Introduction
Breast cancer (BC) is one of the most common
malignant tumors in women worldwide and is associated with high
morbidity and mortality rates (1).
It mostly occurs in women between the ages of 55 and 60 years
(2). In 2019, ~268,600 new cases of
BC were diagnosed and 41,760 women died from this disease in US
(3). Accumulating evidence suggests
that the incidence of BC is increasing, and patients diagnosed with
BC are younger (4,5). Triple-negative BC (TNBC) is a unique
and aggressive subtype of BC, accounting for 15–20% of all cases of
BC (6). In addition, TNBC is
associated with high recurrence rate and poor outcomes (6). TNBC is characterized by the absence of
the estrogen receptor, human epidermal growth factor receptor type
2 and progesterone receptor expression (7). Due to the lack of specific early
diagnostic and prognostic biomarkers, the prognosis of TNBC remains
poor (8). Therefore, an urgent
demand currently exists for the development of possible diagnostic
and therapeutic targets to improve the survival rates of patients
with TNBC.
Long non-coding RNAs (lncRNAs) are RNA molecules
that are >200 nucleotides in length and that do not encode
proteins (9). Previous studies have
suggested that lncRNAs can exert various functions in numerous
biological processes, and that they are implicated in the
development and progression of a variety of cancers (10,11).
The lncRNA titin-antisense RNA1 (TTN-AS1) was first found to be a
highly expressed oncogene in esophageal squamous cell carcinoma,
where it can promote cell proliferation and metastasis (12). In addition, a previous study has
reported that TTN-AS1 increases the proliferation and invasive and
migratory abilities of cervical cancer (13), whereas another study demonstrated
that TTN-AS1 can facilitate papillary thyroid cancer tumor
progression (14). However, the
role of TTN-AS1 in TNBC remains poorly understood.
In the present study, clinical TNBC tissue samples
and cell lines (MDA-MB-453 and MDA-MB-231) were used to investigate
the biological function and underlying mechanism of TTN-AS1 in
TNBC. The results from the present study may uncover a novel
therapeutic target for the intervention of TNBC.
Materials and methods
Clinical sample collection
A total of 30 pairs of TNBC and matched adjacent
normal tissues (distance from tumor margin, 2 cm) were collected
from patients with TNBC (age range, 27–79 years; mean age, 52.9±4.1
years) who underwent surgical resection at The Nanjing Maternal and
Child Health Care Hospital (Nanjing, China) between April 2017 and
May 2018. None of the patients received chemotherapy or
radiotherapy prior to the operation. All resected tissues samples
were immediately snap frozen in liquid nitrogen following surgery
and stored at −80°C for further analysis. The present study was
approved by the Ethics Committee of Nanjing Maternal and Child
Health Care Hospital and complied with the principles of the
Helsinki Declaration. All patients provided signed informed
consent.
Cell culture
The human TNBC cell lines MDA-MB-453 and MDA-MB-231
and the normal breast epithelial cell line MCF-10A were obtained
from the Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences. All cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and placed in a
humidified atmosphere under 5% CO2 at 37°C.
Cell transfection
MDA-MB-453 and MDA-MB-231 cells were seeded into
6-well plates at a density of 1×106 cells/well and
transfection was performed when ~90% confluence was reached
following manufacturer's protocol. Short hairpin RNA (shRNA)
targeted against TTN-AS1 (shRNA-TTN-AS1-1 or shRNA-TTN-AS1-2) and a
scrambled shRNA that served as the negative control (shRNA-NC),
were designed and synthesized by Shanghai GenePharma Co., Ltd. The
shRNAs (3 µg) were transfected into the TNBC cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. In
addition, microRNA (miR/miRNA)-211-5p mimic, miR-211-5p inhibitor
and their corresponding negative control (miR-NC) obtained from
Shanghai GenePharma Co., Ltd., were transfected into the cells at a
concentration of 40 nM using Lipofectamine® 3000 as
previously described (15). Cells
were collected 24 h following transfection, and transfection
efficiency was evaluated by reverse transcription-quantitative PCR
(RT-qPCR).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 kit (Sigma-Aldrich; Merck KGaA) was used
to detect cell proliferation. Briefly, transfected cells were
seeded into 96-well plates (3,000 cells/100 µl). After incubation
for 24, 48 or 72 h, 10 µl CCK-8 solution was added to each well and
then cultured for a further 2 h. Absorbance was read at 450 nm
using a microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
Cells in the logarithmic growth phase were seeded
into 6-well plates at the density of 600 cells/well and were
cultured at 37°C for 14 days until colony formation became visible.
Culture medium was regularly changed every 2 days. The colonies
were subsequently fixed with 4% paraformaldehyde for 30 min at room
temperature, stained with 0.5% crystal violet for 10–30 min and
imaged under a fluorescence inversion microscope (Olympus;
magnification, 10×). ImageJ software (version 1.52r; National
Institutes of Health) was used to count colonies.
Transwell assay
The invasive ability of TNBC cells was evaluated
using Transwell assay (pore size, 8.0 µm; Corning Inc.) coated with
Matrigel (BD Biosciences) overnight at 37°C. Briefly, 500 µl
serum-free media containing 2×105 cells were seeded in
the upper chamber, and the lower chamber was filled with media
containing 10% FBS. Invasive cells were stained with crystal violet
48 h after incubation, and were subsequently imaged under a
fluorescence inversion microscope (Olympus; magnification, 100×).
Cell number was analyzed using ImageJ software (version 1.52r;
National Institutes of Health).
Wound healing assay
Cell migration was assessed using a wound healing
assay. Briefly, cells (3×105 cells/well) were seeded
into a 6-well plate and incubated in RPMI-1640 medium containing
10% FBS at 37°C until 80% confluence was reached. Subsequently,
cells were serum-starved for 24 h. A straight scratch was then
introduced in the cell monolayers using a 200 µl plastic pipette
tip. PBS was used to wash the cells and remove any debris.
Following incubation for further 48 h in serum-free medium, the
average distance of cells migrating into the wound was measured
using an fluorescence inversion microscope (Olympus; magnification,
100×).
RNA extraction and RT-qPCR
Total RNA was isolated from TNBC tissue samples and
cell lines using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc) according to the manufacturer's protocol.
