|
1
|
Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW,
Li RQ, Yang KY, Li J, Li M, Law PT, et al: The draft genome,
transcriptome, and microbiome of Dermatophagoides farinae
reveal a broad spectrum of dust mite allergens. J Allergy Clin
Immunol. 135:539–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roberts G, Almqvist C, Boyle R, Crane J,
Hogan SP, Marsland B, Saglani S and Woodfolk JA: Developments in
the field of allergy in 2017 through the eyes of clinical and
experimental allergy. Clin Exp Allergy. 48:1606–1621. 2018.
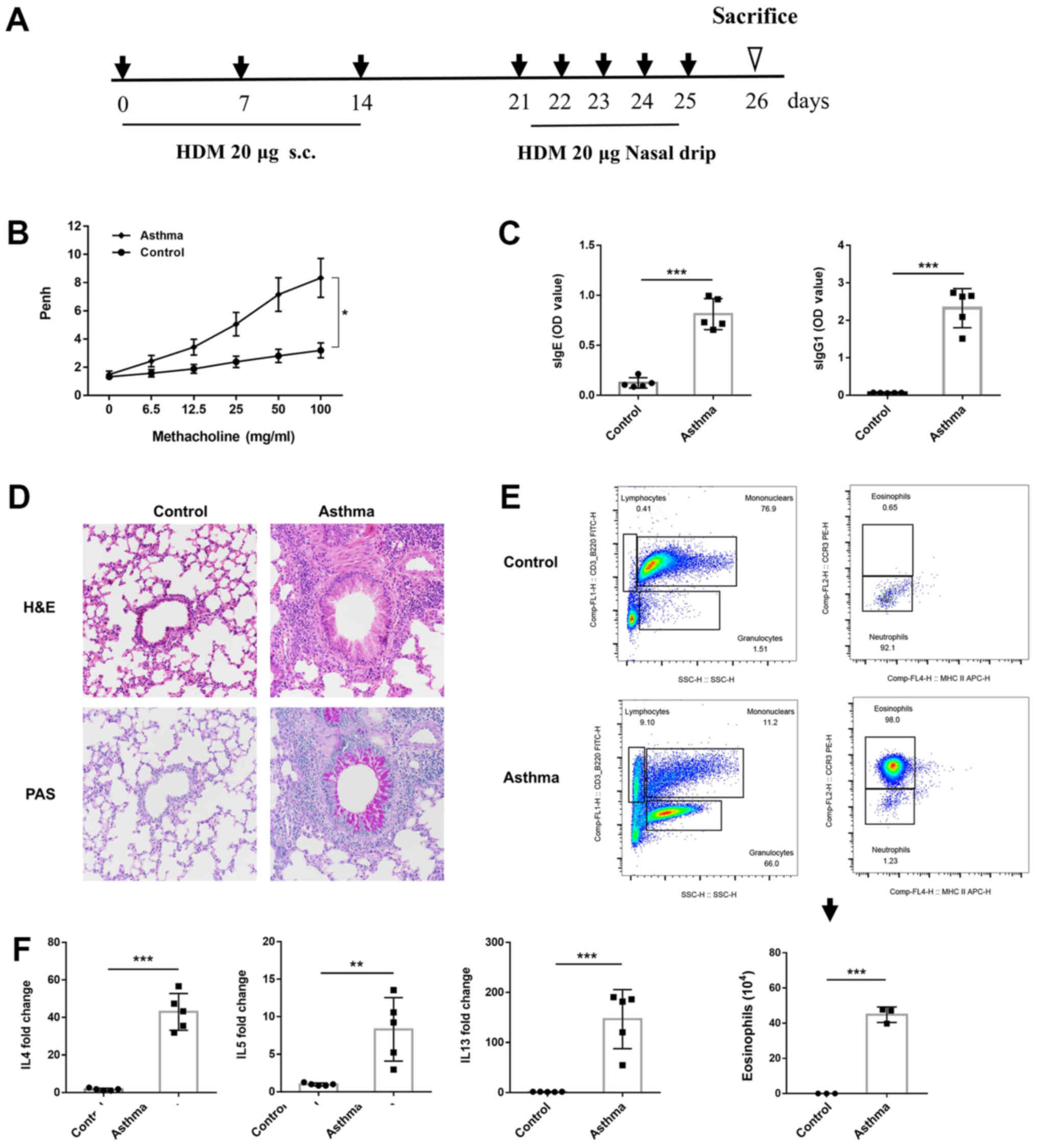
View Article : Google Scholar : PubMed/NCBI
|
|
3
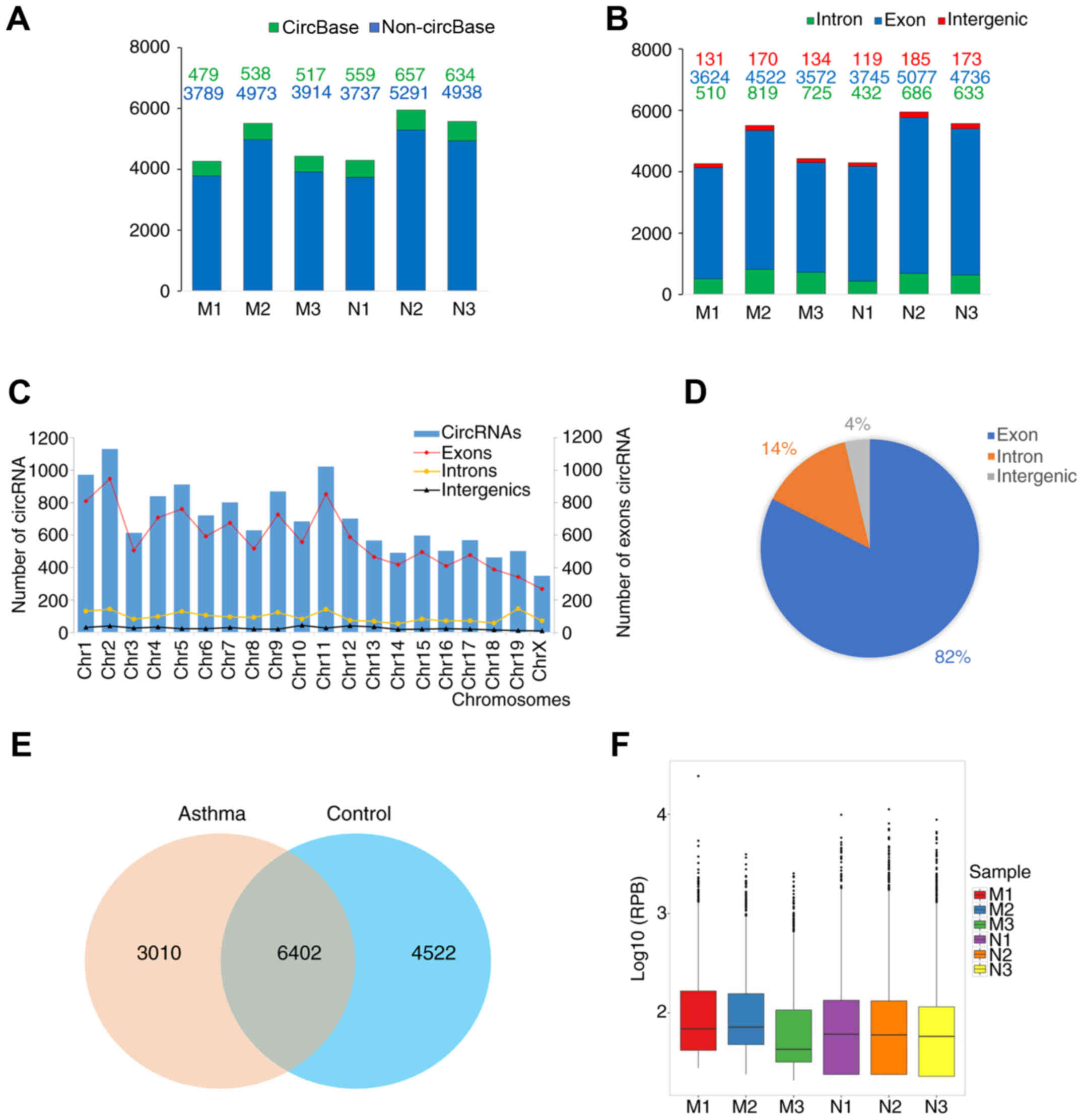
|
Corren J and Ziegler SF: TSLP: From
allergy to cancer. Nat Immunol. 20:1603–1609. 2019. View Article : Google Scholar : PubMed/NCBI
