103 NP cells/well were
seeded into 96-well plates. Following culture for 1, 3, 5 and 7
days, 15 µl MTT solution (Sigma-Aldrich; Merck KGaA) was added to
each well, and the cells were incubated for a further 4 h at 37°C.
Then, 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was
added to dissolve the formazan crystals. Absorbance of the samples
was detected at 492 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Migration assay
Transwell chamber inserts in 24-well plates (pore
size, 8 µm; Corning Life Sciences) were used for NP cell migratory
ability evaluation. Briefly, 4×104 cells in 100 µl
serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.) were
carefully added to the upper chamber, and 600 µl complete DMEM (10%
FBS; Gibco; Thermo Fisher Scientific, Inc.) was added to the lower
chamber in order to induce migration. Following incubation for 20 h
at 37°C, the cells in the upper chamber were gently removed, and
those that had migrated to the lower chamber were fixed with 100%
methanol (Sigma-Aldrich; Merck KGaA) for 10 min and stained with
0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for another 10 min
at room temperature. Finally, stained cells in three random visual
fields (magnification, ×200) were captured under a light microscope
and counted with ImageJ software (version 1.8.0; National
Institutes of Health).
Microarray analysis
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and sent to
Shanghai Biotechnology Corporation to complete the microarray
analysis. The RNeasy micro kit (Qiagen GmbH) and RNase-Free DNase
set (Qiagen GmbH) were used to purify the total RNA according to
the manufacturer's instructions. The purified RNA was labeled and
amplified by Low Input Quick Amp Labeling kit (Agilent
Technologies, Inc.) according to the manufacturer's instructions.
Subsequently, 1.65 µg Cy3-labeled cRNA of each sample was loaded
onto a custom microarray for hybridization, and microarray scanning
was performed on an Agilent Microarray Scanner (Agilent
Technologies, Inc.). The resulting data were extracted with Feature
Extraction software 10.7 (Agilent Technologies, Inc.), and the raw
data were processed using the R statistical software package
(version 3.4.1) (21). DE lncRNAs
were filtered following the criteria absolute fold-change [FC
(abs)] ≥2 and P<0.05.
Prediction of lncRNA targets
It has been suggested that lncRNAs regulate gene
expression in the cis- and trans-regulatory manner (22,23).
In the present study, genes transcribed within 10 kb upstream or
downstream of the DE lncRNAs were considered as cis-targets.
LncRNAs and their potential cis-targets were paired and visualized
using the UCSC genome browser (genome.ucsc.edu/) (24). For trans-regulation prediction, two
criteria were used for screening: i) Sequence complementarity of DE
lncRNAs and potential mRNA targets, and ii) the complementary
energy between the two sequences was ≤-30. BLAST software (National
Center for Biotechnology Information) was used for the first
screening, while RNAplex software (bioinf.uni-leipzig.de/Software/RNAplex/) was used to
decide on the trans-acting targets by the calculation of
complementary energy.
Bioinformatics analyses
Gene Ontology (GO) enrichment analysis (geneontology.org) (25) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis (genome.jp/kegg/) (26) were performed to assess the
potential roles of the lncRNA targets. Gene number ≥2 and P<0.05
were used as thresholds to screen relevant GO terms and KEGG
pathways.
Cell transfection
NP cells from the control group were seeded in
6-well plates (2×105 cells/well). When confluence
reached ~80%, the cells were transfected with 4 µg
pCDH-CMV-MCS-EF1-GFP-T2A-Puro plasmid (Hanbio Biotechnology Co.,
Ltd.) encoding the full length of lnc-HRK-2:1 or equivalent empty
vector using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Transfection was performed for 48 h at 37°C, and the
resulting cells were harvested for RT-qPCR validation. NP cells
transfected with the plasmid or empty vector were termed
lnc-HRK-2:1 or negative control (NC) groups, respectively.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
software (SPSS Inc.). Data are presented as the mean ± standard
deviation. Unpaired Student's t-test was used to analyze the
differences between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Phenotypic differences of control and
degenerated NP cells
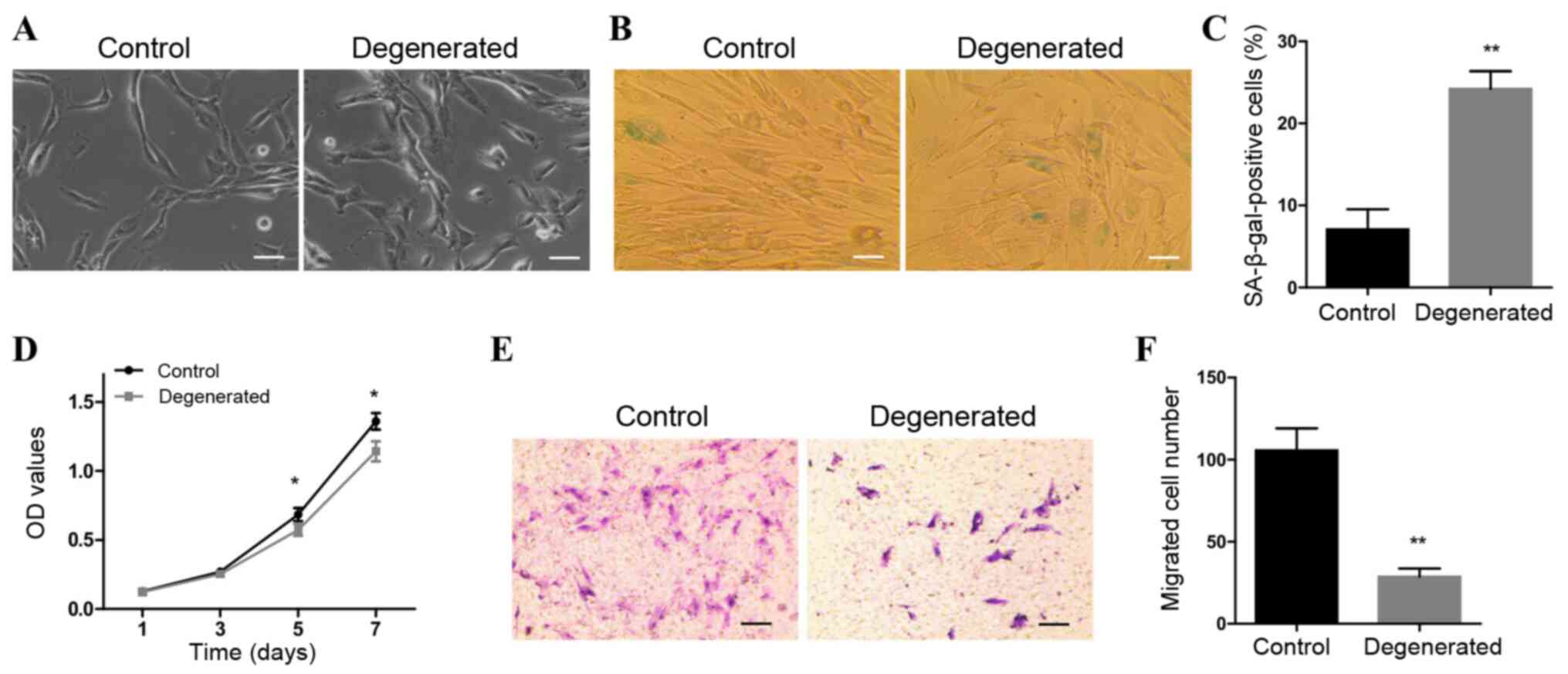
As presented in Fig.
