Introduction
Atherosclerosis is the leading cause of disability
and death worldwide, resulting in ~17.3 million deaths, which is
expected to exceed 23.6 million each year by 2030 (1–3).
Atherosclerosis is the pathological basis of the majority of
cardiovascular diseases and poses a serious threat to human health
(4). After cardiovascular and
cerebrovascular diseases, peripheral arterial disease (PAD) is the
third most common cause of morbidity among individuals with
atherosclerosis (5). PAD
frequently occurs in the arteries of the lower extremities and
occlusive atherosclerosis impairs the blood supply to the lower
extremities (6). Accumulating
evidence has revealed that a variety of long non-coding RNAs
(lncRNAs) participate in the onset and progress of atherosclerosis,
and are involved in multiple pathological processes and signaling
pathways, suggesting that lncRNAs may serve a vital role in
atherosclerosis (7–12). For example, silencing the lnc RNA
myocardial infarction associated transcript inhibits endothelial
cell proliferation, migration and tube formation (13). Another example is that
lncRNA-DYNLRB2-2 not only accelerates cholesterol efflux, but also
decreases the levels of cellular inflammatory cytokines [such as
interleukin (IL)-6, IL-1β and TNF-α] under hyperlipidemic stress
(14).
lncRNAs, a class of non-coding RNAs, contain >200
nucleotides and were previously considered to be transcriptional
noise (15,16); however, the extensive functions of
lncRNAs have gradually been identified. lncRNAs contribute to a
wide range of vital processes, such as chromatin remodeling,
genomic imprinting, dosage compensation effects, gene expression,
post-transcriptional modification of mRNA and regulation of
proteins (17,18). lncRNAs provide a novel layer of
regulation in the mechanism underlying atherosclerosis (8–11).
However, the full repertoire of the physiological and pathological
functions of lncRNAs has not been identified.
lncRNA WEE2 antisense RNA 1 (WEE2-AS1) is the
antisense RNA of the gene encoding WEE1 homolog 2 (WEE2) (19). Although the sequence of lncRNA
WEE2-AS1 has been characterized, its subcellular localization and
biological function are not completely understood (19). M-phase or maturation promoting
factor (MPF) activation is a prerequisite for the cell cycle
transition from the G2 phase to the M phase. WEE2
protein inhibits MPF activity by phosphorylating the Tyr15 residue
of its regulatory subunit, cyclin dependent kinase 1 (CDK1)
(20–22). At present, the identified role of
WEE2 is primarily based on the findings of germ cell research
(23,24). Antisense RNAs may exert positive or
negative influences on sense mRNAs at the post-transcriptional,
processing and mRNA stability levels (25–31).
Therefore, it was hypothesized that altering the expression level
of lncRNA WEE2-AS1 may affect the expression of WEE2, leading to
alterations in WEE2-related functions, especially those associated
with the cell cycle.
Vascular endothelial cells are the primary
regulators for maintaining vascular stability. Endothelial
dysfunction, which is the initiation event of a series of
pathological cascades in atherosclerosis, serves a crucial role in
the pathogenesis of the disease, especially in the initial stage
(1,2). When exposed to various
atherosclerosis risk factors (for example, hyperlipidemia,
hypertension, shear stress and diabetes), the function of vascular
endothelial cells is impaired, leading to certain pathological
alterations, including inhibition of endothelial cell viability and
alterations to the cell cycle (3,5). It
has also been suggested that atherosclerosis is a response of the
vascular wall to endothelial damage (6). Nitric oxide is an effective
vasodilator and anti-inflammatory substance, and endothelial
dysfunction inhibits the production of nitric oxide, leading to a
loss of inhibition of vascular smooth muscle cell (VSMC)
proliferation (32). Extensive
proliferation of VSMCs can cause thickening of blood vessel walls,
which narrows the vascular lumen (33). Endothelial cells, which also
experience dysfunction, express a large number of adhesion
molecules, which can recruit and bind inflammatory cells to
exacerbate vascular damage (34).
Local inflammation induced by endothelial injury promotes plaque
formation and thrombosis (35–40).
A large number of lncRNAs are expressed in endothelial cells and
serve crucial regulatory roles (41). For instance, antisense non-coding
RNA CDKN2B antisense RNA 1 in the INK4 locus preserves the normal
phenotype of endothelial cells by inhibiting the expression of
Kruppel-like factor 2 (11).
Therefore, it was hypothesized that lncRNA WEE2-AS1 may be
associated with endothelial dysfunction.
In the present study, the expression level of lncRNA
WEE2-AS1 in an ASO group and a normal control group was measured.
The function and molecular mechanism underlying lncRNA WEE2-AS1
during the regulation of the cell cycle were also assessed. In
addition, whether antisense lncRNA WEE2-AS1 could influence the
expression of the corresponding protein-coding gene was
investigated.
Materials and methods
Sample acquisition
The present study was approved by the Ethical
Committee of the First Affiliated Hospital of Sun Yat-sen
University and was conducted in accordance with the Declaration of
Helsinki. The participants or their guardian provided written
informed consent before the tissue and blood samples were
obtained.
Arterial samples were obtained from the main artery
of amputated lower limbs at The First Affiliated Hospital of Sun
Yat-sen University (China) between October 2015 and May 2017. A
total of 5 atherosclerotic samples were obtained from patients with
severe lower extremity ASO who had undergone amputation (4 male
patients and 1 female patient; mean age, 67.60 years; age range,
59–75 years). All patients with severe lower extremity ASO were
diagnosed with arteriosclerosis obliterans according to the
guidelines issued by the European Society of Cardiology in 2011
(42). A total of 5 healthy
arterial samples were obtained from donors without atherosclerosis
who had undergone amputation (3 male donors and 2 female donors;
mean age, 52.20 years; age range, 42–62 years) at The First
Affiliated Hospital of Sun Yat-sen University between October 2015
and May 2017.
In addition, blood samples were collected from
patients with severe lower extremity ASO (mean age, 65.93 years;
age range, 53–80 years; 9 male patients and 6 female patients) and
healthy subjects (mean age, 66.13 years; age range, 52–82 years; 9
male patients and 6 female patients). Blood samples were obtained
from the superficial veins of the upper limbs at The First
Affiliated Hospital of Sun Yat-sen University (China) between
January 2015 and November 2017. The blood samples were collected
into tubes containing EDTA as an anticoagulant, and were
centrifuged at 3,000 × g for 10 min at 4°C. The upper plasma was
transferred to the EP tube and immediately frozen at −80°C until
RNA extraction.
Human umbilical cords were obtained from healthy
women (mean age, 30.14 years; age range, 25–35 years) post-delivery
at The First Affiliated Hospital of Sun Yat-sen University between
June 2015 and March 2017.
Isolation and culture of human
umbilical vein endothelial cells (HUVECs)
Human umbilical cords were obtained from healthy
post-delivery women and endothelial cells were immediately isolated
from the umbilical veins as previously described (43). HUVECs were cultured in Endothelial
Cell Basal Medium (EBM-2; Lonza Group, Ltd.) supplemented with 5%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and endothelium growth
medium kit (Lonza Group, Ltd.). Cells were cultured at 37°C in a
humidified environment with 5% CO2. The medium was
changed every 3 days. Passage 1–10 cells were used for subsequent
experiments.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from HUVECs and plasma
samples (250 µl) using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and TRIzol® LS reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), respectively. An
equal volume of plasma sample was used as the internal control
(44). The ultraviolet absorbance
of each sample at a wavelength of 260 and 280 nm was measured to
assess RNA purity and concentration, respectively. Total RNA was
reverse transcribed into cDNA using the PrimeScript™ RT reagent kit
(Takara Bio, Inc.) according to the manufacturer's instructions.
Subsequently, qPCR was performed using SYBR-Green PCR Master Mix
(Roche Diagnostics GmbH) according to the manufacturer's
instructions. The thermocycling conditions used for qPCR consisted
of pre-incubation followed by 40 amplification cycles. Each cycle
comprised 95°C for 10 sec, 60°C for 20 sec, and 72°C for 20 sec.
The following primers were used for qPCR: lncRNA WEE2-AS1 forward,
5′-AGAAATCACCAACCGGCTCA-3′ and reverse, 5′-GAACTTCGCTTTCCCCCTGT-3;
and GAPDH forward, 5′-GCACCGTCAAGGCTGAGAGAAC-3 and reverse,
5′-TGGTGAAGACGCCAGTGGA-3′. The qPCR products were resolved by
agarose gel (1%) electrophoresis to confirm the specificity of the
primers. mRNA expression levels were quantified using the
2−∆∆Cq method (45) and
normalized to the internal reference gene GAPDH.
Hematoxylin and eosin (H&E)
staining
Artery tissue samples were immediately fixed in 4%
paraformaldehyde at room temperature for 1 day, embedded in
paraffin and cut into 4-µm thick sections. The sections were
mounted on slides and heated at 62°C for ~4 h. Subsequently, the
slides were deparaffinized, air-dried and stained with hematoxylin
at room temperature for ~5 min. The slides were washed in running
water, rinsed with ammonia water and washed again in running water.
After counterstaining with eosin at room temperature for ~5 min,
the slides were dehydrated using ascending ethanol concentrations
(95–100%). The coverslips were positioned using a xylene-based
mountant. Slides were observed under a light microscope and scanned
by the Panoramic MIDI scanner (3DHISTECH Kft.). Images were
captured using CaseViewer (Version 2.0; 3DHISTECH Kft.) at ×35 or
×200 magnification.
Oil Red O Staining
Artery tissue samples preserved in liquid nitrogen
were cut into 4-µm thick sections and mounted onto slides. The
slides were air-dried at room temperature and fixed in 4%
paraformaldehyde for 15 min at room temperature. The slides were
rinsed in distilled water and air-dried. Subsequently, the slides
were stained with Oil Red O solution for 8–10 min at room
temperature and rinsed in distilled water. Slides were
differentiated in 75% ethanol for ~2 sec at room temperature,
washed in distilled water and stained with hematoxylin for 3–5 min
at room temperature. Subsequently, the slides were rinsed in
running water for 3 min and mounted using glycerin jelly. Slides
were observed under a light microscope and scanned by the Panoramic
MIDI scanner. Images were captured using CaseViewer at ×35 or ×200
magnification.
