Introduction
Wound healing is a complex physiological process. In
the clinic setting, enhancing wound healing has long been a goal of
physicians. Patients with certain conditions are particularly
susceptible to chronic, non-healing wounds, such as those of
diabetic wounds (1). In addition,
tending to chronic and intractable wounds increases the cost of
healthcare (2). Therefore,
elucidating the mechanism underlying wound healing so that this
process is accelerated is currently necessary and urgently
sorted.
Circulating fibrocytes (CFs), also termed peripheral
blood fibrocytes, are bone-marrow derived mesenchymal stem cells
that account for ~1% of the total number of mononuclear cells in
the blood (3). Chen et al
(4) previously reported that
fibrocytes accelerate wound healing by stimulating cell
proliferation, re-epithelialization and angiogenesis in a mouse
diabetic model. Notably, a previous study demonstrated that CFs can
differentiate into pericytes, which are indispensable components of
new blood vessels (5).
Additionally, fibrocytes have been documented to induce an
angiogenic phenotype in cultured endothelial cells (6) and contribute to the stabilization of
newly-formed vessels during angiogenesis (5). Recruitment of fibrocytes into wound
areas forms the basis of their functions (4). Thymic stromal lymphopoietin has been
reported to function in airway remodeling by promoting CF
recruitment to the lungs in mice subjected to chronic allergen
exposure (7). In addition,
contribution of chemokines chemokine ligand (CCL)5, CCL11 and CCL24
in the recruitment of fibrocytes to the airway in severe asthma has
also been previously demonstrated (8). Aside from asthma, fibrocyte numbers
have also been observed to be increased in the circulating blood in
patients with necrotizing enterocolitis, where they may be
recruited to the inflammatory intestinal tract through the CXC
chemokine receptor (CXCR) 4-CXC chemokine ligand (CXCL) 12 axis
(9). Platelet-derived growth
factor (PDGF)-BB-PDGF receptor-β biological axis has also been
reported to serve a role in the transmigration of fibrocytes into
fibrotic lungs (10). Although the
mechanism underlying the transmigration of fibrocytes has been
explored in numerous studies, an insufficient number studies
investigated the mechanism associated with the transmigration of
fibrocytes during the wound healing process. In particular,
chemokines that participate in the recruitment of fibrocytes during
wound healing remain poorly understood. Based on the data from gene
chip analysis (Pang, unpublished data), it was hypothesized that a
specific chemokine secreted by endothelial cells in newly-formed
blood vessels during wound healing may mediate the chemotaxis of
fibrocytes towards endothelial cells in the wound through a cognate
receptor expressed on fibrocytes.
The present study aimed to explore the role of
chemokines secreted by human umbilical vein endothelial cells
(HUVECs) in mediating the transmigration of CFs. Gene chip analysis
was first applied to measure the expression levels of chemokines in
HUVECs that were co-cultured with CFs in a permeable Transwell
system or cultured alone. Based on these data from this gene chip,
it was hypothesized that some chemokines may be involved in
mediating the transmigration of fibrocytes toward HUVECs. Further
transmigration assay experiments were performed to identify the
functional role of screened chemokines and its corresponding
receptor. The present study demonstrated that the chemokine ligand
15 (CCL15) and its receptor chemokine receptor 1 (CCR1) are
involved in mediating the recruitment of fibrocytes to HUVECs,
which may provide a new target for accelerating angiogenesis, which
can in turn accelerate wound healing.
Materials and methods
Cell isolation and culture
Human CFs were isolated and purified as previously
described (11). Briefly, human
CFs were isolated from leukapheresis packs (6 donors; male; age
range, 20–40 years old) provided by the Xijing Hospital Blood
Center (Xi'an, China) using the Histopaque®−1077 (cat.
no. 10771; Sigma-Aldrich; Merck KGaA) density gradient method. The
leukapheresis sample was mixed with PBS (1:1), before the diluted
sample was layered over Histopaque®−1077 (2:1) and
centrifuged at 800 × g for 20 min at room temperature. The
peripheral blood mononuclear cell (PBMC) layer was acquired by
gentle aspiration, which was washed with PBS and centrifuged at
1,200 × g for 5 min at room temperature three times. The PBMC layer
was resuspended in DMEM (HyClone; Cytiva) supplemented with 4.0 mM
L-glutamine, 4,500 mg/l glucose, 100 U/ml penicillin-G and 100 U/ml
streptomycin. PBMCs were then seeded into six-well plates at a
density of 1×107 cells/ml and 24-well plates at a
density of 5×105 cells/ml and cultured at 37°C with 5%
CO2. After 7 days of culture, non-adherent cells were
removed from the culture dishes and CFs were retained, and the
media was replaced. After 14 days of culture, the resultant
enriched human fibrocyte populations were >95% pure based on
collagen I and CD34 immunohistochemical staining, which was
performed as described previously (11).
HUVECs were purchased from ScienCell Research
Laboratories (cat. no. 8000). HUVECs were cultured in endothelial
cell medium supplemented with 5% FBS (cat. no. 1001; ScienCell
Research Laboratories), 4 mM L-glutamine, 100 U/ml penicillin-G,
100 U/ml streptomycin and 1% endothelial cell growth supplement
(v/v; cat. no. 1052; ScienCell Research Laboratories) at 37°C with
5% CO2. HUVECs used in the present study were from
passages 3–4. The experimental protocol was approved by the Ethics
Committee for Experimentation of the Fourth Military Medical
University (Shaanxi, China).
Cell co-culture system
Purified CFs and HUVECs were separately cultured in
six-well plates for 2 days at 37°C with 5% CO2 prior to
co-culture. In the co-culture system, the HUVECs were suspended in
endothelial cell medium (ScienCell Research Laboratories, Inc.) at
a density 1×105 cells/ml and added into the lower
chamber before the CFs were suspended in DMEM at a density
1×105 cells/ml and added into the upper Transwell
inserts (cat. no. 140640; Thermo Scientific; Thermo Fisher
Scientific, Inc.). HUVECs and CFs co-cultured for 24 h at 37°C were
then extracted for RNA microarray analysis or used for Transwell
migration experiments. HUVECs and CFs that were co-cultured for 48
h were detached for reverse transcription-quantitative (RT-q) PCR,
western blotting and ELISA. Separately cultured HUVECs and CFs in
six-well plates were used as the control groups.
Gene chip microassay
Gene chip microarray analysis was performed using
HumanWG-6_V3 (Illumina, Inc.) according to the manufacturer's
protocol. HUVECs were lysed using TRIzol® (Thermo Fisher
Scientific, Inc.) after 24 h co-culture. Total RNA was then
extracted using the TRIzol® reagent, followed by
purification and DNase I treatment using a Qiagen RNeasy mini kit
(Qiagen GmbH) according to the manufacturer's protocol. An Agilent
Bioanalyzer (Agilent Technologies GmbH) was used for quality
control. Biotinylated circular (c)RNA was prepared with the Ambion
MessageAmp kit (cat. no. AM1819; Thermo Scientific; Thermo Fisher
Scientific, Inc.) for Illumina arrays according to the
manufacturer's protocol. Labeled cRNA was hybridized to the probes
on the chip and washed. The results were scanned by a SureScan
Microarray scanner (cat. no. G4900DA; Agilent Technologies, Inc,)
according to the manufacturer's protocol. The data were normalized
using quantile normalization by GenomeStudio v2.0 (Illumina, Inc.).