|
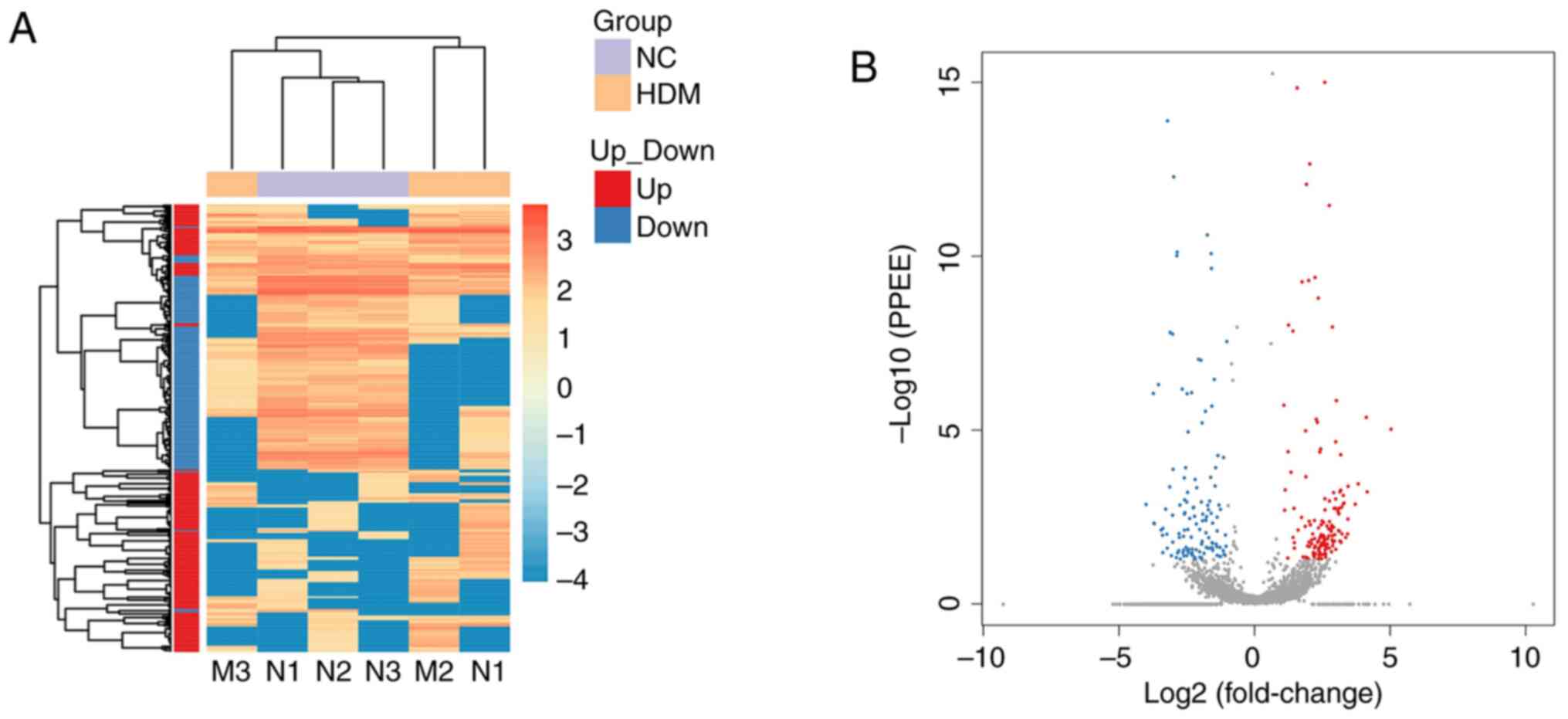
|
4
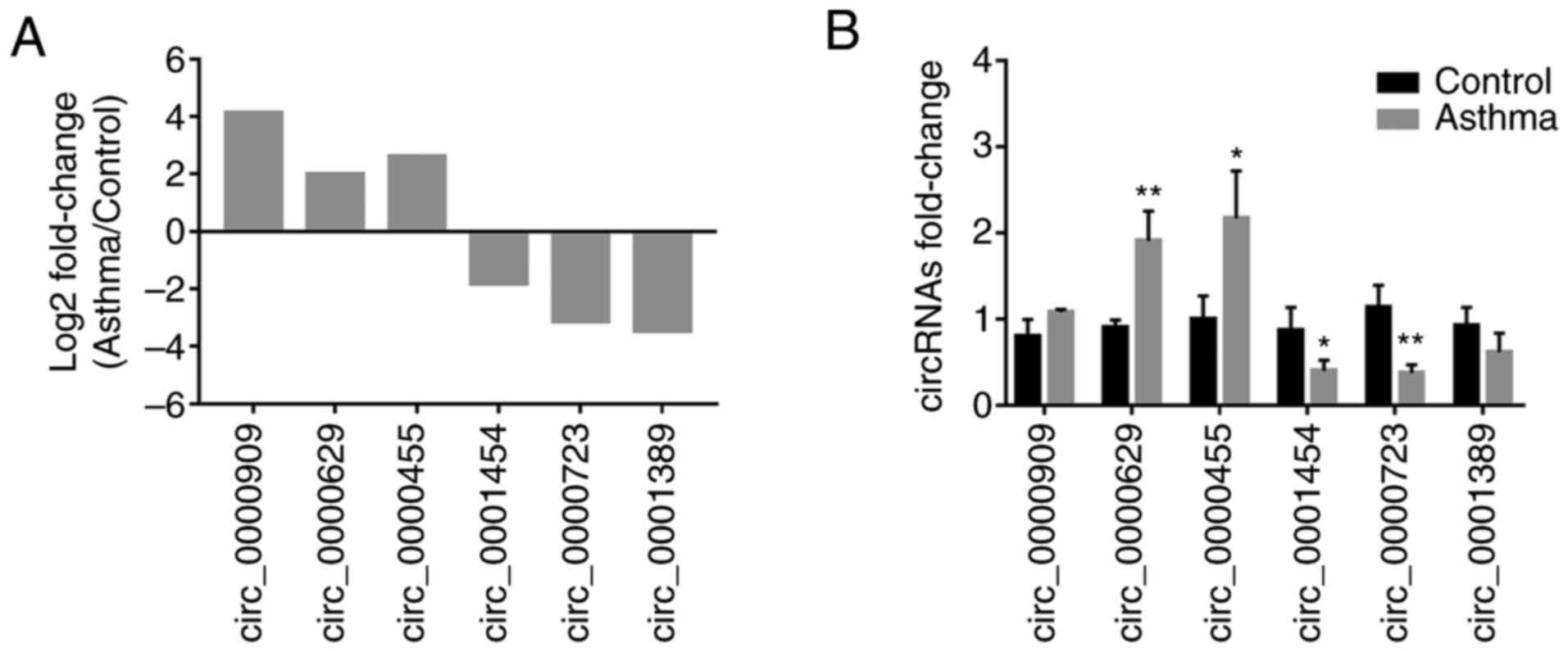
|
Ortiz RA and Barnes KC: Genetics of
allergic diseases. Immunol Allergy Clin North Am. 35:19–44. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gomez JL: Epigenetics in Asthma. Curr
Allergy Asthma Rep. 19:562019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dai X, Zhang S and Zaleta-Rivera K: RNA:
Interactions drive functionalities. Mol Biol Rep. 47:1413–1434.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sonkoly E, Janson P, Majuri ML, Savinko T,
Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al:
miR-155 is overexpressed in patients with atopic dermatitis and
modulates T-cell proliferative responses by targeting cytotoxic T
lymphocyte-associated antigen 4. J Allergy Clin Immunol.