Fluorescence in situ hybridization
(FISH)
The sequence of the lncWEE2-AS1 probe used for FISH
was 5′-GCCCGCTTCTTGCACATCTTACTC-3′. The lncWEE2-AS1 probe, which
was labeled with Cy3, was synthesized by Servicebio. Artery tissue
samples were immediately fixed in 4% paraformaldehyde at room
temperature for 1 day, and embedded in paraffin. The artery tissue
samples imbedded in paraffin wax were cut into 4-µm thick sections.
The sections were mounted on slides and baked. At room temperature,
the slides were deparaffinized at room temperature as follows: 100%
xylene for 15 min, 100% xylene for 15 min, 100% ethanol for 5 min,
100% ethanol for 5 min, 85% ethanol for 5 min, 75% ethanol for 5
min and diethyl pyrocarbonate-treated water for 5 min.
Subsequently, the slides were air-dried. The slides were boiled in
antigen retrieval buffer (citrate) for 10 min and cooled naturally.
The tissues were digested with protease K (Servicebio, Inc.) at
37°C for 30 min. After washing with pure water, the slides were
washed three times in PBS for 5 min each time. After preheating to
the hybridization temperature, the probes (8 ng/µl) were applied to
the slides and incubated at 37°C overnight. Post-hybridization
washes were performed using sodium chloride-sodium citrate buffer
at room temperature for 10 min. The slides were then incubated with
DAPI for 10 min at room temperature to stain the nuclei and then
mounted with anti-fade fluorescence mountant. Images were obtained
using a fluorescence microscope at ×400 or ×900 magnification and
analyzed using ImageJ software (version 6.0; National Institutes of
Health).
For FISH using cells, the cell suspension
(1-2×104 cells) was added to a slide and cultured at
37°C for 24 h. The supernatant was removed and the slides were
washed twice with PBS, and then incubated in 4% formaldehyde for 20
min at room temperature. The slides were washed three times with
PBS. The subsequent steps were as performed as aforementioned from
the protease K digestion step.
Construction of a protein-protein
network
The protein-protein interaction network was
constructed and mapped by analyzing data obtained from Kyoto
Encyclopedia of Genes and Genomes (genome.jp/kegg/), Pfam
(pfam.xfam.org/), Search Tool for the Retrieval of Interacting
Genes/Proteins (string-db.org/) and Gene Ontology
(geneontology.org/) databases (46–48).
Immunofluorescence
Slides containing cells were prepared as described
above (up to the 4% formaldehyde incubation step). The slides were
washed with PBS and blocked with 3% bovine serum albumin
(Servicebio, Inc.) for 30 min at room temperature. The slides were
incubated at 4°C overnight with the following primary antibodies:
Mouse anti-platelet-endothelial cell adhesion molecule-1 (CD31;
1:3,000; cat. no. 3528; Cell Signaling Technology, Inc.) and rabbit
anti-von Willebrand Factor (vWF; 1:1,000; cat. no. GB11020;
Servicebio, Inc.). Following washing in PBS three times for 5 min
each, the slides were incubated with fluorophore-tagged secondary
antibodies for 50 min at room temperature in the dark. The
secondary antibodies included FITC-conjugated Goat Anti-Mouse
(1:200; cat. no. GB22301; Servicebio, Inc.) and Anti-Rabbit IgG
(1:200; cat. no. GB22303; Servicebio, Inc.). Subsequently, the
slides were incubated with DAPI for 10 min at room temperature and
mounted with anti-fade fluorescence reagent. Stained slides were
observed using a fluorescence microscope. Slides were scanned by
the Panoramic MIDI scanner. Images were captured using CaseViewer
at ×200 or ×900 magnification.
Small interfering (si)RNA
transfection
Cells were seeded into 6-well plates and incubated
for 24 h. At 60% confluence, cells were transfected for 10 h with
50 nmol/l lncRNA WEE2-AS1 siRNA (siR-lnc WEE2-AS1) or the negative
control (NC) siRNA (siR-NC) using Lipofectamine® RNAiMAX
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). To knockdown
lncRNA WEE2-AS1, two siRNAs (Suzhou GenePharma Co., Ltd.) were used
with the following sequences: siR-lnc WEE2-AS1#1,
5′-GCCCAUCACAUUUCUCAUUTT-3′; and siR-lnc WEE2-AS1#2:
5′-GCAGCAAGCGACGUUCUUATT-3′. The siR-NC (Suzhou GenePharma Co.,
Ltd.) used as the negative control had the following sequence:
5′-UUCUCCGAACGUGUCACGUTT-3′.
Lentivirus preparation and
infection
Lentiviruses expressing lncRNA WEE2-AS1 (lv-lnc
WEE2-AS1) and negative control (lv-lnc WEE2-AS1-NC) sequences were
prepared by GeneCopoeia, Inc. Cells were infected with lv-lnc
WEE2-AS1 or lv-lnc WEE2-AS1-NC according to the manufacturer's
protocol. Following infection with lv-lnc WEE2-AS1 or lv-lnc
WEE2-AS1-NC at 37°C for 10 h, HUVECs were cultured in selection
medium containing puromycin. Stably infected cells were cultured in
EBM-2 containing puromycin.
Cell viability assay
Cell suspensions (100 µl per well) were added to a
96-well plate. Following culture at 37°C for 72 h, cell viability
was measured using a Cell Counting Kit-8 (CCK-8) assay (Dojindo
Molecular Technologies, Inc.). Briefly, 10 µl CCK-8 reagent was
added to each well and incubated at 37°C for 2 h. The absorbance of
each well was measured at a wavelength of 450 nm using a
spectrophotometer.
Cell cycle analysis
Cells were seeded at 1×105 cells/well in
6-well plates. Following siRNA transfection or lentivirus
infection, flow cytometry was performed to assess the cell cycle
distribution. Following trypsinization, cells were centrifuged at
100 × g for 3 min at room temperature. Then cells were washed with
PBS. The cell concentration was adjusted to 1×106
cells/ml. Subsequently, cells were fixed in 70% cold ethanol at 4°C
overnight. Fixed cells were washed with PBS, centrifuged at 100 × g
for 3 min at room temperature and resuspended. Subsequently, cells
(1×106/ml) were stained using RNase and propidium iodide
buffer (Nanjing KeyGen Biotech Co., Ltd.) for 60 min at room
temperature in the dark. Cell cycle distribution was assessed using
a CytoFLEX flow cytometer (Beckman Coulter, Inc.) and analyzed
using ModFit LT software (version 4.1; Verity Software House,
Inc.).
Western blotting
Total protein was extracted from HUVECs using lysis
buffer (Nanjing KeyGen Biotech Co., Ltd.) and quantified using a
BCA protein assay kit (Nanjing KeyGen Biotech Co., Ltd.). Equal
amounts of protein (40-50 µg) were separated via 8–12% SDS-PAGE and
transferred to PVDF membranes (EMD Millipore). The membranes were
blocked in TBS containing 5% non-fat dry milk for ≥1 h at room
temperature. Subsequently, the membranes were incubated overnight
at 4°C with the following primary antibodies: Anti-cell division
cycle 25B (CDC25B; 1:1,000; cat. no. 9525; Cell Signaling
Technology, Inc.), anti-cyclin dependent kinase 1 (CDK1; 1:1,000;
cat. no. ab32594; Abcam), anti-myelin transcription factor 1 (MYT1;
1:1,000; cat. no. 4282; Cell Signaling Technology, Inc.), anti-CDK1
(phospho Y15; 1:1,000; cat. no. ab47594; Abcam), anti-WEE2 (1:500;
cat. no. ab138162; Abcam), β-actin (1:100,000; cat. no. AC026;
ABclonal Biotech Co., Ltd.) and GAPDH (1:100,000; cat. no. AC036;
ABclonal Biotech Co., Ltd.). Following primary incubation, the
membranes were incubated with a horseradish peroxidase-conjugated
anti-rabbit IgG (1:2,000; cat. no. 7074; Cell Signaling Technology,
Inc.) secondary antibody for 2 h at room temperature.
Immunoreactive proteins were visualized using enhanced
chemiluminescence (EMD Millipore) and photographed using an
Amersham Imager 600 (Cytiva). Protein expression levels were
quantified using ImageJ software (version 6.0; National Institutes
of Health) with β-actin and GAPDH as the loading controls.
Statistical analysis
Data are expressed as the mean ± SD. Statistical
analyses were performed using SPSS software (version 21.0; IBM
Corp.). The paired Student's t-test was used to compare differences
between two groups. One-way ANOVA followed by Tukey's post hoc test
was used to compare differences among multiple groups. P<0.05
was considered to indicate a statistically significant difference.
All experiments were performed in triplicate.
Results
lncRNA WEE2-AS1 expression levels are
upregulated in ASO plasma and artery samples
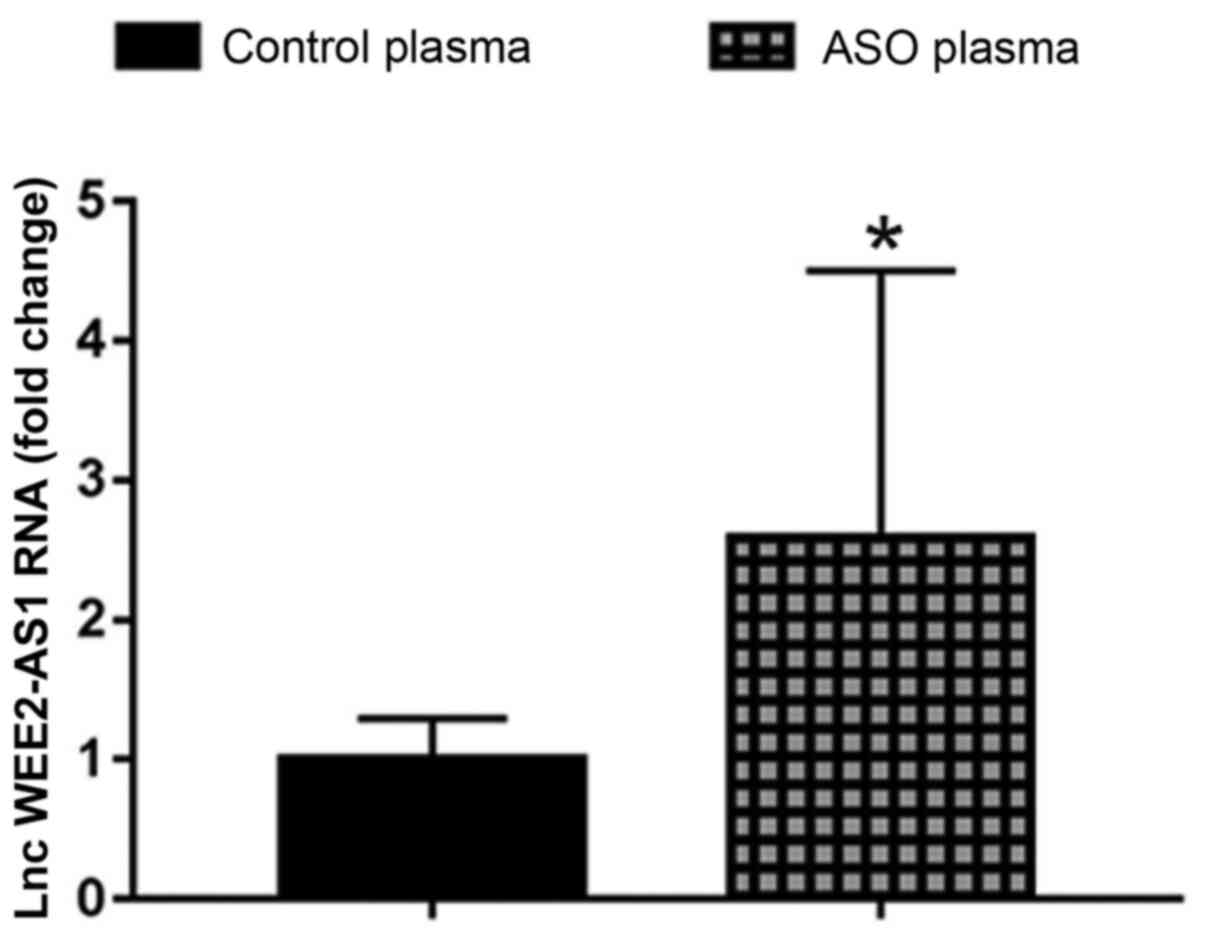
RT-qPCR was performed to evaluate the expression
levels of lncRNA WEE2-AS1 in plasma samples obtained from patients
with ASO and healthy control subjects. The results indicated that
the expression level of lncRNA WEE2-AS1 was significantly
upregulated in the ASO group compared with the healthy control
group (Fig. 1).