All data have been deposited into the Gene Expression Omnibus
database and are available with the accession number GSE108626.
RT-qPCR
The total RNA of CFs and HUVECs was isolated using a
Takara MiniBEST Universal RNA Extraction Kit (cat. no. 9767; Takara
Bio, Inc.) according to the manufacturer's protocol. Reverse
transcription was performed with PrimeScript™ RT Master Mix (cat.
no. RR036A; Takara Bio, Inc.) according to the manufacturer's
protocol. qPCR was subsequently performed using TB Green™ Premix Ex
Taq II (cat. no. RR820A; Takara Bio, Inc.). The sequences of
primers used were as follows: CCL15 forward,
5′-CTCTCCTGCCTCATGCTTGT-3′ and reverse, 5′-CAGCAGCAAAGTGAAAGCTG-3′;
CCL2 forward, 5′-CCCCAGTCACCTGCTGTTAT-3′ and reverse,
5′-AGATCTCCTTGGCCACAATG-3′; CXCL8 forward,
5′-CTGCGCCAACACAGAAATTA-3′ and reverse,
5′-TGAATTCTCAGCCCTCTTCAA-3′; CCR1 forward,
5′-CTGGTTGGAAACATCCTGGT-3′ and reverse, 5′-GGAAGCGTGAACAGGAAGAG-3′;
CCR3 forward, 5′-TGGCGGTGTTTTTCATTTTC-3′ and reverse,
5′-CCGGCTCTGCTGTGGAT-3′; and GAPDH forward,
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′.
All primers were obtained from Sangon Biotech Co., Ltd. The PCR
amplifications were performed in a CFX Connect Real-Time System
(Bio-Rad Laboratories, Inc.) under standard cycling conditions in
accordance with the manufacturer's protocol. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for min; followed by 40 cycles of denaturation
at 95°C for 15 sec, annealing and elongation at 60°C for 1 min;
then a final extension at 68°C for 3 min. Expression levels were
quantified using the 2−ΔΔCq method (12) and normalized to GAPDH. Data were
analyzed with CFX Manager software version 2.1 (Bio-Rad
Laboratories, Inc.).
ELISAs
The concentrations of chemokines in single culture
or co-culture supernatants were detected by ELISAs. CFs and HUVECs
were co-cultured as aforementioned. The medium volume in each
chamber was 2 ml. The media were not changed during this
experiment. After a 48-h culture, the culture medium of the HUVECs
was collected. The medium samples were centrifuged at 600 × g for
10 min at room temperature and the supernatant was stored at −80°C
for subsequent experiments. The supernatants were diluted 1:2 in
serum-free endothelial cell medium before ELISAs were performed
with human CCL2/monocyte chemoattractant protein-1 (cat. no. DCP00;
R&D Systems, Inc.), interleukin-8/CXCL8 (cat. no. D8000C;
R&D Systems, Inc.) and macrophage inflammatory protein 5/CCL15
(cat. no. ab100598; Abcam) ELISA kits, according to the
manufacturers' protocols. Data were acquired by a Tecan Sunrise™
microplate absorbance reader (Tecan Group, Ltd.) and normalized
based on cell numbers. The experiment was repeated three times.
Chemokine blocking experiment
The polyclonal antibodies used to block the
corresponding chemokines were rabbit polyclonal anti-CCL2 (1:10;
cat. no. 25542-1-AP; ProteinTech Group, Inc.), rabbit polyclonal
anti-CCL15 (1:10; cat. no. ab221040; Abcam) and rabbit polyclonal
anti-CXCL8 (1:10; cat. no. ab7747; Abcam). The addition of the
antibodies to HUVECs cultured in the co-culture system was
performed at 37°C for 24 h with 5% CO2 prior to further
experiments. Control groups were added with the same amount of TE
buffer (pH 9.0), which is the solvent of these antibodies only.
Western blotting analysis
Western blotting analysis was performed following
standard procedures. Briefly, T-PER™ Tissue Protein Extraction
reagent (Thermo Fisher Scientific, Inc.) was used to extract total
protein from cells, and the protein content was determined using
Bradford assay. Protein samples (30–40 µg/lane) were separated with
8% SDS-PAGE and transferred onto PVDF membranes. After blocking
with 5% non-fat milk in TBS-0.05% Tween-20 for 1 h at room
temperature, the PVDF membranes were incubated with anti-CCR1 mouse
monoclonal (1:1,000; cat. no. ab129002; Abcam) and anti-actin mouse
monoclonal (1:1,000; cat. no. ab11003; Abcam) primary antibodies
for 3h at 37°C and then incubated with a horseradish
peroxidase-conjugated rabbit polyclonal anti-mouse IgG-H&L
secondary antibody (1:2,000; cat. no. ab6728; Abcam) for 1 h at
room temperature. Protein bands were visualized with ECL solution
and a ChemiDoc XRS+ system (Bio-Rad Laboratories, Inc.). The
densities of the specific bands were further analyzed with ImageJ
software v1.53a (National Institutes of Health).
CCL15 and CCR1 overexpression
293T cells (American Type Culture Collection) were
kept in DMEM supplemented with 10% FBS, 1% glutamine and 1% P/S
antibiotics (Thermo Fisher Scientific, Inc.) at 37°C and 5%
CO2. Human CCL15 and CCR1 genes were cloned into
pLenti-CMV-EGFP-3FLAG vectors (OBiO Technology Corp., Ltd.). Using
the calcium phosphate transfection protocol (13), 293T cells were transfected with
lentiviral vectors (4.81×108 TU/ml CCL15;
8.50×108 TU/ml CCR1; 8.41×108 TU/ml empty
vector) together with packaging vectors pMD2-VSVG and pPAX2 (OBiO
Technology Corp., Ltd.). Lentiviral particles prepared by
transfecting the 293T cells empty lentiviral particle vector
together with packaging vectors were used as negative controls.
Lentiviruses were harvested 48 h post transfection and concentrated
by ultracentrifugation at 3,000 × g for 2 h at 4°C. HUVECs and CFs
were then infected with the lentiviral particles at a multiplicity
of infection (MOI) of 40 in the presence of 8 mg/ml polybrene
(Sigma-Aldrich; Merck KGaA). The cells were harvested 3–4 days
after transfection for subsequent experimentation.
CCL15 and CCR1 knockdown
CCL15 small interfering (si)RNA CCL15-homo-667
(5′-CCAGUAGUUCUGAACAGCUTT-3′) was synthesized by Sangon Biotech Co.