PrimeScript™ RT reagent Kit (Takara Bio, Inc.) was then used to
produce the cDNA following manufacturer's protocol. Subsequently,
qPCR was performed using Power SYBR® Green Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Pre-denaturation at 95°C for 10
min, denaturation at 95°C for 15 sec, annealing at 60°C for 1 min,
for a total of 40 cycles. Sequences of the gene-specific primers
used in this study were as follows: TTN-AS1 forward,
5′-CGATACCATTGAACACGCTGC-3′ and reverse, 5′-GGTTGAGGGTCCCAGTG-3′;
miR-211-5p forward, 5′-ACACTCCAGCTGGGCAAGTAGCATCAACTA-3′ and
reverse, 5′-TGGTGTCGTGGAGTCG-3′; GAPDH forward,
5′-TGTGGGCATCAATGGATTTGG-3′ and reverse,
5′-ACACCATGTATTCCGGGTCAAT-3′; and U6 forward,
5′-TCTGCTCCTATCCCAATTACCTG-3′ and reverse,
5′-ACTCCCGGATCTCTTCTAAGTTG-3′ which were all synthesized by
Guangzhou RiboBio Co., Ltd. GAPDH and U6 were used as internal
controls for TTN-AS1 and miRNA-211-5p, respectively. Relative mRNA
expression levels of TTN-AS1 and miRNA-211-5p were determined using
the 2−ΔΔCq method (16).
Western blotting
Total protein in TNBC tissue samples and cell lines
was isolated using RIPA reagent buffer (Beyotime Institute of
Biotechnology) at 4°C, and the bicinchoninic acid protein assay kit
(Beyotime Institute of Biotechnology) was used to determine protein
concentration. Proteins (40 µg) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes. The membranes were blocked in 5%
skimmed milk and incubated with specific primary antibodies
overnight at 4°C. Primary antibodies included the anti-matrix
metalloproteinase 2 (MMP2) (cat. no. 40994S; 1:1,000), anti-MMP9
(cat. no. 13667T; 1:1,000) and the loading control anti-GAPDH (cat.
no. 5174T; 1:1,000), all from Cell Signaling Technology, Inc. After
rinsing with TBS-0.2% Tween-20 three times, the membranes were
incubated with a goat anti-rabbit HRP-conjugated secondary antibody
(cat. no. 7074S; 1:3,000; Cell Signaling Technology, Inc.) at room
temperature for 1.5 h. The blots were visualized using the Odyssey
Infrared Imaging System (LI-COR Biosciences) and subsequently
quantified using ImageJ software (version 1.52r; National
Institutes of Health).
Luciferase reporter assay
Potential target genes of lncRNA TTN-AS1 were
predicted using an online bioinformatics software (http://starbase.sysu.edu.cn/) (17,18),
and the potential interaction between miR-211-5p and TTN-AS1 was
determined using luciferase reporter assay. Luciferase reporter
vectors encoding the wild-type (WT) or mutant (MUT) TTN-AS1
3′-untranslated region (3′-UTR) were first designed. To perform the
luciferase reporter assay, cells were first co-transfected with
TTN-AS1 3′-UTR-WT or TTN-AS1 3′-UTR-MUT and miR-211-5p mimic or
miR-NC, following which luciferase activity was detected using a
Dual-Luciferase® Reporter Assay System (Promega
Corporation) 48 h after transfection. Renilla luciferase
activity was used as control.
Statistical analysis
All data were presented as the means ± standard
deviation and analyzed using GraphPad Prism 6 (GraphPad Software,
Inc.). Pearson's correlation coefficient was applied to assess the
correlation between the expression of TTN-AS1 and miR-211-5p in
TNBC tissues and adjacent normal tissues. A paired t-test was used
for comparisons between TNBC and matched adjacent tissues. One-way
analysis of variance followed by Tukey's post hoc test was used to
evaluate differences among different groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
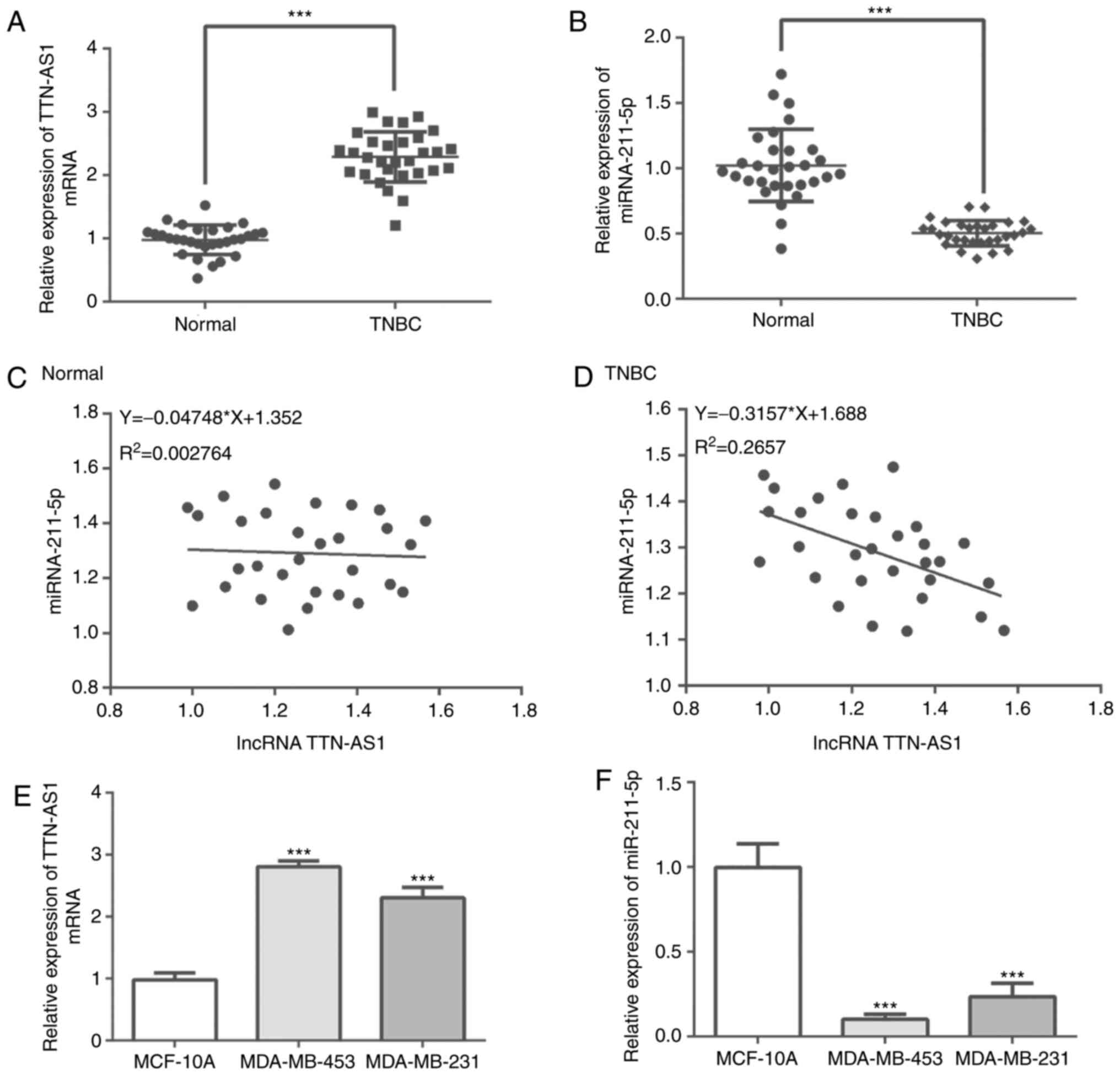
TTN-AS1 expression is upregulated,
whereas miR-211-5p expression is downregulated in TNBC tissues and
TNBC cells
RT-qPCR was performed to evaluateTTN-AS1 and
miR-211-5p expression in human TNBC tissues. The results
demonstrate that TTN-AS1 expression was significantly increased,
whereas the expression of miR-211-5p was significantly decreased in
TNBC tissues compared with adjacent tissues (Fig. 1A and B). Pearson's correlation
coefficients analysis was performed to determine the correlation
between TTN-AS1 and miR-211-5p expression in matched adjacent
tissues and TNBC tissues. The results demonstrated that there was
no correlation between TTN-AS1 and miR-211-5p expression in normal
tissues, whereas miR-211-5p expression was negatively correlated
with TTN-AS1 expression in TNBC tissues (F=−0.3157; P<0.05;
Fig. 1C and D). In addition,
significantly higher expression of TTN-AS1 and lower miR-211-5p
expression were observed in MDA-MB-453 and MDA-MB-231 cells
compared with those in MCF-10A cells (Fig. 1E and F). These results indicated
that TTN-AS1 expression was upregulated and miR-211-5p expression
was downregulated in TNBC tissues and TNBC cells compared with
controls.
