Introduction
Hearing loss ranks fourth among the principal causes
of disability worldwide and leads to impaired communication, social
isolation and reduced quality of life (1). Hearing loss is a common sensory
disorder that results from genetic and environmental factors,
including genetic mutations, ototoxic drugs, noise exposure and
ageing (2). These physicochemical
or pathological factors could induce the damage or loss of human
inner ear hair cells (3).
Currently, the most common therapy for sensorineural hearing loss
is hearing rehabilitation with hearing devices; however, the sound
quality perceived is not as good as with the original cochlea
(4). Sensory hair cells are located
in the organ of Corti and act as mechanosensory cells (5). Mammalian auditory hair cells cannot
self-regenerate; therefore, the hearing loss caused by hair cell
loss is permanent (6). Regeneration
of cochlear hair cells has been considered a promising treatment
approach for noise-induced sensorineural hearing loss and
age-related hearing loss (7,8). It
was demonstrated that cochlear progenitor cells (CPCs) can
differentiate into either sensory hair cells or supporting cells,
whose number is the determinant factor of the final length of the
cochlea (9), indicating that
manipulation of progenitor cells could be considered as a key point
for hair cell regeneration. Further investigation on the mechanism
of CPC proliferation is therefore required.
Any dysregulation of microRNA (miRNA) in the cochlea
is likely to damage the structure of the auditory system and cause
hearing loss (10). For instance,
it has been reported that an increase in miR-34a level is
associated with increased hearing threshold and a larger loss of
hair cells in the cochlea (11).
However, the impact of dysregulated miRNAs on the biological
functions of CPCs remains unclear. In retinal progenitors, miR-125
has been reported to be expressed, but its expression is altered
over the developmental period (12). Furthermore, miR-125 significantly
suppresses the proliferation of cardiac progenitor cells during
hypoxia (13). However, the
expression and function of miR-125 in CPCs remain unclear. It has
been reported that cyclin-dependent kinase 2 (CDK2) activity could
determine whether eukaryotic cells enter the next cell cycle or
enter a transient G0-like state at the end of mitosis (14), suggesting the pivotal role of CDK2
in cell biological functions. Further investigating the effects of
the miR-125/CDK2 axis on the biological process of CPCs is
therefore required.
The present study aimed to identify the expression
levels and function of miR-125 in CPCs and to elucidate the
potential mechanism by which miR-125 regulates the proliferation of
CPCs. Identifying the role and underlying mechanism of miR-125 in
CPC proliferation may provide novel insights towards the
regeneration of hair cells and treatment of hearing loss.
Materials and methods
Ethical statement
Animal experiments were approved by the local Ethics
Committee of Hunan Provincial People's Hospital. All animals were
cared for according to the Guide for the Care and Use of Laboratory
Animals [National Institutes of Health (NIH); version 8, 2011].
Animals
Neonatal rats (0–3 days old) were purchased from
Shanghai Model Organisms Center, Inc. and housed under specific
pathogen-free conditions at humidity of 60–65%.
Isolation and culture of CPCs
After being intraperitoneally anaesthetized by 2%
pentobarbital sodium (40 mg/kg), the neonatal rats were sterilized
in 75% ethanol and sacrificed by decapitation. The bilateral
temporal bones were obtained and then placed in precooled (4°C)
°normal saline. The cochlear and spiral ligaments were removed by
microdissection. After being washed with Hank's balanced salt
solution (HBSS; HyClone; GE Healthcare Life Sciences) twice,
cochlear tissues were cut into pieces (0.5 mm3). Then,
the tissues were digested by 0.125% tryptase at 37°C for 20 min,
during which the tissues were flipped every 5 min using a
fire-polished Pasteur pipet. Subsequently, 10% foetal calf serum
(FCS, HyClone; GE Healthcare Life Sciences) was added for 5 min to
terminate the digestion at room temperature. Eventually, samples
were centrifuged at 111 × g for 5 min at room temperature. The
supernatant was discarded and the pellets were washed with HBSS
twice. DMEM/F12 (HyClone; GE Healthcare Life Sciences) containing
B27 (1:50) and N2 (1:100; both Sigma-Aldrich; Merck KGaA) was used
to resuspend cells that were filtered using a 100-mesh copper net
(pore size, 70 µm). A single cell suspension was prepared using
DMEM/F12 supplemented with epidermal growth factor (20 ng/ml),
basic fibroblast growth factor (20 ng/ml) and penicillin (100 U/ml)
(PeproTech China). Living cells (5×105 ml) were placed
in 24-well plates and cultured at 37°C in a humidified incubator
containing 5% CO2. The culture medium was replaced every
other day. Cells were passaged every 5 days at a density of
5×105/ml. Cells used for in vitro differentiation
were subjected to suspension culture at 37°C for 3 days. Then, the
culture medium was replaced with the aforementioned culture medium
containing 10% FCS. Afterwards, the cells were further cultured at
37°C for 12 days to induce differentiation.
In vitro differentiation of rat
CPCs
The 3rd generation of CPCs was cultured for 4 days,
then the CPCs were centrifuged at 88 × g for 10 min at room
temperature. Supernatant was removed and cells were resuspended in
DMEM/F12 containing 10% FCS. Cells (1×104) were
incubated in culture dishes that contained polylysine-encased
coverslips at 37°C with 5% CO2 for 12 h. Once cells had
adhered, the supernatant was discarded and the medium was replaced.
Half of the medium was replaced every other day until the seventh
day. The medium consisted of DMEM/F12 solution, 100 U/ml
penicillin-streptomycin, 2% B27 and 2% N2.
Cell proliferation evaluation by MTT
assay
Cells (1×106/ml) in different growth
phases were seeded in a 96-well plate (100 µl/well) and incubated
at 37°C with 5% CO2. Then, cells were incubated with 10
µl of 5 mg/ml MTT (Beyotime Institute of Biotechnology) for 4 h at
37°C. Subsequently, the MTT solution was discarded and 20% sodium
dodecyl sulphate (100 µl/well) was added to the cells for 4 h at
37°C with 5% CO2 to terminate the reaction. The
absorbance was read at 570 nm using a microplate reader (SpectraMax
190; Molecular Devices, LLC).