1A, control NP cells were long in shape with small nuclei. The
majority of degenerative NP cells were morphologically similar to
the control cells, whereas a minority were short shuttle-like,
polygonal or rounded in shape. SA-β-gal staining results indicated
that the number of SA-β-gal-positive cells was significantly higher
in the degenerated group compared with that in the control group
(Fig. 1B and C). In addition, the
abilities of the cells to grow and migrate were significantly
decreased in degenerated NP cells compared with those of the
control cells (Fig. 1D-F). These
results indicated that degenerative NP cells exhibited increased
cellular senescence and reduced growth and migratory abilities.
Identification of DE lncRNAs in
degenerated NP cells
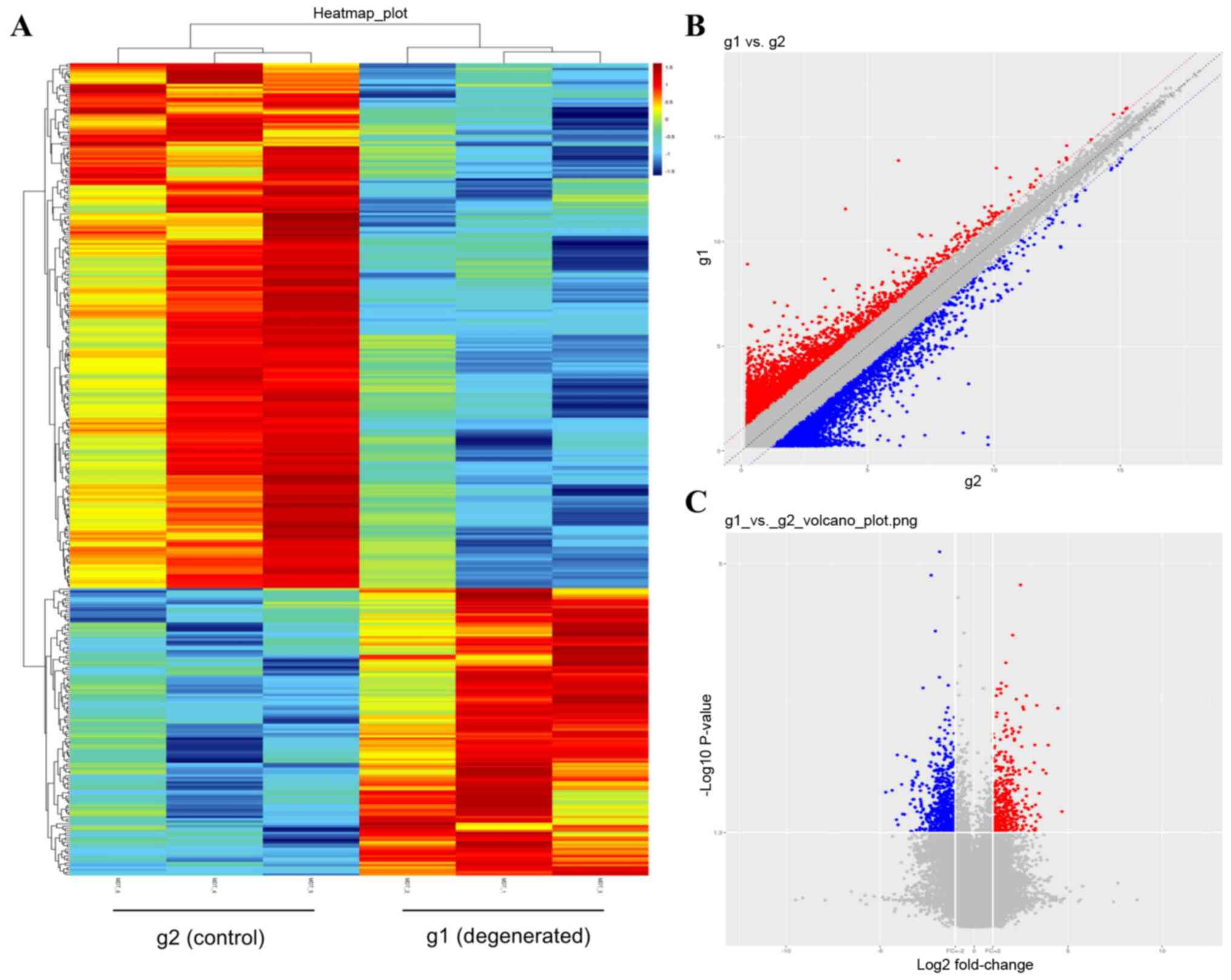
LncRNA microarray analysis screened and identified
353 DE lncRNAs that satisfied the FC≥2 and P<0.05 thresholds
between the control and degenerated NP cells (Table SI). Heatmaps (Fig. 2A) provided a clear outline of
lncRNA expression in each sample, and the general expression trends
of lncRNAs from three independent assays in each group were
relatively consistent. Among the 353 DE lncRNAs, 228 were
downregulated and 125 were upregulated, presented as blue and red
dots, respectively, in Fig. 2C.
The most upregulated lncRNA was NR_026812, and its FC was close to
the FC (abs) of ENST00000438810, which was the most downregulated
lncRNA. The top 10 upregulated and top 10 downregulated lncRNAs are
listed in Tables II and III.
 | Table II.Top 10 upregulated lncRNAs in
degenerated nucleus pulposus cells. |
Table II.
Top 10 upregulated lncRNAs in
degenerated nucleus pulposus cells.
| No. | LncRNA | P-value | FC | Database |
|---|
| 1 | NR_026812 | 0.0257 | 25.531 | RefSeq |
| 2 |
ENST00000438158 | 0.0010 | 21.957 |
ENSEMBL_GENCODE |
| 3 | NR_122111 | 0.0031 | 15.407 | RefSeq |
| 4 |
ENST00000454588 | 0.0069 | 12.723 |
ENSEMBL_GENCODE |
| 5 | lnc-HRK-2:1 | 0.0009 | 10.085 | lncipedia |
| 6 | lnc-MFAP5-3:2 | 0.0353 | 9.782 | lncipedia |
| 7 | NR_031707 | 0.0467 | 9.433 | RefSeq |
| 8 | NR_105061 | 0.0031 | 9.382 | RefSeq |
| 9 |
lnc-ST8SIA5-1:2 | 0.0073 | 7.804 | lncipedia |
| 10 | lnc-HFE2-2:1 | 0.0486 | 7.482 | lncipedia |
 | Table III.Top 10 downregulated lncRNAs in
degenerated nucleus pulposus cells. |
Table III.
