Introduction
Non-small-cell lung cancer (NSCLC) is a disease in
which malignant cells proliferate in the tissues of the lung. It
accounts for ~85% of all lung cancer cases, and is the leading
cause of cancer-associated mortality worldwide (1). There are several types NSCLC depending
on the type of cancer cell, including squamous cell carcinoma
(squamous cell origin), large cell carcinoma (numerous types of
undifferentiated cells) and adenocarcinoma (cells that line the
alveoli) (2). It is well known that
smoking is a major risk factor for the development of NSCLC
(3). Additionally, there are
numerous modes of NSCLC treatment, including surgery, radiation
therapy, chemotherapy, targeted therapy, laser therapy,
photodynamic therapy, cryosurgery and radiosensitizers (4); however, due to limitations in
diagnosis, the majority of patients with NSCLC are diagnosed at
stages III and IV, for whom existing treatments are not curative
(5). Therefore, there is an urgent
need for more efficient therapeutic approaches for NSCLC
treatment.
MicroRNAs (miRNAs) are a group of small non-coding
RNAs comprising 22–24 nucleotides (6). miRNAs specifically bind to the 3′
untranslated region (3′ UTR) of target mRNAs, leading to mRNA
degradation and abnormal levels of protein expression (7). miRNAs are differentially expressed in
tumor and normal tissues, and function as oncogenes or tumor
suppressors (8). Numerous miRNAs
have been reported to serve key roles in the pathogenesis of NSCLC
(9). miR-34b and miR-520h have been
suggested to be key factors in the regulation of NSCLC, and miR-22
has been reported as an oncogene and novel biomarker (9,10). The
tumor suppressor properties of miR-654-3p have been investigated in
papillary thyroid cancer and miR-654-3p has also been suggested as
a potential biomarker for the tumorigenicity of VERO cells
(11,12). It has been demonstrated that the
expression of miR-654-3p differed in NSCLC tissues from normal
tissues (9).
Ras protein activator like 2 (RASAL2) gene encodes
the enzyme, RasGTPase-activating protein nGAP (13). RasGTPases are essential components
of signaling pathways that propagate signals from cell surface
receptors to regulate a variety of cellular processes, including
cell cycle progression, cell survival, actin cytoskeletal
organization, cell polarity and movement (14). It was recently demonstrated that
RASAL2 serves roles as a tumor and metastasis suppressor by
inhibiting the proliferative and metastatic abilities of
nasopharyngeal carcinoma cells (15).
In the present study, bioinformatics research using
TargetScan, revealed that RASAL-2 was a target of miR-654-3p. The
present study aimed to investigate the effects of miR-654-3p on the
viability and apoptosis of NSCLC cells by targeting RASAL2, which
may have potential as a novel therapeutic target for the treatment
of NSCLC.
Materials and methods
Patients and tissues
Tumor tissues were collected from 45 patients with
NSCLC between August 2016 and August 2017 at Linyi Central Hospital
(Linyi, China). The clinicopathological features (including gender,
age, tumour stage, and tumor, node and metastasis staging) of
patients with NSCLC enrolled in our study were presented in
Table I. The tumor and the
paracancerous tissues were fixed in 10% formalin for 48 h at room
temperature. Tumor and non-tumor samples were confirmed by
pathological examination. No patients received chemotherapy or
radiotherapy prior to surgery. In addition, the present study was
approved by the ethics committee of Linyi Central Hospital, and
written informed consent was obtained from all patients prior to
enrolment.
 | Table I.Expression of miR-645 and RASAL2 in
the tissues of patients with lung carcinoma. |
Table I.
Expression of miR-645 and RASAL2 in
the tissues of patients with lung carcinoma.