Cell transfection
After incubation for 2 days, CPCs were transfected
with miR-125 mimic (50 nM), miR-125 inhibitor (50 nM), small
interfering (si)-CDK2 (2 µg) or the corresponding negative controls
(NC mimic, NC inhibitor or si-NC) or were co-transfected with
miR-125 inhibitor + si-CDK2 (all Shanghai GenePharma Co., Ltd.)
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The
sequences were as follows: miR-125 mimic, forward
5′-ACAAGUCAGGUUCUUGGGACCU-3′, reverse 3′-UGUUCAGUCCAAGAACCCUGGA-5′;
mimic NC, forward 5′-ACACGUCAGCAUUAACUCCUUG-3′, reverse
3′-UGUGCAGUCGUAAUUGAGGAAC-5′; miR-125 inhibitor
5′-UGUUCAGUCCAAGAACCCUGGA-3′; inhibitor NC,
5′-ACUGCCAUCUAUCUCGGAACGA-3′; si-CDK2, GGTGTACCCAGTACTGCCA;
si-NCGACTTCATAAGGCGATGC. Cell transfection was performed for 48 h
at room temperature, after which cells were cultured at 37°C for 48
h before subsequent experiments.
Immunofluorescence
Cells were collected and resuspended in culture
medium containing 10% FCS. Cells were then seeded onto
polylysine-treated coverslips at a density of 1×105 and
incubated at 5% CO2 and 37°C. Following incubation for
24 h, the cells used for identification of CPCs and differentiation
were fixed with 4% paraformaldehyde for 20 min at room temperature
and washed with PBS three times. Subsequently, cells were incubated
with 1% Triton X-100 on ice for 5 min, washed with PBS three times
and blocked with 5% bovine serum albumin (cat. no. ST025; Beyotime
Institute of Biotechnology) diluted in PBS. The blocking buffer was
discarded and cells were incubated with primary antibodies against
nestin (1:100; cat. no. MAB353; EMD Millipore), BrdU (1:500; cat.
no. MAB4072; EMD Millipore) and myosin VIIA (1:100; cat. no.
Ab150386; Abcam) at 4°C overnight. Subsequently, cells were washed
with PBS three times and incubated with Cy3-labelled secondary
antibody IgG (1:100; Sigma-Aldrich; Merck KGaA) at 37°C for 30 min.
The nuclei were stained with DAPI (5 µg/ml; cat. no. C1002;
Beyotime Institute of Biotechnology) at room temperature for 5 min.
After being washed with PBS three times, cells were visualized and
imaged under a fluorescence microscope (magnification, ×400;
Olympus BX51; Olympus Corporation).
Cell cycle analysis
Cells were washed with ice-cold PBS three times and
centrifuged at 1,000 × g at room temperature for 5 min. Cells were
suspended in ice-cold PBS and fixed in absolute ethanol for 30 min
at 4°C. The ethanol was discarded after incubation and cells were
washed with PBS to remove the residual ethanol. Subsequently, cells
were resuspended in PBS with 3 µl RNAse (cat. no. ST578; Beyotime
Institute of Biotechnology) and incubated at 37°C for 30 min.
Eventually, cells were stained with propidium iodide (PI; 50 µg/ml;
Sigma-Aldrich; Merck KGaA) for 30 min at 37°C. Cell cycle
distribution was detected by flow cytometry and data were analyzed
using CellQuest Pro software version 5.1 (FACSCalibur; Becton,
Dickinson and Company).
Reverse transcription quantitative
(RT-q) PCR
CPCs were lysed in 1 ml TRIzol® (Thermo
Fisher Scientific, Inc.), and total RNA was extracted according to
the manufacturer's instructions. Following RNA quantification, cDNA
was obtained by reverse transcription using BeyoRT™ II and cDNA
synthesis kit (cat. no. D7168M; Beyotime Institute of
Biotechnology), and then subjected to RT-qPCR using a Quanti Fast
SYBR® Green PCR kit (Qiagen, Inc.). The procedures were
performed as follows: 40 cycles of pre-degradation at 95°C for 2
min, degradation at 95°C for 10 sec, annealing at 60°C for 40 sec
and extension at 72°C for 20 sec. Each experiment was repeated
three times. The relative expression levels were normalized to
endogenous controls GAPDH or U6 (for endogenous normalization for
miRNA) and were expressed as 2−ΔΔCq (15). The sequences of the primers used are
presented in Table I.
 | Table I.Sequences of the primers used for
reverse transcription quantitative PCR. |
Table I.
Sequences of the primers used for
reverse transcription quantitative PCR.
| Name | Primer sequence
(5′→3′) |
|---|
| miR-125 |
|
|
Forward | TCCAGGGTTCTTGGAC |
|
Reverse |
GCAGGGTCCGAGGTATTC |
| CDK2 |
|
|
Forward |
TGCCCTTTCACTGCCTATGG |
|
Reverse |
GAGGAAAGCCAAGACCCACA |
| PCNA |
|
|
Forward |
CTCCTCATCCTTGCGTCCTCATAT |
|
Reverse |
GAGGCACTTGGCAATGTATTCGATAT |
| Nestin |
|
|
Forward |
ATCTACACATACACGGGTTCCA |
|
Reverse |
TTCTTCTTCTCCTCCTCATTCA |
| U6 |
|
|
Forward |
CTCGCTTCGGCAGCACA |
|
Reverse |
AACGCTTCACGAATTTGCGT |
| GAPDH |
|
|
Forward |
TCTTGTGCAGTGCCAGCCT |
|
Reverse |
TGAGGTCAATGAAGGGGTCG |
Western blotting
CPCs were lysed using RIPA lysis buffer (Beyotime
Institute of Biotechnology) on ice and the proteins were quantified
using bicinchoninic acid (Beyotime Institute of Biotechnology).