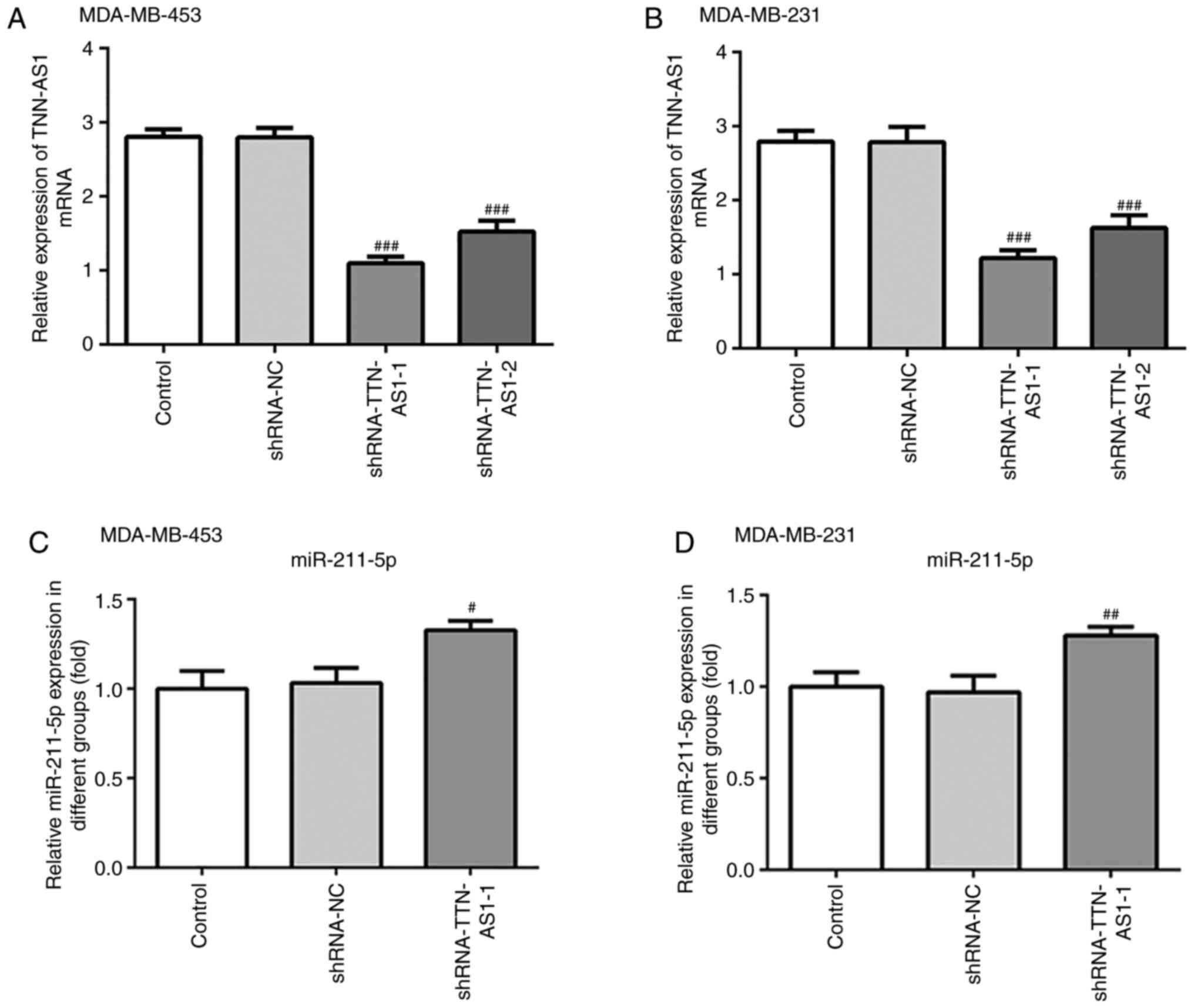
TTN-AS1 knockdown increases the
expression of miR-211-5p in TNBC cells
To investigate the regulatory relationship between
TTN-AS1 and miR-211-5p, TTN-AS1 shRNA-1 and shRNA-2 were
transfected into both MDA-MB-453 and MDA-MB-231 cells to knockdown
TTN-AS1 expression. As presented in Fig. 2A and B, the expression levels of
TTN-AS1 in both MDA-MB-453 and MDA-MB-231 cells were significantly
decreased following transfected with shRNA compared with shRNA-NC.
Since transfection with shRNA-1 exhibited the greater silencing
effect, TTN-AS1 shRNA-1 was used for subsequent experiments. The
expression of miR-211-5p was first evaluated following TTN-AS1
knockdown. The depletion of TTN-AS1 expression induced the
upregulation of miR-211-5p in MDA-MB-453 and MDA-MB-231 cells
(Fig. 2C and D). These data
suggested that TTN-AS1 may affect the expression of miR-21-5p in
TNBC cells.
