Introduction
Osteoporosis is a common bone disease that mainly
affects the elderly, and is characterized by low bone density and
microarchitectural deterioration of bone tissue (1–3).
Osteoporosis is associated with an excessive replacement of
osteoblasts with osteoclasts (4).
As a result, studying of the osteogenic differentiation of bone
marrow-derived mesenchymal stem cells (BMSCs) has emerged as the
main direction in the exploration of the pathogenesis of
osteoporosis (5). Since
chondrocyte development and bone marrow adipogenesis are based on
osteoblast formation (6,7), osteogenic differentiation of BMSCs is
favored to osteoblast maturation and contributes to osteoporosis
prevention (8,9). The pivotal role of bone morphogenetic
protein 2 (BMP2), one of the most well-characterized proteins of
the BMP family, in promoting bone formation has been well
documented, indicating that factors that regulate BMP2 expression
may be considered as important modulators in bone formation.
1,25(OH)2D3, the most active
vitamin D metabolite, is a pleiotropic hormone. Through binding to
its intra-nuclear receptor, vitamin D receptor (VDR),
1,25(OH)2D3 has numerous regulatory effects,
including calcium homeostasis, cell proliferation and
differentiation (10,11). Notably, it is widely accepted that
vitamin D is important for bone growth as its deficiency can lead
to osteomalacia and rickets (12).
Nevertheless, the effects of vitamin D on bone formation remains
under debate. In a previous study, Erben et al (13,14)
reported that 1,25(OH)2D3 administration
evidently enhanced new bone remodeling in vivo. However,
Sooy et al (15) found that
the knockdown of VDR improved osteogenic potential in vitro.
Additionally, Yamaguchi and Weitzmann (16) revealed that high-dose
administration of 1,25(OH)2D3 significantly
inhibited the mineralization of osteoblasts. Fu et al
(11) further reported that
1,25(OH)2D3 was able to reduce BMP2
expression in BMSCs. However, the mechanisms underlying the action
of 1,25(OH)2D3 in osteogenic differentiation
suppression remain largely unclear.
The canonical Wnt/β-catenin pathway has been
identified to serve an important role in the regulation of the
osteogenic differentiation of BMSCs (17). Under normal conditions, β-catenin
protein remains at a low level without Wnt ligands through
phosphorylation and ubiquitination-mediated degradation. However,
β-catenin is released from the ‘degradation complex’, which
contains Axin1/2, APC, casein kinase 1 and glycogen synthase kinase
3β, when Wnt ligands bind to their receptors, thus resulting in the
stabilization of β-catenin and its accumulation in the cytoplasm
and nucleus. However, to date, the roles of β-catenin in the
osteogenic differentiation of stem cells remain paradoxical. For
instance, Liu et al (18)
reported that activation of β-catenin repressed the osteogenic
differentiation of periodontal ligament stem cells, a new
population of MSCs. By contrast, the studies by Monroe et al
(17) and Mo et al
(19) demonstrated that the
osteogenic differentiation of BMSCs depends on Wnt/β-catenin
signaling activation. Therefore, whether Wnt/β-catenin signaling is
involved in the 1,25(OH)2D3-mediated
osteogenic differentiation and the exact role of this signaling
need to be elucidated.
In the present study, the aim was to explore the
mechanism underlying the 1,25(OH)2D3-mediated
suppression of osteogenic differentiation in vitro by
recruiting BMSCs.
Materials and methods
BMSC isolation, culture and
identification
In total, three Sprague-Dawley (SD) rats (140±10 g),
aged 4-weeks-old, were purchased from Better Biotechnology Co.,
Ltd. (Nanjing, Jiangsu) and were maintained under specific
pathogen-free conditions at 20–26°C with 55±5% humidity in 12 h
light/dark cycle with ad libitum access to food and water.
After 1 week of accommodation, the rats were humanely euthanized
via cervical dislocation. Next, BMSCs were isolated from the femurs
and tibias of the SD rats according to the method described in a
previous study (20). All animal
experiment protocols were approved by the Review Committee for the
Use of Human or Animal Subjects of the Shanghai Jiao Tong
University (Shanghai, China).
Subsequent to repeated flushing, the BMSCs were
cultured in culture medium containing 89% Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.), and kept at 37°C with 5% CO2. The culture medium
was refreshed twice per week. BMSCs at the 3rd passage were used in
all the experiments.
To identify the BMSCs, cells at the 3rd passage were
trypsinized, collected and subjected to flow cytometry analysis
(CytoFLEX; Beckman Coulter Commercial Enterprise, Inc.) with
antibodies against CD34 (1:50; cat. no. ab81289; Abcam), CD45
(1:10; cat. no. 554878; BD Bioscience, CD44 (1:10; cat. no. 550974;
BD Bioscience) and CD90 (1:10; cat. no. 561973; BD Bioscience).
Cells identified as BMSCs negatively expressed CD34 and CD45, and
positively expressed CD44 and CD90 (19). The results were analyzed using the
FlowJo software (version 7.6; FlowJo LLC, Inc.).
Osteogenic induction and cell
treatments
For osteogenic induction, BMSCs (1×106
cells/well) were seeded into 6-well plates and cultured in the
complete culture medium supplemented with osteogenic induction
medium (OIM) containing 10−7 M dexamethasone, 10 mM
β-glycerophosphate and 50 µM ascorbate-2-phosphate (all purchased
from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 14 days
(21). Next, the cells were
collected and subjected to ossification assessment.
For 1,25(OH)2D3 treatment,
BMSCs were incubated with 1, 5, 10, 20 or 50 nM of
1,25(OH)2D3 (Sigma-Aldrich; Merck KGaA)
dissolved in ethanol for 48 h. Equivalent volume of 100% ethanol
was used in the negative control group. Furthermore, XAV-939
(Selleck Chemicals, Shanghai, China) treatment was used to inhibit
Wnt/β-catenin signaling in BMSCs at a concentration of 10 µM for 1
h, with equal volume of DMSO (Beyotime Institute of Biotechnology)
as a negative control. For co-treatment of
1,25(OH)2D3 and XAV-939 (MedChemExpress,
Inc.), the cells were first treated with 10 µM XAV-939 for 1 h at
37°C, followed by treatment with 10 nM
1,25(OH)2D3 for 48 h at 37°C.
Stable transfection cell lines
In order to induce upregulation of BMP2 and Smad1
levels in BMSCs, overexpressing lentivirus vectors targeting the
rat BMP2 (OE-BMP2) and Smad1 (OE-Smad1) genes were designed and
synthesized by GenePharma Co., Ltd. Briefly, the BMSCs
(5×105) were seeded in 6-well plates and cultured at
37°C overnight. Next, the cells were infected with OE-BMP2,
OE-Smad1 or OE-NC (serving as the negative control vector) using 5
µg/ml polybrene (Hanbio Biotechnology Co., Ltd.), followed by
incubation with 100 µg/ml G418 or 7 µg/ml puromycin for 14 days to
select the stably infected cell lines, respectively.
Small interfering RNAs (siRNAs)
Three siRNAs targeting the rat β-catenin gene
(si-β-catenin) and a negative control vector (si-NC) were purchased
from OriGene Technologies, Inc. (Rockville, MD, USA; cat. no.