Ltd. HUVECs were transfected with either si-CCL15 or nonsense siRNA
(cat. no. 4404021; Silencer™ Negative Control No. 1 siRNA; Thermo
Fisher Scientific, Inc.) using Lipofectamine® 2000 (cat.
no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.) at a
concentration of 50 nM according to the manufacturer's protocol.
Small harpin (sh)RNA of human CCR1 (5′-CCTACAATTTGACTATACTT-3′) or
control shRNA (targeting sequence 5′-TTCTCGAACGTGTCACGT-3′) was
cloned into the pLKD-CMV-Puro-U6-shRNA vector (OBiO Technology
Corp., Ltd.), following which recombinant lentiviruses were
generated as aforementioned. CFs were infected with the lentiviral
particles at MOI=40 in the presence of 8 mg/ml polybrene
(Sigma-Aldrich; Merck KGaA). The cells were harvested 3–4 days
after transfection for subsequent experimentation.
Transwell migration assay
HUVECs were suspended in endothelial cell medium at
a density of 1×105 cells/ml and plated into the lower
chamber. The CFs were suspended in DMEM at a density of
1×105 cells/ml and plated into the upper chamber of the
Transwell inserts (cat. no. 140640; Thermo Scientific; Thermo
Fisher Scientific, Inc.). After 24 h of co-culture at 37°C, the CFs
were fixed with 4% formaldehyde for 30 min at room temperature
(cat. no. DF0133; Beijing Leagene Biotech Co., Ltd.) and stained
with 0.1% crystal violet (cat. no. C0121; Beyotime Institute of
Biotechnology) for 5 min at room temperature. The number of CFs
that migrated through the membrane towards the HUVECs and attached
on the underside of the membrane was then counted under a light
microscope (magnification, ×20) in three randomly selected fields
of view per chamber to obtain an average count.
Statistical analysis
All experiments were repeated three times. The data
were expressed as the mean ± SEM. One-way ANOVA was used to compare
differences between > two groups of data. Dunnett's post hoc
test was performed for the multiple comparison of two groups vs.
the control group. Tukey's post hoc test was performed for multiple
comparisons of > two groups. An unpaired two-tailed Student's
t-test was performed to compare the difference between two groups.
Statistical analysis was performed using SPSS v19.0 software (IBM
Corp.) and figures were plotted using GraphPad Prism 8.0 software
(GraphPad Software, Inc.).
Results
CCL15 is a vital chemokine that
mediates the transmigration of CFs toward HUVECs
Previous studies have demonstrated that fibrocytes
migrate towards vascular endothelial cells in vitro
(5,6,12,14).
To study the mechanism underlying the tropism of CFs for HUVECs, a
Transwell co-culture system was used to mimic the in vivo
communication between endothelial cells and fibrocytes. HUVECs were
subjected to gene chip microarray analysis after co-culture with
CFs or after culturing alone. The results demonstrated that the
expression levels of the chemokines CCL2, CCL15 and CXCL8 were
significantly increased in the HUVECs co-cultured with CFs compared
with those in HUVECs cultured alone (Fig. 1A and B). Additionally, ELISA
results demonstrated that the concentrations of CCL2, CCL15 and
CXCL8 were significantly higher in the HUVECs co-cultured with CFs
compared with those in HUVECS cultured alone (Fig. 1C).
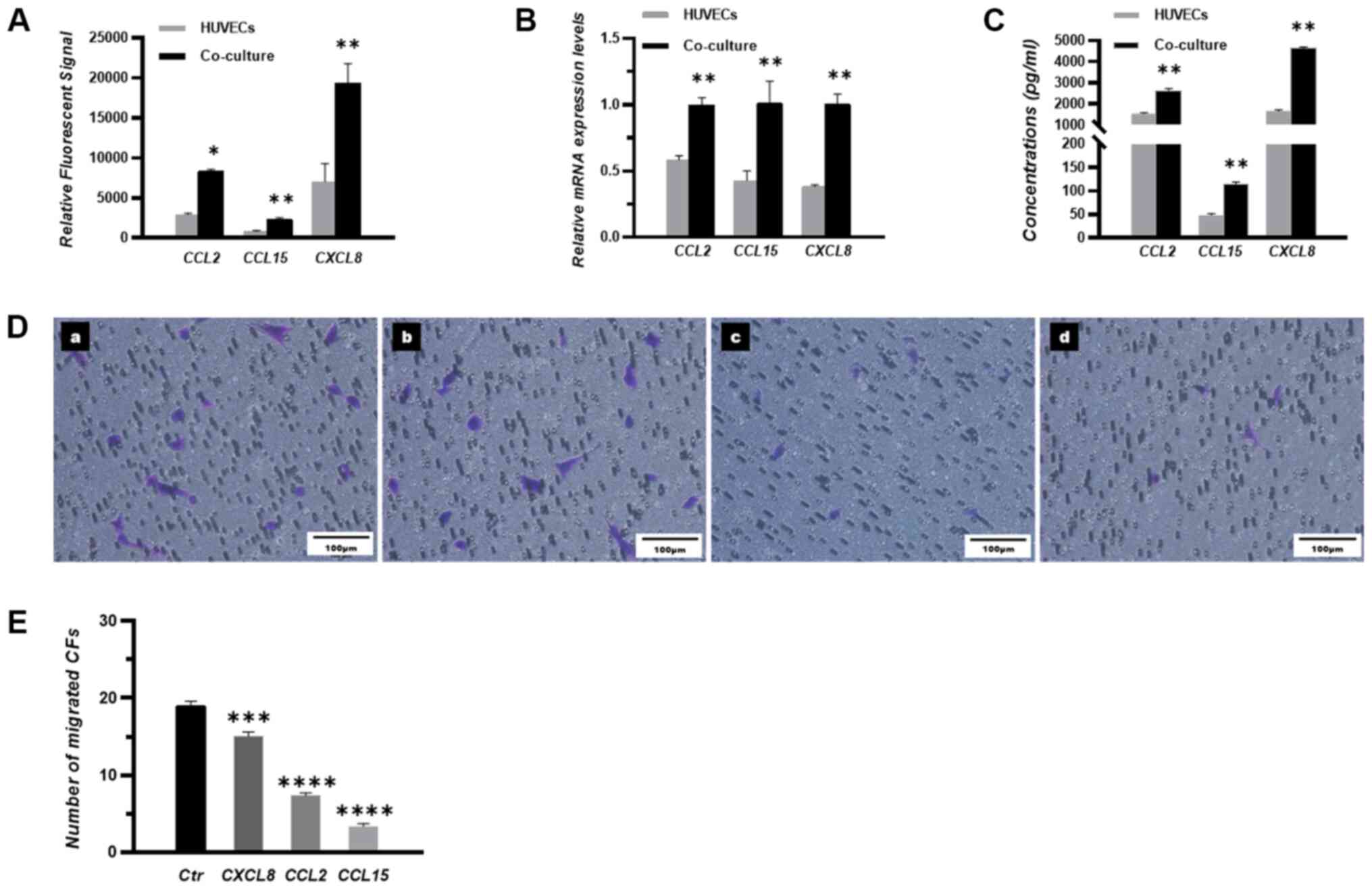 | Figure 1.CCL15 is a vital chemokine that
mediates the transmigration of CFs toward HUVECs. (A) Expression
levels of the three chemokines in mono-cultured and co-cultured
HUVECs were measured by gene chip microarray analysis and expressed
as relative fluorescent signals. (B) Relative mRNA expression
levels of CCL2, CCL15 and CXCL8 in the mono-cultured HUVECs and
co-cultured HUVECs were measured by reverse
transcription-quantitative PCR. (C) Concentrations of CCL2, CCL15
and CXCL8 in the medium of mono-cultured HUVECs and co-cultured
HUVECs were measured by ELISA. (D) Migratory CFs were stained and
imaged under a microscope. (D-a) Control group, (D-b) CXCL8
blocking group, (D-c) CCL2 blocking group and (D-d) CCL15 blocking
group. n=3. Scale bar, 100 µm. (E) The number of migrated CFs in
each group. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001 vs. HUVECs (parts A-C) or Ctr (part E). HUVEC,
human umbilical vein endothelial cell; CFs, circulating fibrocytes;
Ctr, control; CCL, chemokine ligand; CXCL, CXC chemokine
ligand. |
The transmigration of CFs toward HUVECs was next
assessed using Transwell assay. Polyclonal antibodies were used to
evaluate the effects of CCL2, CCL15 and CXCL8 on modulating the
transmigration of CFs. Notably, a fewer number of migrated CFs was
observed following the addition of the CCL15 antibody compared with
that following the addition of CCL2 or CXCL8 antibodies (Fig. 1D and E).