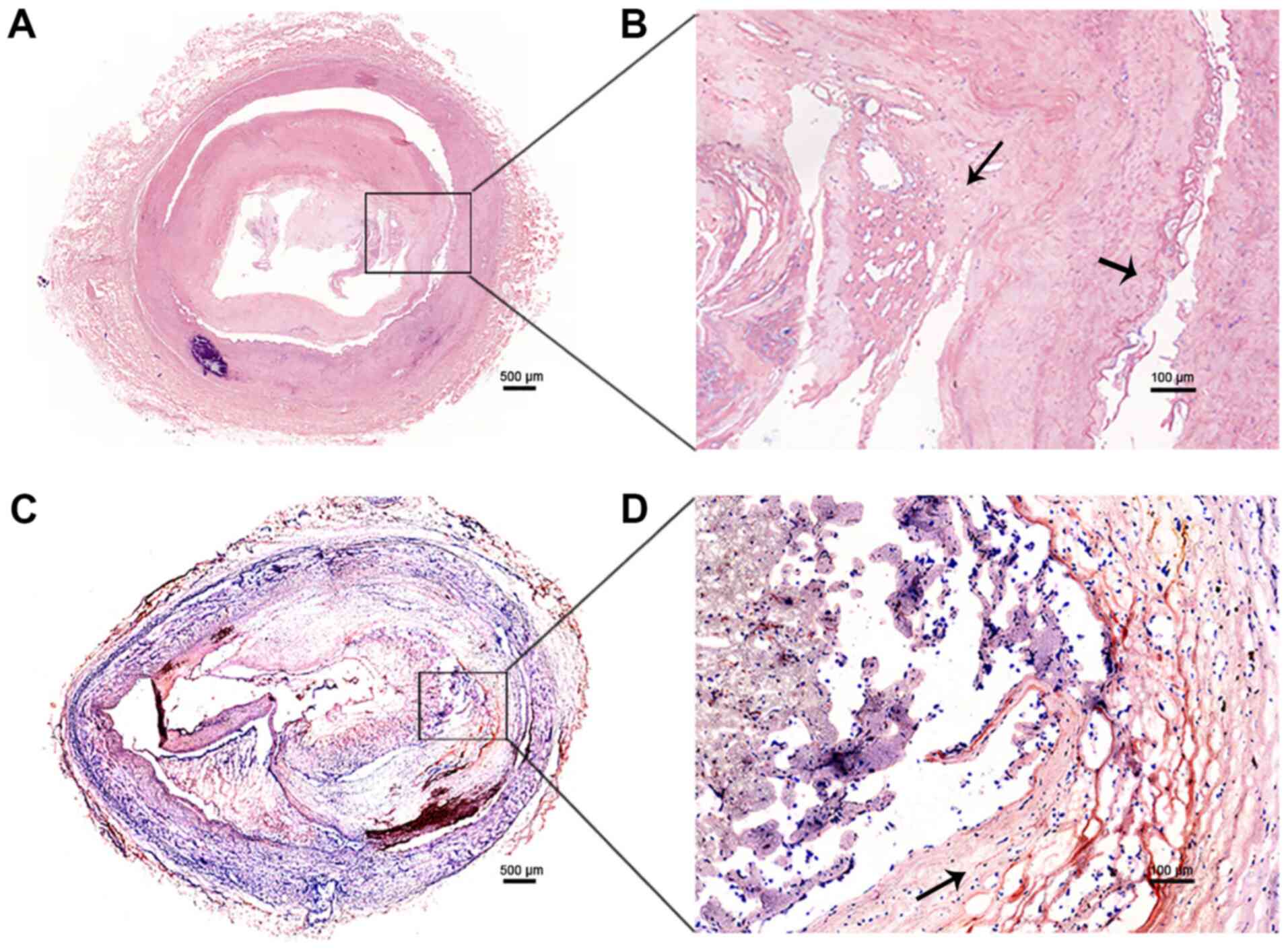
The expression level of lncRNA WEE2-AS1 was further
evaluated between tissue samples obtained from 5 patients with ASO
and 5 healthy subjects. The results of H&E (Fig. 2A and B) and Oil Red O staining
(Fig. 2C and D) indicated that the
ASO tissue samples displayed pathological alterations consistent
with ASO, including thickening and hardening of the blood vessel
walls, artery stenosis, occlusion, endothelial cell injury, VSMC
proliferation, atheromatous plaques, lipid deposition and rupture
of the fiber cap. By contrast, the results of H&E staining
showed no pathological alterations in the healthy artery tissue
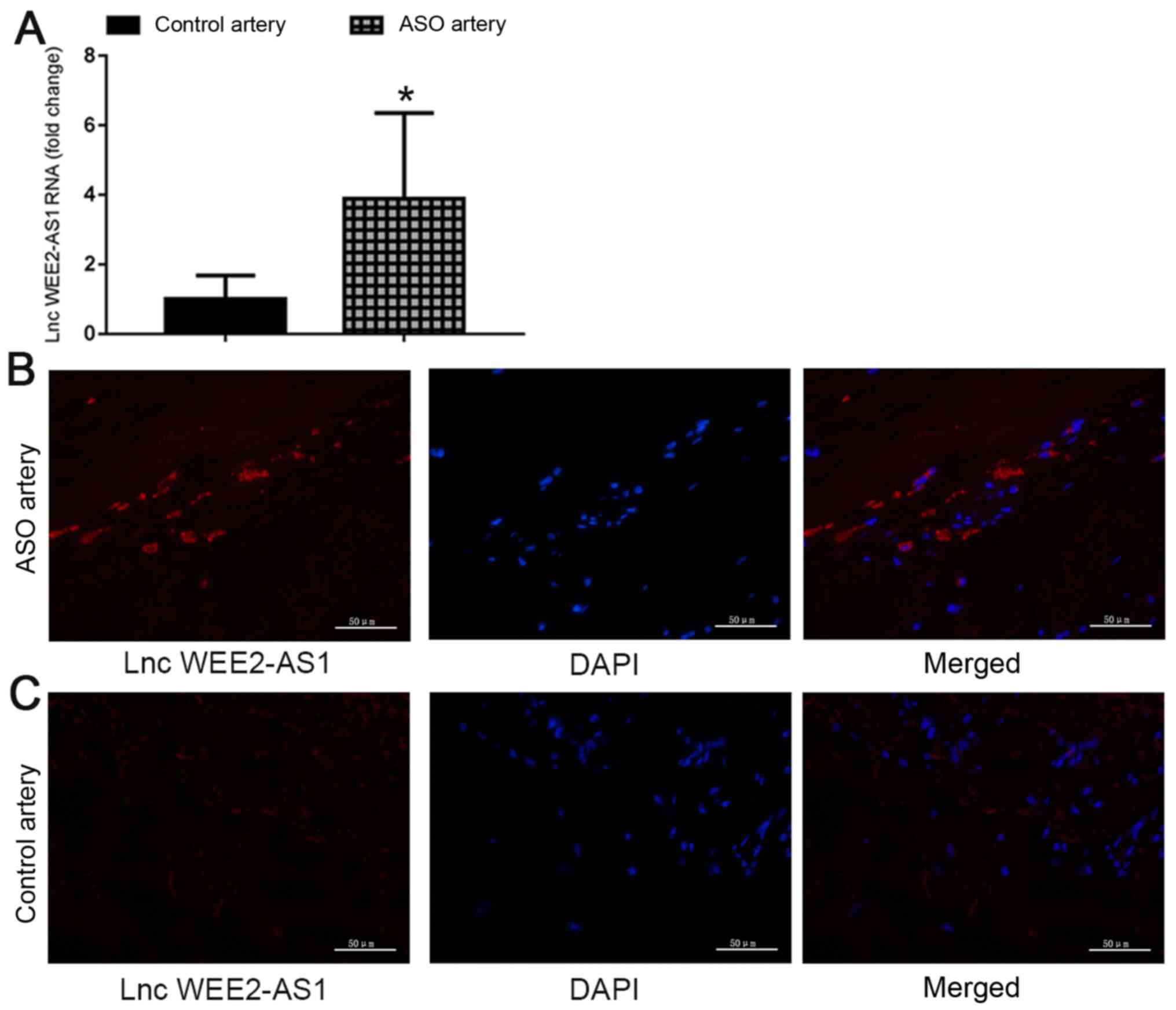
samples. The FISH analysis results suggested that, compared with
healthy control arteries, the signal intensity for lncRNA WEE2-AS1
in the ASO artery group was significantly higher (Fig. 3). Collectively, the results
suggested that the expression level of lncRNA WEE2-AS1 was
significantly upregulated in plasma and artery tissue samples
obtained from patients with ASO compared with healthy control
subjects, which indicated that there might be an association