SR500644). The siRNAs (si-1, si-2 and si-3) were applied to knock
down β-catenin expression in BMSCs, using Lipofectamine®
2000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Following 6 h of
cell transfection, the medium was replaced with fresh DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% FBS. The
transfection efficiency was detected by using reverse
transcription-quantitative PCR (RT-qPCR) and western blotting
assays after 24 and 48 h of cell transfection. The siRNA sequences
used were: si-1, 5′-CCAGCAAATCATGCGCCTT-3′; si-2,
5′-GCTGCATAATCTCCTGCTA-3′; and si-3, 5′-CCACTAATGTCCAGCGCTT-3′.
RT-qPCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the
concentration was measured using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). Next, the RNA was reversely transcribed into complementary
DNA (cDNA) using a PrimeScript™ RT reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China). Next, qPCR analysis was
performed with SYBR GreenER™ qPCR SuperMix (Thermo Fisher
Scientific, Inc.). The qPCR conditions involved pre-denaturation at
95°C for 5 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min.
The relative expressions of mRNAs were calculated by using the
2−ΔΔCq method (22).
The primer sequences used in this experiment were as follows: GAPDH
forward, 5′-AGTGCCAGCCTCGTCTCATA-3′, and reverse,
5′-GATGGTGATGGGTTTCCCGT-3′; BMP2 forward,
5′-GGACATGGTTGTGGAGGGTT-3′, and reverse,
5′-TGTTTTCCCAACTTATTTTCGTAGA-3′; Msh homeobox 2 (Msx2) forward,
5′-GGAGATTGCAAGAGGGCGTA-3′, and reverse,
5′-GGGCTAGCTGACTGTGTTGT-3′; Runt-related transcription factor 2
(Runx2) forward, 5′-CGCCTCACAAACAACCACAG-3′, and reverse,
5′-TCACTGCACTGAAGAGGCTG-3′; β-catenin forward,
5′-ATCATTCTGGCCAGTGGTGG-3′, and reverse,
5′-GACAGCACCTTCAGCACTCT-3′; Smad1 forward,
5′-TCAATAGAGGAGATGTTCAAGCAGT-3′, and reverse,
5′-GGTGGTAGTTGCAGTTCCGA-3′.
Western blotting
Total protein was extracted from the BMSCs using
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology), while the nuclear and cytoplasmic
proteins were extracted from the cells using a CelLytic™ NuCLEAR™
Extraction kit (Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. Following the assessment of protein
concentrations with a BCA kit (Thermo Fisher Scientific, Inc.), 25
µg protein from each sample was subjected to 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, followed by transfer
onto PVDF membranes (Thermo Fisher Scientific, Inc.). The membranes
were then blocked with 5% non-fat milk for 1 h at room temperature
and probed overnight at 4°C with the following primary antibodies:
GAPDH (1:10,000 dilution; ProteinTech), BMP2 (1:1,000 dilution;
cat. no. ab14933; Abcam), Runx2 (1:1,000 dilution; cat. no.
ab76956; Abcam), Msx2 (1:2,000 dilution; cat. no. HPA005652;
Sigma-Aldrich), osteocalcin (OCN; 1:2,000 dilution; cat. no.
ab93876; Abcam), osteopontin (OPN; 1:2,000 dilution; cat. no.
ab8448; Abcam), β-catenin (1:2,500 dilution; cat. no. 9562; Cell
Signaling Technology), disheveled-1 (Dvl-1; 1:2,000 dilution; cat.
no. D3570; Sigma-Aldrich), Smad1 (1:1,000 dilution; cat. no. 9743;
Cell Signaling Technology), tubulin (1:1,000 dilution; cat. no.
2148; Cell Signaling Technology) and histone (1:5,000 dilution;
cat. no. ab1791; Abcam). Subsequently, incubation with a
horseradish peroxidase-conjugated secondary antibody (1:5,000; cat.
no. SA00001-1/ SA00001-2; ProteinTech) was performed for 1 h at
room temperature. GAPDH, histone and tubulin were used as the
internal references for the total, nuclear and cytoplasmic protein
expression, respectively. The band signals were enhanced by
chemiluminescence (ECL reagent; EMD Millipore, Billerica, MA, USA)
and visualized with a FluorChem Q system (SelectScience, Waltham,
MA, USA). Protein expression quantification was performed using
ImageJ software (version 1.48; National Institutes of Health,
Bethesda, MD, USA).
Alkaline phosphatase (ALP)
activity
To assess ALP activity, BMSCs at a density of
1×105 cells/well were seeded into 24-well plates and
cultured for 14 days with OIM. Next, the ALP activity was measured
with an ALP activity kit (Shanghai Suer Biological Technology Co.,
Ltd., Shanghai, China) according to the manufacturer's protocol. In
brief, cells were lysed with RIPA lysis buffer on ice, and then the
lysates were mixed with a working solution for 15 min at 37°C. The
optical density values at 520 nm were subsequently measured, and
the ALP activity was normalized to the total intracellular protein
concentration.
In order to assess whether
1,25(OH)2D3 affected the osteogenic
differentiation of BMSCs, after 12 days of incubation with OIM,
BMSCs (5×105/well in 6-well plates) were cultured in OIM
containing 1,25(OH)2D3 (10 nM) for a further
48 h and the ALP activity was determined. In addition, to assess
the effects of BMP2 and Smad1 on osteogenic differentiation, the
OE-BMP2/OE-Smad1 stable expressing cells were cultured in OIM for
14 days, followed by ALP detection.
Alizarin red-S staining
Cells at a density of 5×106/well in
6-well plates treated with 1,25(OH)2D3 (10
nM) for 48 h or OE-BMP2/OE-Smad1 stable expression cell lines were
washed with PBS and then fixed with 75% ethanol at 4°C for 30 min.
Next, the cells were stained with Alizarin red-S (40 mM, pH 6.2) at
room temperature for a total of 30 min. Any excess stain was
removed by distilled water, and images of the plates were captured
to visualize cell calcification. Quantification was performed using
Image Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Cell Counting Kit-8 assay (CCK-8)
A CCK-8 assay was used to detect cell proliferation.
In brief, BMSCs were seeded in 96-well plates at a density of
3×103 cells/well and cultured at 37°C overnight.
Subsequently, the cells were subjected to different treatments,
including 1,25(OH)2D3 (10 nM)/ethanol (10 nM)
and/or XAV-939 (10 µM)/DMSO (10 µM). Following incubation at 37°C
for 48 h, the cell culture medium was replaced with 10 µl CCK-8
reagent (Beyotime Institute of Biotechnology) and 90 µl fresh
medium, and incubated at 37°C for another 4 h. The absorbance at
450 nm was finally measured with a plate reader (model 680; Bio-Rad
Laboratories, Inc.).
Luciferase gene reporter assay
The fragment of rat BMP2 promoter region C that
contains the VDR binding sites was amplified by PCR using the
followed primers: Forward primer, 5′-ATTTGCCCTAAACTCGGGCATCTG-3′,
and reverse primer, 5′-TTCGTCCCGAGCTGCCAAT-3′ (11). Next, the fragment was cloned into
the pGL3 promoter vector (Promega Corporation) containing a SV40
promoter upstream of the luciferase gene. The pSV40-BMP2-Luciferase
vector was then transfected into the BMSCs cells with or without 10
nM 1,25(OH)2D3 treatment. Cells were
harvested 48 h after the aforementioned treatments, and the
relative luciferase activities were measured using the Dual-Glo
luciferase assay kit (Promega Corporation).