| Factors | Case | Low miR-645 [n
(%)] | High miR-645 [n
(%)] | χ2 | P-value | Low RASAL2 [n
(%)] | High RASAL2 [n
(%)] | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
| Male | 34 (75.56%) | 18 (40.00) | 16 (35.56) | 2.200 | 0.138 | 19 (42.22) | 15 (33.33) | 1.267 | 0.260 |
|
Female | 11 (24.44%) | 3 (6.67) | 8 (17.78) |
|
| 4 (8.89) | 7 (15.56) |
|
|
| Age (years) |
|
|
|
|
|
|
|
|
|
|
<60 | 8 (17.78%) | 2 (4.44) | 6 (13.33) | 1.835 | 0.176 | 3 (6.67) | 5 (11.11) | 0.721 | 0.396 |
| ≥60 | 37 (82.22%) | 19 (42.22) | 18 (40.00) |
|
| 20 (44.44) | 17 (37.78) |
|
|
| Tumor size (cm) |
|
|
|
|
|
|
|
|
|
| ≥5 | 18 (40.00%) | 12 (26.67) | 6 (13.33) | 4.821 | 0.028 | 13 (28.89) | 5 (11.11) | 5.351 | 0.021 |
|
<5 | 27 (60.00%) | 9 (20.00) | 18 (40.00) |
|
| 10 (22.22) | 17 (37.78) |
|
|
| Stage (NSCLC) |
|
|
|
|
|
|
|
|
|
| I,
II | 30 (66.67%) | 12 (26.67) | 18 (40.00) | 1.607 | 0.205 | 14 (31.11) | 16 (35.56) | 0.712 | 0.399 |
| III,
IV | 15 (33.33%) | 9 (20.00) | 6 (13.33) |
|
| 9 (20.00) | 6 (13.33) |
|
|
| Metastasis |
|
|
|
|
|
|
|
|
|
| No | 32 (71.11%) | 11 (24.44) | 21 (46.67) | 6.724 | 0.010 | 12 (26.67) | 20 (44.44) | 8.213 | 0.004 |
|
Yes | 13 (28.89%) | 10 (22.22) | 3 (6.67) |
|
| 11 (24.44) | 2 (4.44) |
|
|
Cell line
A549 cells were purchased from the Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China) and
cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in 5% CO2 in
a humidified incubator.
Cell transfection
The cells were divided into three groups: Control,
negative control (NC) and miR-654-3p mimics. The cells in the
control group were incubated in medium without treatment; cells in
the NC group were transfected with miR-NC mimics, and cells in
miR-654-3p mimics group were transfected with miR-654-3p mimics.
All cells were incubated at 37°C in a humidified 5% CO2
atmosphere for 48 h. The miR-654 mimics and control were obtained
from New England Biolabs, Inc. (Ipswich, MA, USA). The cells were
seeded in triplicate in 24-well plates and transfected with 500 ng
miR-654-3p mimics or miR-NC mimics using 2.5 µl
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
After 6 h post-transfection, the medium was replaced with fresh
medium containing 10% FBS. The sequences of the transfected mimics
were the following: miR-654 mimics forward,
5′-UGGUGGGCCGCAGAACAUGUGC-3′; miR-654 mimics reverse,
5′-ACAUGUUCUGCGGCCCACGAAU-3′; NC mimics forward,
5′-UUCUCCGAACGUGUCACGUUU-3′; NC mimics reverse,
5′-ACGUGACACGUUCGGAGAAUU-3′.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was employed to determine miR-654-3p and
mRNA expression levels. An RNeasy Mini kit (Qiagen GmbH, Hilden,
Germany) was used to isolate RNA from cells or tissues according to
the manufacturer's protocol, and theconcentration of RNA was
determined using a NanoDrop 2000 spectrophotometer (NanoDrop;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). An M-MLV
reverse transcriptase cDNA synthesis kit (Thermo Fisher Scientific)
was applied to synthesize cDNA via RT; samples were incubated at
43°C for 30 min, 97°C for 5 min and 5°C for 5 min. The PrimeScript™
RT-PCR kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used
for qPCR. The thermocycling conditions were as follows: 95°C for 6
min (denaturation), 94°C for 30 sec (initiation), 60°C for 30 sec
(annealing), and 75°C for 1.5 min (elongation) for 36 cycles. U6
was used to normalize the expression of miR-654-3p and GAPDH was
used as an internal reference for mRNA expression. This experiment
was performed in triplicate. Expression was quantified using the
2−ΔΔCq method (16). The
sequences of the primers used are as follows: miR-654-3p, forward:
5′-GGGATGTCTGCTGACCA-3′; reverse: 5′-CAGTGCGTGTCGTGGA-3′; U6,
forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse:
5′-AACGCTTCACGAATTTGCGT-3′; Bcl-2-associated × protein (Bax),
forward: 5′-CACCAGCTCTGAACAGATCATGA-3′, reverse:
5′-TCAGCCCATCTTCTTCCAGATGT-3′; B cell lymphoma-2 (Bcl-2), forward:
5′-CACCCCTGGCATCTTCTCCTT-3′, reverse: 5′-AGCGTCTTCAGAGACAGCCAG-3′;
RASAL2, forward: 5′-TGTTCTGTCCTTGAGCCAGT-3′, reverse:
5′-TCCACCTCAGACATCACCAA-3′; and GAPDH, forward:
5′-GCACCACCAACTGCTTAGC-3′, reverse:
5′-GGCATGGACTGTGGTCATGAG-3′.