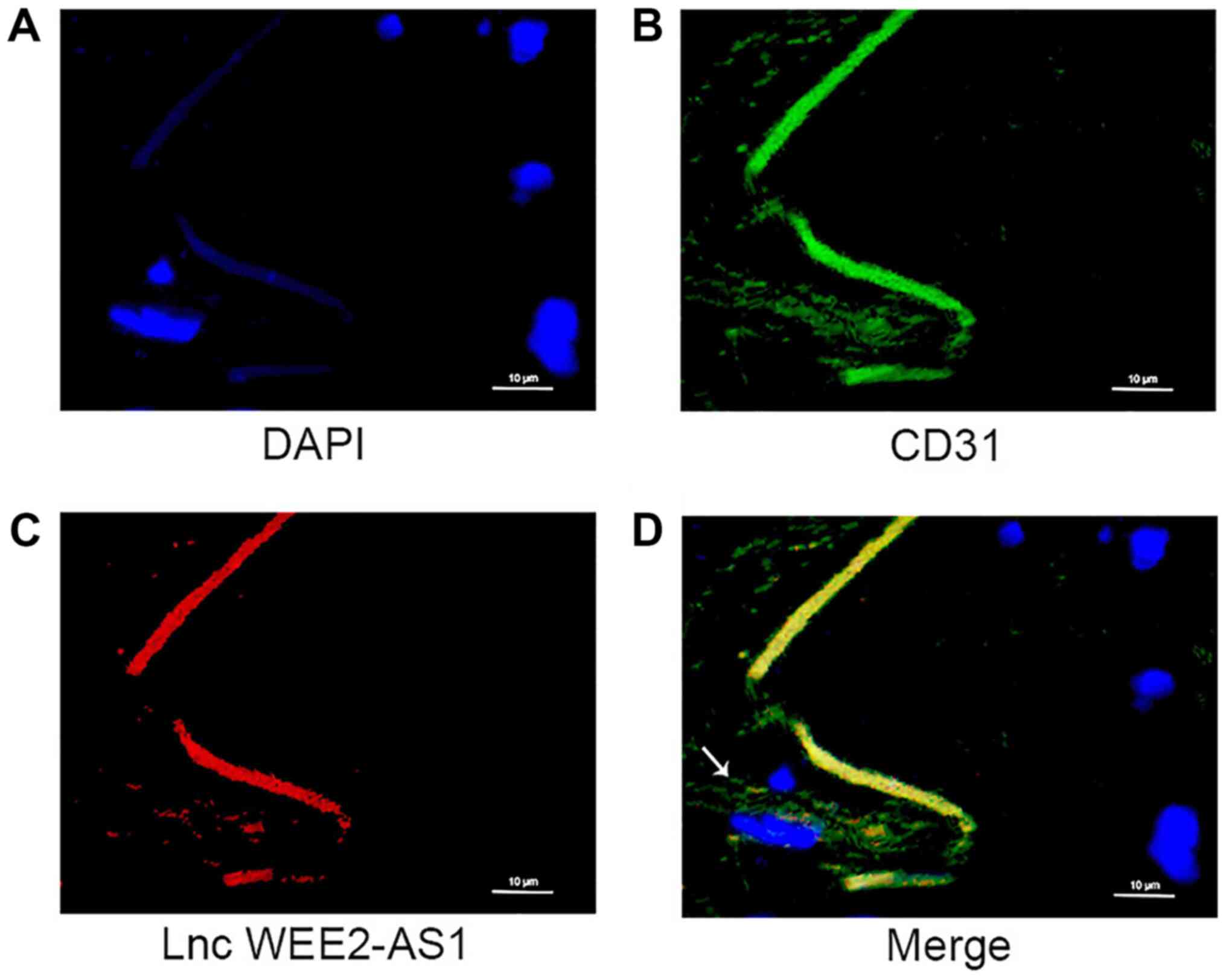
between the expression level of lncRNA WEE2-AS1 and ASO. Moreover,
FISH and immunofluorescence assays indicated that lncRNA WEE2-AS1
co-localized with CD31 in endothelial cells, which suggested that
lncRNA WEE2-AS1 was expressed by human arterial endothelial cells
(Fig. 4).
Subcellular localization of lncRNA
WEE2-AS1 in HUVECs
HUVECs are one of the most important cell models for
the in vitro study of alterations to endothelial cell
function in ASO (49,50); therefore, the present study
investigated the functions of lncRNA WEE2-AS1 in HUVECs.
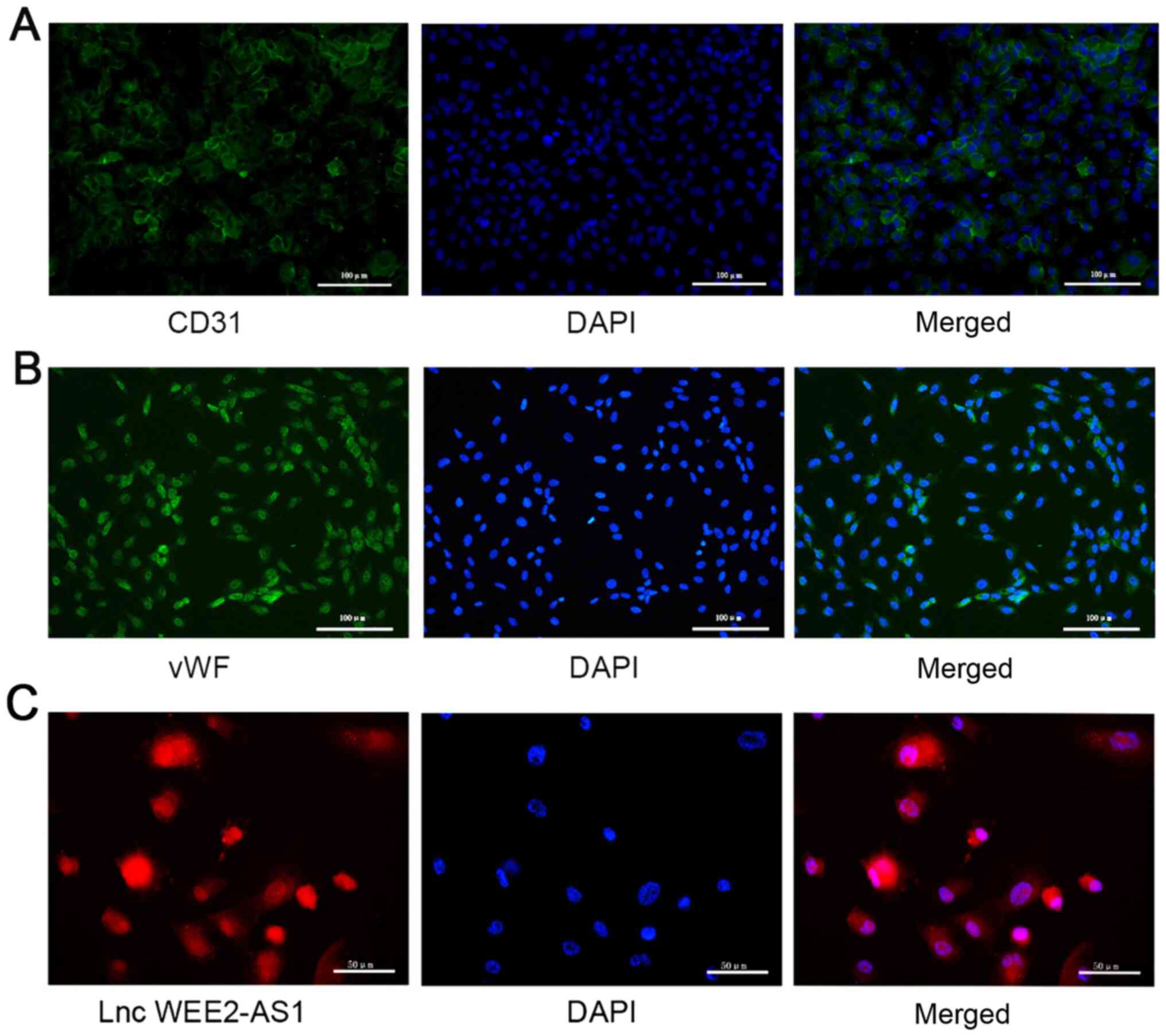
Immunofluorescence was performed to characterize cells isolated
from human umbilical veins. The cells stained positively for
endothelial cell markers, including CD31 and vWF (Fig. 5A and B) (49,51,52),
which indicated that the isolated cells were HUVECs.
FISH was performed to investigate the subcellular
localization of lncRNA WEE2-AS1 in HUVECs. The results indicated
that lncRNA WEE2-AS1 was expressed in the cytoplasm and nuclei of
HUVECs, implying that it may participate in a variety of
physiological and pathological processes (Fig. 5C).