Top 10 downregulated lncRNAs in
degenerated nucleus pulposus cells.
| No. | LncRNA | P-value | FC | FC (abs) | Database |
|---|
| 1 |
ENST00000438810 | 0.0139 | 0.039 | 25.883 |
ENSEMBL_GENCODE |
| 2 | NR_026576 | 0.0131 | 0.049 | 20.496 | RefSeq |
| 3 | lnc-GABRE-2:1 | 0.0333 | 0.057 | 17.458 | lncipedia |
| 4 |
lnc-TMEM229B-2:1 | 0.0043 | 0.059 | 16.935 | lncipedia |
| 5 |
lnc-RP11-383H13.1.1–4:1 | 0.0391 | 0.061 | 16.502 | lncipedia |
| 6 |
ENST00000518064 | 0.0109 | 0.069 | 14.465 |
ENSEMBL_GENCODE |
| 7 | lnc-TUBB2A-5:1 | 0.0279 | 0.072 | 13.947 | lncipedia |
| 8 | lnc-HOXD3-1:6 | 0.0282 | 0.077 | 12.997 | lncipedia |
| 9 | lnc-MT1G-2:3 | 0.0045 | 0.082 | 12.210 | lncipedia |
| 10 | NR_125920 | 0.0202 | 0.095 | 10.500 | RefSeq |
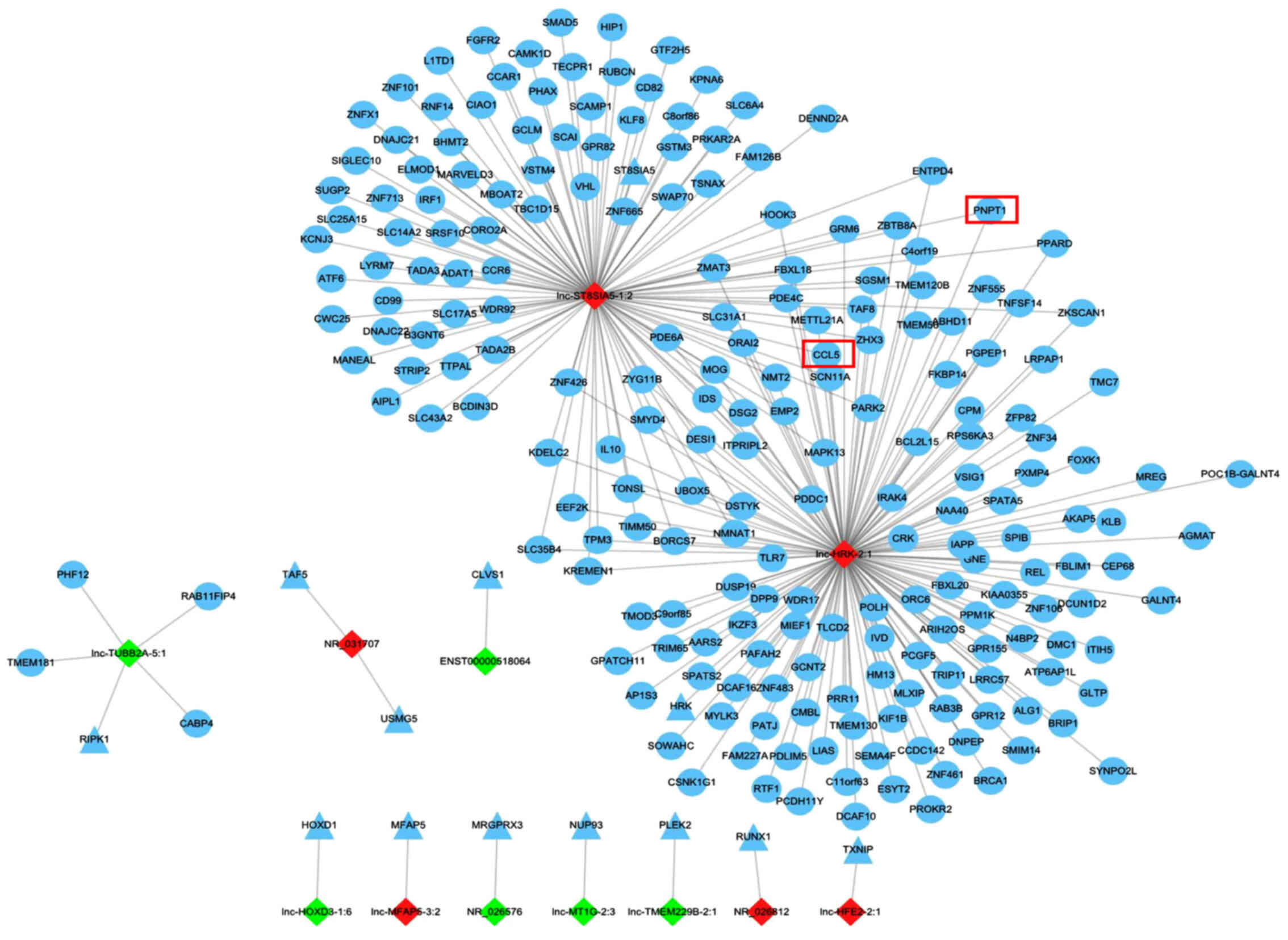
Prediction of DE lncRNA targets
For improved understanding of the unknown lncRNAs,
the present study predicted the potential cis- and trans-targets of
the DE lncRNAs, which may mediate the expression of nearby genes
located on the same chromosome and genes located on other
chromosomes, respectively. As a result, a total of 251 cis- and
2,170 trans-regulatory targets were identified (Tables SII and SIII), and targets of the top 20 DE
lncRNAs were included in a lncRNA target network (Fig. 3). Among the 20 lncRNAs, 12 had cis-
or trans-targets. Of note, targets of lnc-ST8SIA5-1:2 and
lnc-HRK-2:1, sharing almost one-third of the trans-regulatory
targets, were abundant compared with the limited targets of the
other 10 lncRNAs.