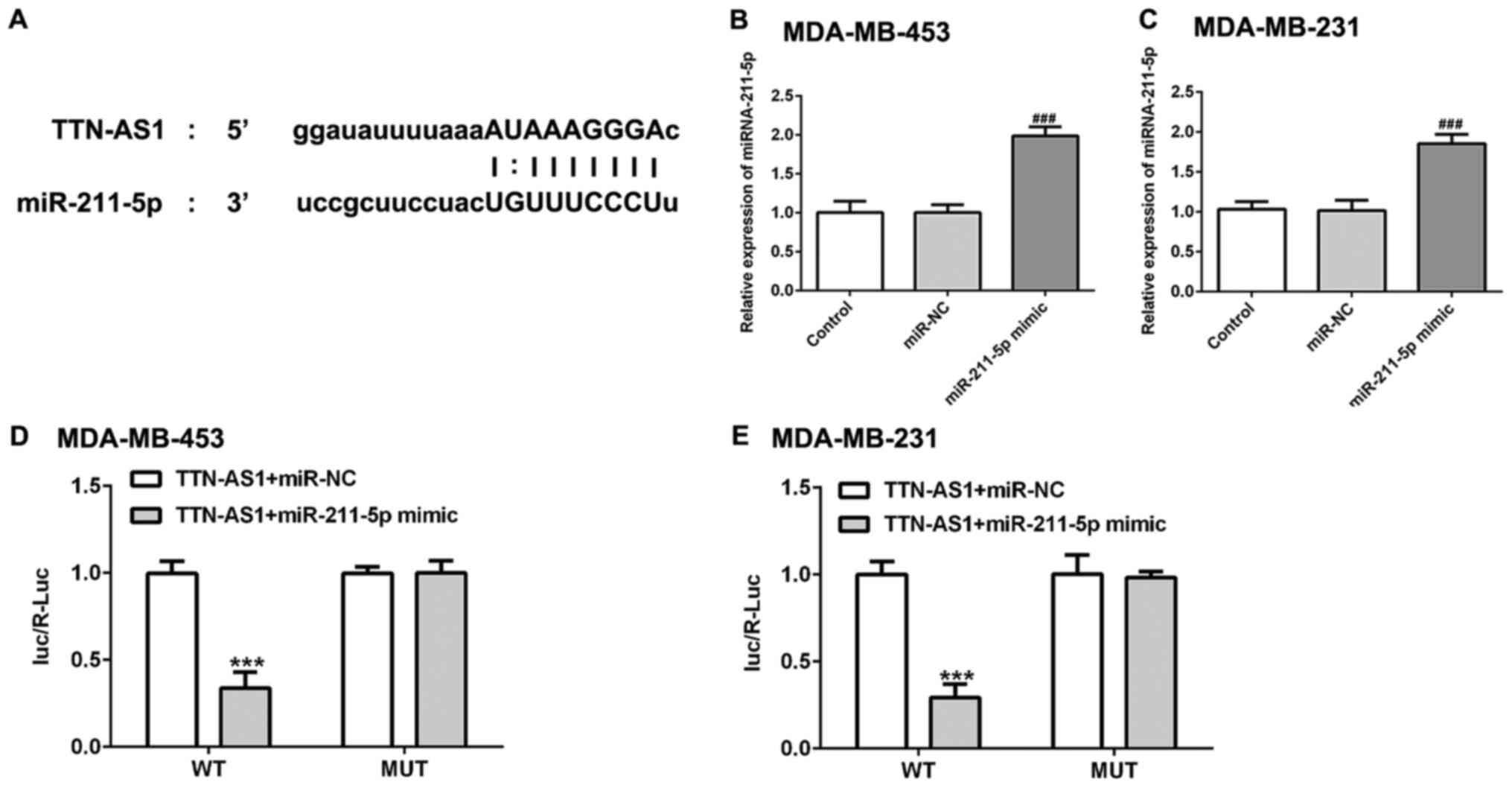
miR-211-5p is a direct target of
TTN-AS1
Using an online bioinformatics program (http://starbase.sysu.edu.cn/), it was found that
miR-211-5p was a potential target of TTN-AS1 (Fig. 3A). Then, miR-211-5p was
overexpressed by transfection with miR-211-5p mimic. As presented
in Fig. 3B and C, the expression
level of miR-211-5p was significantly upregulated in miR-211-5p
mimic group compared with the miR-NC group. Subsequently,
dual-luciferase assay was used to verify this potential
relationship. Luciferase activity was found to be decreased
following co-transfection with the miR-211-5p mimic and lncRNA
TTN-AS1-WT (Fig. 3D and E),
suggesting that miR-211-5p may be a direct target of TTN-AS1.
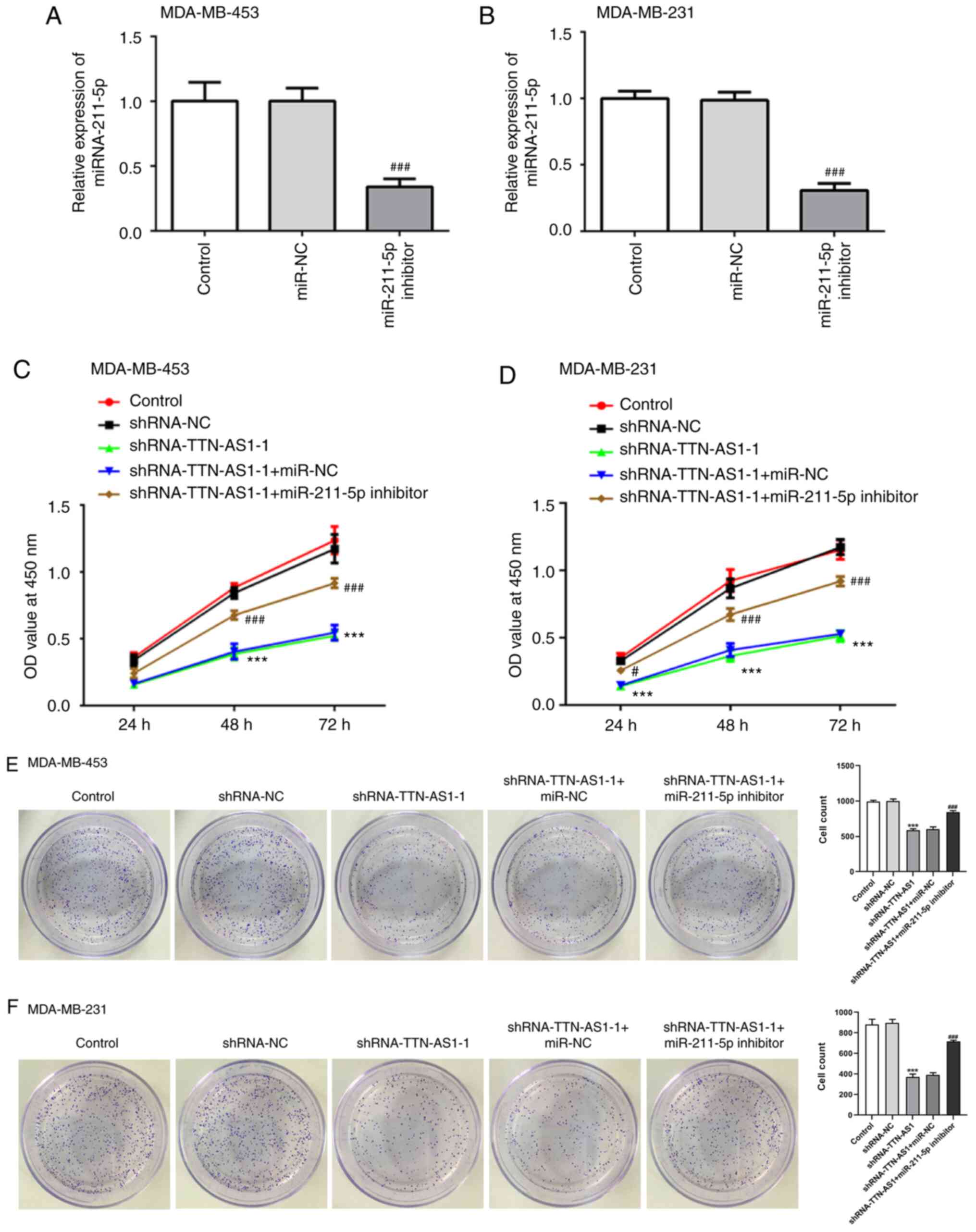
Transfection with the miR-211-5p
inhibitor reverses the inhibitory impact of TTN-AS1 knockdown on
TNBC cell proliferation
To explore the regulatory relationship between
TTN-AS1 and miR-211-5p in TNBC, a miR-211-5p inhibitor was
transfected into MDA-MB-453 and MDA-MB-231 cells, and RT-qPCR was