Proteins (50 µg/lane) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes. Subsequently, the membranes were
blocked with Tris buffer containing 5% non-fat milk at room
temperature for 1 h and incubated with primary antibodies against
GAPDH (1:10,000; cat. no. ab181602; Abcam), CDK2 (1:1,000; cat. no.
ab32147; Abcam), proliferating cell nuclear antigen (PCNA; 1:1,000;
ab92552; Abcam), nestin (1:1,000; cat. no. ab6142; Abcam) and
β-catenin (1:5,000; cat. no. ab32572; Abcam) at 4°C overnight.
Membranes were washed with PBST (10% Tween-20) three times and were
incubated with goat anti-rabbit IgG (1:5,000; cat. no. CW0103S;
CoWin Biosciences) or goat anti-rat IgG (1:2,000; cat. no.
ab205719; Abcam) secondary antibodies at room temperature for 30
min. Membranes were washed with PBST four times and enhanced
chemiluminescence reagent (cat. no. CW0049S; CoWin Biosciences) was
used to detect the signal on the membrane using a chemiluminescence
imaging system (GE Healthcare). ImageJ software (version 1.46; NIH)
was used to analyze protein bands.
Dual luciferase reporter assay
TargetScan (targetscan.org/vert_72/) was used to predict the
binding sites between miR-125 and CDK2. According to the
prediction, wild-type (wt) and mutant (mut)-type sequences of the
binding sites between miR-125 and CDK2 were synthesized and cloned
into the reporter vectors (pGL3-Basic, Promega Corporation), which
were named mut-CDK2 and wt-CDK2. 293T cells (American Type Culture
Collection) were co-transfected with mut-CDK2 or wt-CDK2 and
miR-125 mimic, miR-125 inhibitor, NC mimic or NC inhibitor
(GenePharma, Shanghai, China). OPTI-MEM (49 µl) was pipetted onto
24-well plates to dilute 1 µl Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and the final volume
was 50 µl. After 48 h transfection, luciferase activity was
detected using a Lucifer Reporter analytic system (Promega
Corporation) by comparison with Renilla luciferase
activity.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 7.0 (GraphPad Software, Inc.). Comparisons between two groups
were measured by Student's t-test, whereas comparisons among
multiple groups were conducted by one-way analysis of variance
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Evaluation of the CPC in vitro
differentiation model
After culture, cells that were isolated from the
cochleae of neonatal rats formed progenitor spheres with different
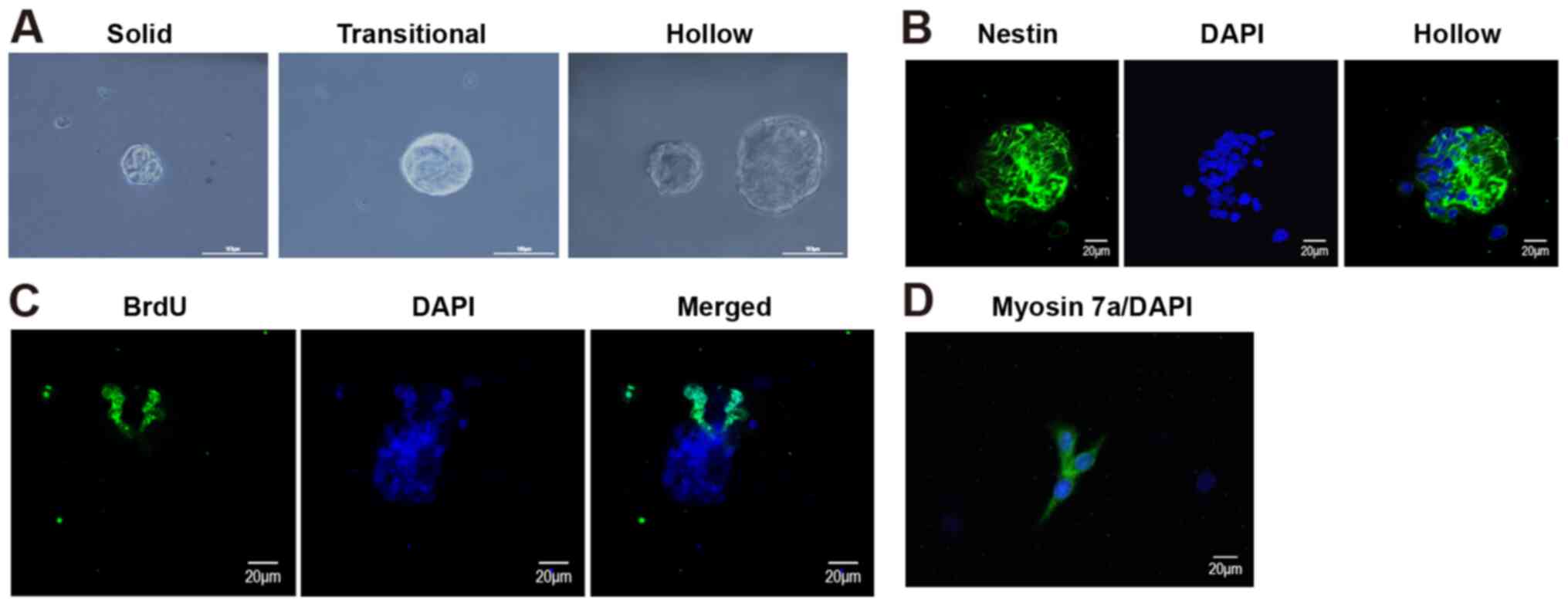
morphologies, including solid, transitional and hollow spheres
(Fig. 1A). Furthermore, a fraction
of CPCs was positive for nestin and BrdU. As a marker of neural
stem cells, nestin was mainly expressed in the cytoplasm (Fig. 1B), and BrdU, which is a marker for
mitosis, was mainly expressed in the nuclei (Fig. 1C). To identify the directional
differentiation cell capacity, in vitro directional
differentiation was induced for 12 days. Immunofluorescence was
used to detect the expression of the hair cell marker myosin VII,
and the results suggested that myosin VII was expressed in the
isolated cells (Fig. 1D), which
confirmed that the isolated cells had the potential to
differentiate into hair cells.