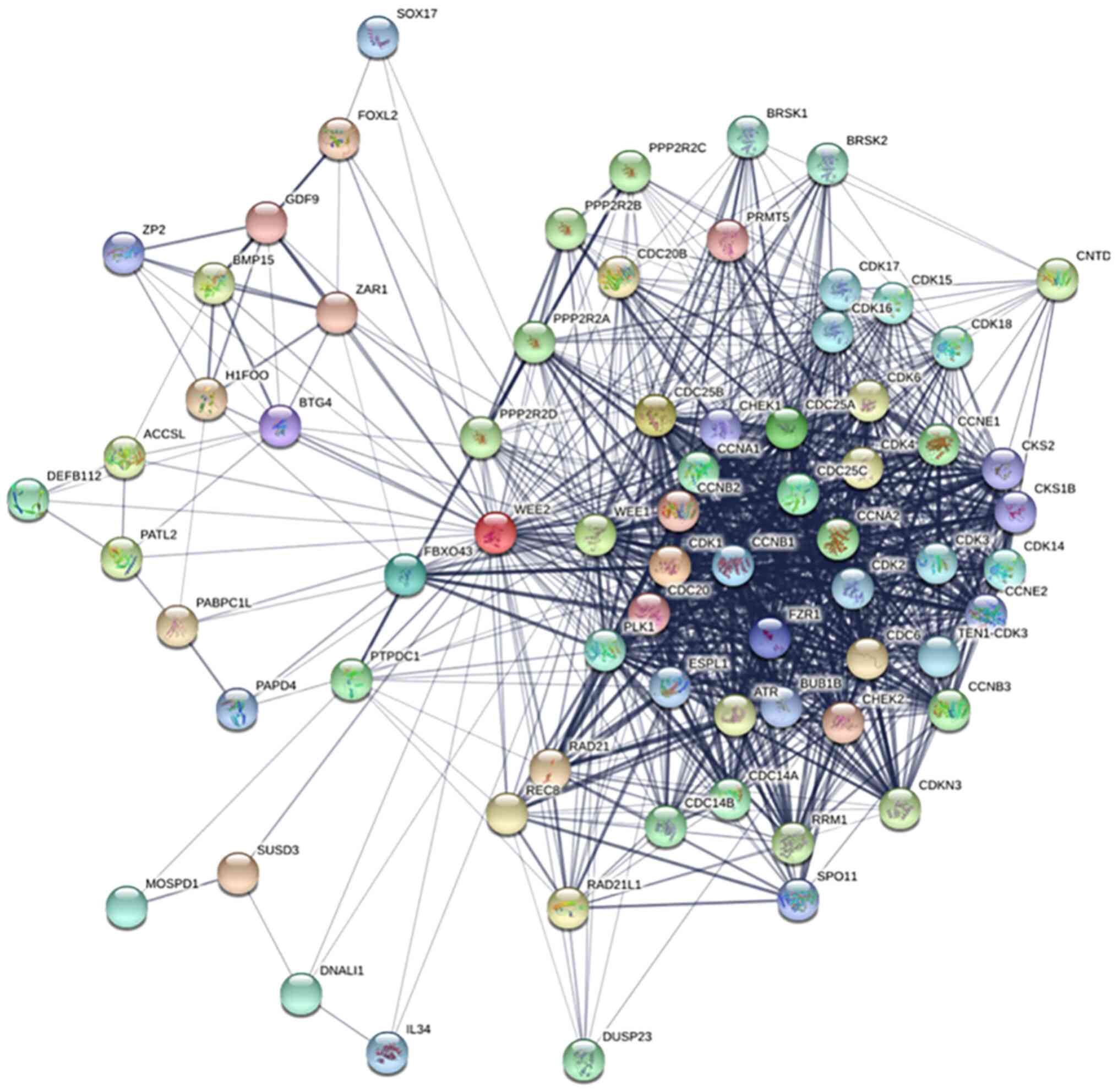
Bioinformatic analysis of the WEE2
protein interaction network
lncRNA WEE2-AS1 is the antisense RNA of the WEE2
gene (19). The protein encoded by
the sense strand has important significance for the prediction of
antisense lncRNAs, which indicates a potential association between
the biological functions of the antisense and sense strands
(53–55). To explore the biological function
of lncRNA WEE2-AS1, a protein-protein interaction network of WEE2
was constructed and mapped (Fig.
6). Analysis of the network suggested that WEE2 participated in
several critical pathways, including cell viability, mitotic cell
cycle and the G2/M checkpoint.
lncRNA WEE2-AS1 regulates HUVEC
viability and cell cycle progression
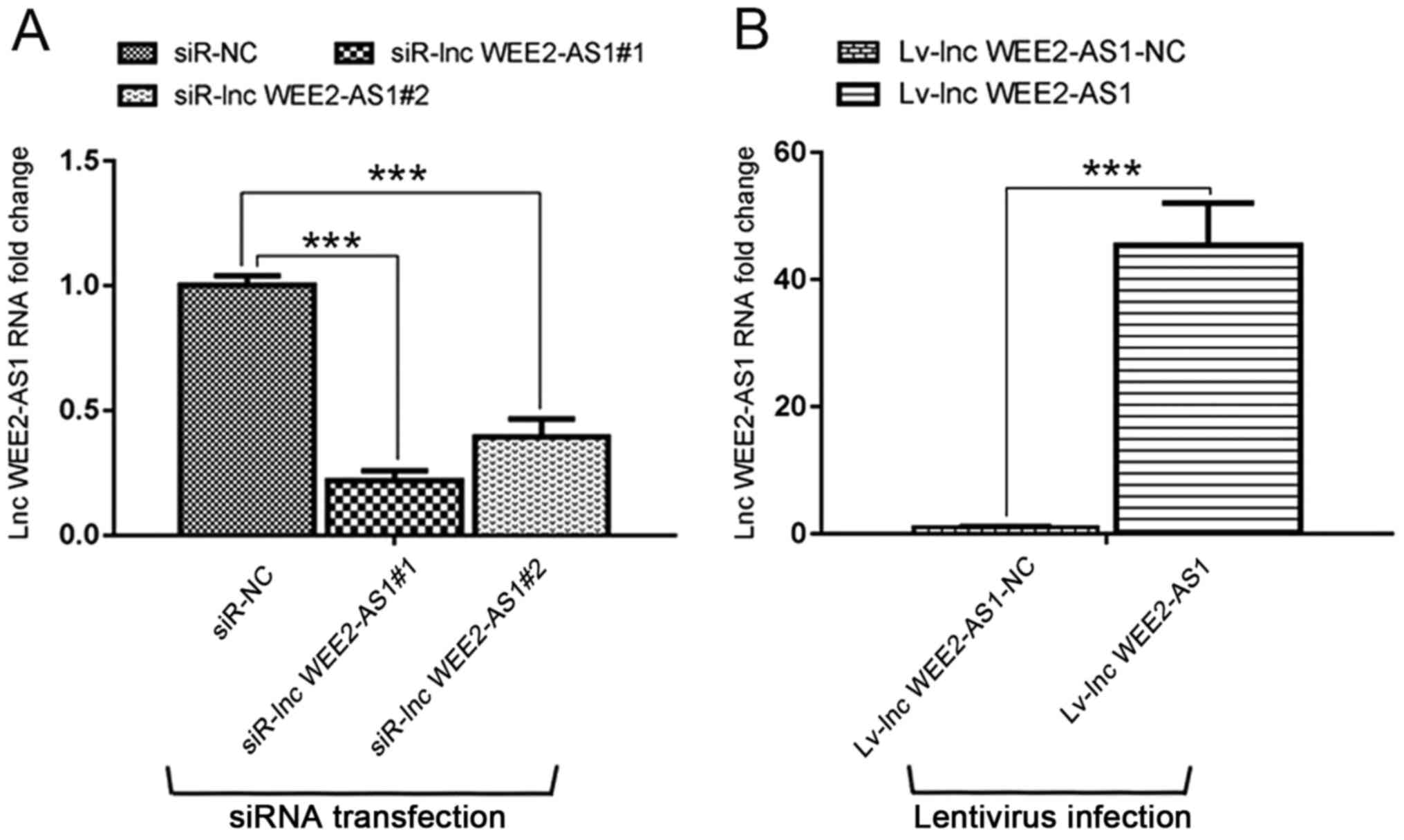
RT-qPCR was performed to evaluate the effect of
lncRNA WEE2-AS1 knockdown and overexpression on HUVECs. Compared
with the corresponding negative control groups, lncRNA WEE2-AS1
expression levels were significantly reduced by siRNA transfection
and significantly increased by lentivirus infection. The results
indicated that the siRNA transfection and lentivirus infection were
successful (Fig. 7A and B).
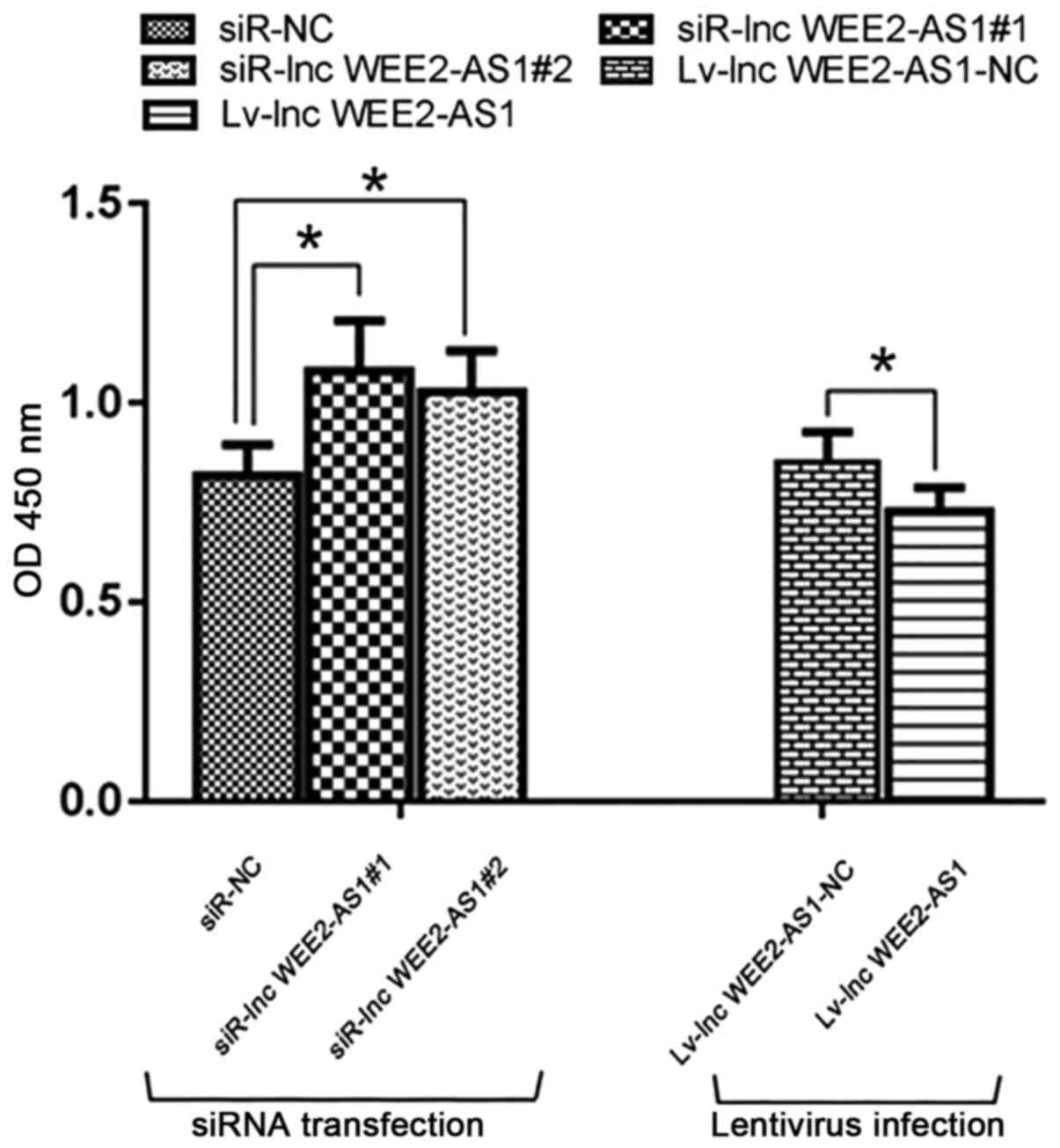
The CCK-8 assay was conducted to explore the effect
of lncRNA WEE2-AS1 on vascular endothelial cell viability. lncRNA
WEE2-AS1 knockdown significantly enhanced cell viability compared
with the negative control (siR-NC) group, whereas lncRNA WEE2-AS1
overexpression significantly inhibited cell viability compared with
the negative control (lv-lnc WEE2-AS1-NC) group (Fig. 8).