Bioinformatics analyses of predicted
lncRNA targets
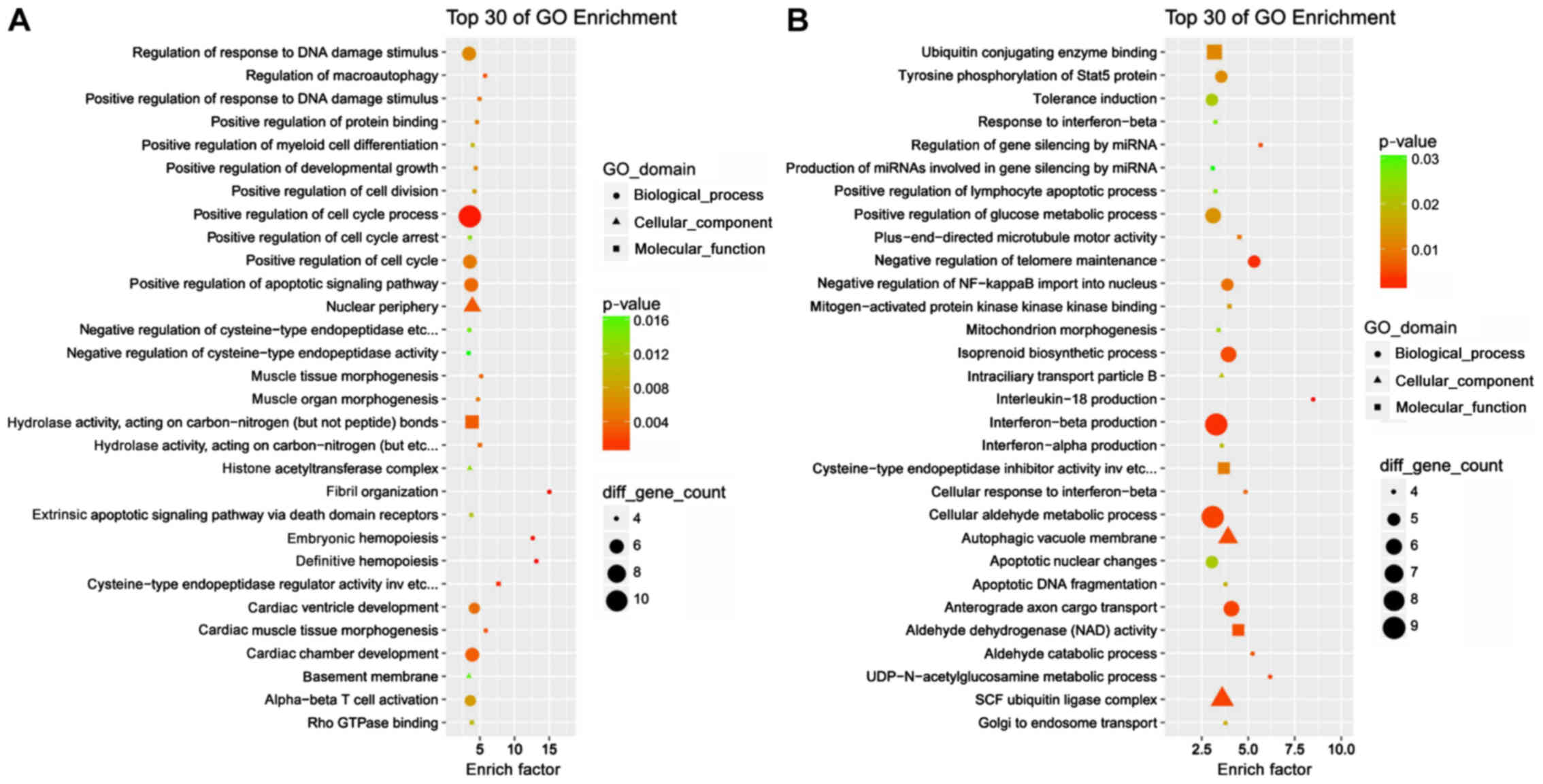
Bioinformatics analyses of the cis- and
trans-regulatory targets were subsequently performed. GO term
analysis demonstrated that the majority of the cis- and
trans-acting targets were enriched in ‘cellular processes’ and
‘binding’ terms (Figs. S1 and
S2). The top 30 GO enrichment
results provided more details. Cis-acting targets were involved in
the ‘regulation of response to DNA damage stimulus’, ‘positive
regulation of cell cycle processes’ and ‘positive regulation of
cell cycle arrest, whereas trans-acting targets were associated
with ‘interferon-β production’ and ‘cellular aldehyde metabolic
process’ (Fig. 4), which may
underlie the involvement of the DE lncRNAs in cellular senescence.
KEGG pathway analysis demonstrated that the cis-regulatory targets
were implicated in ‘apoptosis’ and ‘proteasome’ pathways, and the
trans-regulatory targets were enriched in ‘lysosome’ and
‘glycosylphosphatidylinositol (GPI)-anchor biosynthesis’ (Figs. S3 and S4).
RT-qPCR validation of
senescence-associated lncRNAs and targets
To screen cellular senescence-associated targets of
lncRNAs with high FC, key words such as ‘aging’ and ‘senescence’
were retrieved in GO and KEGG enrichment analyses, and 15 targets
(Table IV) were identified among
the trans-targets in GO enrichment. Next, the 15 targets were
searched in the network of the top 20 lncRNA targets (Fig. 3) to screen the coincident and
cis-acting targets C-C motif chemokine ligand 5 (CCL5) and
polyribonucleotide nucleotidyltransferase 1 (PNPT1), and its two
lncRNAs, lnc-ST8SIA5-1:2 and lnc-HRK-2:1 were screened out. Then,
the expression levels of the two lncRNAs and their two targets were
detected. RT-qPCR results demonstrated that the two lncRNAs were
significantly upregulated in degenerative NP cells compared with
the control cells, consistent with microarray analysis results
(Fig. 5A). In addition, the
expression levels of the two targets were enhanced in the
degeneration group compared with those in the control group
(Fig. 5A).