Identification of CPC
proliferation
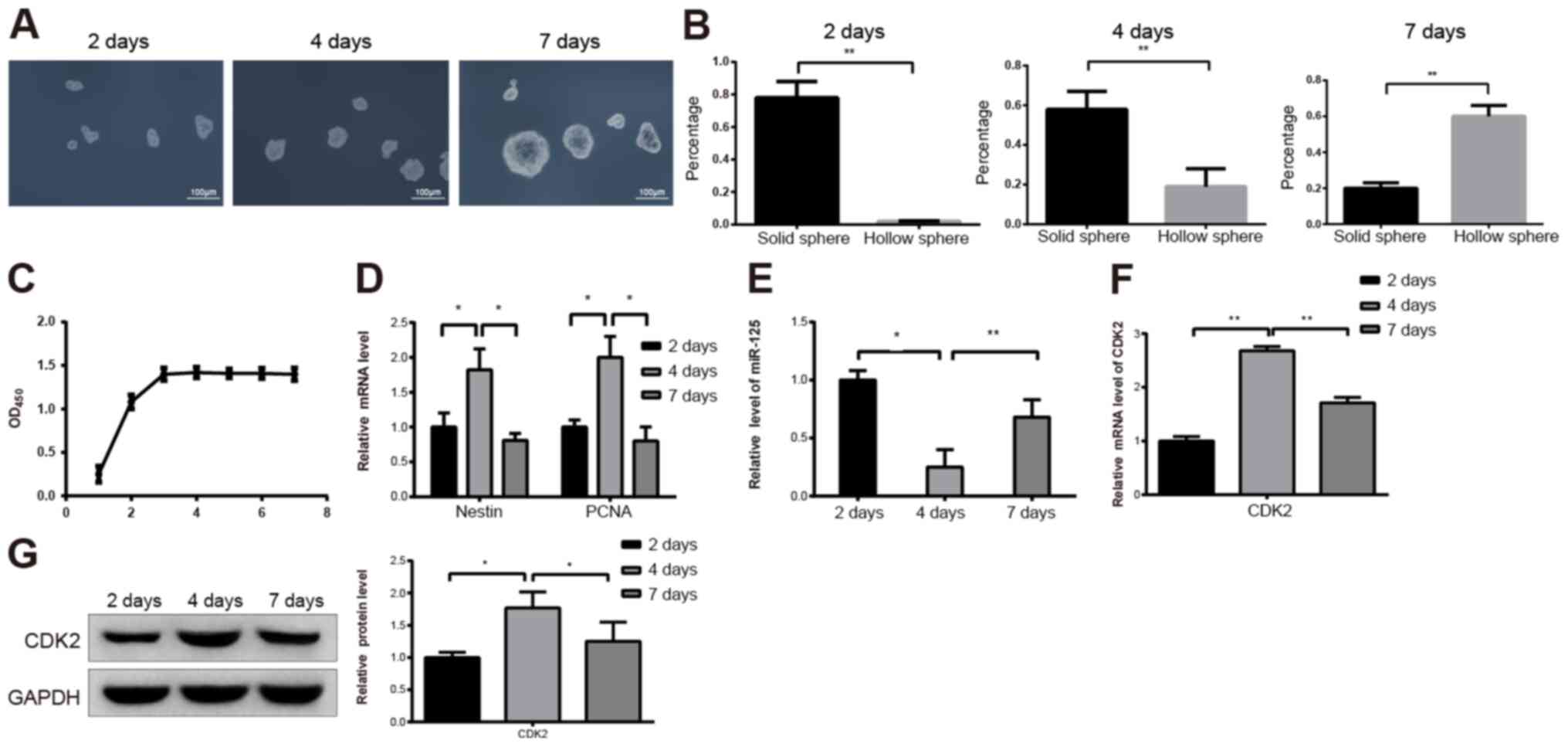
The three types of progenitor spheres had different
proportions depending on culture durations. For spheres cultured
for 2, 4 and 7 days, the size of the progenitor spheres gradually
increased in a time-dependent manner (Fig. 2A). During this time, the proportion
of solid spheres decreased and the proportion of hollow spheres
increased (Fig. 2B). CPCs in
different growth phases were subjected to MTT to measure their
proliferation abilities. The results demonstrated that CPCs were in
the exponential phase after incubation for 1–3 days, and the cells
reached a plateau after 3 days of incubation (Fig. 2C; P<0.05). The results from
RT-qPCR demonstrated that the expression levels of nestin and PCNA
were significantly increased and decreased after 4 and 7 days,
respectively (Fig. 2D;
P<0.05).
To investigate the role of miR-125 in CPC
proliferation, miR-125 expression was detected by RT-qPCR. In the
spheres cultured for 4 days, the expression of miR-125 was
significantly lower than that in the spheres cultured for 2 days
(Fig. 2E; P<0.05). Furthermore,
in the spheres cultured for 7 days, miR-125 expression was higher
than that in the spheres cultured for 4 days (Fig. 2E; P<0.01). In addition, results
from RT-qPCR demonstrated that CDK2 expression level was
significantly increased in spheres cultured for 4 days compared
with spheres cultured for 2 days (Fig.
2F; P<0.01). In addition, CDK2 expression level decreased
significantly in the spheres cultured for 7 days compared with
spheres cultured for 4 days (Fig.
2F; P<0.01). These findings were confirmed by western
blotting (Fig. 2G; P<0.05).