CCL15 mediates the transmigration of
CFs toward HUVECs in vitro
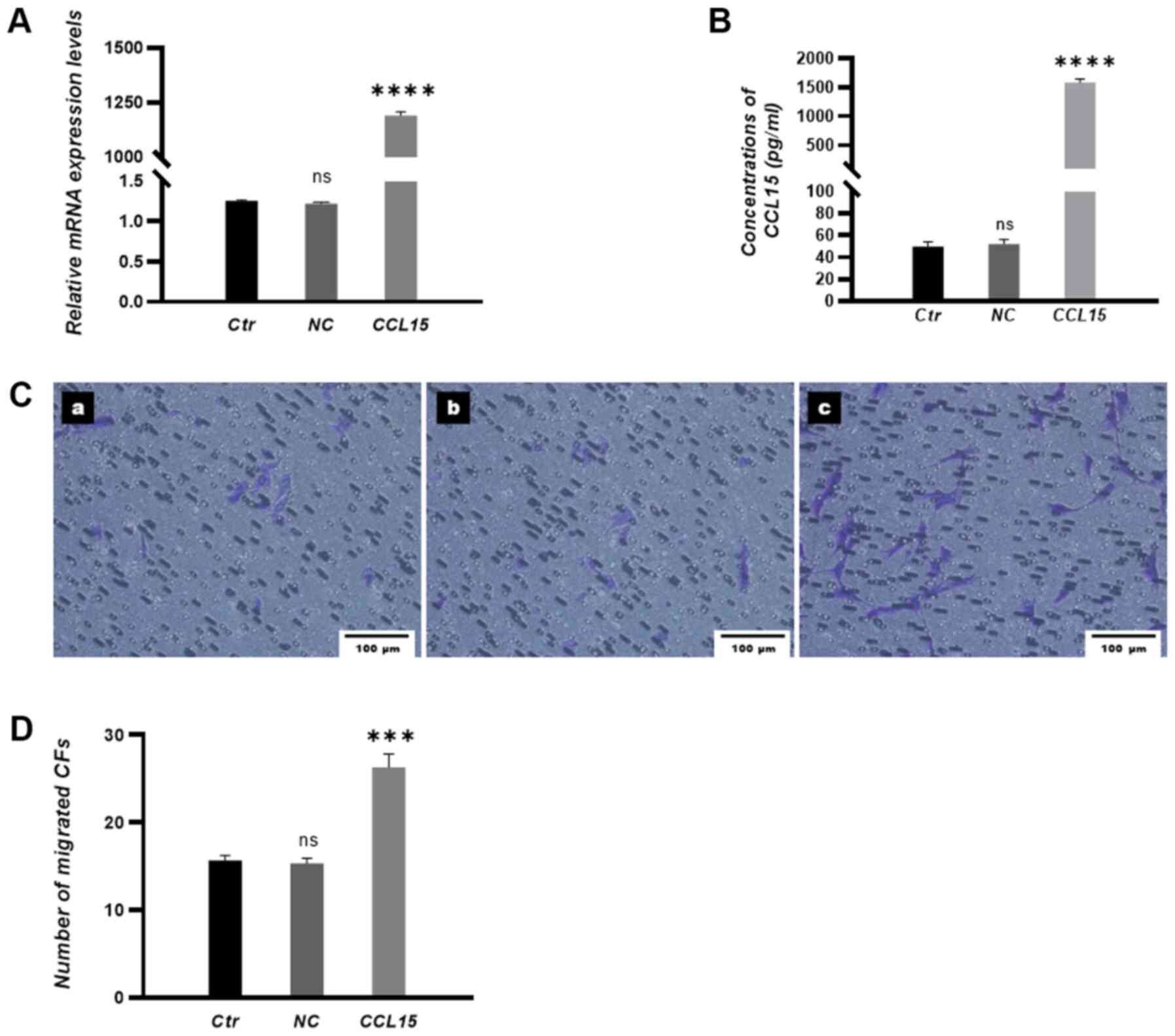
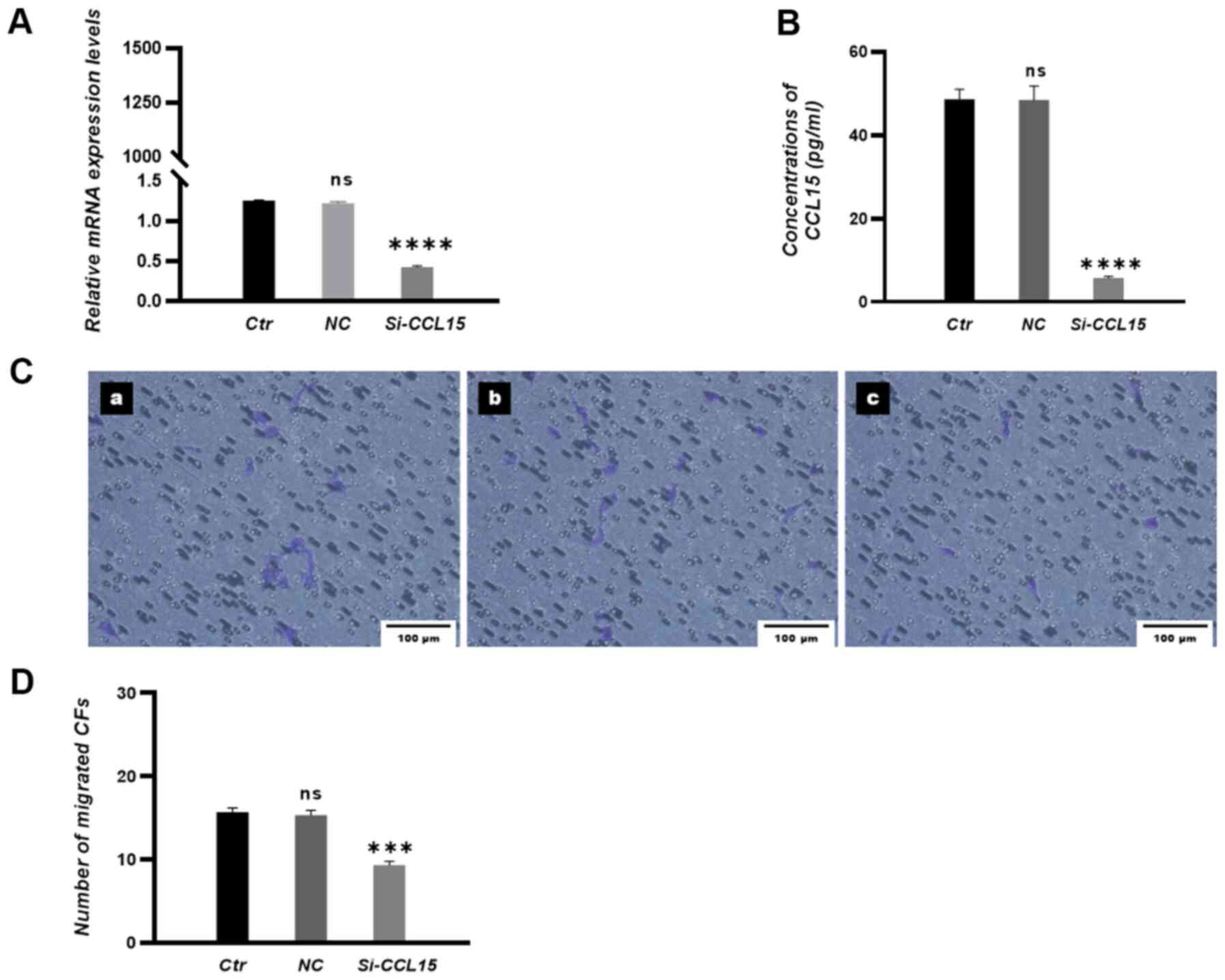
To investigate the chemotactic role of CCL15, CCL15
in HUVECs were either overexpressed or silenced before transfection
efficiency was examined by RT-qPCR and ELISA. The results
demonstrated that the expression levels of CCL15 were significantly
increased in the CCL15 overexpression group (Fig. 2A and B) and significantly decreased
in the si-CCL15 group (Fig. 3A and
B) compared with those in negative control (NC) groups.
Subsequently, CCL15 overexpression in HUVECs significantly promoted
the migration of CFs towards HUVECs (Fig. 2C and D), whilst knocking down CCL15
expression in HUVECs significantly attenuated the transmigration of
CFs towards HUVECs in co-culture conditions (Fig. 3C and D).