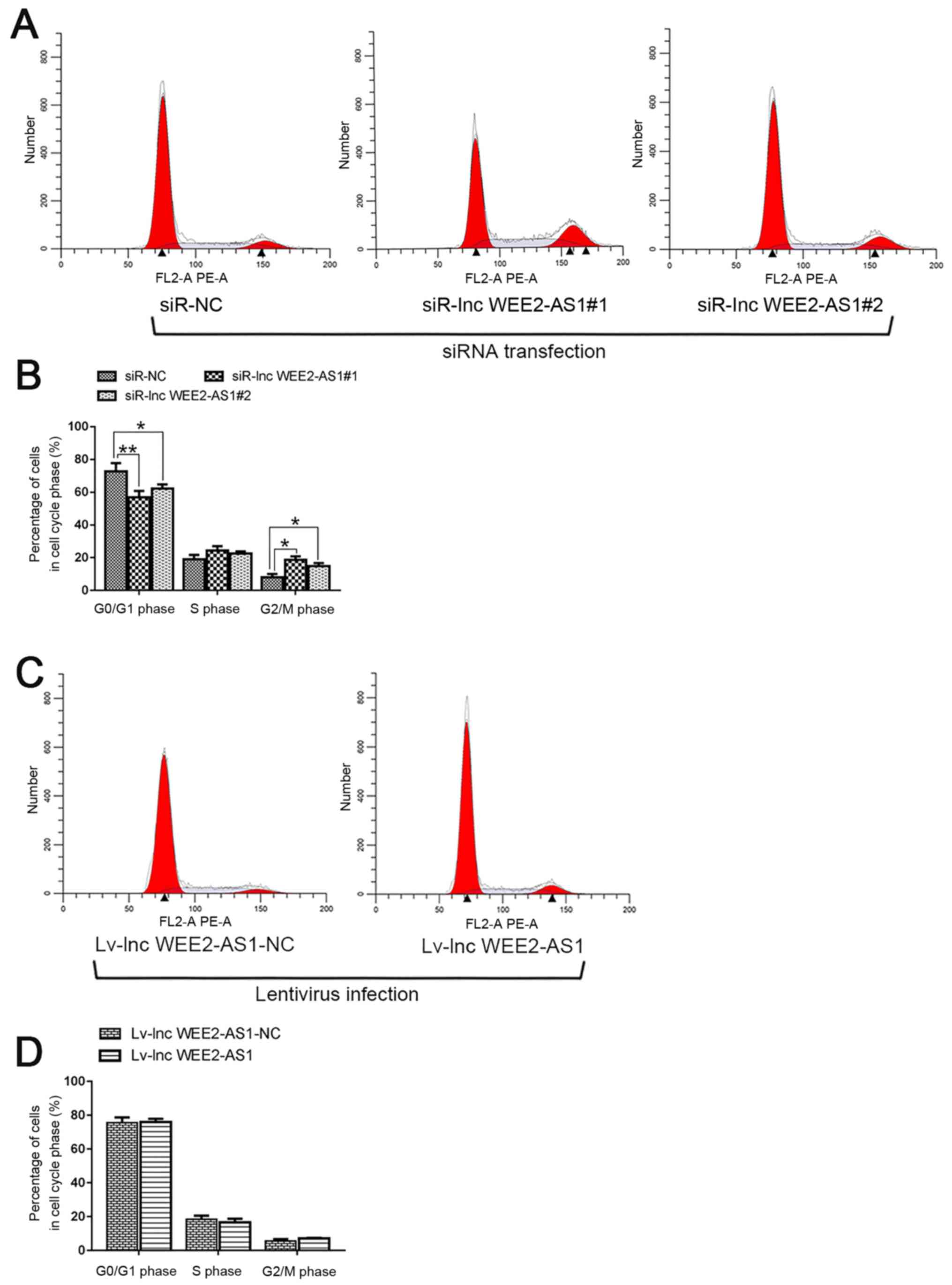
To investigate the function of lncRNA WEE2-AS1
during mitosis, the cell cycle distribution was assessed via flow
cytometry. The results indicated that lncRNA WEE2-AS1 knockdown
significantly decreased the proportion of cells in the
G0/G1 phase, but significantly increased the
proportion of cells in the G2/M phase compared with the
negative control (siR-NC) group (Fig.
9A and B). However, no significant difference in the proportion
of cells in the different cell cycle phases was detected between
lncRNA WEE2-AS1-overexpression cells and negative control (lv-lnc
WEE2-AS1-NC) cells (Fig. 9C and
D).
lncRNA WEE2-AS1 modulates WEE2
expression and MPF activity
Antisense lncRNAs are widespread in human cells and
have emerged as regulators of the corresponding sense strand genes
(56). However, the mechanism
underlying how antisense lncRNAs affect the expression of the
neighboring coding gene remains unclear. Therefore, whether lncRNA
WEE2-AS1 regulated the corresponding sense gene expression was
investigated. The western blotting results indicated that lncRNA
WEE2-AS1 knockdown significantly decreased WEE2 protein expression
levels compared with the negative control (siR-NC) group, whereas
lncRNA WEE2-AS1 overexpression significantly increased WEE2 protein
expression levels compared with the negative control (lv-lnc
WEE2-AS1-NC) group (Fig.
10A).
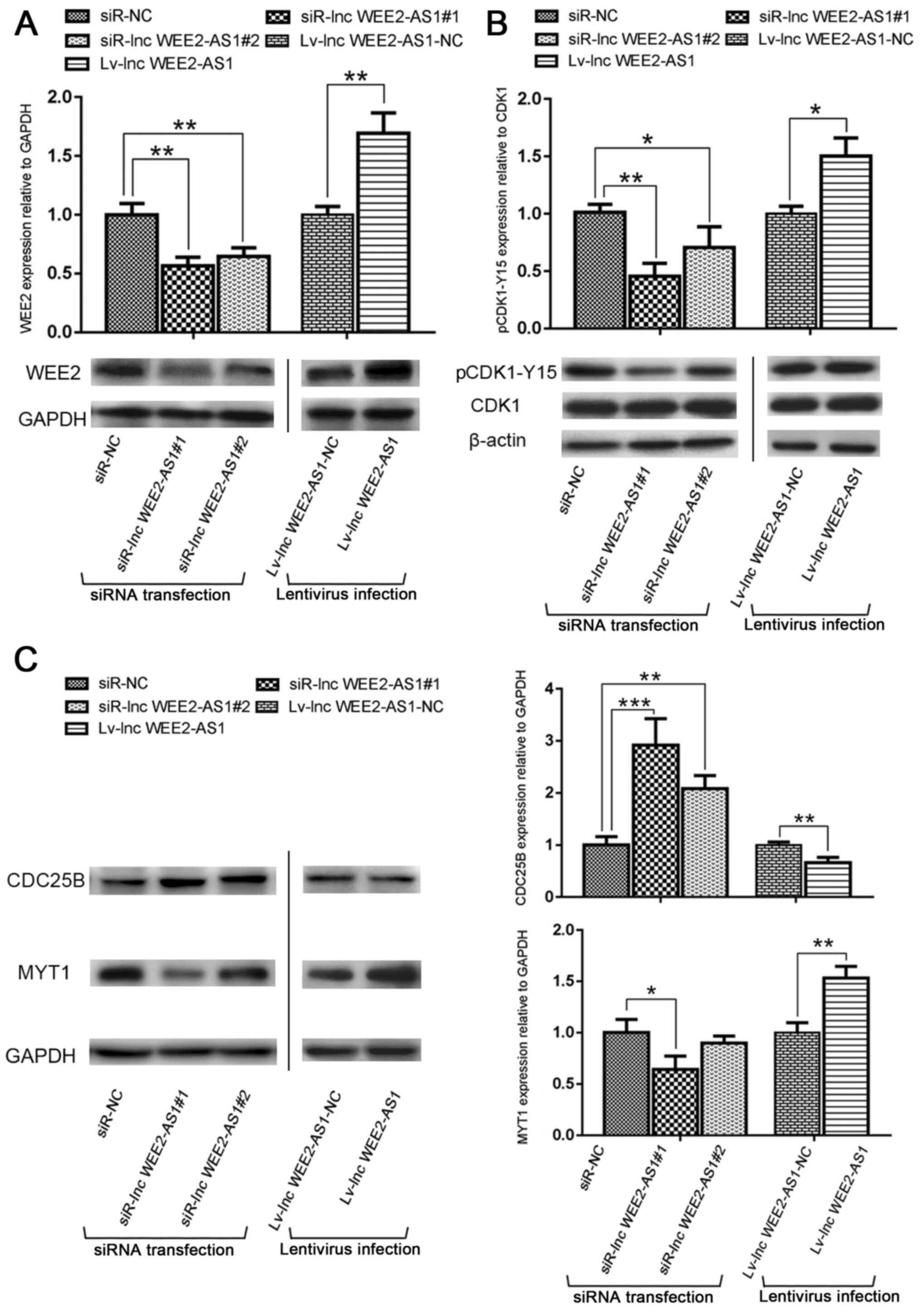 | Figure 10.Western blotting was performed to
assess the expression of G2/M transition-related proteins. The
effects of lncRNA WEE2-AS1 knockdown and overexpression on (A)
WEE2, (B) the ratio of pCDK1-Y15 to CDK1, (C) CDC25B and MYT1
protein expression levels were determined by western blotting.