Inhibitory effect of miR-125 on CPC
proliferation
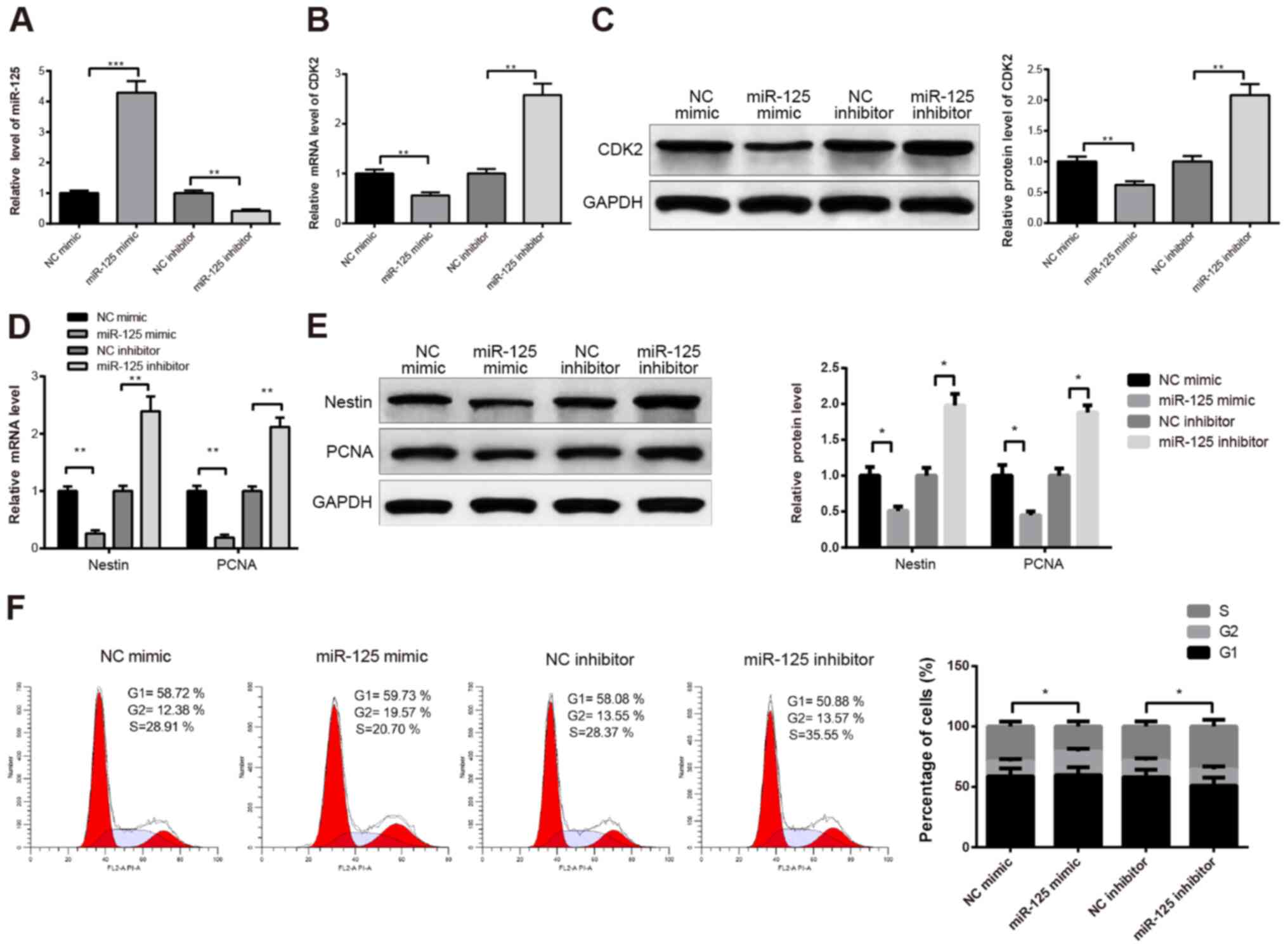
To further explore the effect of miR-125 on CPC
proliferation, CPCs cultured for 2 days were transfected with
miR-125 mimic or miR-125 inhibitor. As confirmed by RT-qPCR,
miR-125 expression was elevated in the miR-125 mimic group compared
with the NC mimic group, and the expression of miR-125 was
decreased in the miR-125 inhibitor group compared with the NC
inhibitor group (Fig. 3A;
P<0.001 and P<0.01). Furthermore, the expression level of
CDK2 was significantly decreased in the miR-125 mimic group
compared with the NC mimic group (Fig.
3B; P<0.01), and significantly increased in the miR-125
inhibitor group compared with NC inhibitor group (Fig. 3B; P<0.01). These results were
confirmed by western blotting (Fig.
3C). These findings suggested that miR-125 could negatively
regulate CDK2. Furthermore, the results from RT-qPCR demonstrated
that nestin and PCNA were significantly downregulated in the
miR-125 mimic group compared with the NC mimic group, and
significantly upregulated in the miR-125 inhibitor group compared
with the NC inhibitor group (Fig.
3D; P<0.01). These data were confirmed by western blotting
(Fig. 3E; P<0.05). The detection
of the cell cycle by flow cytometry demonstrated that miR-125
inhibition increased the number of cells in the S phase, and that
miR-125 overexpression had the opposite effect (Fig. 3F; P<0.05). Taken together, these
findings suggested that miR-125 may inhibit CPC proliferation.
miR-125 exerts its regulatory effect
on CPCs via CDK2
TargetScan (targetscan.org/vert_72/) results demonstrated that
there were binding sites between miR-125 and CDK2 (Fig. 4A). According to these results,
wt-CDK2 and mut-CDK2 were constructed, and 293T cells were
co-transfected with wt-CDK2 or mut-CDK2 and miR-125 mimic or
miR-125 inhibitor. The results from dual luciferase reporter assay
demonstrated that miR-125 mimic significantly decreased the
luciferase activity of 293T cells transfected with wt-CDK2
(P<0.01), and that miR-125 mimic did not alter the luciferase
activity of 293T cells transfected with mut-CDK2 compared with the
NC mimic group (Fig. 4B).
Subsequently, transfection with miR-125 inhibitor significantly
increased the luciferase activity of 293T cells transfected with
wt-CDK2 (P<0.01), whereas the luciferase activity of 293T cells
transfected with mut-CDK2 was not changed by the miR-125 inhibitor
compared with the NC inhibitor (Fig.
4B).