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was performed as previously described
(23). Briefly, the crosslinked
chromatins were immunoprecipitated with antibodies against histone
H3 dimethylated lysine 9 (H3K9me2; cat. no. ab1220; Abcam) and
acetyl-histone H3 (histone H3-Ac; cat. no. ab4729; Abcam). The
enrichment of the specific amplified region was analyzed by
RT-qPCR. Normal IgG was used as a negative control. The primers for
amplifying the fragments of the BMP2 promoter in region C were as
follows: Forward, 5′-CGCCCCGCCCCGCCCCG-3′, and reverse,
5′-ATTTGCCCTAAACTCGGGCATCTG-3′ (11).
Immunoprecipitation (IP)
An IP assay was used to evaluate the interaction
between Dvl-1 and Smad1 proteins. Briefly, cells were lysed in 5 ml
lysis buffer (containing 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5%
Nonidet P40 and protease inhibitor cocktail) for 30 min at 4°C.
After 1 h of incubation with 50 µl Dynabeads protein A (Invitrogen;
Thermo Fisher Scientific, Inc.), the supernatants containing 200 µg
proteins were incubated with antibody against Smad1 (2 mg; cat. no.
9743; Cell Signaling Technology) at 4°C overnight. Next, the
Dynabeads were washed with lysis buffer for five times, followed by
resuspension in SDS-PAGE loading buffer and assessment by western
blotting using antibody against Dvl-1 (1:2,000; cat. no. D3570;
Sigma-Aldrich; Merck KGaA).
Statistical analysis
Each experiment was repeated at least three times,
and the data are represented as the mean ± standard deviation. Data
analysis was performed with SPSS software, version 21.0 (IBM
Corporation, Armonk, NY, USA), using the Student's t-test for
comparisons between two groups, or one-way analysis of variance
followed by Bonferroni post-hoc test for comparisons of more than
two groups. P<0.05 was considered to denote a statistically
significant difference.
Results
BMP2 upregulation reverses the
1,25(OH)2D3-induced inhibition of osteogenic
differentiation
The flow cytometry results demonstrated that the
isolated cells positively expressed CD44 and CD90, while they
negatively expressed CD34 and CD45 (Fig. 1A), suggesting that BMSCs were
successfully isolated. The present study then attempted to explore
whether BMP2 was involved in
1,25(OH)2D3-mediated osteogenesis inhibition
of BMSCs. First, stable overexpression of BMP2 was induced in the
BMSCs, and the expression of BMP2 was found to be significantly
elevated at the mRNA and protein levels in the OE-BMP2 group,
compared with the OE-NC group (Fig. 1B
and C). Next, the ALP activity of cells treated with
1,25(OH)2D3 was assessed. Compared with the
negative control (ethanol) group, 1,25(OH)2D3
treatment decreased ALP activity in a dose-dependent manner
(Fig. 1D). Significant reduction
in ALP activity was observed at a minimum concentration of 10 nM
(Fig. 1D), and thus this
concentration was selected for use in subsequent assays. No evident
changes in BMSC viability were observed when cells were treated
with 1,25(OH)2D3 or/and overexpressed BMP2
(Fig. 1E).
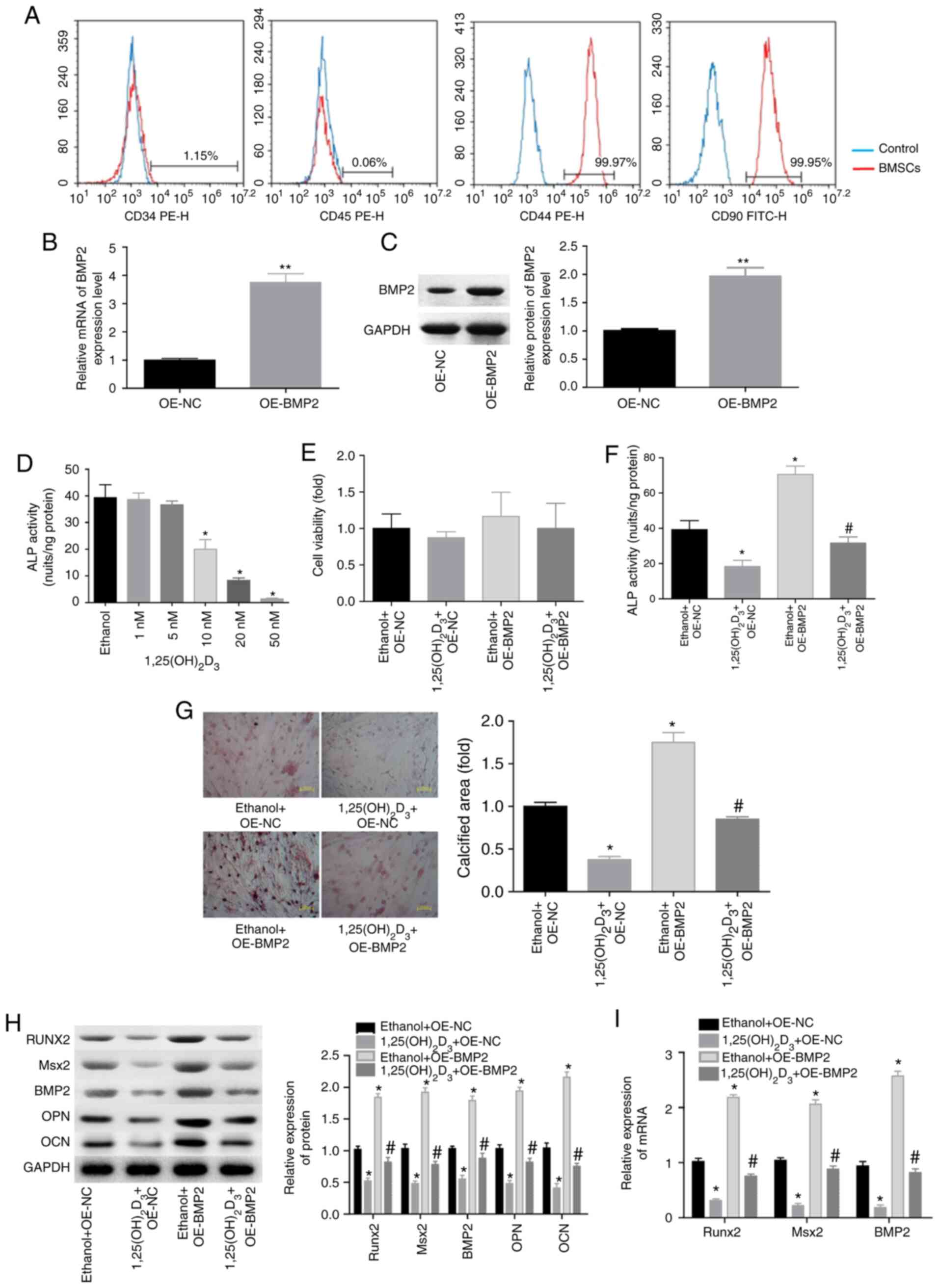 | Figure 1.1,25(OH)2D3
repressed the osteogenic differentiation of BMSCs via BMP2
downregulation. (A) Flow cytometry was conducted to test the
proportion of CD34+, CD44+, CD45+
and CD80+ in the isolated BMSCs. (B) mRNA and (C)
protein levels of BMP2, determined by RT-qPCR and western blotting
subsequent to stable infection of BMSCs with OE-BMP2 or OE-NC
(n=3). (D) The effect of different concentrations of
1,25(OH)2D3 (1, 5, 10, 20 or 50 nM) on ALP
activity in BMSCs, with ethanol serving as a negative control
(n=3). (E) Cell viability determined by Cell Counting Kit-8 assay
and (F) ALP activity in BMSCs treated with
1,25(OH)2D3 and/or OE-BMP2. (G) Alizarin
red-S was used to assess the calcified nodules of BMSCs treated
with 1,25(OH)2D3 and/or OE-BMP2.