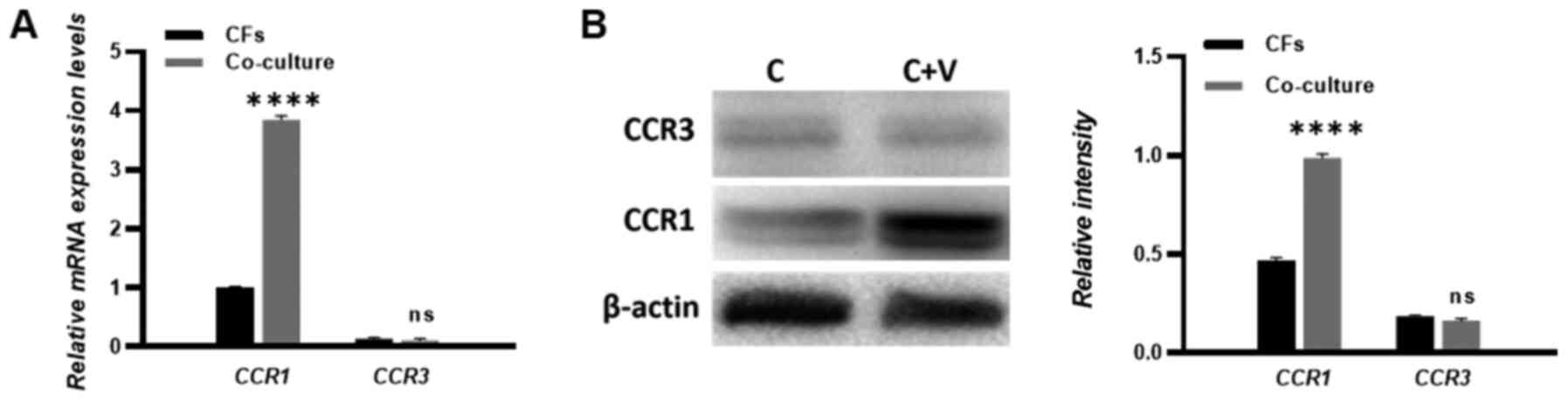
CCR1 mediates the migration of CFs
toward HUVECs in response to CCL15 in vitro
The CCL15-CCR1 chemokine axis has been previously
reported to modulate the migration and accumulation of numerous
cell types (15,16). Therefore, to investigate whether
CFs were recruited by HUVECs through the CCL15-CCR1 axis, the
expression of homologous CCL15 receptors CCR1 and CCR3 was examined
in CFs. RT-qPCR and western blotting were performed to measure the
expression levels of CCR1 and CCR3. The results showed that the
expression of CCR1 was increased in the co-culture compared with
the CF single culture. Meanwhile, the expression of CCR3 could not
be detected in either the single culture CFs or the HUVECs
co-cultured CFs, which suggests a deficiency in CCR3 expression in
the CFs (Fig. 4A and B).