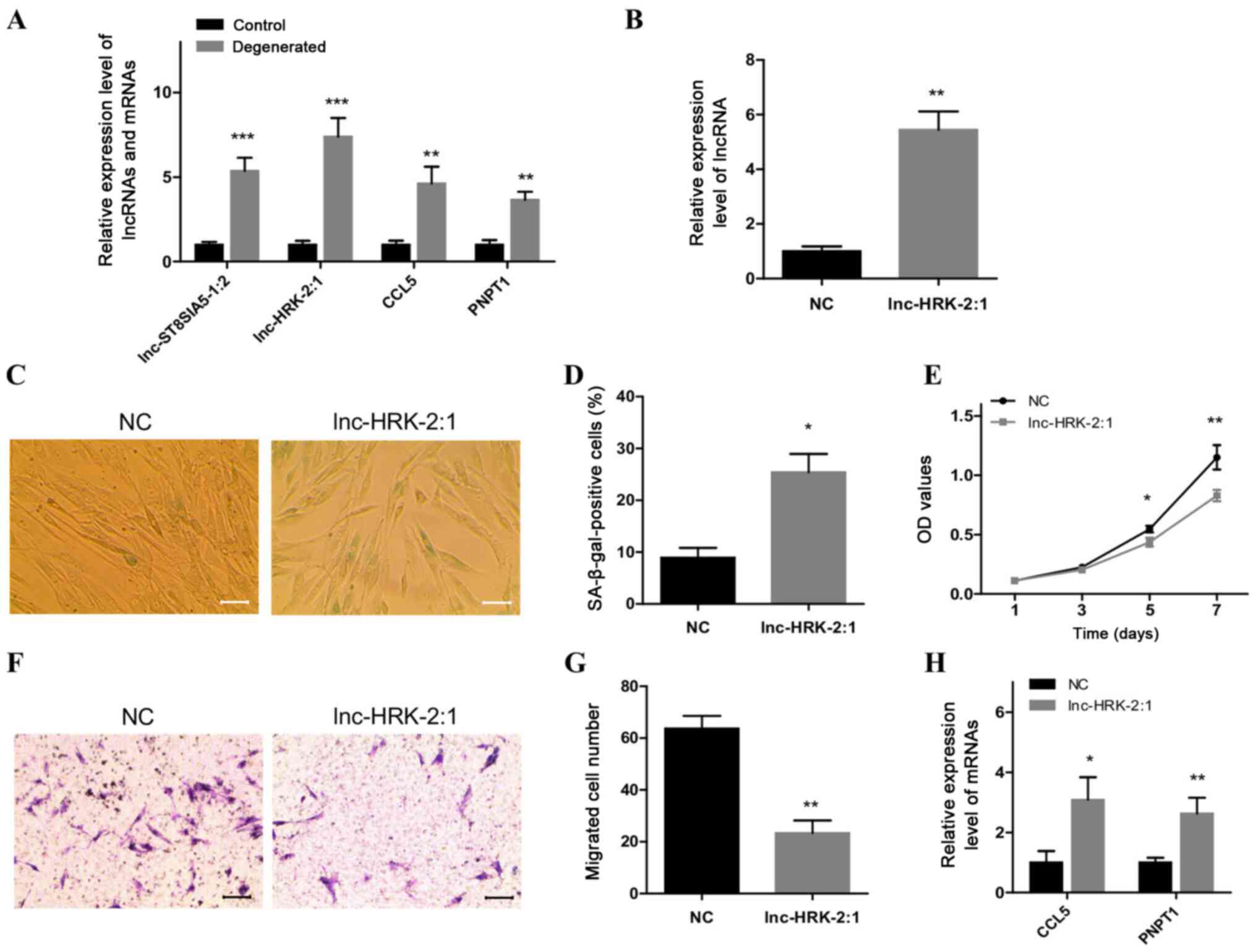 | Figure 5.Overexpression of lnc-HRK-2:1 induces
a senescent phenotype in control NP cells. (A) RT-qPCR analysis of
the expression levels of two candidate lncRNAs, lnc-ST8SIA5-1:2 and
lnc-HRK-2:1, and their targets CCL5 and PNPT1 in degenerated NP
cells. (B) RT-qPCR analysis of the level of lnc-HRK-2:1 after
overexpression of lnc-HRK-2:1 in control NP cells. (C and D)
SA-β-gal staining analysis of senescent cells in
lnc-HRK-2:1-overexpressing NP cells. Scale bar, 50 µm. (E) MTT
assays were used to evaluate the role of lnc-HRK-2:1 on cell
viability. (F and G) Transwell assays were performed to detect the
role of lnc-HRK-2:1 in cell migratory ability. Scale bar, 100 µm.
(H) RT-qPCR analysis of the expression levels of CCL5 and PNPT1 in
control NP cells overexpressing lnc-HRK-2:1. *P<0.05,
**P<0.01 and ***P<0.001 vs. control. NP, nucleus pulposus;
OD, optical density; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; lnc-, long non-coding RNA; CCL5,
C-C motif chemokine ligand 5; PNPT1, polyribonucleotide
nucleotidyltransferase 1; SA-β-gal, senescence-associated
β-galactosidase. |
 | Table IV.Aging- and senescence-associated
target genes. |
Table IV.
Aging- and senescence-associated
target genes.
| GO ID | Term | Type | Count | Genes |
|---|
| 0007569 | Cell aging | BP | 8 | ERCC1 LIMS1 SMC5
PRELP ZKSCAN3 TERF2 ATM PNPT1 |
| 0090398 | Cellular
senescence | BP | 4 | TERF2 SMC5 ZKSCAN3
PNPT1 |
| 0007568 | Aging | BP | 15 | CCL5 ERCC1 SMC5 MOG
PRELP ZKSCAN3 TERF2 P2RY2 PNPT1 NQO1 IL15 CASP2 LIMS1 STAT3
ATM |
Overexpression of lnc-HRK-2:1 induces
a senescent phenotype of control NP cells
Lnc-HRK-2:1 was selected for further study due to
its high expression levels in degenerative NP cells. Lnc-HRK-2:1
was overexpressed in control NP cells, which was validated by
RT-qPCR (Fig. 5B). SA-β-gal
staining results demonstrated that the numbers of senescent cells
were significantly increased in control NP cells following the
overexpression of lnc-HRK-2:1 (Fig. 5C
and D). In addition, suppressed growth and migratory abilities
were also observed in the lnc-HRK-2:1-overexpressing NC cells
compared with those in the NC group (Fig. 5E-G). These altered phenotypes
suggested a senescence-inducing effect of lnc-HRK-2:1 on phenotypic
transition in control NP cells. The expression levels of the two
target genes were also detected, and the results revealed
significant upregulation of CCL5 and PNPT1 expression levels in
control NP cells following overexpression of lnc-HRK-2:1 compared
with those in the NC group (Fig.
5H).
Discussion
Generally, aging is regarded as a dependent variable
to investigate the effects of senescence- or aging-mediated
mechanisms for IVD degeneration (1,9,27).
However, the onset of IVD degeneration is not solely dependent on
aging factors. Interactions between external stimuli and aging or
external stimuli-induced cellular senescence are similar to genuine
pathological circumstances (5).
The present study aimed to screen lncRNAs involved in cellular
senescence in IVD degenerative samples to provide a realistic
pathological development mechanism.
To exclude the factor of aging, young patients of a
similar age were selected as sources of NP tissues in the present
study. For the control group, NP tissues were collected from
patients who had suffered car accidents, who commonly experience
IVD protrusion (28,29). In a vehicle accident, the pressure
on the lumbar joints can be abruptly increased, resulting in joint
compression, fiber ring rupture and NP herniation, and may cause
spinal compression and further motor deficit if herniated NP
tissues are not resected in a timely manner (29). In the present study, only the IVD
degeneration of patients confirmed to be grade I was used as
control samples.