performed to examine the transfection efficiency. miR-211-5p
expression was significantly downregulated following transfection
with the miR-211-5p inhibitor in the two TNBC cell lines (Fig. 4A and B). Cell proliferation was then
measured using CCK-8 (Fig. 4C and
D) and colony formation (Fig. 4E
and F) assays. The results demonstrated that depletion of
TTN-AS1 levels decreased the cell viability and colony numbers of
both MDA-MB-453 and MDA-MB-231 cells compared with the shRNA-NC
group, which was reversed by transfection with the miR-211-5p
inhibitor. These findings suggested that TTN-AS1 may regulate TNBC
cell proliferation by targeting miR-211-5p.
Transfection with the miR-211-5p
inhibitor inhibits the effects of TTN-AS1 knockdown on the
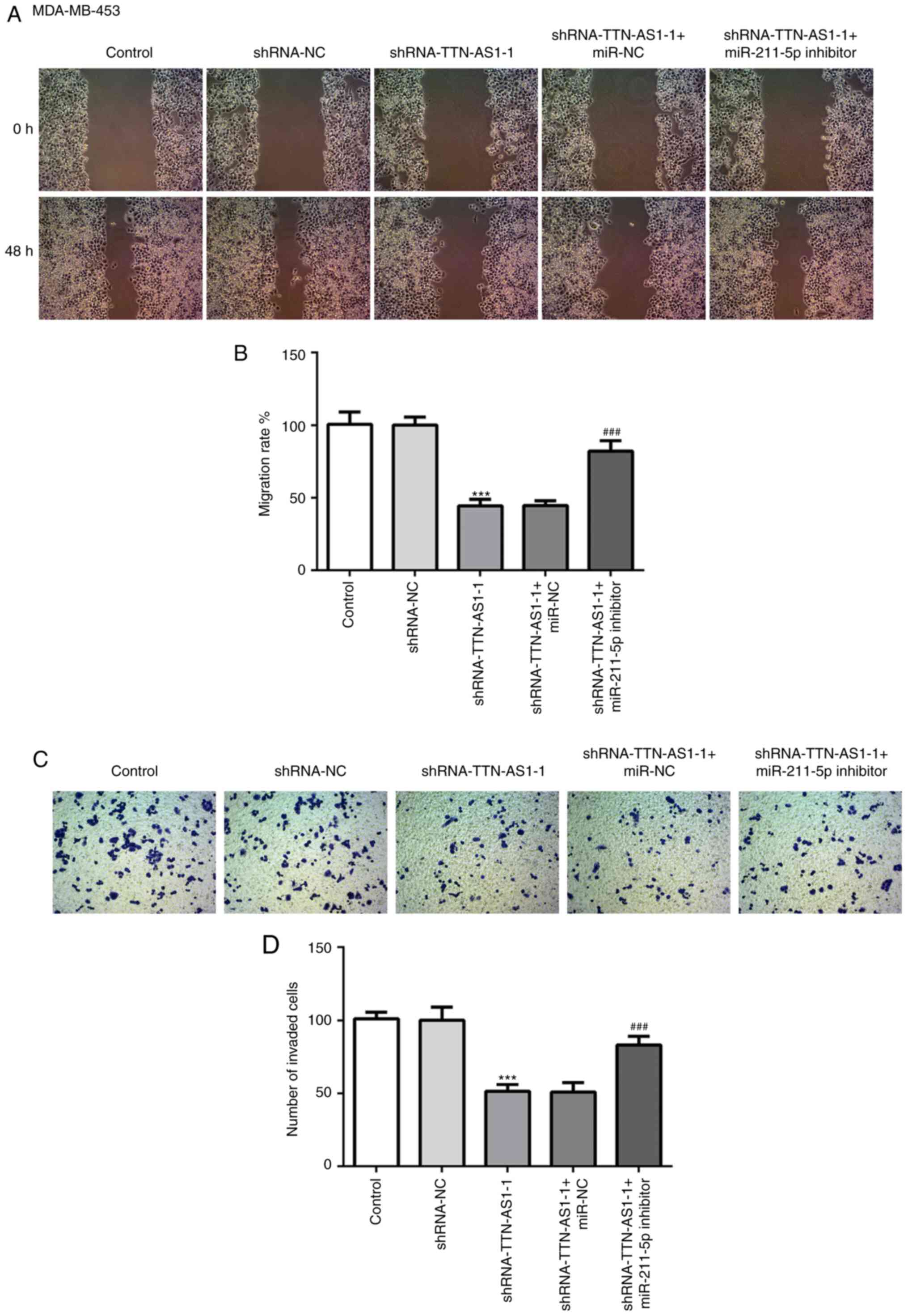
migratory and invasive abilities of TNBC cells
To understand the role of TTN-AS1 in invasion and
migration, Transwell and wound healing assays were performed with
TNBC cells. The results of the wound healing assay demonstrated
that transfection with the TTN-AS1 shRNA-1 decreased MDA-MB-453
cell migratory ability compared with cells transfected with