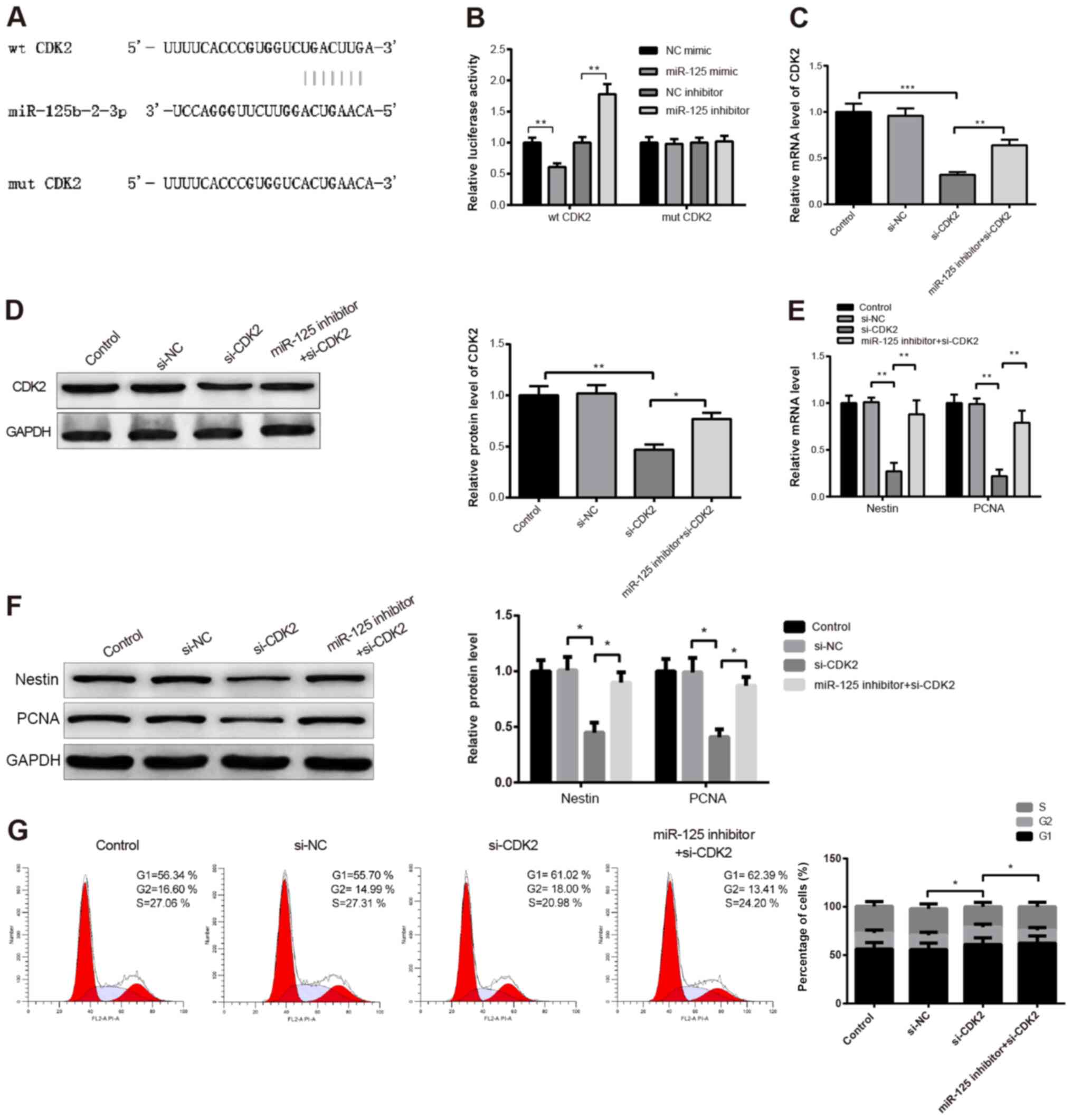 | Figure 4.miR-125 regulated CPC proliferation by
downregulating CDK2. (A) Following CPC transfection with miR-125
mimic or miR-125 inhibitor, the potential binding sites between
miR-125 and CDK2 were predicted. (B) Detection of luciferase
activity of 293T cells transfected with wt-CDK2 or mut-CDK2. mRNA
and protein expression of CDK2 was determined by (C) RT-qPCR and
(D) western blotting in CPCs co-transfected with miR-125 inhibitor
and si-CDK2. mRNA and protein expression of nestin and PCNA was
measured by (E) RT-qPCR and (F) western blotting. (G) Cell cycle
analysis was detected by flow cytometry. All data were expressed as
the means ± standard deviation. *P<0.05, **P<0.01
***P<0.001. CPCs, cochlear progenitor cells; PCNA, proliferating
cell nuclear antigen; RT-qPCR, reverse transcription quantitative;
si, small interfering; mut, mutant; wt, wild type; NC, negative
control; CDK2, cyclin-dependent kinase 2. |
To test the interactions between miR-125 and CDK2,
CPCs cultured for 2 days were co-transfected with miR-125 inhibitor
and si-CDK2. The detection of the transfection efficiency of
si-CDK2 by RT-qPCR and western blotting revealed a satisfactory
CDK2 knockdown in CPCs (Fig. 4C and
D; *P<0.05, **P<0.01, ***P<0.001). Furthermore,
miR-125 inhibitor upregulated CDK2 expression (Fig. 4C and D; *P<0.05, **P<0.01,
***P<0.001). In addition, the expression levels of nestin and
PCNA were significantly decreased following CDK2 knockdown compared
with control group (Fig. 4E;
P<0.01), whereas they were significantly increased in the
miR-125 inhibitor + si-CDK2 group compared with the si-CDK2 group
(Fig. 4E; P<0.01). Similar
results were observed by western blotting (Fig. 4F; P<0.05). In addition, the
number of cells in the S phase was decreased in the si-CDK2 group
compared with the control group, and was increased in the miR-125
inhibitor + si-CDK2 group compared with the si-CDK2 group (Fig. 4G; P<0.05). Taken together, these
findings suggested that miR-125 may inhibit CPC proliferation by
downregulating CDK2.
Discussion
CPCs have been considered as the best candidates for
hair cell regeneration (16). In
the past decade, much attention has been given to the regeneration
of hair cells from CPCs to rescue hearing loss. For example, a
previous study isolated progenitors with high Lgr6 expression
levels from transgenic mice and found that high Lgr6 expression
elevated the population of progenitor cells, increasing therefore
hair cell generation (17). The
present study described the role and mechanism of miR-125 in CPC
proliferation. Exploring the mechanisms underlying CPC
proliferation may account for the treatment of hearing loss
following hair cell damage.