NP cells isolated from the two groups investigated
in the present study were morphologically similar, and only a
limited number of the degenerated NP cells were rounded. Generally,
IVD cells undergo morphological changes from initially being
spindle-shaped and turning rounded during aging and degeneration
(30). Similar morphological
alterations were also confirmed in NP cells in a rat aging model
(9). Thus, the limited number of
rounded NP cells from the degeneration group in the present study
suggested that slight cellular senescence was occurring in patient
NP tissues, which may be attributed to the lower ages and moderate
level of IVD degeneration of these patients. Cell phenotyping
results revealed that degenerated NP cells exhibited increased
cellular senescence and decreased growth and migratory abilities.
These phenotypic changes were consistent with previous studies
involving models of aging and clinical cases (9,31).
LncRNAs are a class of molecules that serve roles in
regulating gene expression and have been demonstrated to be
involved in diverse physiological and pathogenic effects in the
endocrine, reproductive, metabolic, immune, nervous and
cardiovascular systems (32). The
present study identified 353 DE lncRNAs between NP cells cultured
from moderate and low grade IVD degeneration. The total number of
DE lncRNAs identified in the present study was not as substantial
as the 1,806 DE lncRNAs from NP tissues identified previously
(33). The considerable difference
the in number of identified DE lncRNAs may have occurred due to
sample differences. First, the number of DE lncRNAs in NP tissues
sourced from cases with high grade IVD degeneration is undoubtedly
higher compared with that of tissues from patients with moderate
IVD degeneration. Second, lncRNAs in NP tissues not only include
lncRNAs from NP cells, but also those from certain inflammatory or
immune cells gathered at IVD degenerative sites, as well as
vesicles secreted from various cell types, which would hinder the
study of lncRNA mechanisms as it is difficult to distinguish
specific sources of lncRNAs. The selection of NP cells rather than
tissues as well as cases with moderate-rather than high-grade IVD
degeneration may help to eliminate interference by other factors
and enable the identification of lncRNAs involved in NP cellular
phenotype alterations in early IVD degeneration.
LncRNA functions can be broadly classified into
those that act on cis-targets, affecting the expression of
neighboring genes, and those remote from transcription sites that
operate on trans-targets (23,32).
In the present study, prediction tools identified 251 cis- and
2,170 trans-acting targets, and bioinformatics analyses provided an
improved understanding of these targets. For example, proteasome
subunit α 3 (PSMA3), proteasome 26S subunit-2C non-ATPase 6
(PSMD6), inhibitor of DNA binding 2–2C HLH protein (ID2) and G2 and
S-phase expressed 1 (GTSE1) were enriched in ‘positive regulation
of cell cycle arrest’, whereas NSE4 homolog A-2C SMC5-SMC6 complex
component (NSMCE4A), TP53 induced glycolysis regulatory phosphatase
(TIGAR), acidic residue methyltransferase 1 (ARMT1), scaffolding
protein involved in DNA repair (SPIDR), family with sequence
similarity 175 member A (FAM175A) and tumor protein-2C
translationally-controlled 1 (TPT1) were enriched in ‘regulation of
response to DNA damage stimulus’. Cell cycle arrest as well as DNA
damage are two important alterations identified as mechanisms
involved when IVDs undergo aging and degeneration (34,35).
The majority of the targets identified in the present study were
involved in either cell cycle arrest or DNA damage responses in
other cells; however, the role of these targets in IVD degeneration
is still unknown. Thus, these targets and their associated lncRNAs
may provide potential choices for studies involving new targets and
molecular mechanisms in IVD degeneration.
Next, cellular senescence-associated targets of
lncRNAs with high FC (CCL5 and PNPT1, and their lncRNAs
lnc-ST8SIA5-1:2 and lnc-HRK-2:1) were screened. RT-qPCR validation
indicated that the two lncRNAs and two targets were all
significantly upregulated in degenerative NP cells, which suggested
that certain relationships existed between the two lncRNAs and
increased cellular senescence in degenerated IVD NP cells. In
addition, overexpression of lnc-HRK-2:1 in the control NP cells
induced a senescent phenotype with an elevated percentage of
senescent cells, and reduced growth and migratory abilities, and
the expression levels of CCL5 and PNPT1 were also significantly
enhanced, which indicated that lnc-HRK-2:1-mediated the senescence
of NP cells is a key mechanism for the development of IVD
degeneration.
CCL5, also termed ‘regulated upon activation, normal
T cell expressed and secreted’, is a member of the pro-inflammatory
chemokine family and serves a crucial role in immune cell
chemotaxis (36). Although there
is no previous evidence of association between CCL5 and cellular
senescence in the process of IVD degeneration, elevated expression
levels of CCL5 were detected in patients with degenerated IVD,
discogram-positive painful IVD tissue, as well as normal NP cells
treated with interleukin-1β compared with their corresponding
controls (37–39). In addition, high levels of CCL5
secretion were also observed in aged theca-interstitial cells,
senescent melanoma cells and a human fibroblast cell line (AG04382)
from an aged donor (40,41). Human polynucleotide phosphorylase
(hPNPaseold-35), the protein encoded by the PNPT1 gene,
is an evolutionarily conserved 3′,5′-exoribonuclease involved in
regulating various physiological processes including the
maintenance of mitochondrial homeostasis, aging-related
inflammation and mitochondrial RNA import (42–44).
Previous studies have demonstrated that overexpression of PNPT1 in
cancer and normal cells results in distinctive growth inhibition
with a characteristic senescent-like phenotype (45,46).
These results suggested that enhanced CCL5 and PNPT1 expression
levels in NP cells from IVD degenerated samples as well as
lnc-HRK-2:1 overexpressing cells in the present study maybe
implicated in cellular senescence-associated IVD degeneration.
As demonstrated by the present microarray results,
lnc-HRK-2:1 is located on chromosome 12 with a transcript size of
2,020 bp; reports regarding lnc-HRK-2:1 are lacking. The present
study, for the first time, uncovered some of the roles of
lnc-HRK-2:1 in IVD degeneration. CCL5 and PNPT1 were the predicted
trans-targets of lnc-HRK-2:1, and overexpression of lnc-HRK-2:1
resulted in distinctive upregulation of the two targets, which may
be due to direct trans-actions or indirect regulators. The first
identified typical mechanism of trans-regulation was that involving
the lncRNA HOTAIR acting as a scaffold that coordinates the
recruitment of chromatin-modifying complexes to the HOXD locus,
thus regulating gene expression (23), which provides an ideal basis for
the future study of potential lnc-HRK-2:1 trans-acting
mechanisms.