126:581–589.e1-e20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Knolle MD, Chin SB, Rana BMJ, Englezakis
A, Nakagawa R, Fallon PG, Git A and McKenzie ANJ: MicroRNA-155
protects group 2 innate lymphoid cells from apoptosis to promote
type-2 immunity. Front Immunol. 9:22322018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Daniel E, Roff A, Hsu MH, Panganiban R,
Lambert K and Ishmael F: Effects of allergic stimulation and
glucocorticoids on miR-155 in CD4+ T-cells. Am J Clin
Exp Immunol. 7:57–66. 2018.PubMed/NCBI
|
|
10
|
Zhang H, Nestor CE, Zhao S, Lentini A,
Bohle B, Benson M and Wang H: Profiling of human CD4+
T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1.
J Allergy Clin Immunol. 132:1005–1008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang PP, Sun J and Li W: Genome-wide
profiling reveals atrial fibrillation-related circular RNAs in
atrial appendages. Gene. 728:1442862020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vidal AF, Sandoval GT, Magalhães L, Santos
SE and Ribeiro-dos-Santos Â: Circular RNAs as a new field in gene
regulation and their implications in translational research.
Epigenomics. 8:551–562. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dhamija S and Menon MB: Non-coding
transcript variants of protein-coding genes-what are they good for?
RNA Biol. 15:1025–1031. 2018.PubMed/NCBI
|
|
16
|
Cates EC, Fattouh R, Wattie J, Inman MD,
Goncharova S, Coyle AJ, Gutierrez-Ramos JC and Jordana M:
Intranasal exposure of mice to house dust mite elicits allergic
airway inflammation via a GM-CSF-mediated mechanism. J Immunol.
173:6384–6392. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valentine H, Williams WO and Maurer KJ:
Sedation or inhalant anesthesia before euthanasia with
CO2 does not reduce behavioral or physiologic signs of
pain and stress in mice. J Am Assoc Lab Anim Sci. 51:50–57.
2012.PubMed/NCBI
|
|
18
|
Van Rijt LS, Kuipers H, Vos N, Hijdra D,
Hoogsteden HC and Lambrecht BN: A rapid flow cytometric method for
determining the cellular composition of bronchoalveolar lavage
fluid cells in mouse models of asthma. J Immunol Methods.
288:111–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang E, Liu X, Tu W, Do DC, Yu H, Yang L,
Zhou Y, Xu D, Huang SK, Yang P, et al: Benzo(a)pyrene facilitates
dermatophagoides group 1 (Der f 1)-induced epithelial cytokine
release through aryl hydrocarbon receptor in asthma. Allergy.
74:1675–1690. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Soneson C and Delorenzi M: A comparison of
methods for differential expression analysis of RNA-seq data. BMC
Bioinformatics. 14:912013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Glažar P, Papavasileiou P and Rajewsky N:
CircBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila.
Genome Biol. 5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miller JD: The role of dust mites in
allergy. Clin Rev Allergy Immunol. 57:312–329. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakagome K and Nagata M: Involvement and
possible role of eosinophils in asthma exacerbation. Front Immunol.
9:22202018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kirschvink N, Vincke G, Onclinx C, Peck MJ
and Gustin P: Comparison between pulmonary resistance and penh in
anaesthetised rats with tracheal diameter reduction and after
carbachol inhalation. J Pharmacol Toxicol Methods. 51:123–128.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakaya M, Dohi M, Okunishi K, Nakagome K,
Tanaka R, Imamura M, Baba S, Takeuchi N, Yamamoto K and Kaga K:
Noninvasive system for evaluating allergen-induced nasal
hypersensitivity in murine allergic rhinitis. Lab Invest.