*P<0.05, **P<0.01 and ***P<0.001 vs. siR-NC or lv-lnc
WEE2-AS1-NC. lncRNA/lnc, long non-coding RNA; WEE2-AS1, WEE2
antisense RNA 1; WEE2, WEE1 homolog 2; p, phosphorylated; CDK1-Y15,
cyclin dependent kinase 1-phosphorylated Y15; CDC25B, cell division
cycle 25B; MYT1, myelin transcription factor 1; siR, small
interfering RNA; NC, negative control; lv, lentivirus. |
MPF activation is a prerequisite for the cell cycle
transition from the G2 phase to the M phase, and is
regulated by a variety of cell cycle-related proteins via both
negative and positive loops (57).
WEE2 inhibits MPF activity by phosphorylating the Tyr15 residue of
CDK1 to regulate the cell cycle (20,21).
To identify the regulatory mechanism underlying lncRNA WEE2-AS1,
the effect of lncRNA WEE2-AS1 expression on MPF activity was
investigated. The western blotting results indicated that lncRNA
WEE2-AS1 knockdown significantly decreased the expression level of
MYT1 and the phosphorylation of CDK1, but increased the expression
level of CDC25B compared with the negative control (siR-NC) group.
By contrast, lncRNA WEE2-AS1 overexpression displayed the opposite
effect on CDK1 phosphorylation, and the expression of MYT1 and
CDC25B (Fig. 10B and C).
Therefore, the western blotting results indicated an association
between lncRNA WEE2-AS1 and the regulation of cell viability.
Discussion
The molecular mechanism underlying atherosclerosis
is not completely understood; however, numerous studies have
indicated that lncRNAs serve pivotal roles in the pathological
processes of atherosclerosis (7–12).
The present study suggested that lncRNA WEE2-AS1
expression levels were significantly upregulated in the plasma and
artery tissue samples of patients with ASO compared with healthy
control subjects, which indicated that lncRNA WEE2-AS1 may be
involved in ASO. The results also suggested that lncRNA WEE2-AS1
was expressed in the cytoplasm and nuclei of human vascular
endothelial cells, which indicated that lncRNA WEE2-AS1 may be
involved in a variety of physiological and pathological
processes.
Endothelial dysfunction can lead to alterations to
synthetic and secretory functions, causing disruption of the local
microbalance, which represents the beginning of atherosclerosis.
Identifying factors that control endothelial dysfunction is
essential for the treatment of atherosclerosis (58). Increasing evidence suggests that
various lncRNAs are involved in endothelial dysfunction. For
instance, silencing the lncRNA myocardial infarction associated
transcript inhibits endothelial cell proliferation, migration and
tube formation (13). Abnormal
alterations to the viability and cell cycle progression of
endothelial cells are important features of atherosclerosis.
Therefore, whether lncRNA WEE2-AS1 could affect endothelial cell
viability was investigated in the present study. lncRNA WEE2-AS1
knockdown significantly promoted endothelial cell viability,
whereas lncRNA WEE2-AS1 overexpression significantly inhibited
endothelial cell viability compared with the negative control
groups. Cell cycle dysregulation is implicated in various diseases,
including atherosclerosis. Therefore, the effect of lncRNA WEE2-AS1
on the cell cycle was assessed. lncRNA WEE2-AS1 knockdown
significantly decreased the proportion of cells in the
G0/G1 phase and significantly increased the
proportion of cells in the G2/M phase compared with the
negative control group. However, there was no significant
difference in the proportion of cells in the different cell cycle
phases between lncRNA WEE2-AS1-overexpression cells and negative
control cells. lncRNAs commonly contain multiple splice variants
(59); however, the present study
only overexpressed the single consensus gene model of lncRNA
WEE2-AS1 via lentivirus transfection, which may provide an
explanation for the minimal effects of lncRNA WEE2-AS1
overexpression on cell viability and the cell cycle compared with
lncRNA WEE2-AS1 knockdown. Moreover, the results also indicated
that only overexpressing the single consensus gene model of lncRNA
WEE2-AS1 via lentivirus transfection makes it difficult to assess
the full extent of its biological functions. The transcription and
activation of lncRNAs involves complex post-transcriptional
processing, modification and regulation processes (60); therefore, the current understanding
of these potential mechanisms remains in its infancy. Further
studies are required to identify the splice variant of lncRNA
WEE2-AS1 that is most closely related to the viability and cell
cycle of endothelial cells, as well as the underlying
mechanisms.
In eukaryotic cells, MPF is an indispensable inducer
for entry into the mitosis phase of the cell cycle. MPF is a
complex comprised of a catalytic subunit, Cyclin B, and a
regulatory subunit, CDK1 (20–22).
WEE2 and MYT1 perturb the G2/M transition via specific
phosphorylation of residues of CDK1 (61,62).
In opposition to the inhibitory phosphorylation of WEE2 and MYT1,
the stimulatory dephosphorylation of CDC25B restores MPF activity
(63,64). Once activated MPF reaches the
threshold for G2/M transition, cells are triggered to
undergo mitosis (65,66). The balance between WEE2 and CDC25B
participates in the regulation of MPF activity and maintains the
orderly progression of mitosis (67,68).
The present study investigated the regulatory role of lncRNA
WEE2-AS1 on cell viability and the cell cycle. lncRNA WEE2-AS1 was
expressed in the nuclei and cytoplasm of vascular endothelial
cells, and increased the synthesis of WEE2 protein, which indicated
that antisense lncRNA WEE2-AS1 might positively regulate the
expression of WEE2 encoded by the sense strand gene and could be
involved in gene transcription and post-transcriptional regulation.
Furthermore, the results indicated that lncRNA WEE2-AS1 knockdown
significantly decreased WEE2, MYT1 and pCDK1 expression levels, but
increased CDC25B expression levels compared with the negative
control group. By contrast, lncRNA WEE2-AS1 displayed the opposite
effects on protein expression. The results also suggested that
lncRNA WEE2-AS1 may modulate the G2/M transition of the
cell cycle and serve a role in cell cycle progression. A large
number of antisense lncRNAs have been identified to upregulate the
expression of their sense gene. For example, PDCD4-AS1 and PDCD4
are positively correlated (22).
The results of the present study implied that lncRNA WEE2-AS1
regulated the expression of WEE2 gene. Although a series of models
have been proposed to illustrate how lncRNAs regulate sense RNA,
there is no convincing molecular explanation. Further research is
required to reveal the molecular mechanism underlying lncRNA
WEE2-AS1-mediated regulation of WEE2 expression. lncRNAs can
regulate gene transcription in the nucleus or the activity of other
noncoding and coding RNAs in the cytoplasm (69–72).
Moreover, lncRNAs can form secondary, tertiary and even more
complex and higher order structures, which can tether functionally
interacting or unrelated proteins together (73,74).
Thus, further research is required to determine the molecular
mechanisms underlying lncRNA WEE2-AS1-mediated regulation of
crucial proteins in the G2/M transition pathway. Hu
et al (75) demonstrated
that lncRNA WEE2-AS1 accelerated hepatocellular carcinoma cell
proliferation in hepatitis B virus-related HCC. Therefore, whether
the role of lncRNA WEE2-AS1 depends on the specificity of tissue
and cell type requires further investigation (75).
In conclusion, the present study indicated that
lncRNA WEE2-AS1 expression was significantly upregulated in the
plasma and artery tissue samples of patients with ASO compared with
healthy control subjects. Moreover, lncRNA WEE2-AS1 may regulate
endothelial cell viability via the G2/M transition
pathway of the cell cycle. The results of the present study may
further the current understanding of the molecular mechanism
underlying ASO and may aid with the identification of specific
probes and targeted drugs for the diagnosis and treatment of
ASO.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81070258, 81270378,
81370368, 81670441, 81600336, 81200231 and 81300237), the Guangdong
Department of Finance (grant nos. 2010901 and 2014SC104), the
Guangzhou Science and Technology Plan Projects (grant no.
2015B090903064), the Ministry of Health of China (Tube) Key Project
of Hospital Clinic (grant no. 254003) and the Pearl River S&T
Nova Program of Guangzhou (grant nos. 201610010050 and
201806010006).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW, WL, BJ and CZ conceived and designed the study.
BJ performed the majority of the experiments, analyzed the data and
drafted the manuscript. SW, WL and CZ revised the manuscript. RW,
JM and RL performed some experiments and analyzed some the data.
ZL, RW, JC, MW and WW participated in the collection of clinical
specimens and performed some experiments. SW supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the First Affiliated Hospital of Sun Yat-sen
University [approval no. (2013)70] and conducted in accordance with
the 1964 Helsinki declaration and its later amendments or
comparable ethical standards. All participants or their guardians
provided written informed consent before the tissue and blood
samples were obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO), .
Cardiovascular disease: Global atlas on cardiovascular disease
prevention and control. WHO; Geneva: 2011
|
|
2
|
Smith SC Jr, Collins A, Ferrari R, Holmes
DR Jr, Logstrup S, McGhie DV, Ralston J, Sacco RL, Stam H, Taubert
K, et al: Our time: A call to save preventable death from
cardiovascular disease (heart disease and stroke). J Am Coll
Cardiol. 60:2343–2348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laslett LJ, Alagona P Jr, Clark BA III,
Drozda JP Jr, Saldivar F, Wilson SR, Poe C and Hart M: The
worldwide environment of cardiovascular disease: Prevalence,
diagnosis, therapy, and policy issues: A report from the American
College of Cardiology. J Am Coll Cardio. 60 (Suppl 25):S1–S49.
2012. View Article : Google Scholar
|
|
4
|
Solanki A, Bhatt LK and Johnston TP:
Evolving targets for the treatment of atherosclerosis. Pharmacol
Ther. 187:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fowkes FG, Rudan D, Rudan I, Aboyans V,
Denenberg JO, McDermott MM, Norman PE, Sampson UK, Williams LJ,
Mensah GA and Criqui MH: Comparison of global estimates of
prevalence and risk factors for peripheral artery disease in 2000
and 2010: A systematic review and analysis. Lancet. 382:1329–1340.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sean MC and Peter RV: Peripheral arterial
disease. Heart Lung Circ. 27:427–432. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Turner AW, Wong D, Khan MD, Dreisbach CN,
Palmore M and Miller CL: Multi-omics approaches to study long
non-coding RNA function in atherosclerosis. Front Cardiovasc Med.
6:92019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Zheng L, Wang Q and Hu YW: Emerging
roles and mechanisms of long noncoding RNAs in atherosclerosis. Int
J Cardiol. 228:570–582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Voellenkle C, Garcia-Manteiga JM, Pedrotti
S, Perfetti A, De Toma I, Da Silva D, Maimone B, Greco S, Fasanaro
P, Creo P, et al: Implication of long noncoding RNAs in the
endothelial cell response to hypoxia revealed by RNA-sequencing.