Imbalances in ECM metabolism serve an important role
in IVD degeneration (47). To
identify lncRNAs potentially involved in ECM metabolism,
ECM-related GO and KEGG enrichment analyses were performed, and a
total of 12 targets were observed in the GO term ‘extracellular
matrix organization’. Among these, ADAM metallopeptidase with
thrombospondin type 1 motif 5 (ADAMTS5) and
microfibrillar-associated protein 5 (MFAP5) were of interest.
ADAMTS5 is one of the major proteolytic enzymes involved in ECM
degradation and serves a vital role in this process (48,49).
MFAP5 is an ECM glycoprotein that has been demonstrated to be
involved in ECM remodeling (50).
Thus, it was inferred that lncRNAs corresponding to ADAMTS5 and
MFAP5, NONHSAT081552 and lnc-MFAP5-3:2, may also serve certain
roles in ECM metabolism during IVD degeneration.
There were certain limitations in the present study.
First, the sample size (three samples per group) of NP tissues used
for microarray analysis was small; an increased sample size may be
beneficial for the identification of additional valuable lncRNAs.
Second, the expression levels of lncHRK-2:1 have been confirmed at
the cellular level, whereas the location and expression levels of
lncHRK-2:1 in clinical IVD tissue is unclear, and further studies
may clarify this.
In summary, the present study demonstrated that
increased lnc-HRK-2:1 expression levels promoted the senescence of
NP cells in the development of IVD degeneration, which may be
attributed to the upregulation of CCL5 and PNPT1. In addition,
hundreds of DE lncRNAs were identified, and thousands of potential
DE lncRNA targets were predicted in the present study. The
corresponding bioinformatic analyses conducted in the present study
may also provide diverse perspectives to elucidate the mechanisms
underlying IVD degeneration.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was financially supported by the Guangxi
Provincial Natural Science Foundation (grant no. JJBA40179).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FT and YW worked on the conception and design of the
study. DL and DH analyzed and interpreted the data. JL, LL and HC
performed the experiments and collected the data. All authors
contributed to preparation of the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Review
Board of Liuzhou Traditional Chinese Medicine Hospital, and written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang F, Cai F, Shi R, Wang XH and Wu XT:
Aging and age related stresses: A senescence mechanism of
intervertebral disc degeneration. Osteoarthritis Cartilage.
24:398–408. 2016.PubMed/NCBI
|
|
2
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 88 (Suppl 2):S21–S24. 2006.
|
|
3
|
Colombier P, Clouet J, Hamel O, Lescaudron
L and Guicheux J: The lumbar intervertebral disc: From embryonic
development to degeneration. Joint Bone Spine. 81:125–129.
2014.PubMed/NCBI
|
|
4
|
Sampara P, Banala RR, Vemuri SK, Av GR and
Gpv S: Understanding the molecular biology of intervertebral disc
degeneration and potential gene therapy strategies for
regeneration: A review. Gene Ther. 25:67–82. 2018.PubMed/NCBI
|
|
5
|
Feng C, Liu H, Yang M, Zhang Y, Huang B
and Zhou Y: Disc cell senescence in intervertebral disc
degeneration: Causes and molecular pathways. Cell Cycle.
15:1674–1684. 2016.PubMed/NCBI
|
|
6
|
Le Maitre CL, Freemont AJ and Hoyland JA:
Accelerated cellular senescence in degenerate intervertebral discs:
A possible role in the pathogenesis of intervertebral disc
degeneration. Arthritis Res Ther. 9:R452007.PubMed/NCBI
|
|
7
|
Gruber HE, Ingram JA, Norton HJ and Hanley
EN Jr: Senescence in cells of the aging and degenerating
intervertebral disc: Immunolocalization of senescence-associated
beta-galactosidase in human and sand rat discs. Spine (Phila Pa
1976). 32:321–327. 2007.PubMed/NCBI
|
|
8
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013.PubMed/NCBI
|
|
9
|
Cheng S, Li X, Lin L, Jia Z, Zhao Y, Wang
D, Ruan D and Zhang Y: Identification of aberrantly expressed genes
during aging in rat nucleus pulposus cells. Stem Cells Int.
2019:27852072019.PubMed/NCBI
|
|
10
|
Ben-Porath I and Weinberg RA: The signals
and pathways activating cellular senescence. Int J Biochem Cell
Biol. 37:961–976. 2005.PubMed/NCBI
|
|
11
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation Centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991.PubMed/NCBI
|
|
12
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012.PubMed/NCBI
|
|
13
|
Cossu AM, Mosca L, Zappavigna S, Misso G,
Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and
Boccellino M: Long Non-coding RNAs as important biomarkers in
laryngeal cancer and other head and neck tumours. Int J Mol Sci.
20:34442019.
|
|
14
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as Heracles in autophagy.
Autophagy. 13:1004–1024. 2017.PubMed/NCBI
|
|
15
|
Li X, Lou Z, Liu J, Li H, Lei Y, Zhao X
and Zhang F: Upregulation of the long noncoding RNA lncPolE
contributes to intervertebral disc degeneration by negatively
regulating DNA polymerase epsilon. Am J Transl Res. 11:2843–2854.
2019.PubMed/NCBI
|
|
16
|
Wei R, Chen Y, Zhao Z, Gu Q and Wu J:
LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via
targeting the Notch signaling pathway. J Cell Physiol.
234:22163–22171. 2019.PubMed/NCBI
|
|
17
|
Xiao Y, Hu J and Yin W: Systematic
Identification of Non-coding RNAs. Adv Exp Med Biol. 1094:9–18.
2018.PubMed/NCBI
|
|
18
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001.PubMed/NCBI
|
|
19
|
Tang X, Richardson WJ, Fitch RD, Brown CR,
Isaacs RE and Chen J: A new non-enzymatic method for isolating
human intervertebral disc cells preserves the phenotype of nucleus
pulposus cells. Cytotechnology. 66:979–986. 2014.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
21
|
Team RC: R: A language and environment for
statistical computing. R Foundation for Statistical Computing;
Vienna, Austria. Computing: 2013
|
|
22
|
Petazzi P, Sandoval J, Szczesna K, Jorge
OC, Roa L, Sayols S, Gomez A, Huertas D and Esteller M:
Dysregulation of the long non-coding RNA transcriptome in a Rett
syndrome mouse model. RNA Biol. 10:1197–1203. 2013.PubMed/NCBI
|
|
23
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI
|
|
24
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002.PubMed/NCBI
|
|
25
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI
|
|
26
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI
|
|
27
|
Zhang YG, Sun ZM, Liu JT, Wang SJ, Ren FL
and Guo X: Features of intervertebral disc degeneration in rat's
aging process. J Zhejiang Univ Sci B. 10:522–527. 2009.PubMed/NCBI
|
|
28
|
Khan AA, Mahmood S, Saif T and Gul A:
Spinal cord injury without radiographic abnormality (SCIWORA) in
adults: A report of two cases. J Pak Med Assoc. 67:1275–1277.