Magnification, ×200. (H) Western blotting of the protein levels of
BMP2, Runx2, Msx2, OPN and OCN in BMSCs with different treatments.
(I) RT-qPCR analysis of the mRNA levels of Runx2, BMP2 and Msx2 in
differently treated BMSCs (n=3). *P<0.05 and **P<0.01, vs.
corresponding control group (OE-NC, ethanol or ethanol + OE-NC
group); #P<0.05 vs.
1,25(OH)2D3+OE-NC group. BMSCs, bone
marrow-derived mesenchymal stem cells; BMP2, bone morphogenetic
protein 2; ALP, alkaline phosphatase; Runx2, Runt-related
transcription factor 2; Msx2, Msh homeobox 2; OPN, osteopontin;
OCN, osteocalcin; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; OE, overexpressing vector; NC, negative
control. |
When cells were treated with
1,25(OH)2D3 (10 nM) for 48 h, the ALP
activity and calcified area of BMSCs were significantly decreased,
whereas BMP2 overexpression rescued these results (Fig. 1F and G). Furthermore,
1,25(OH)2D3 treatment significantly decreased
the expression levels of ossification-associated proteins,
including BMP2, OPN, OCN, Runx2 and Msx2 (Fig. 1H), and reduced the mRNA levels of
BMP2, Runx2 and Msx2 in BMSCs (Fig.
1I). However, these effects of
1,25(OH)2D3 were all neutralized when BMP2
was overexpressed in BMSCs (Fig. 1H
and I). These results indicated that
1,25(OH)2D3 repressed the osteogenic
differentiation of BMSCs via downregulating BMP2 expression.
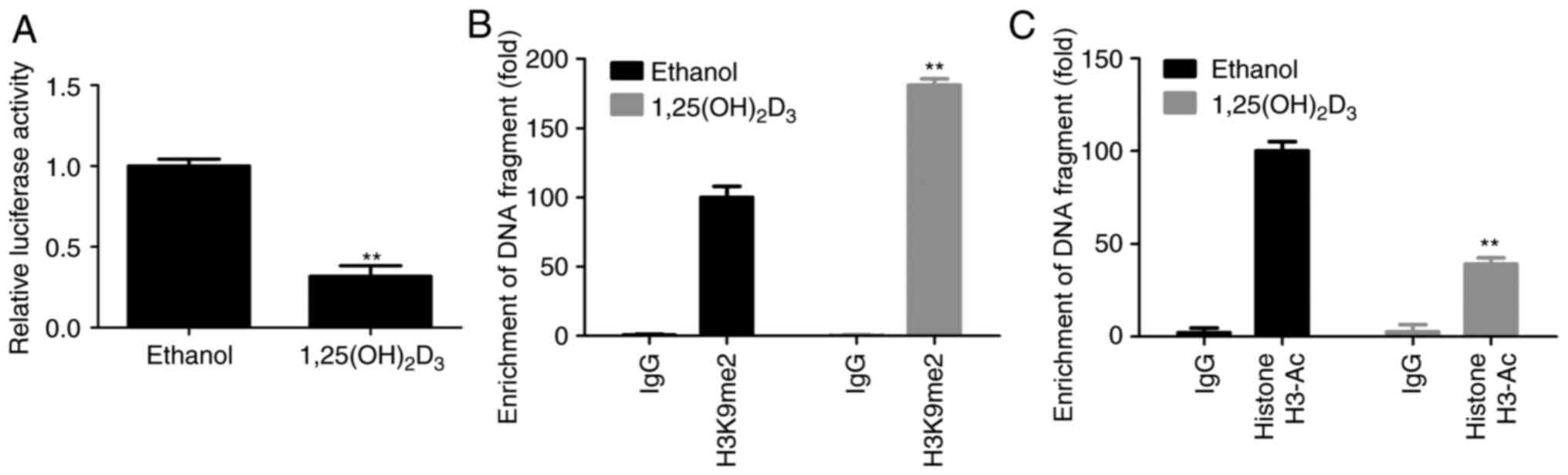
1,25(OH)2D3
decreases BMP2 expression through DNA methylation and histone
modification
Next, the current study explored the mechanism
underlying 1,25(OH)2D3-induced BMP2
downregulation in BMSCs. Compared with the control (ethanol) group,
1,25(OH)2D3 treatment significantly decreased
the activity of the pSV40-BMP2-Luciferase promoter (Fig. 2A), and enhanced the combination
between BMP2 promoter and H3K9me2 (Fig. 2B), while it weakened the
interaction between BMP2 promoter and histone H3-Ac (Fig. 2C). These results suggested that
1,25(OH)2D3 downregulated BMP2 expression at
the epigenetic level.
1,25(OH)2D3
represses the osteogenic differentiation through activating
β-catenin signaling
Next, the study explored the role of β-catenin
signaling in 1,25(OH)2D3-induced osteogenesis
repression. The results revealed that
1,25(OH)2D3 treatment enhanced the nuclear
expression level of β-catenin and decreased its cytoplasmic
expression level, whereas BMP2 overexpression reversed the effects
of 1,25(OH)2D3 (Fig. 3A). These findings suggested that
the activation of β-catenin signaling may be involved in the
1,25(OH)2D3-induced repression of the
osteogenic differentiation of BMSCs. To further explore the role of
β-catenin, the inhibitor of β-catenin, XAV-939, was recruited to
repress β-catenin levels. The results demonstrated that XAV-939
treatment had no evident influence in BMSC viability (Fig. 3B). XAV-939 significantly blunted
the role of 1,25(OH)2D3 in the inhibition of
ALP activity (Fig. 3C) and in the
reduction of the calcified area (Fig.
3D), as well as reversed the
1,25(OH)2D3-induced decrease in the
expression levels of Runx2, Msx2, BMP2, OPN and OCN (Fig. 3E). The aforementioned findings
illustrated that 1,25(OH)2D3 repressed the
osteogenic differentiation of BMSCs through activating β-catenin
signaling.