86:917–926. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Verheijden KA, Henricks PA, Redegeld FA,
Garssen J and Folkerts G: Measurement of airway function using
invasive and non-invasive methods in mild and severe models for
allergic airway inflammation in mice. Front Pharmacol. 5:1902014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kong DH, Kim YK, Kim MR, Jang JH and Lee
S: Emerging roles of vascular cell adhesion molecule-1 (VCAM-1) in
immunological disorders and cancer. Int J Mol Sci. 19:10572018.
View Article : Google Scholar
|
|
35
|
Alexis NE, Soukup J, Nierkens S and Becker
S: Association between airway hyperreactivity and bronchial
macrophage dysfunction in individuals with mild asthma. Am J
Physiol Lung Cell Mol Physiol. 280:L369–L375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fitzpatrick AM, Holguin F, Teague WG and
Brown LA: Alveolar macrophage phagocytosis is impaired in children
with poorly controlled asthma. J Allergy Clin Immunol.
121:1372–1378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang Z, Zhang Q, Thomas CM, Chana KK,
Gibeon D, Barnes PJ, Chung KF, Bhavsar PK and Donnelly LE: Impaired
macrophage phagocytosis of bacteria in severe asthma. Respir Res.
15:722014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kowal K, Żebrowska E and Chabowski A:
Altered sphingolipid metabolism is associated with asthma phenotype
in house dust mite-allergic patients. Allergy Asthma Immunol Res.
11:330–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rybak-Wolf A, Stottmeister C, Glazar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yan J, Zhang X, Sun S, Yang T, Yang J, Wu
G, Qiu Y, Yin Y and Xu W: miR-29b reverses T helper 1 cells/T
helper 2 cells imbalance and alleviates airway eosinophils
recruitment in OVA-induced murine asthma by targeting inducible
co-stimulator. Int Arch Allergy Immunol. 180:182–194. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nakano T, Inoue Y, Shimojo N, Yamaide F,
Morita Y, Arima T, Tomiita M and Kohno Y: Lower levels of
hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells
of pediatric patients with asthma. J Allergy Clin Immunol.
132:1224–1227.e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gonzalo JA, Tian J, Delaney T, Corcoran J,
Rottman JB, Lora J, Al-garawi A, Kroczek R, Gutierrez-Ramos JC and
Coyle AJ: ICOS is critical for T helper cell-mediated lung mucosal
inflammatory responses. Nat Immunol. 2:597–604. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee CG, Link H, Baluk P, Homer RJ,
Chapoval S, Bhandari V, Kang MJ, Cohn L, Kim YK, McDonald DM and
Elias JA: Vascular endothelial growth factor (VEGF) induces
remodeling and enhances TH2-mediated sensitization and inflammation
in the lung. Nat Med. 10:1095–1103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Feng MJ, Shi F, Qiu C and Peng WK:
MicroRNA-181a, −146a and −146b in spleen CD4+ T
lymphocytes play proinflammatory roles in a murine model of asthma.
Int Immunopharmacol. 13:347–353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qiu YY, Zhang YW, Qian XF and Bian T:
miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate
Th1/Th2 balance in asthma through the combinatorial regulation of
Runx3. Am J Transl Res. 9:3184–3199. 2017.PubMed/NCBI
|
|
47
|
Lu TX and Rothenberg ME: Diagnostic,
functional, and therapeutic roles of microRNA in allergic diseases.
J Allergy Clin Immunol. 132:3–13; quiz 14.2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee SH, Jeong HM, Choi JM, Cho YC, Kim TS,
Lee KY and Kang BY: Runx3 inhibits IL-4 production in T cells via
physical interaction with NFAT. Biochem Biophys Res Commun.
381:214–217. 2009. View Article : Google Scholar : PubMed/NCBI
|