2017.PubMed/NCBI
|
|
29
|
Amelot A, Bouazza S, George B, Orabi M and
Bresson D: Anterior extrusion of fusion cage in posttraumatic
cervical disk disease. J Neurol Surg A Cent Eur Neurosurg.
76:168–171. 2015.PubMed/NCBI
|
|
30
|
Zhao CQ, Wang LM, Jiang LS and Dai LY: The
cell biology of intervertebral disc aging and degeneration. Ageing
Res Rev. 6:247–261. 2007.PubMed/NCBI
|
|
31
|
Jeong SW, Lee JS and Kim KW: In vitro
lifespan and senescence mechanisms of human nucleus pulposus
chondrocytes. Spine J. 14:499–504. 2014.PubMed/NCBI
|
|
32
|
Sun M and Kraus WL: From discovery to
function: The expanding roles of long noncoding RNAs in physiology
and disease. Endocr Rev. 36:25–64. 2015.PubMed/NCBI
|
|
33
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014.PubMed/NCBI
|
|
34
|
Zhan S, Wang K, Song Y, Li S, Yin H, Luo
R, Liao Z, Wu X, Zhang Y and Yang C: Long non-coding RNA HOTAIR
modulates intervertebral disc degenerative changes via
Wnt/β-catenin pathway. Arthritis Res Ther. 21:2012019.PubMed/NCBI
|
|
35
|
Vo NV, Hartman RA, Patil PR, Risbud MV,
Kletsas D, Iatridis JC, Hoyland JA, Le Maitre CL, Sowa GA and Kang
JD: Molecular mechanisms of biological aging in intervertebral
discs. J Orthop Res. 34:1289–1306. 2016.PubMed/NCBI
|
|
36
|
Pattappa G, Peroglio M, Sakai D, Mochida
J, Benneker LM, Alini M and Grad S: CCL5/RANTES is a key
chemoattractant released by degenerative intervertebral discs in
organ culture. Eur Cell Mater. 27:124–136. 2014.PubMed/NCBI
|
|
37
|
Kepler CK, Markova DZ, Dibra F, Yadla S,
Vaccaro AR, Risbud MV, Albert TJ and Anderson DG: Expression and
relationship of proinflammatory chemokine RANTES/CCL5 and cytokine
IL-1β in painful human intervertebral discs. Spine (Phila Pa 1976).
38:873–880. 2013.PubMed/NCBI
|
|
38
|
Grad S, Bow C, Karppinen J, Luk KD, Cheung
KM, Alini M and Samartzis D: Systemic blood plasma CCL5 and CXCL6:
Potential biomarkers for human lumbar disc degeneration. Eur Cell
Mater. 31:1–10. 2016.PubMed/NCBI
|
|
39
|
Liu W, Liu D, Zheng J, Shi P, Chou PH, Oh
C, Chen D, An HS and Chee A: Annulus fibrosus cells express and
utilize C-C chemokine receptor 5 (CCR5) for migration. Spine J.
17:720–726. 2017.PubMed/NCBI
|
|
40
|
Shen L, Chen Y, Cheng J, Yuan S, Zhou S,
Yan W, Liu J, Luo A and Wang S: CCL5 secreted by senescent
theca-interstitial cells inhibits preantral follicular development
via granulosa cellular apoptosis. J Cell Physiol. 234:22554–22564.
2019.PubMed/NCBI
|
|
41
|
Eyman D, Damodarasamy M, Plymate SR and
Reed MJ: CCL5 secreted by senescent aged fibroblasts induces
proliferation of prostate epithelial cells and expression of genes
that modulate angiogenesis. J Cell Physiol. 220:376–381.
2009.PubMed/NCBI
|
|
42
|
Sarkar D and Fisher PB: Polynucleotide
phosphorylase: An evolutionary conserved gene with an expanding
repertoire of functions. Pharmacol Ther. 112:243–263.
2006.PubMed/NCBI
|
|
43
|
Sarkar D, Lebedeva IV, Emdad L, Kang DC,
Baldwin AS Jr and Fisher PB: Human polynucleotide phosphorylase
(hPNPaseold-35): A potential link between aging and inflammation.
Cancer Res. 64:7473–7478. 2004.PubMed/NCBI
|
|
44
|
Sokhi UK, Das SK, Dasgupta S, Emdad L,
Shiang R, DeSalle R, Sarkar D and Fisher PB: Human polynucleotide
phosphorylase (hPNPaseold-35): Should I eat you or not-that is the
question? Adv Cancer Res. 119:161–190. 2013.PubMed/NCBI
|
|
45
|
Sarkar D, Leszczyniecka M, Kang DC,
Lebedeva IV, Valerie K, Dhar S, Pandita TK and Fisher PB:
Down-regulation of Myc as a potential target for growth arrest
induced by human polynucleotide phosphorylase (hPNPaseold-35) in
human melanoma cells. J Biol Chem. 278:24542–24551. 2003.PubMed/NCBI
|
|
46
|
Chan I, Lebedeva IV, Su ZZ, Sarkar D,
Valerie K and Fisher PB: Progression elevated gene-3 promoter
(PEG-Prom) confers cancer cell selectivity to human polynucleotide
phosphorylase (hPNPase(old-35))-mediated growth suppression. J Cell
Physiol. 215:401–409. 2008.PubMed/NCBI
|
|
47
|
Wang WJ, Yu XH, Wang C, Yang W, He WS,
Zhang SJ, Yan YG and Zhang J: MMPs and ADAMTSs in intervertebral
disc degeneration. Clin Chim Acta. 448:238–246. 2015.PubMed/NCBI
|
|
48
|
Banala RR, Vemuri SK, Dar GH, Palanisamy
V, Penkulinti M, Surekha MV, Gurava Reddy AV, Nalam MR and Subbaiah
G: Efficiency of dual siRNA-mediated gene therapy for
intervertebral disc degeneration (IVDD). Spine J. 19:896–904.
2019.PubMed/NCBI
|
|
49
|
Lu L, Hu J, Wu Q, An Y, Cui W, Wang J and
Ye Z: Berberine prevents human nucleus pulposus cells from
IL1-β-induced extracellular matrix degradation and apoptosis by
inhibiting the NF-κB pathway. Int J Mol Med. 43:1679–1686.
2019.PubMed/NCBI
|
|
50
|
Vaittinen M, Kolehmainen M, Ryden M,
Eskelinen M, Wabitsch M, Pihlajamaki J, Uusitupa M and Pulkkinen L:
MFAP5 is related to obesity-associated adipose tissue and
extracellular matrix remodeling and inflammation. Obesity (Silver
Spring). 23:1371–1378. 2015.PubMed/NCBI
|