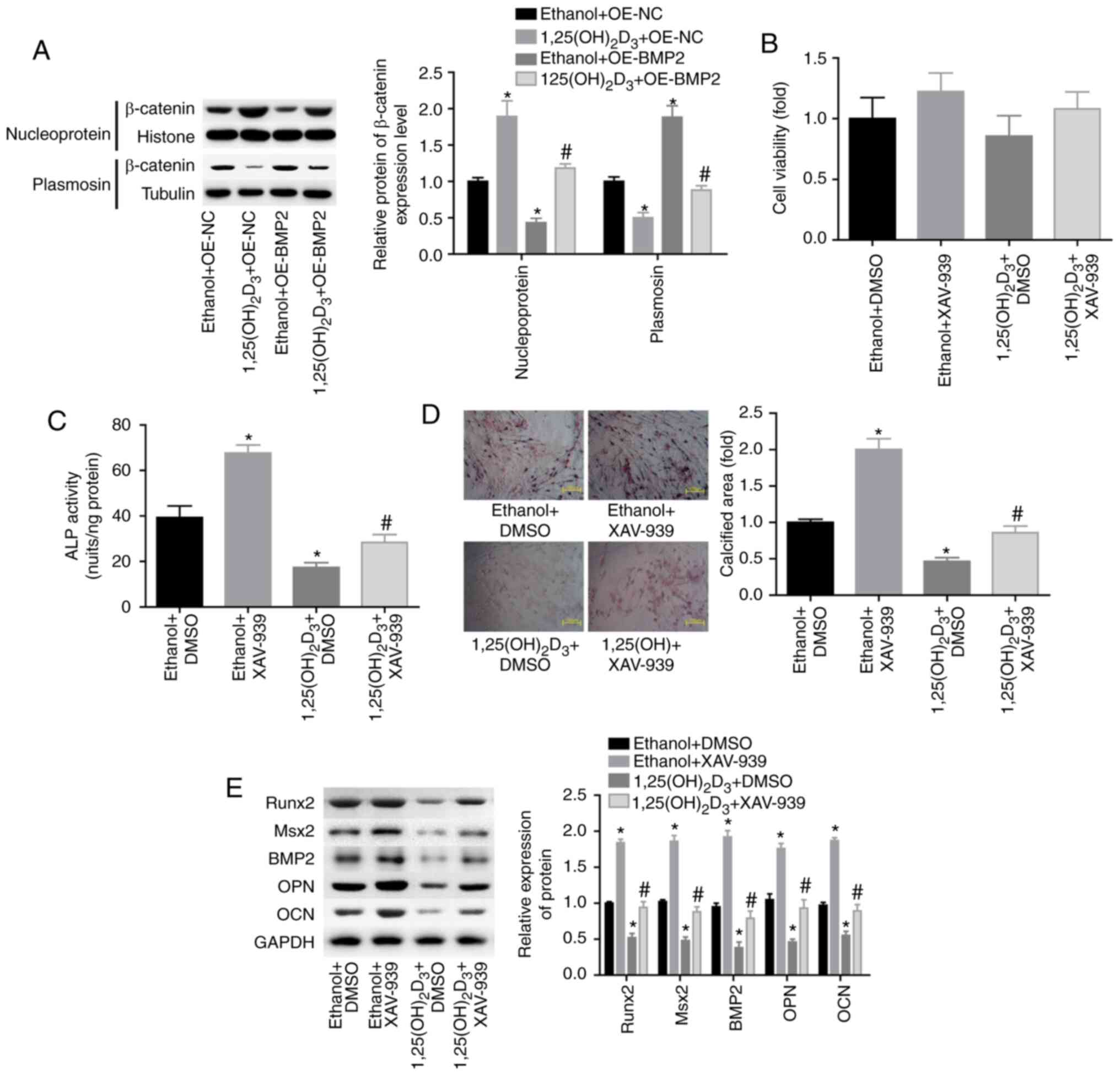 | Figure 3.Inhibition of β-catenin signaling
rescued the repression of the osteogenic differentiation of BMSCs
induced by 1,25(OH)2D3. (A) Western blotting
of the expression of β-catenin in the nucleus and cytoplasm of
BMSCs treated with ethanol plus OE-NC or OE-BMP2, or with
1,25(OH)2D3 (10 nM) plus OE-NC or OE-BMP2.
*P<0.05 vs. ethanol + OE-NC group, #P<0.05 vs.
1,25(OH)2D3 + OE-NC group. (B) Cell viability
determined by Cell Counting Kit-8 assay and (C) ALP activity in
differently treated BMSCs (ethanol plus DMSO or XAV-939, and
1,25(OH)2D3 plus DMSO or XAV-939). (D)
Alizarin red-S was used to assess the calcified nodules of BMSCs.
Magnification, ×200. (E) Western blotting of the protein expression
levels of BMP2, Runx2, Msx2, OPN and OCN after BMSCs were treated
with ethanol plus DMSO or XAV-939, and with
1,25(OH)2D3 plus DMSO or XAV-939. (n=3). C-E,
*P<0.05 vs. ethanol + DMSO group; #P<0.05 vs.
1,25(OH)2D3 + DMSO group. BMSCs, bone
marrow-derived mesenchymal stem cells; BMP2, bone morphogenetic
protein 2; ALP, alkaline phosphatase; Runx2, Runt-related
transcription factor 2; Msx2, Msh homeobox 2; OPN, osteopontin;
OCN, osteocalcin; OE, overexpressing vector; NC, negative
control. |
1,25(OH)2D3
represses the osteogenic differentiation through impairing the
interaction between Dvl-1 and Smad1 proteins via β-catenin
As Wnt/β-catenin signaling closely interacts with
BMP signaling pathway (24), the
present study further explored the role of the interaction between
β-catenin and BMP2 signaling pathways in the
1,25(OH)2D3-mediated inhibition of the
osteogenic differentiation of BMSCs. Following transfection with
siRNAs targeting the β-catenin gene, si-2 exhibited the highest
knockdown efficiency among the three siRNAs at the mRNA and protein
levels (Fig. 4A and B); therefore,
si-2 was selected for use in further experiments. IP assay revealed
that 1,25(OH)2D3 treatment evidently reduced
the crosstalk between Dvl-1 and Smad1 protein, while knockdown of
β-catenin significantly impaired this effect (Fig. 4C). This suggested that the weakness
in Dvl-1 and Smad1 interaction may serve a vital role in
1,25(OH)2D3-induced osteogenic
differentiation repression. To further explore this interaction,
BMSCs were transfected with OE-Smad1 to upregulate Smad1
expression. As shown in Fig. 4D and
E, the mRNA and protein expression levels of Smad1 were
significantly increased when BMSCs were transfected with OE-Smad1,
while it had no evident influence on cell viability (Fig. 4F). In contrast to the
1,25(OH)2D3 treatment, Smad1 upregulation
significantly increased the ALP activity and calcified area in
BMSCs, and impaired the effect of 1,25(OH)2D3
on osteogenic differentiation repression (Fig. 4G and H). These findings
demonstrated that 1,25(OH)2D3 repressed the
osteogenic differentiation of BMSCs through regulating the
interaction between BMP and β-catenin signaling pathways.