negative control. Subsequently, the migratory ability was recovered
following co-transfection with the miR-211-5p inhibitor (Fig. 5A and B). Similar results were
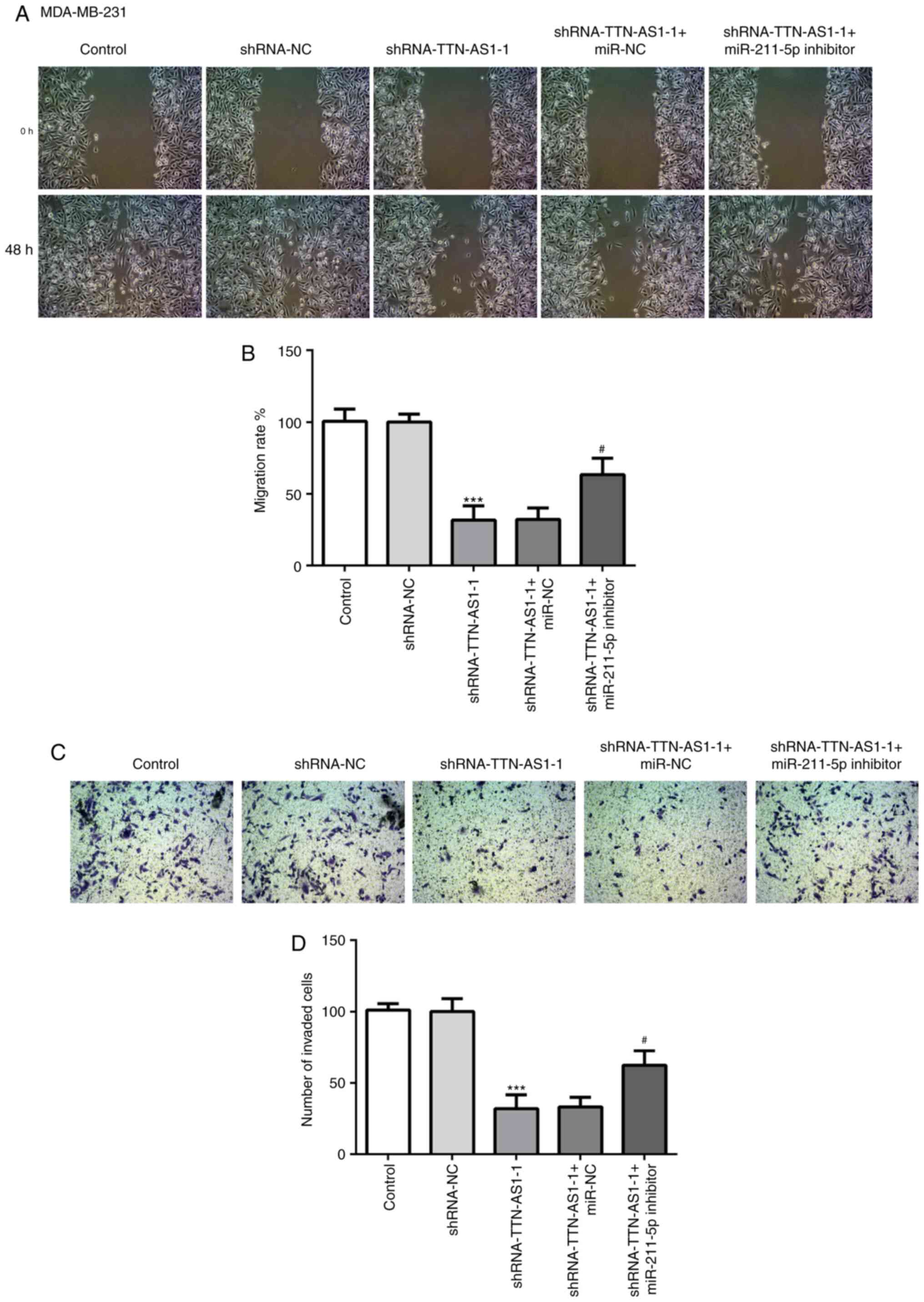
observed from the cell invasion assays (Fig. 5C and D). In addition, changes in
cell migratory (Fig. 6A and B) and
invasive (Fig. 6C and D) abilities
in the MDA-MB-231 cell line following transfection with TTN-AS1
shRNA-1 and/or the miR-211-5p inhibitor presented similar results
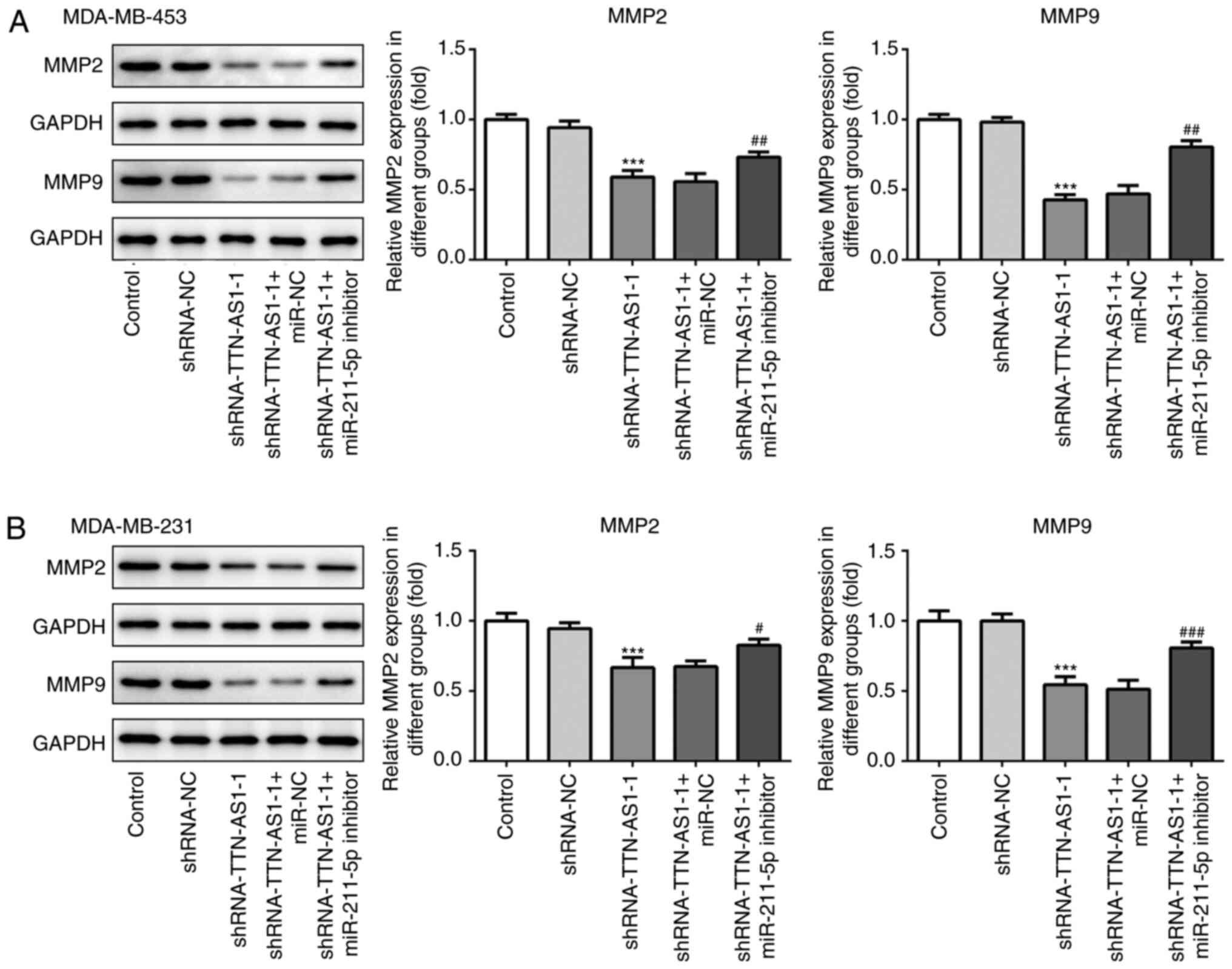
as those obtained with MDA-MB-453 cells. Subsequently, the protein
expression of matrix metalloproteinase (MMP)-2 and MMP-9, which are
associated with migration, were assessed by western blotting. As
presented in Fig. 7A and B, TTN-AS1
knockdown significantly downregulated the expression of MMP-2 and
MMP-9 compared with cells transfected with shRNA-NC, which was
reversed by co-transfection with the miR-211-5p inhibitor. These
observations suggested that TTN-AS1 may regulate TNBC cell invasive
and migratory abilities by targeting miR-211-5p.