Sci Rep. 6:241412016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Zhu H and Ge J: Long noncoding RNA:
Recent updates in atherosclerosis. Int J Biol Sci. 12:898–910.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou T, Ding JW, Wang XA and Zheng XX:
Long noncoding RNAs and atherosclerosis. Atherosclerosis.
248:51–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weirick T, Militello G and Uchida S: Long
non-coding RNAs in endothelial biology. Front Physiol. 9:5222018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: LncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu YW, Yang JY, Ma X, Chen ZP, Hu YR, Zhao
JY, Li SF, Qiu YR, Lu JB, Wang YC, et al: A lincRNA-
DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway
is essential for the regulation of cholesterol homeostasis. J Lipid
Res. 55:681–697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bhartiya D and Scaria V: Genomic
variations in non-coding RNAs: Structure, function and regulation.
Genomics. 107:59–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gentiluomo M, Crifasi L, Luddi A, Locci D,
Barale R, Piomboni P and Campa D: Taste receptor polymorphisms and
male infertility. Hum Reprod. 32:2324–2331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morgan DO: Principles of CDK regulation.
Nature. 374:131–134. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okamoto K and Sagata N: Mechanism for
inactivation of the mitotic inhibitory kinase Wee1 at M phase. Proc
Natl Acad Sci USA. 104:3753–3758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakanishi M, Ando H, Watanabe N, Kitamura
K, Ito K, Okayama H, Miyamoto T, Agui T and Sasaki M:
Identification and characterization of human Wee1B, a new member of
the Wee1 family of Cdk-inhibitory kinases. Genes Cells. 5:839–847.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanna CB, Yao S, Patta MC, Jensen JT and
Wu X: WEE2 is an oocyte-specific meiosis inhibitor in rhesus
macaque monkeys. Biol Reprod. 82:1190–1197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sang Q, Li B, Kuang Y, Wang X, Zhang Z,
Chen B, Wu L, Lyu Q, Fu Y, Yan Z, et al: Homozygous mutations in
WEE2 cause fertilization failure and female infertility. Am J Hum
Genet. 102:649–657. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and Computationally
predicted microRNA targets on Long noncoding RNAs. Nucleic Acids
Res. 41((Database Issue)): D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu KP, Zhang CL and Ma XL: Antisense
lncRNA FOXF1-AS1 promotes migration and invasion of osteosarcoma
cells through the FOXF1/MMP-2/-9 pathway. Int J Biol Sci.
13:1180–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pelechano V and Steinmetz LM: Gene
regulation by antisense transcription. Nat Rev Genet. 14:880–893.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Villegas VE and Zaphiropoulos PG:
Neighboring gene regulation by antisense long non-coding RNAs. Int
J Mol Sci. 16:3251–3266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jadaliha M, Gholamalamdari O, Tang W,
Zhang Y, Petracovici A, Hao Q, Tariq A, Kim TG, Holton SE, Singh
DK, et al: A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS
Genet. 14:e10078022018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choy JC, Granville DJ, Hunt DW and McManus
BM: Endothelial cell apoptosis: Biochemical characteristics and
potential implications for atherosclerosis. J Mol Cell Cardiol.
33:1673–1690. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mantella LE, Quan A and Verma S:
Variability in vascular smooth muscle cell stretch-induced
responses in 2D culture. Vasc Cell. 7:72015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mudau M, Genis A, Lochner A and Strijdom
H: Endothelial dysfunction: The early predictor of atherosclerosis.
Cardiovasc J Afr. 23:222–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vanhoutte PM, Shimokawa H, Feletou M and
Tang EH: Endothelial dysfunction and vascular disease-a 30th
anniversary update. Acta Physiol (Oxf). 219:22–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deng L, Bradshaw AC and Baker AH: Role of
noncoding RNA in vascular remodelling. Curr Opin Lipidol.
27:439–448. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Q, Kim YR, Vikram A, Kumar S, Kassan M,
Gabani M, Lee SK, Jacobs JS and Irani K: P66Shc-induced
MicroRNA-34a causes diabetic endothelial dysfunction by
downregulating sirtuin1. Arterioscler Thromb Vasc Biol.
36:2394–2403. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yin Y, Li X, Sha X, Xi H, Li YF, Shao Y,
Mai J, Virtue A, Lopez-Pastrana J, Meng S, et al: Early
hyperlipidemia promotes endothelial activation via a
caspase-1-sirtuin 1 pathway. Arterioscler Thromb Vasc Biol.
35:804–816. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leung SW and Vanhoutte PM:
Endothelium-dependent hyperpolarization: Age, gender and blood
pressure, do they matter? Acta Physiol (Oxf). 219:108–123. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stott JB, Barrese V and Greenwood IA: Kv7
channel activation underpins EPAC-dependent relaxations of rat
arteries. Arterioscler Thromb Vasc Biol. 36:2404–2411. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singh KK, Mantella LE, Pan Y, Quan A,
Sabongui S, Sandhu P, Teoh H, Al-Omran M and Verma S: A global
profile of glucose-sensitive endothelial-expressed long non-coding
RNAs. Can J Physiol Pharmacol. 94:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
European Stroke Organisation, Tendera M,
Aboyans V, Bartelink ML, Baumgartner I, Clément D, Collet JP,
Cremonesi A, De Carlo M, Erbel R, et al: ESC Guidelines on the
diagnosis and treatment of peripheral artery diseases: Document
covering atherosclerotic disease of extracranial carotid and
vertebral, mesenteric, renal, upper and lower extremity arteries:
The Task Force on the Diagnosis and Treatment of Peripheral Artery
Diseases of the European Society of Cardiology (ESC). Eur Heart J.
32:2851–2906. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bai Y, Zhang M and Bian F: Culture and
identification of human umbilical vein endothelial cells in vitro
using Trypsin digestion method. Chin J Tissue Eng Res.
16:2695–2698. 2012.
|
|
44
|
Chu M, Wu R, Qin S, Hua W, Shan Z, Rong X,
Zeng J, Hong L, Sun Y, Liu Y, et al: Bone marrow-derived
MicroRNA-223 works as an endocrine genetic signal in vascular
endothelial cells and participates in vascular injury from kawasaki
disease. J Am Heart Assoc. 6:e0048782017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
46
|
Raman K: Construction and analysis of
protein-protein interaction networks. Autom Exp. 2:22010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sardiu ME and Washburn MP: Building
protein-protein interaction networks with proteomics and
informatics tools. J Biol Chem. 286:23645–23651. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Miryala SK, Anbarasu A and Ramaiah S:
Discerning molecular interactions: A comprehensive review on
biomolecular interaction databases and network analysis tools.
Gene. 642:84–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bruno B, Arnaud B, Nelly B and Michel V: A
protocol for isolation and culture of human umbilical vein
endothelial cells. Nat Protoc. 2:481–485. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cao Y, Gong Y, Liu L, Zhou Y, Fang X,
Zhang C, Li Y and Li J: The use of human umbilical vein endothelial
cells (HUVECs) as an in vitro model to assess the toxicity of
nanoparticles to endothelium: A review. J Appl Toxicol.
37:1359–1369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li L and Guo PS: CD31: Beyond a marker for
endothelial cells. Cardiovasc Res. 94:3–5. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zanetta L, Marcus SG, Vasile J, Dobryansky
M, Cohen H, Eng K, Shamamian P and Mignatti P: Expression of Von
Willebrand factor, an endothelial cell marker, is up-regulated by
angiogenesis factors: A potential method for objective assessment
of tumor angiogenesis. Int J Cancer. 85:281–288. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Numata K and Kiyosawa H: Genome-wide
impact of endogenous antisense transcripts in eukaryotes. Front
Biosci (Landmark Ed). 17:300–315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen LL and Carmichael GG: Decoding the
function of nuclear long non-coding RNAs. Curr Opin Cell Biol.
22:357–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Katayama S, Tomaru Y, Kasukawa T, Waki K,
Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et
al: Antisense transcription in the mammalian transcriptome.
Science. 309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Takeo K: Entry into mitosis: A solution to
the decades-long enigma of MPF. Chromosoma. 124:417–428. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qaradakhi T, Apostolopoulos V and Zulli A:
Angiotensin (1–7) and alamandine: Similarities and differences.
Pharmacol Res. 111:820–826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Malumbres M and Barbacid M: Mammalian
cyclin-dependent kinases. Trends Biochem Sci. 30:630–641. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Booher RN, Holman PS and Fattaey A: Human
Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not
Cdk2 activity. J Biol Chem. 272:22300–22306. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Morgan DO: The cell cycle: Principles of
control. New Science Press; London: 2007
|
|
64
|
Burrows AE, Sceurman BK, Kosinski ME,
Richie CT, Sadler PL, Schumacher JM and Golden A: The C. elegans
Myt1 ortholog is required for the proper timing of oocyte
maturation. Development. 133:697–709. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Baldin V, Cans C, Knibiehler M and
Ducommun B: Phosphorylation of human CDC25B phosphatase by
CDK1-cyclin A triggers its proteasome-dependent degradation. J Biol
Chem. 272:32731–32734. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lindqvist A, Rodriguez-Bravo V and Medema
RH: The decision to enter mitosis: Feedback and redundancy in the
mitotic entry network. J Cell Biol. 185:193–202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Branzei D and Foiani M: Regulation of DNA
repair throughout the cell cycle. Nat Rev Mol Cell Biol. 9:297–308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nilsson I and Hoffmann I: Cell cycle
regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res.
4:107–114. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kornfeld JW and Brüning JC: Regulation of
metabolism by long, non-coding RNAs. Front Genet. 5:572014.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang
J and Tian W: Molecular mechanisms and function prediction of long
noncoding RNA. ScientificWorldJournal. 2012:5417862012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cao J: The functional role of long
non-coding RNAs and epigenetics. Biol Proced Online. 16:112014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX
and Hong W: The ways of action of long non-coding RNAs in cytoplasm
and nucleus. Gene. 547:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
St Laurent G, Wahlestedt C and Kapranov P:
The landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hu Z, Huang P, Yan Y, Zhou Z, Wang J and
Wu G: Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes
hepatocellular carcinoma proliferation and invasion. Biochem
Biophys Res Commun. 508:79–86. 2019. View Article : Google Scholar : PubMed/NCBI
|