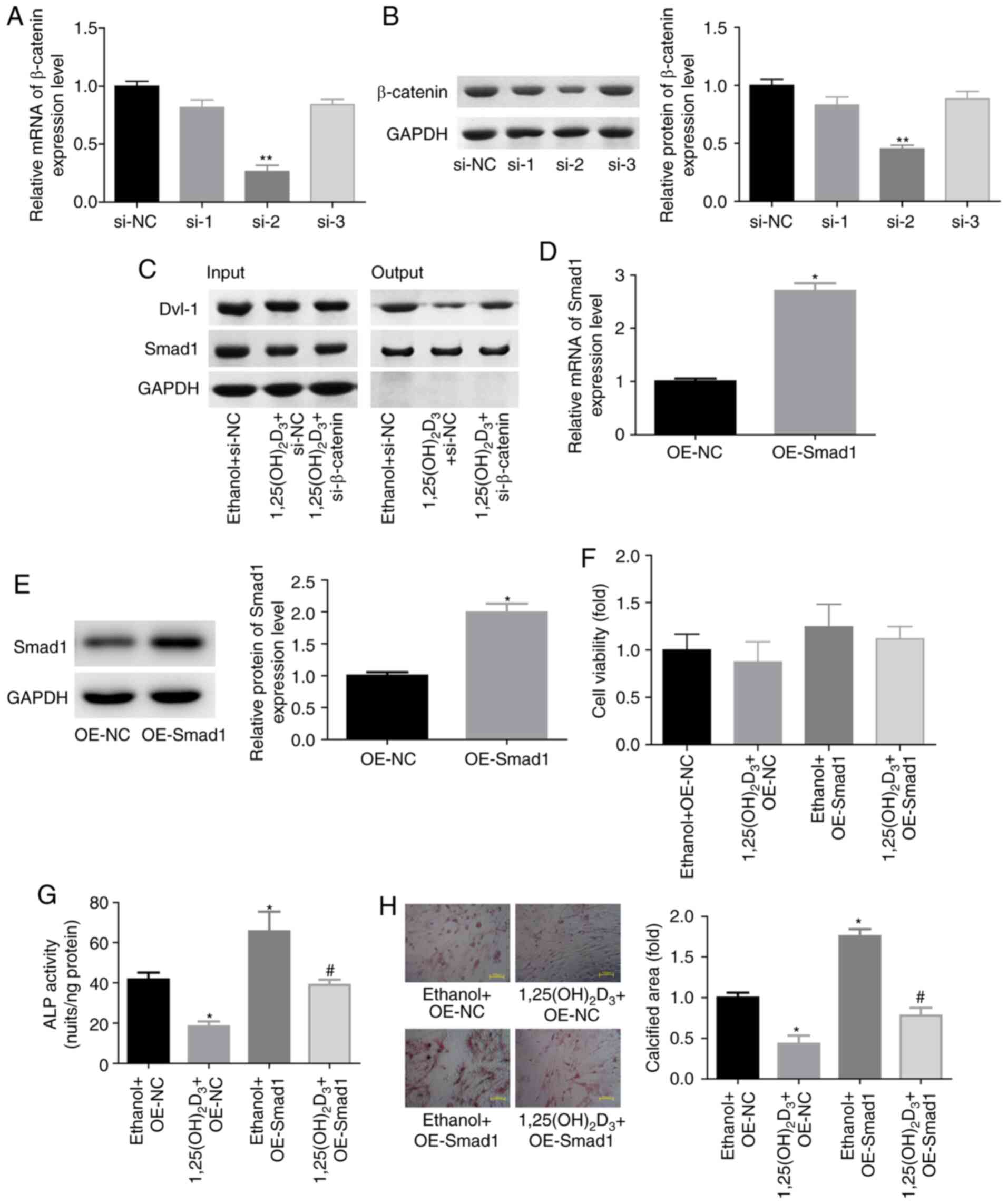 | Figure 4.
1,25(OH)2D3/β-catenin repressed the
osteogenic differentiation of BMSCs through impairing the
interaction between Dvl-1 and Smad1 proteins. (A) mRNA and (B)
protein levels of β-catenin in BMSCs transfected with siRNAs were
assessed by RT-qPCR and western blotting, respectively. **P<0.01
vs. si-NC group. (C) Immunoprecipitation assay, evaluating the
crosstalk between Dvl-1 and Smad1 protein in BMSCs treated with
1,25(OH)2D3 (10 nM, 48 h) with or without
si-β-catenin transfection. (D) mRNA and (E) protein expression
levels of Smad1 in BSMCs stably infected with OE-Smad1 or OE-NC
were determined by RT-qPCR and western blotting, respectively.
*P<0.05 vs. OE-NC group. (F) BMSC viability determined by Cell
Counting Kit-8 assay, and (G) ALP activity in BMSCs. *P<0.05 vs.
ethanol + OE-NC group, #P<0.05 vs.
1,25(OH)2D3 + OE-NC group. (H) Alizarin red-S
was used to assess the calcified nodules of BMSCs. Magnification,
×200. *P<0.05 vs. ethanol + OE-NC group, #P<0.05
vs. 1,25(OH)2D3 + OE-NC group. (n=3). BMSCs,
bone marrow-derived mesenchymal stem cells; Dvl-1, disheveled-1;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; si-/siRNA, small interfering RNA; ALP, alkaline
phosphatase; OE, overexpressing vector; NC, negative control. |
Discussion
The imbalance in bone formation and bone resorption
induced by the inactivity of osteoblasts and the hyperactivation of
osteoclasts is considered as the main reason of osteoporosis
(21,25). Therefore, the presence of a
sufficient number of osteoblasts is beneficial for osteoporosis
prevention or even treatment (26). In the present study, the effects of
BMPs and Wnt/β-catenin signaling on
1,25(OH)2D3-mediated repression of the
osteogenic differentiation of BMSCs was investigated. The study
results demonstrated that 1,25(OH)2D3
treatment induced a significant inhibition in the osteogenic
differentiation of BMSCs, including reduced the expression levels
of several ossification markers (27), including Runx2, Msx2, BMP2, OPN and
OCN. Furthermore, 1,25(OH)2D3 decreased ALP
activity and the calcified area of BMSCs, and these effects were
induced by downregulating BMP2 and activating β-catenin
signaling.
It is well documented that Wnt/β-catenin and BMP
signaling pathways are essential for BMSCs to differentiate into
osteoblasts (17,28). To further comprehend the role of
β-catenin and BMP signaling pathways in
1,25(OH)2D3-induced osteogenic
differentiation repression, the present study initially explored
the effects of 1,25(OH)2D3 on the expression
of BMP2, which serves as an osteogenic activation factor through
stimulating osteoblast differentiation and osteogenic nodule
formation (28,29). The current study results indicated
that 1,25(OH)2D3 negatively regulated BMP2
expression, not only at the transcriptional level, but also at the
translational level. The decrease in the expression levels of
ossification markers (Runx2, Msx2, OPN, OCN), ALP activity and the
calcified area of BMSCs induced by
1,25(OH)2D3 treatment were rescued when BMP2
expression was upregulated, suggesting that BMP2 repression was
strongly implicated in the
1,25(OH)2D3-induced repression of the
osteogenic differentiation of BMSCs.