To determine the mechanism of CPC differentiation
into hair cells, cells were isolated from the cochleae of neonatal
rats and further examined. BrdU, a thymine analogue, can be
incorporated into new DNA during DNA synthesis (S phase) (18), suggesting that BrdU-expressing cells
have proliferation ability. Nestin is a class VI intermediate
filament protein associated with pluripotency in pluripotent stem
cells, which is abundantly expressed in the developing central
nervous system and downregulated in proliferative areas of the
dentate gyrus and subventricular zone in adults (19). Nestin-expressing cells present a
stem- or progenitor-like character. In the present study, cells
isolated from the cochleae of neonatal rats were characterized by
observing the number of cells positive for BrdU and nestin. The
results demonstrated that nestin was mainly expressed in the
cytoplasm whereas BrdU was expressed in the nuclei. Further results
suggested that isolated cells had the potential to differentiate
into hair cells, as evidenced by the expression of myosin VIIA,
which normally functions in the cochlear hair cells of the inner
ear. Furthermore, the results from the present study demonstrated
that CPC proliferation gradually decreased in a time-dependent
manner.
miRNAs have integral roles in regulating cell
proliferation, differentiation and maturation in addition to the
cell fate determination of stem cells (20). Numerous miRNAs, such as miR-124 and
miR-182, have been implicated in inner ear development and hair
cell fate by modulating downstream mRNA expression (21,22).
However, the roles of miRNAs in mediating CPC proliferation remain
unclear. The inhibitory effects of miR-125 family members on cell
proliferation have been reported in various types of cell,
including colorectal cancer cells, cardiomyocytes and osteoblasts
(23–25). In the present study, miR-125 was
found to be downregulated and eventually upregulated in the
progenitor spheres. Furthermore, overexpression of miR-125 in CPCs
decreased the levels of nestin and PCNA as well as the number of
cells in the S phase. A previous study demonstrated that NEUROG1
overexpression can inhibit the proliferation of otic progenitors by
decreasing CDK2 expression (26).
In the present study, the expression of CDK2 was increased and then
decreased in CPCs, suggesting the involvement of CDK2 in CPC
proliferation. In addition, miR-125 upregulation decreased the
expression of CDK2 and CDK2 expression was upregulated by the
introduction of miR-125 inhibition into CPCs. To determine the
mechanism of miR-125 on CPC proliferation, the relationship between
miR-125 and CDK2 was investigated. A dual luciferase reporter assay
demonstrated that miR-125 could negatively target CDK2. The results
also revealed that miR-125 inhibition reversed the suppressive
effect of CDK2 knockdown on CPC proliferation. Taken together, this
study demonstrated that miR-125 inhibited CPC proliferation by
downregulating CDK2. This study did not investigate the role of the
miR-125/CDK2 axis in other biological processes of CPCs, such as
differentiation, although results confirmed that isolated CPCs had
the potential to differentiate into hair cells and that CPC
proliferation was attenuated after incubation for 7 days. Previous
studies have reported the essential roles of miR-125 and CDK2 in
progenitor or stem cell differentiation (27–29).
The function of miR-125/CDK2 axis in CPC differentiation requires
further investigation.
In summary, the results from the present study
suggested that CPCs may have the potential to differentiate into
hair cells. In addition, miR-125 inhibited CPC proliferation by
negatively targeting CDK2. Hearing loss treatment based on
progenitor or stem cell strategies could therefore be considered;
however, the mechanism of CPC proliferation needs to be further
investigated. This study attempted to explain the molecular
mechanisms of miRNA-125 on the regeneration of hair cells.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China for Distinguished Young Scholars (grant
no. 81800921), Natural Science Foundation of Hunan Province (grant
no. 2020JJ5303), the Foundation of Health and Family Planning
Commission of Hunan Province (grant no. C2017033) and Hunan
Provincial Technology Innovation-Oriented Projects (grant no.
2018SK50705).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TP conceptualized and designed the experiments and
supervised the study. JJP performed the experiments. GYM, ZQT, BL
and EZ analyzed the data. BL and EZ revised the manuscript
critically for important intellectual content. TP and JJP wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the Ethics
Committee of Hunan Provincial People's Hospital (approval no.
2019S47).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cunningham LL and Tucci DL: Hearing loss
in adults. N Engl J Med. 377:2465–2473. 2017. View Article : Google Scholar
|
|
2
|
Kim YR, Baek JI, Kim SH, Kim MA, Lee B,
Ryu N, Kim KH, Choi DG, Kim HM, Murphy MP, et al: Therapeutic
potential of the mitochondria-targeted antioxidant MitoQ in
mitochondrial-ROS induced sensorineural hearing loss caused by Idh2
deficiency. Redox Biol. 20:544–555. 2019. View Article : Google Scholar
|
|
3
|
Tang M, Yan X, Tang Q, Guo R, Da P and Li
D: Potential application of electrical stimulation in stem
cell-based treatment against hearing loss. Neural Plast.
2018:95063872018. View Article : Google Scholar
|
|
4
|
Lee MY and Park YH: Potential of gene and
cell therapy for inner ear hair cells. Biomed Res Int.
2018:81376142018. View Article : Google Scholar
|
|
5
|
Roccio M, Perny M, Ealy M, Widmer HR,
Heller S and Senn P: Molecular characterization and prospective
isolation of human fetal cochlear hair cell progenitors. Nat
Commun. 9:40272018. View Article : Google Scholar
|
|
6
|
Lin SCY, Thorne PR, Housley GD and
Vlajkovic SM: Purinergic signaling and aminoglycoside ototoxicity:
The opposing roles of P1 (Adenosine) and P2 (ATP) receptors on
cochlear hair cell survival. Front Cell Neurosci. 13:2072019.
View Article : Google Scholar
|
|
7
|
Revuelta M, Santaolalla F, Arteaga O,
Alvarez A, Sanchez-Del-Rey A and Hilario E: Recent advances in
cochlear hair cell regeneration-A promising opportunity for the
treatment of age-related hearing loss. Ageing Res Rev. 36:149–155.
2017. View Article : Google Scholar
|
|
8
|
Youm I and Li W: Cochlear hair cell
regeneration: An emerging opportunity to cure noise-induced
sensorineural hearing loss. Drug Discov Today. 23:1564–1569. 2018.