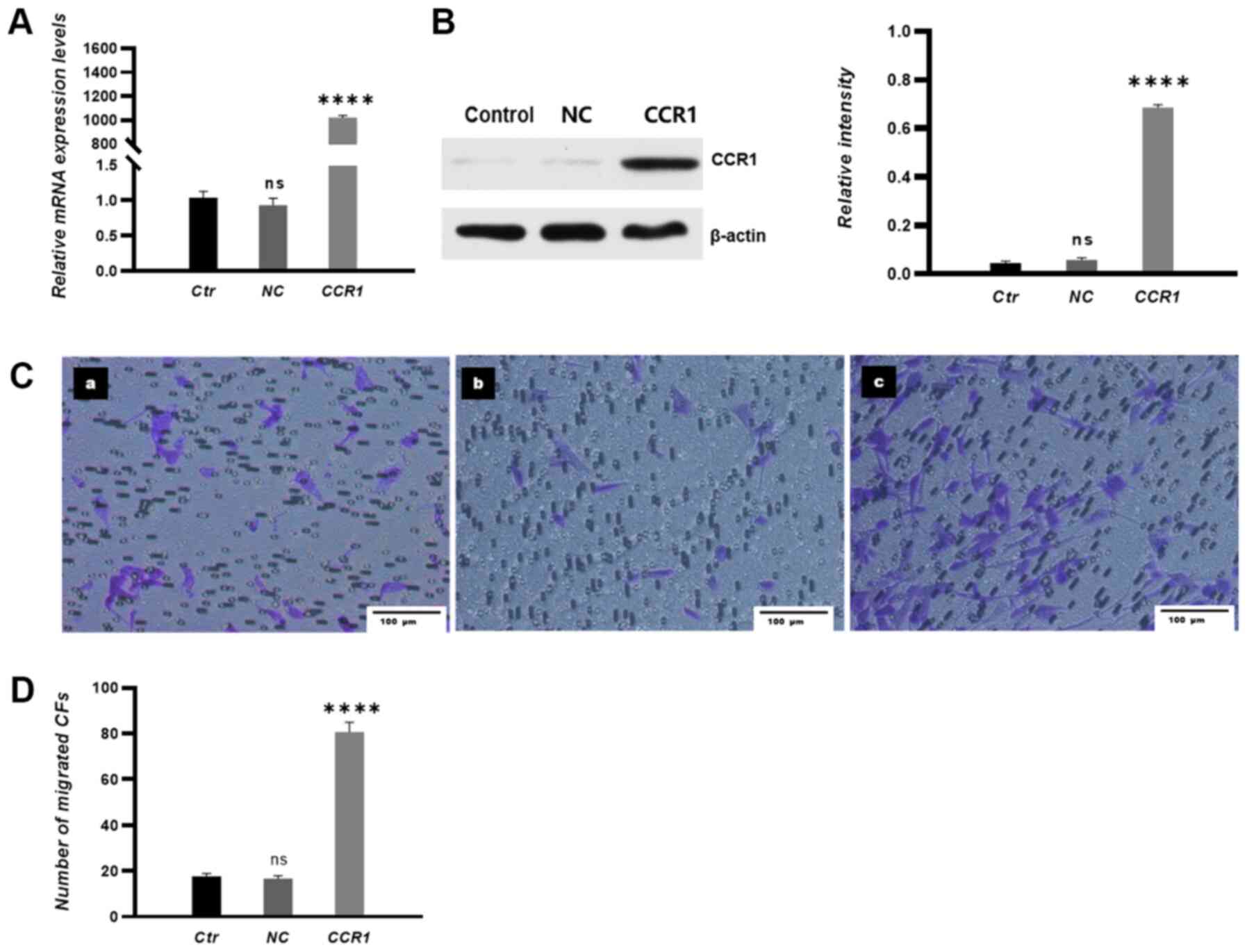
To verify if the CCL15-induced CF transmigration was
due to interactions between CCL15 and CCR1, CCR1 expression levels
were then manipulated. Transfection efficiency of the CCR1
overexpression vector was examined by RT-qPCR and western blotting.
The results demonstrated that the expression levels of CCR1 mRNA
and proteins were significantly increased in CFs in the CCR1
overexpression group compared with those in CFs in NC groups
(Fig. 5A and B). CCR1
overexpression in CFs significantly promoted the migration of CFs
towards HUVECs in co-culture conditions (Fig. 5C and D).
The transfection efficiency of CCR1 shRNA in CFs was
examined by RT-qPCR and western blotting. The results demonstrated
that the expression levels of CCR1 in CFs were significantly
decreased in the sh-CCR1 group compared with those in CFs in NC
groups (Fig. 6A and B). In
addition, knocking down CCR1 expression significantly attenuated
the migration of CFs towards HUVECs in co-culture conditions
(Fig. 6C and D).
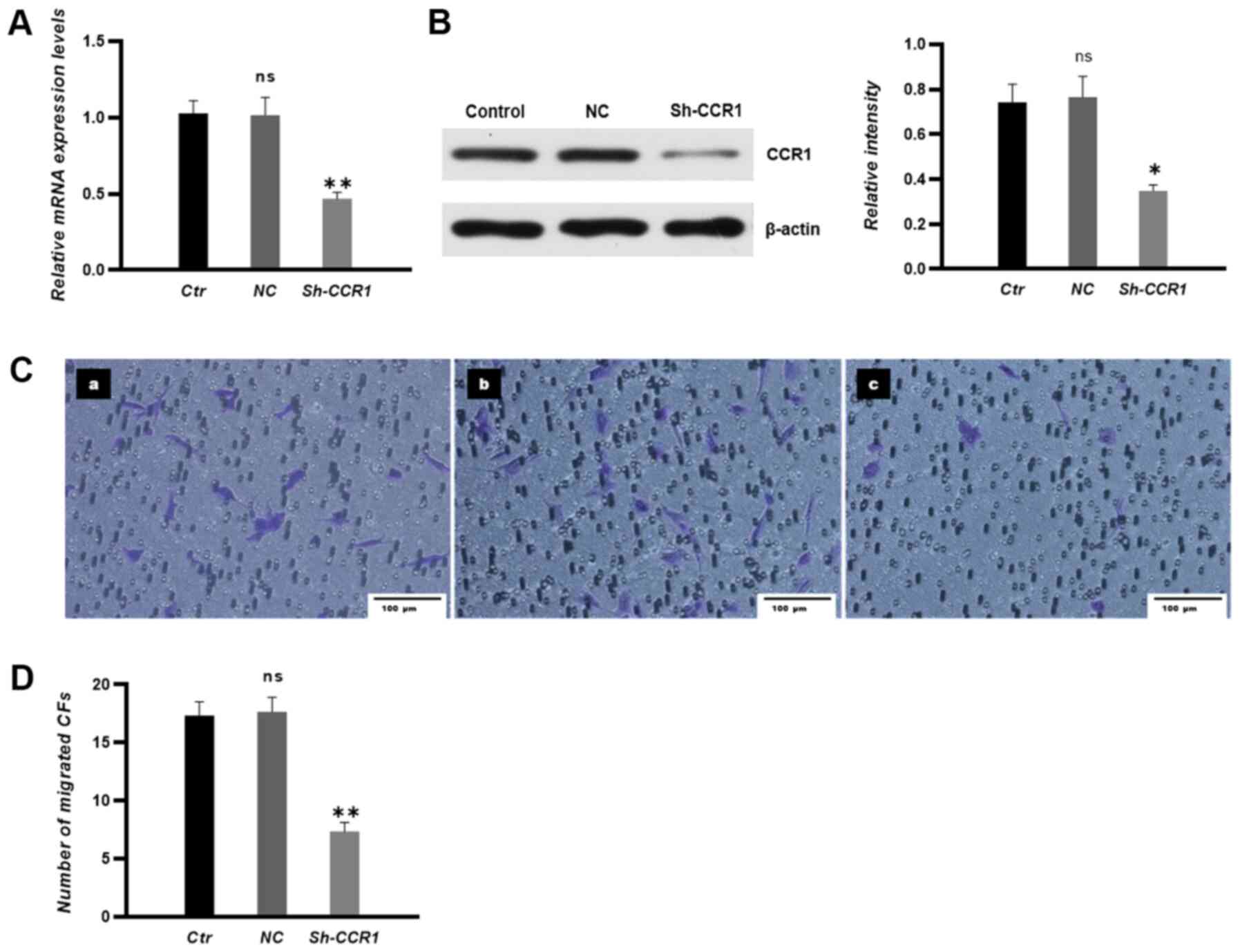 | Figure 6.CCR1 knockdown in CFs attenuates the
migration of CFs towards human umbilical vein endothelial cells
in vitro. (A) mRNA and (B) protein expression levels in each
group were measured by reverse transcription-quantitative PCR and
western blotting, respectively. (C) Migrated CFs were stained and
imaged under a microscope. (C-a) Control group, (C-b) NC group and
(C-c) short hairpin-CCR1 group. n=3. Scale bar, 100 µm. (D) The
number of migrated CFs. Untransfected CFs were used in Ctr groups
and non-sense shRNA transfected CFs were used in the NC groups.