Previous evidence demonstrated that the
1,25(OH)2D3 role depends on its combination
with its receptor VDR. Once 1,25(OH)2D3 binds
to VDR, this receptor will heterodimerize with the retinoid X
receptor and translocate into the nucleus to combine with vitamin
D3 responsive elements (VDREs) in the promoter regions of the
target genes of VDR, leading to upregulation or downregulation of
gene transcription (30). Tagami
et al (31) revealed that
certain corepressors were able to block the VDRE in VDR target
genes and deacetylate histones in the absence of
1,25(OH)2D3, suggesting that
1,25(OH)2D3 can regulate target gene
expression through VDREs. Notably, a study by Fu et al
(11) demonstrated that inhibition
of histone deacetylase using trichostatin A and DNA
methyltransferase using 5-aza-29-deoxycytidine increased BMP2
expression. In addition, 1,25(OH)2D3
treatment significantly increased the levels of H3K9me2 and reduced
the acetylation of histone H3 at the same BMP2 promoter region in
UMR-106 cells (11). Consistently,
the present study also demonstrated that
1,25(OH)2D3 treatment decreased the
acetylation of histone H3 and increased the expression of its
methylated modification H3K9me3, suggesting that
1,25(OH)2D3 negatively regulates BMP2
expression at the epigenetic level.
The present study further investigated the effects
of the 1,25(OH)2D3/BMP2 axis on Wnt/β-catenin
signaling activation. It was observed that
1,25(OH)2D3 treatment evidently enhanced the
nuclear accumulation of β-catenin protein, while BMP2
overexpression reversed this effect, suggesting that
1,25(OH)2D3 activated β-catenin signaling
through downregulating BMP2. However, the results of the present
study were the opposite from those reported by Larriba et al
(32), who indicated that
1,25(OH)2D3 functioned as a multilevel
repressor of Wnt/β-catenin signaling in cancer cells, particularly
in colon cancer. This divergence may be caused by the different
cell contents and different usage concentrations. In detail, both
Palmer et al (33) and
Larriba et al (34) used a
1,25(OH)2D3 concentration of 10−7
M in colon cancer cells, while the present study applied a
concentration of 10−8 M
1,25(OH)2D3 in BMSCs. Furthermore, in the
current study, it was observed that inhibition of β-catenin
signaling with XAV-939 significantly weakened the effects of
1,25(OH)2D3 on the inhibition of ALP activity
and calcification formation, indicating that
1,25(OH)2D3 inhibited the differentiation of
BMSCs to osteoblasts through activating β-catenin signaling
pathway. Although a number of studies have demonstrated that
enhanced Wnt/β-catenin signaling promotes bone formation (35), there are also researchers who
reported the opposite effect, that is, activation of the
Wnt/β-catenin pathway weakens osteogenic differentiation (36,37).
Increasing evidence has suggested that
Wnt/β-catenin signaling closely interacts with the BMP signaling
pathway. For instance, Haramis et al (24) demonstrated that inhibition of
BMP2/4 signaling in the intestine increased polyp formation with
β-catenin upregulation. Derfoul et al (38) further reported that BMP2 reversed
Wnt3a-induced inhibition of OCN and OPN expression in C3H10T1/2
cells. In addition, BMP2 markedly reduced Wnt3a-induced β-catenin
nuclear accumulation and BMSC proliferation through enhancing the
binding of Smad1 and Dvl-1, which is required for β-catenin
activation (39–41). As reported in the present study,
repression of β-catenin signaling by XAV-939 treatment neutralized
the 1,25(OH)2D3-mediated reduction in BMP2
expression, suggesting that 1,25(OH)2D3
negatively regulated BMP2 expression via activating β-catenin.
Furthermore, BMP2 was able to enhance the interaction between Smad1
and Dvl-1, and promote the activation of β-catenin (39); it can thus be speculated that
1,25(OH)2D3 may modulate the interaction
between Dvl-1 and Smad1 via β-catenin. In accordance with our
predictions, it was observed that knockdown of β-catenin
neutralized the 1,25(OH)2D3-mediated decrease
in the interaction between Dvl-1 and Smad1 proteins. This result
suggested that the weakness in the interaction between Dvl-1 and
Smad1 proteins may be involved in
1,25(OH)2D3-mediated repression of osteogenic
differentiation, which was further confirmed by the ALP activity
detection and Alizarin red-S staining assays in BMSCs with Smad1
overexpression.
In conclusion, the present study revealed that
1,25(OH)2D3 inhibited the differentiation of
BMSCs into osteoclast-like cells through inactivating BMPs and
activating Wnt/β-catenin signaling. The study provides a deeper
understanding on the mechanisms of vitamin D in the inhibition of
osteogenic differentiation, as well as reconsiders the role of
vitamin D in osteoporosis treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
GC contributed to the design of the study and
revised the manuscript. XH performed the experiments and data
analysis, and wrote the manuscript. NZ and YW performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Experiments in this study involving animals were
approved by the Review Committee for the Use of Human or Animal
Subjects of Shanghai Jiao Tong University and were performed in
accordance with the National Institutes of Health Guidelines on the
Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Raisz LG: Pathogenesis of osteoporosis:
Concepts, conflicts, and prospects. J Clin Invest. 115:3318–3325.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanis JA, McCloskey EV, Johansson H and
Oden A: Approaches to the targeting of treatment for osteoporosis.
Nat Rev Rheumatol. 5:425–431. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hayashi M, Nakashima T, Yoshimura N,
Okamoto K, Tanaka S and Takayanagi H: Autoregulation of osteocyte
Sema3A orchestrates estrogen action and counteracts bone aging.
Cell Metab. 29:627–637.e625. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu Y, Newman H, Shen L, Sharma D, Hu G,
Mirando AJ, Zhang H, Knudsen E, Zhang GF, Hilton MJ and Karner CM:
Glutamine metabolism regulates proliferation and lineage allocation
in skeletal stem cells. Cell Metab. 29:966–978 e964. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shoback D: Update in osteoporosis and
metabolic bone disorders. J Clin Endocrinol Metab. 92:747–753.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosen CJ and Bouxsein ML: Mechanisms of
disease: Is osteoporosis the obesity of bone? Nat Clin Pract
Rheumatol. 2:35–43. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosen CJ, Ackert-Bicknell C, Rodriguez JP
and Pino AM: Marrow fat and the bone microenvironment:
Developmental, functional, and pathological implications. Crit Rev
Eukaryot Gene Expr. 19:109–124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan Y, Hanai JI, Le PT, Bi R, Maridas D,
DeMambro V, Figueroa CA, Kir S, Zhou X, Mannstadt M, et al:
Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell
Metab. 25:661–672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren R, Ocampo A, Liu GH and Izpisua
Belmonte JC: Regulation of stem cell aging by metabolism and
epigenetics. Cell Metab. 26:460–474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jurutka PW, Whitfield GK, Hsieh JC,
Thompson PD, Haussler CA and Haussler MR: Molecular nature of the
vitamin D receptor and its role in regulation of gene expression.
Rev Endocr Metab Disord. 2:203–216. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fu B, Wang H, Wang J, Barouhas I, Liu W,
Shuboy A, Bushinsky DA, Zhou D and Favus MJ: Epigenetic regulation
of BMP2 by 1,25-dihydroxyvitamin D3 through DNA methylation and
histone modification. PLoS One. 8:e614232013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
DeLuca HF: The vitamin D story: A
collaborative effort of basic science and clinical medicine. FASEB
J. 2:224–236. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Erben RG, Scutt AM, Miao D, Kollenkirchen
U and Haberey M: Short-term treatment of rats with high dose
1,25-dihydroxyvitamin D3 stimulates bone formation and increases
the number of osteoblast precursor cells in bone marrow.