Discussion
Accumulating evidence has demonstrated that lncRNAs
participate in the tumorigenesis process. For instance, lncRNA long
intergenic non-protein coding RNA, p53 induced transcript was
reported to suppress miR-523-3p and hamper retinoblastoma
progression by upregulating Dickkopf-1 (19). lncRNA differentiation antagonizing
non-protein coding RNA promotes the growth and metastasis of oral
squamous cell carcinoma cells by altering miR-216a-5p expression
(20). In addition, lncRNAs have
been reported to serve as pivotal regulators in the initiation and
progression of a number of malignancies, including TNBC (21–23).
The lncRNA TTN-AS1 is a newly identified lncRNA; the increased
expression of which was first reported in esophageal squamous cell
carcinoma (12). TTN-AS1 has been
demonstrated to accelerate the proliferation and invasive and
migratory abilities of cervical cancer cells, in addition to
facilitating the progression of papillary thyroid cancer tumors
(13,14). The present study demonstrated that
TTN-AS1 was overexpressed in TNBC tissues and cell lines compared
with normal adjacent tissues and the normal breast epithelial cell
line, respectively. In addition, TTN-AS1 knockdown inhibited the
proliferation and invasive and migratory abilities of TNBC cells by
targeting miR-211-5p expression, which may provide a potentially
novel insight into the development of therapeutic strategies for
clinical intervention of TNBC.
It has been reported that lncRNAs can modulate gene
expression by sponging specific miRNAs, indirectly regulating gene
expression at the post-transcriptional level (24). Therefore, bioinformatics predictions
were performed using the TargetScan online tool, which predicted
miR-211-5p to be a direct target of TTN-AS1. This was verified
using luciferase reporter assays in the present study. Increasing
evidence suggested that miR-211-5p functions as a tumor suppressor
in numerous malignancies, including renal cell carcinoma, papillary
thyroid cancer and tongue cancer (15,25,26).
Notably, emerging evidence supports the notion that miR-211-5p
expression is downregulated in TNBC tissues and cells, and the
upregulation of miR-211-5p expression can inhibit the progression
of TNBC (27). To understand the
regulatory relationship between TTN-AS1 and miR-211-5p, TTN-AS1
shRNA was transfected into MDA-MB-453 and MDA-MB-231 cells,
following which TNBC cell proliferation was measured using CCK-8
and colony formation assays. It was found that TTN-AS1 knockdown
inhibited the proliferation of TNBC cells, which was reversed
following transfection with miR-211-5p. Excessive proliferation of
cancer cells can accelerate the progression of tumorigenesis and
development (28). These findings
suggested therefore that TTN-AS1 may regulate TNBC proliferation by
targeting miR-211-5p.
Cancer cell migration and invasion are pivotal
processes of metastasis, which is the primary cause of
cancer-associated mortality (29).
During cancer development, the migratory and invasive abilities of
cancer cells are increased, resulting in cancer cell migration to
distant sites throughout the body, where they can become secondary
tumors (30). It has previously
been suggested that the poor prognosis associated with TNBC is
associated with enhanced invasive and migratory abilities of TNBC
cells (31). The present study
demonstrated that TTN-AS1 knockdown inhibited the invasive and
migratory abilities of both MDA-MB-453 and MDA-MB-231 cells, which
was reversed by transfection with the miR-211-5p inhibitor. MMPs,
including MMP-2 and MMP-9, are major components that are implicated
in invasion and migration, and the upregulation of these proteins
is closely associated with the aggressiveness and metastasis of
TNBC (32–34). In the present study, decreased MMP-2
and MMP-9 expression was observed following TTN-AS1 knockdown,
which was reversed following transfection with the miR-211-5p
inhibitor. These findings suggested that TTN-AS1 may regulate TNBC
invasive and migratory abilities by targeting miR-211-5p.
The present study investigated the role of TTN-AS1
in TNBC and the potential underlying mechanisms. TTN-AS1 was found
to be significantly upregulated in TNBC tissues and cell lines,
suggesting that TTN-AS1 may be considered as a significant
biomarker in the evaluation of patients with TNBC prognosis. In
addition, TTN-AS1 was found to regulate TNBC proliferation and
invasive and migratory abilities by targeting miR-211-5p, which may
provide novel insights into the mechanism of TNBC and help the
development of novel therapeutic interventions.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ES and XL interpreted the data and performed the
experiments. CL and KL collected the data, searched the literature
and designed the study. ES wrote the manuscript and KL revised the
manuscript. All authors read and approval the final manuscript.
Ethics approval and consent to
participate
All patients in this study signed the written
informed consents. This study was approved by the Ethics Committee
of Nanjing Maternal and Child Health Care Hospital and complied
with the principles of the Helsinki Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zagouri F, Sergentanis TN, Tsigginou A,
Dimitrakakis C, Zografos GC, Dimopoulos MA and Psaltopoulou T:
Female breast cancer in Europe: Statistics, diagnosis and treatment
modalities. J Thorac Dis. 6:589–590. 2014.PubMed/NCBI
|
|
2
|
Woolston C: Breast cancer. Nature.