*P<0.05, **P<0.01 vs. NC; ns vs. Ctrl. CFs, circulating
fibrocytes; Ctr, control; NC, negative control; sh, short hairpin;
CCR, chemokine receptor; ns, non-significant. |
Discussion
Chemokines involved in the transmigration of
fibrocytes in fibrotic diseases have been widely studied (8,17,18).
However, chemokines that regulate the transmigration of fibrocytes
during wound healing remain poorly understood. In the present
study, chemokines secreted by HUVECs were screened using a gene
chip, following which changes in their expression with or without
CFs co-culture were compared. To the best of our knowledge, the
present study was the first to report that endothelial cell-derived
CCL15 modulated the migration of CFs toward HUVECs. CCR1 and CCR3
are two homologous receptors for CCL15. Notably, the present study
demonstrated that CFs only expressed CCR1 when co-cultured with
HUVECs. In addition, the present study demonstrated that
overexpression of CCL15 in HUVECs or overexpression of CCR1 in CFs
promoted the transmigration of CFs towards HUVECs, whilst knockdown
of CCL15 or CCR1 attenuated the migration of CFs towards HUVECs.
These results suggest that the CCL15-CCR1 axis serves a vital role
in mediating the chemoattraction of CFs towards endothelial cells
in vitro.
The use of permeable Transwell co-culture systems to
explore the interactions between two cell types are widely used
(19,20). The present study co-cultured CFs
and HUVECs to mimic the angiogenesis process during wound healing
and measured the expression levels of chemokines in HUVECs with or
without co-culture with CFs. Gene chip analysis performed in the
present study demonstrated that the chemokines CCL2, CCL15 and
CXCL8 exhibited the largest fold-changes in expression. Therefore,
the effects of CCL2, CCL15 and CXCL8 on CF migration were compared
using transmigration assays. The results revealed a potentially
vital role for CCL15 in regulating the migration of CFs.
A previous study identified that CCL15 can stimulate
chemotactic endothelial cell migration and differentiation, which
confirmed the in vitro and in vivo angiogenic
activity of CCL15 (21). CCL15 has
also been observed to increase the adhesion of human monocytes to
endothelial cells under static and shear stress conditions
(22). Results from the present
study suggested that endothelial cell-derived CCL15 serves an
important role in modulating the transmigration of CFs, which is an
additional function of CCL15. The results of the present study
suggested that the chemokine CCL15 may participate in the
angiogenesis process by promoting CF migration, which are the
precursors of pericytes (11).
Furthermore, the present study also confirmed the expression of
CCR1 in CFs. The CCL15-CCR1 axis between endothelial cells and
fibrocytes is important. A previous study reported that the
CCL15-CCR1 interaction forms a complex tumor-promoting inflammatory
microenvironment in human hepatocellular carcinoma (23). Unfortunately, in the present study,
the mechanism downstream of the CCL15-CCR1 axis was not explored
further, which may have other functions in addition to modulating
CF migration. Therefore, future studies should focus on exploring
the CCL15-CCR1 axis in mediating the differentiation of fibrocytes
and the activation of downstream signaling pathways related to
other functions of fibrocytes.
Chemokines that modulate the transmigration of CFs
have been previously explored. CCL2 has been previously found to
mediate fibrocyte migration to the lung during asthma, whilst
CX3CL1 and CXCL2 can also regulate the transmigration of CFs
(18,24). In the present study, although gene
chip resulted in the identification of several chemokines a
limitation of the present study is that only three chemokines were
selected based on their fold-changes in expression. Therefore, the
roles of other chemokines identified by gene chip analysis in
mediating fibrocyte migration during angiogenesis may have been
overlooked. In addition to transmigration, differentiation of
fibrocytes into pericytes is equally vital in the process of
angiogenesis (24). However, the
present study did not examine the differentiation ratio in
fibrocytes after co-culture with HUVECs. Whether the CCL15-CCR1
axis is involved in CF differentiation of CFs require further
study.
In conclusion, to the best of our knowledge, the
present study identified an important role of CCL15 in mediating
the recruitment of fibrocytes by endothelial cells for the first
time. The present study also confirmed the expression of CCR1 in
fibrocytes and determined the regulatory role of the CCL15-CCR1
axis during the recruitment process of circulating fibrocytes.
Acknowledgements
The authors would like to thank Professor Xueyong Li
(Department of Plastic Surgery, Tangdu Hospital, Fourth Military
Medical University) for guiding the present research.
Funding
This study was supported by the National High
Technology Research and Development Program of China (863 Program;
grant no. SS2015AA020313) and the National Nature Science
Foundation of China (grant no. 81701909).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL and NP performed the experiments and wrote the
manuscript. XW and LX analyzed the data and revised the manuscript.
RH and XX performed the experiments. XuL conceived and designed the
study, prepared the figures and revised the manuscript. JL and XiL
analyzed the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee for
Experimentation of the Fourth Military Medical University (approval
no. TDLL-20170129; Jan 2017).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cho H, Blatchley MR, Duh EJ and Gerecht S:
Acellular and cellular approaches to improve diabetic wound
healing. Adv Drug Deliver Rev. 146:267–288. 2019.
|
|
2
|
Jones RE, Foster DS and Longaker MT:
Management of chronic wounds-2018. JAMA. 320:1481–1482.