Endocrinology. 138:4629–4635. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Erben RG, Bromm S and Stangassinger M:
Therapeutic efficacy of 1alpha,25-dihydroxyvitamin D3 and calcium
in osteopenic ovariectomized rats: Evidence for a direct anabolic
effect of 1alpha,25-dihydroxyvitamin D3 on bone. Endocrinology.
139:4319–4328. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sooy K, Sabbagh Y and Demay MB:
Osteoblasts lacking the vitamin D receptor display enhanced
osteogenic potential in vitro. J Cell Biochem. 94:81–87. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamaguchi M and Weitzmann MN: High dose
1,25(OH)2D3 inhibits osteoblast mineralization in vitro. Int
J Mol Med. 29:934–938. 2012.PubMed/NCBI
|
|
17
|
Monroe DG, McGee-Lawrence ME, Oursler MJ
and Westendorf JJ: Update on Wnt signaling in bone cell biology and
bone disease. Gene. 492:1–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu N, Shi S, Deng M, Tang L, Zhang G, Liu
N, Ding B, Liu W, Liu Y, Shi H, et al: High levels of β-catenin
signaling reduce osteogenic differentiation of stem cells in
inflammatory microenvironments through inhibition of the
noncanonical Wnt pathway. J Bone Miner Res. 26:2082–2095. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mo J, Yang R, Li F, He B, Zhang X, Zhao Y,
Shen Z and Chen P: Geraniin promotes osteogenic differentiation of
bone marrow mesenchymal stem cells (BMSCs) via activating
β-catenin: A comparative study between BMSCs from normal and
osteoporotic rats. J Nat Med. 73:262–272. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gnecchi M and Melo LG: Bone marrow-derived
mesenchymal stem cells: Isolation, expansion, characterization,
viral transduction, and production of conditioned medium. Methods
Mol Biol. 482:281–294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li P, Kong J, Chen Z, Huang S, Lv G, Wei
B, Wei J, Jing K, Quan J and Chu J: Aloin promotes osteogenesis of
bone-marrow-derived mesenchymal stem cells via the ERK1/2-dependent
Runx2 signaling pathway. J Nat Med. 73:104–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshida M, Ishimura A, Terashima M,
Enkhbaatar Z, Nozaki N, Satou K and Suzuki T: PLU1 histone
demethylase decreases the expression of KAT5 and enhances the
invasive activity of the cells. Biochem J. 437:555–564. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haramis AP, Begthel H, van den Born M, van
Es J, Jonkheer S, Offerhaus GJ and Clevers H: De novo crypt
formation and juvenile polyposis on BMP inhibition in mouse
intestine. Science. 303:1684–1686. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
You L, Pan L, Chen L, Gu W and Chen J:
MiR-27a is Essential for the Shift from osteogenic differentiation
to adipogenic differentiation of mesenchymal stem cells in
postmenopausal osteoporosis. Cell Physiol Biochem. 39:253–265.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tella SH and Gallagher JC: Prevention and
treatment of postmenopausal osteoporosis. J Steroid Biochem Mol
Biol. 142:155–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou W, Greenblatt MB, Brady N, Lotinun S,
Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al: The
microtubule-associated protein DCAMKL1 regulates osteoblast
function via repression of Runx2. J Exp Med. 210:1793–1806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang EA, Rosen V, D'Alessandro JS, Bauduy
M, Cordes P, Harada T, Israel DI, Hewick RM, Kerns KM, LaPan P, et
al: Recombinant human bone morphogenetic protein induces bone
formation. Proc Natl Acad Sci USA. 87:2220–2224. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harris SE, Bonewald LF, Harris MA,
Sabatini M, Dallas S, Feng JQ, Ghosh-Choudhury N, Wozney J and
Mundy GR: Effects of transforming growth factor beta on bone nodule
formation and expression of bone morphogenetic protein 2,
osteocalcin, osteopontin, alkaline phosphatase, and type I collagen
mRNA in long-term cultures of fetal rat calvarial osteoblasts. J
Bone Miner Res. 9:855–863. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leyssens C, Verlinden L and Verstuyf A:
Antineoplastic effects of 1,25(OH)2D3 and its analogs in breast,
prostate and colorectal cancer. Endocr Relat Cancer. 20:R31–R47.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tagami T, Lutz WH, Kumar R and Jameson JL:
The interaction of the vitamin D receptor with nuclear receptor
corepressors and coactivators. Biochem Biophys Res Commun.
253:358–363. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Larriba MJ, González-Sancho JM, Barbáchano
A, Niell N, Ferrer-Mayorga G and Muñoz A: Vitamin D is a multilevel
repressor of Wnt/b-catenin signaling in cancer cells. Cancers
(Basel). 5:1242–1260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pálmer HG, González-Sancho JM, Espada J,
Berciano MT, Puig I, Baulida J, Quintanilla M, Cano A, de Herreros
AG, Lafarga M and Muñoz A: Vitamin D(3) promotes the
differentiation of colon carcinoma cells by the induction of
E-cadherin and the inhibition of beta-catenin signaling. J Cell
Biol. 154:369–387. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Larriba MJ, Valle N, Pálmer HG,
Ordóñez-Morán P, Alvarez-Díaz S, Becker KF, Gamallo C, de Herreros
AG, González-Sancho JM and Muñoz A: The inhibition of
Wnt/beta-catenin signalling by 1alpha,25-dihydroxyvitamin D3 is
abrogated by Snail1 in human colon cancer cells. Endocr Relat
Cancer. 14:141–151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang J, Sonoyama W, Wang Z, Jin Q, Zhang
C, Krebsbach PH, Giannobile W, Shi S and Wang CY: Noncanonical
Wnt-4 signaling enhances bone regeneration of mesenchymal stem
cells in craniofacial defects through activation of p38 MAPK. J
Biol Chem. 282:30938–30948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo X, Li HX, Liu RX, Wu ZS, Yang YJ and
Yang GS: Beta-catenin protein utilized by Tumour necrosis
factor-alpha in porcine preadipocytes to suppress differentiation.
BMB Rep. 42:338–343. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Daoussis D, Andonopoulos AP and Liossis
SN: Wnt pathway and IL-17: Novel regulators of joint remodeling in
rheumatic diseases. Looking beyond the RANK-RANKL-OPG axis. Semin
Arthritis Rheum. 39:369–383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Derfoul A, Carlberg AL, Tuan RS and Hall
DJ: Differential regulation of osteogenic marker gene expression by
Wnt-3a in embryonic mesenchymal multipotential progenitor cells.
Differentiation. 72:209–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bilic J, Huang YL, Davidson G, Zimmermann
T, Cruciat CM, Bienz M and Niehrs C: Wnt induces LRP6 signalosomes
and promotes dishevelled-dependent LRP6 phosphorylation. Science.
316:1619–1622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niehrs C and Shen J: Regulation of Lrp6
phosphorylation. Cell Mol Life Sci. 67:2551–2562. 2010. View Article : Google Scholar : PubMed/NCBI
|