527:S1012015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Sauer AG, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ai H, Zhou W, Wang Z, Qiong G, Chen Z and
Deng S: MicroRNAs-107 inhibited autophagy, proliferation, and
migration of breast cancer cells by targeting HMGB1. J Cell
Biochem. 2:10022018.
|
|
5
|
Cedolini C, Bertozzi S, Londero AP,
Bernardi S, Seriau L, Concina S, Cattin F and Risaliti A: Type of
breast cancer diagnosis, screening, and survival. Clin Breast
Cancer. 14:235–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Metzger-Filho O, Tutt A, de Azambuja E,
Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA Jr, Ellis
P, et al: Dissecting the heterogeneity of triple-negative breast
cancer. J Clin Oncol. 30:1879–1887. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karaayvaz M, Cristea S, Gillespie SM,
Patel AP, Mylvaganam R, Luo CC, Specht MC, Bernstein BE, Michor F
and Ellisen LW: Unravelling subclonal heterogeneity and aggressive
disease states in TNBC through single-cell RNA-seq. Nat Commun.
9:35882018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Costa FF: Non-coding RNAs: New players in
eukaryotic biology. Gene. 357:83–94. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mou E and Wang H: LncRNA LUCAT1
facilitates tumorigenesis and metastasis of triple-negative breast
cancer through modulating miR-5702. Biosci Rep. 39:BSR201904892019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ai B, Kong X, Wang X, Zhang K, Yang X,
Zhai J, Gao R, Qi Y, Wang J, Wang Z and Fang Y: LINC01355
suppresses breast cancer growth through FOXO3-mediated
transcriptional repression of CCND1. Cell Death Dis. 10:5022019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin CY, Zhang SN, Wang Y, Wang Y, Nice E,
Guo C, Zhang E, Yu L, Li M, Liu C, et al: Functional role of a
novel long noncoding RNA TTN-AS1 in esophageal squamous cell
carcinoma progression and metastasis. Clin Cancer Res. 24:486–498.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen P, Wang R, Yue QF and Hao MM: Long
non-coding RNA TTN-AS1 promotes cell growth and metastasis in
cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun.
503:2956–2962. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui ZH, Luo ZY, Lin ZM, Shi LH, Hong YR
and Yan C: Long non-coding RNA TTN-AS1 facilitates tumorigenesis of
papillary thyroid cancer through modulating the miR-153-3p/ZNRF2
axis. J Gene Med. 21:102019. View
Article : Google Scholar
|
|
15
|
Zhang SY, Ma HY, Zhang DM, Xie S, Wang W,
Li Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9:162018.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang JG, Wang L, Xu XL, et al:
Transcriptome-Based Network Analysis Unveils Eight Immune-Related
Genes as Molecular Signatures in the Immunomodulatory Subtype of
Triple-Negative Breast Cancer. Front. Oncol. 10–19. 2020.PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou XP, Wang YP, Li Q, Ma DH, Nie AQ and
Shen XL: LncRNA linc-PINT inhibits miR-523-3p to hamper
retinoblastoma progression by upregulating dickkopf-1 (DKK1).
Biochem Biophys Res Commun. 530:47–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qu XH, Shi YL, Ma Y, Bao WW, Yang L, Li JC
and Zhang F: LncRNA DANCR regulates the growth and metastasis of
oral squamous cell carcinoma cells via altering miR-216a-5p
expression. Hum Cell. 33:1281–1293. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia Y, Zhou Y, Han H, Li P, Wei W and Lin
NX: lncRNA NEAT1 facilitates melanoma cell proliferation,
migration, and invasion via regulating miR-495-3p and E2F3. J Cell
Physiol. 234:19592–19601. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li CL, Lv YL, Shao CY, Chen C, Zhang T,
Wei Y, Fan H, Lv T, Liu H and Song Y: Tumor-derived exosomal lncRNA
GAS5 as a biomarker for early-stage non-small-cell lung cancer
diagnosis. J Cell Physiol. 234:20721–20727. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo LY, Tang HL, Ling L, Li N, Jia X,
Zhang Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA
activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc
degradation in triple-negative breast cancer. Oncogene.
37:6166–6179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang G, Li H, Sun R, Li P, Yang Z, Liu Y,
Wang Z, Yang Y and Yin C: Long non-coding RNA ZEB2-AS1 promotes the
proliferation, metastasis and epithelial mesenchymal transition in
triple-negative breast cancer by epigenetically activating ZEB2. J
Cell Mol Med. 23:3271–3279. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Quan J, Pan X, He T, Lin C and Lai Y, Chen
P, Zhang Z, Yang S, Wang T and Lai Y: Tumor suppressor miR-211-5p
is associated with cellular migration, proliferation and apoptosis
in renal cell carcinoma. Exp Ther Med. 15:4019–4028.
2018.PubMed/NCBI
|
|
26
|
Liang MH, Jia JL, Chen LL, Wei B, Guan Q,
Ding Z, Yu J, Pang R and He G: LncRNA MCM3AP-AS1 promotes
proliferation and invasion through regulating miR-211-5p/SPARC axis
in papillary thyroid cancer. Endocrine. 65:318–326. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen LL, Zhang ZJ, Yi ZB and Li JJ:
MicroRNA-211-5p suppresses tumour cell proliferation, invasion,
migration and metastasis in triple-negative breast cancer by
directly targeting SETBP1. Br J Cancer. 117:78–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang W, Cui G, Ding M, Yang M and Dai D:
MicroRNA-124-3p.1 promotes cell proliferation through
axin1-dependent wnt signaling pathway and predicts a poor prognosis
of triple-negative breast cancer. J Clin Lab Anal.
3:e232662020.
|
|
29
|
Schroeder A, Heller DA, Winslow MM,
Dahlman JE, Pratt GW, Langer R, Jacks T and Anderson DG: Treating
metastatic cancer with nanotechnology. Nat Rev Cancer. 12:39–50.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang B, Shetti D, Fan C and Wei K:
MiR-29b-3p promotes progression of MDA-MB-231 triple-negative
breast cancer cells through downregulating TRAF3. Biol Res.
52:382019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang Q, Bournazou E, Sansone P, Berishaj
M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, et al:
The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and
metastasis. Neoplasia. 15:848–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kamran MZ, Patil P and Gude RP: Role of
STAT3 in cancer metastasis and translational advances. Biomed Res
Int. 2013:4218212013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie Q, Yang Z, Huang X, Zhang Z, Li J, Ju
J, Zhang H and Ma J: Ilamycin C induces apoptosis and inhibits
migration and invasion in triple-negative breast cancer by
suppressing IL-6/STAT3 pathway. J Hematol Oncol. 12:602019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ning N, Liu SL, Liu XH, Tian Z, Jiang Y,
Yu N, Tan B, Feng H, Feng X and Zou L: Curcumol inhibits the
proliferation and metastasis of melanoma via the
miR-152-3p/PI3K/AKT and ERK/NF-kB signaling pathways. J Cancer.
11:1679–1692. 2020. View Article : Google Scholar : PubMed/NCBI
|