2018.PubMed/NCBI
|
|
3
|
Bucala R, Spiegel LA, Chesney J, Hogan M
and Cerami A: Circulating fibrocytes define a new leukocyte
subpopulation that mediates tissue repair. Mol Med. 1:71–81.
1994.PubMed/NCBI
|
|
4
|
Chen D, Zhao Y, Li Z, Shou K, Zheng X, Li
P, Qi B and Yu A: Circulating fibrocyte mobilization in negative
pressure wound therapy. J Cell Mol Med. 21:1513–1522.
2017.PubMed/NCBI
|
|
5
|
Li J, Tan H, Wang X, Li Y, Samuelson L, Li
X, Cui C and Gerber DA: Circulating fibrocytes stabilize blood
vessels during angiogenesis in a paracrine manner. Am J Pathol.
184:556–571. 2014.PubMed/NCBI
|
|
6
|
Hartlapp I, Abe R, Saeed RW, Peng T,
Voelter W, Bucala R and Metz CN: Fibrocytes induce an angiogenic
phenotype in cultured endothelial cells and promote angiogenesis in
vivo. FASEB J. 15:2215–2224. 2001.PubMed/NCBI
|
|
7
|
Chen Z, Meng P, Li HT, Li M, Yang LF, Yan
Y, Li YT, Zou XL, Wang DY and Zhang TT: Thymic stromal
lymphopoietin contribution to the recruitment of circulating
fibrocytes to the lung in a mouse model of chronic allergic asthma.
J Asthma. 55:975–983. 2018.PubMed/NCBI
|
|
8
|
Isgrò M, Bianchetti L, Marini MA, Bellini
A, Schmidt M and Mattoli S: The C-C motif chemokine ligands CCL5,
CCL11, and CCL24 induce the migration of circulating fibrocytes
from patients with severe asthma. Mucosal Immunol. 6:718–727.
2013.PubMed/NCBI
|
|
9
|
Liu Y, Qingjuan S, Gao Z, Deng C, Wang Y
and Guo C: Circulating fibrocytes are involved in inflammation and
leukocyte trafficking in neonates with necrotizing enterocolitis.
Medicine (Baltimore). 96:e74002017.PubMed/NCBI
|
|
10
|
Aono Y, Kishi M, Yokota Y, Azuma M,
Kinoshita K, Takezaki A, Sato S, Kawanoh, Kishi J, Gotoh, et al:
Role of platelet-derived growth factor/platelet-derived growth
factor receptor axis in the trafficking of circulating fibrocytes
in pulmonary fibrosis. Am J Resp Cell Mol. 51:793–801. 2014.
|
|
11
|
Xueyong L, Shaozong C, Wangzhou L, Yuejun
L, Xiaoxing L, Jing L, Yanli W and Jinqing L: Differentiation of
the pericyte in wound healing: The precursor, the process, and the
role of the vascular endothelial cell. Wound Repair Regen.
16:346–355. 2008.PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
13
|
Sambrook J and Russell DW:
Calcium-phosphate-mediated transfection of cells with
high-molecular-weight genomic DNA. Cold Spring Harbor Protoc.
2006:pdb.prot3872. 2006.
|
|
14
|
Smadja DM, Dorfmüller P, Guerin CL, Bieche
I, Badoual C, Boscolo E, Kambouchner M, Cazes A, Mercier O, Humbert
M, et al: Cooperation between human fibrocytes and endothelial
colony-forming cells increases angiogenesis via the CXCR4 pathway.
Thromb Haemostasis. 112:1002–1013. 2014.
|
|
15
|
Yamamoto T, Kawada K, Itatani Y, Inamoto
S, Okamura R, Iwamoto M, Miyamoto E, Chen-Yoshikawa TF, Hiraih,
Hasegawa S, et al: Loss of SMAD4 promotes lung metastasis of
colorectal cancer by accumulation of CCR1+
Tumor-associated neutrophils through CCL15-CCR1 axis. Clin Cancer
Res. 23:833–844. 2017.PubMed/NCBI
|
|
16
|
Inamoto S, Itatani Y, Yamamoto T,
Minamiguchi S, Hiraih, Iwamoto M, Hasegawa S, Taketo mM, Sakai Y
and Kawada K: Loss of SMAD4 promotes colorectal cancer progression
by accumulation of myeloid-derived suppressor cells through the
CCL15-CCR1 chemokine axis. Clin Cancer Res. 22:492–501.
2016.PubMed/NCBI
|
|
17
|
Singh SR, Sutcliffe A, Kaur D, Gupta S,
Desai D, Saunders R and Brightling CE: CCL2 release by airway
smooth muscle is increased in asthma and promotes fibrocyte
migration. Allergy. 69:1189–1197. 2014.PubMed/NCBI
|
|
18
|
Ishida Y, Kimura A, Nosaka M, Kuninaka Y,
Hemmih, Sasaki I, Kaisho T, Mukaida N and Kondo T: Essential
involvement of the CX3CL1-CX3CR1 axis in bleomycin-induced
pulmonary fibrosis via regulation of fibrocyte and M2 macrophage
migration. Sci Rep. 7:168332017.PubMed/NCBI
|
|
19
|
Bian Y, Du Y, Wang R, Chen N, Du X, Wang Y
and Yuan H: A comparative study of HAMSCs/HBMSCs transwell and
mixed coculture systems. IUBMB Life. 71:1048–1055. 2019.PubMed/NCBI
|
|
20
|
Zhang C, Barrios MP, Alani RM, Cabodi M
and Wong JY: A microfluidic transwell to study chemotaxis. Exp Cell
Res. 342:159–165. 2016.PubMed/NCBI
|
|
21
|
Hwang J, Kim CW, Son KN, Han KY, Lee KH,
Kleinman HK, Ko J, Na DS, Kwon BS, Gho YS and Kim J: Angiogenic
activity of human CC chemokine CCL15 in vitro and in vivo. FEBS
Lett. 570:47–51. 2004.PubMed/NCBI
|
|
22
|
Park KH, Lee TH, Kim CW and Kim J:
Enhancement of CCL15 expression and monocyte adhesion to
endothelial cells (ECs) after hypoxia/reoxygenation and induction
of ICAM-1 expression by CCL15 via the JAK2/STAT3 pathway in ECs. J
Immunol. 190:6550–6558. 2013.PubMed/NCBI
|
|
23
|
Liu LZ, Zhang Z, Zheng BH, Shi Y, Duan M,
Ma LJ, Wang ZC, Dong LQ, Dong PP, Shi JY, et al: CCL15 recruits
suppressive monocytes to facilitate immune escape and disease
progression in hepatocellular carcinoma. Hepatology. 69:143–159.
2019.PubMed/NCBI
|
|
24
|
Harris DA: Inhibiting CXCL12 blocks
fibrocyte migration and differentiation and attenuates
bronchiolitis obliterans in a murine heterotopic tracheal
transplant model. J Thoracic Cardiovasc Surg. 145:854–861.
2013.
|