View Article : Google Scholar
|
|
9
|
Huh SH, Warchol ME and Ornitz DM: Cochlear
progenitor number is controlled through mesenchymal FGF receptor
signaling. Elife. 4:e059212015. View Article : Google Scholar
|
|
10
|
Mittal R, Liu G, Polineni SP, Bencie N,
Yan D and Liu XZ: Role of microRNAs in inner ear development and
hearing loss. Gene. 686:49–55. 2019. View Article : Google Scholar
|
|
11
|
Pang J, Xiong H, Yang H, Ou Y, Xu Y, Huang
Q, Lai L, Chen S, Zhang Z, Cai Y and Zheng Y: Circulating miR-34a
levels correlate with age-related hearing loss in mice and humans.
Exp Gerontol. 76:58–67. 2016. View Article : Google Scholar
|
|
12
|
La Torre A, Georgi S and Reh TA: Conserved
microRNA pathway regulates developmental timing of retinal
neurogenesis. Proc Natl Acad Sci USA. 110:E2362–E2370. 2013.
View Article : Google Scholar
|
|
13
|
Li L, Wang Q, Yuan Z, Chen A, Liu Z, Wang
Z and Li H: LncRNA-MALAT1 promotes CPC proliferation and migration
in hypoxia by up-regulation of JMJD6 via sponging miR-125. Biochem
Biophys Res Commun. 499:711–718. 2018. View Article : Google Scholar
|
|
14
|
Spencer SL, Cappell SD, Tsai FC, Overton
KW, Wang CL and Meyer T: The proliferation-quiescence decision is
controlled by a bifurcation in CDK2 activity at mitotic exit. Cell.
155:369–383. 2013. View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Song YL, Tian KY, Mi WJ, Ding ZJ, Qiu Y,
Chen FQ, Zha DJ and Qiu JH: Decreased expression of TERT correlated
with postnatal cochlear development and proliferation reduction of
cochlear progenitor cells. Mol Med Rep. 17:6077–6083. 2018.
|
|
17
|
Zhang Y, Guo L, Lu X, Cheng C, Sun S, Li
W, Zhao L, Lai C, Zhang S, Yu C, et al: Characterization of
Lgr6+ cells as an enriched population of hair cell
progenitors compared to Lgr5+ cells for hair cell
generation in the neonatal mouse cochlea. Front Mol Neurosci.
11:1472018. View Article : Google Scholar
|
|
18
|
Gao J, Wan F, Tian M, Li Y, Li Y, Li Q,
Zhang J, Wang Y, Huang X, Zhang L and Si Y: Effects of
ginsenoside-Rg1 on the proliferation and gliallike directed
differentiation of embryonic rat cortical neural stem cells in
vitro. Mol Med Rep. 16:8875–8881. 2017. View Article : Google Scholar
|
|
19
|
Takeda H, Dondzillo A, Randall JA and
Gubbels SP: Challenges in cell-based therapies for the treatment of
hearing loss. Trends Neurosci. 41:823–837. 2018. View Article : Google Scholar
|
|
20
|
Hei R, Chen J, Qiao L, Li X, Mao X, Qiu J
and Qu J: Dynamic changes in microRNA expression during
differentiation of rat cochlear progenitor cells in vitro. Int J
Pediatr Otorhinolaryngol. 75:1010–1014. 2011. View Article : Google Scholar
|
|
21
|
Huyghe A, Van den Ackerveken P, Sacheli R,
Prevot PP, Thelen N, Renauld J, Thiry M, Delacroix L, Nguyen L and
Malgrange B: MicroRNA-124 regulates cell specification in the
cochlea through modulation of Sfrp4/5. Cell Rep. 13:31–42. 2015.
View Article : Google Scholar
|
|
22
|
Wang XR, Zhang XM, Du J and Jiang H:
MicroRNA-182 regulates otocyst-derived cell differentiation and
targets T-box1 gene. Hear Res. 286:55–63. 2012. View Article : Google Scholar
|
|
23
|
Yang M, Tang X, Wang Z, Wu X, Tang D and
Wang D: miR-125 inhibits colorectal cancer proliferation and
invasion by targeting TAZ. Biosci Rep. 39:BSR201901932019.
View Article : Google Scholar
|
|
24
|
Li L, Zhang M, Chen W, Wang R, Ye Z, Wang
Y, Li X and Cai C: LncRNA-HOTAIR inhibition aggravates oxidative
stress-induced H9c2 cells injury through suppression of MMP2 by
miR-125. Acta Biochim Biophys Sin (Shanghai). 50:996–1006. 2018.
View Article : Google Scholar
|
|
25
|
Tu XM, Gu YL and Ren GQ: miR-125a-3p
targetedly regulates GIT1 expression to inhibit osteoblastic
proliferation and differentiation. Exp Ther Med. 12:4099–4106.
2016. View Article : Google Scholar
|
|
26
|
Song Z, Jadali A, Fritzsch B and Kwan KY:
NEUROG1 regulates CDK2 to promote proliferation in Otic
progenitors. Stem Cell Reports. 9:1516–1529. 2017. View Article : Google Scholar
|
|
27
|
Kaur S, Abu-Shahba AG, Paananen RO,
Hongisto H, Hiidenmaa H, Skottman H, Seppanen-Kaijansinkko R and
Mannerström B: Small non-coding RNA landscape of extracellular
vesicles from human stem cells. Sci Rep. 8:155032018. View Article : Google Scholar
|
|
28
|
Malgrange B, Knockaert M, Belachew S,
Nguyen L, Moonen G, Meijer L and Lefebvre PP: The inhibition of
Cyclin-dependent kinases induces differentiation of supernumerary
hair cells and Deiters' cells in the developing organ of Corti.
FASEB J. 17:2136–2138. 2003. View Article : Google Scholar
|
|
29
|
Shenoy A, Danial M and Blelloch RH: Let-7
and miR-125 cooperate to prime progenitors for astrogliogenesis.
EMBO J. 34:1180–1194. 2015. View Article : Google Scholar
|