Western blotting
Protein expression in tissues and cells was
investigated by western blotting. Cells and tissue samples were
lysed in radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.) and the total protein concentration was
determined using a Bicinchoninic Acid assay (Thermo Fisher
Scientific, Inc.). GAPDH was used as a loading control; 2 µg
protein was loaded for 15% SDS-PAGE, and then transferred to a
polyvinylidene fluoride membrane (Thermo Fisher Scientific, Inc.).
The membrane was blocked with 5% skimmed milk for 2 h at 37°C, then
incubated with the following primary antibodies: Anti-RASAL-2 (cat.
no. ab216127; 1:5,000; Abcam, Cambridge, UK), anti-Bcl-2 (cat. no.
ab209039; 1:5,000; Abcam), anti-Bax (cat. no. ab32503; 1:5,000;
Abcam) and anti-GAPDH (cat. no. ab181602; 1:10,000; Abcam) for 1 h
at room temperature. The membrane was then incubated with a
horseradish peroxidase-conjugated anti-rabbit antibody (ab6721;
1:5,000; Abcam) for 45 min at room temperature. Subsequently, an
enhanced chemiluminescence western blotting kit (Pierce; Thermo
Fisher Scientific, Inc.) was used to visualise the proteins. The
gray values were obtained using ImageJ image analysis software
(National Institutes of Health, Bethesda, MD, USA). This experiment
was performed in triplicate.
MTT assay
Transfected cells were seededinto 24-well plates
(2×106 cells/ml) and incubated at 37°C with 5%
CO2. Subsequently, an MTT Cell Viability Assay kit
(Sigma-Aldrich; Merck KGaA) was used, according to the
manufacturer's protocols. The optical density was measured at a
wavelength of 570 nm using a microplate reader at 0, 12, 24 and 48
h. The experiment was conducted in triplicate.
Flow cytometry assay
Transfected cells were plated into 24-well plates
(2×106 cells/ml) and incubated at 37°C with 5%
CO2 for 72 h. Then, the cells were treated with
propidium iodide (10 µg/ml; Sigma-Aldrich; Merck KGaA) and Annexin
V-fluorescein isothiocyanate (50 µg/ml; Becton, Dickinson and
Company, Franklin Lakes, NJ, USA) in the dark for 15 min at room
temperature. Subsequently, the stained cells were analyzed using a
FACScanflow cytometer (Becton, Dickinson and Company) and Diva
software (version 8.0; Becton, Dickinson and Company)to determine
the apoptotic rate.
Bioinformatics analysis and luciferase
reporter assay
Bioinformatics analysis using TargetScan 7.1
(http://www.targetscan.org/vert_71/)
revealed that RASAL2 was a target of miR-654-3p. A luciferase
reporter assay was performed to verify whether RASAL2 was a direct
target of miR-654-3p. The cells were seeded in 96-well plates
(2×103 cells/well) and incubated at 37°C with 5%
CO2 for 24 h. Subsequently, RASAL2-3′UTR-wild type (WT)
and RASAL2-3′UTR-mutant (MUT) plasmids (2.5 µg, Addgene, Inc.,
Cambridge, MA, USA), miR-654-3p mimics and miR-NC were transfected
to cells using 2.5 µl Lipofectamine 2000. A
BioLux®Gaussia Luciferase Reporter Assay kit (New
England Biolabs, Inc.) was utilized for the analysis of luciferase
activity, according to the manufacturer's protocols. Luciferase
activity was normalized to that of Renilla luciferase.
Statistical methods
All data are presented as the mean ± standard
deviation and were analyzed using GraphPad Prism version 5.01
(GraphPad Software, Inc., La Jolla, CA, USA). A paired Student's
t-test was used to examine differences between the two groups. An
unpaired Student's t-testwas used to compare the luciferase
activity between NC and miR-654-3p groups. One-way analysis of
variance followed by a Newman-Keuls post-hoc test was used to
examine differences among three groups. The correlation between
miR-654-3p and RASAL2 was analysed using Pearson's correlation
coefficient. A χ2 test was performed to analyze the data
presented in Table I. P<0.05 was
considered to indicate a statistically significant difference.
Results
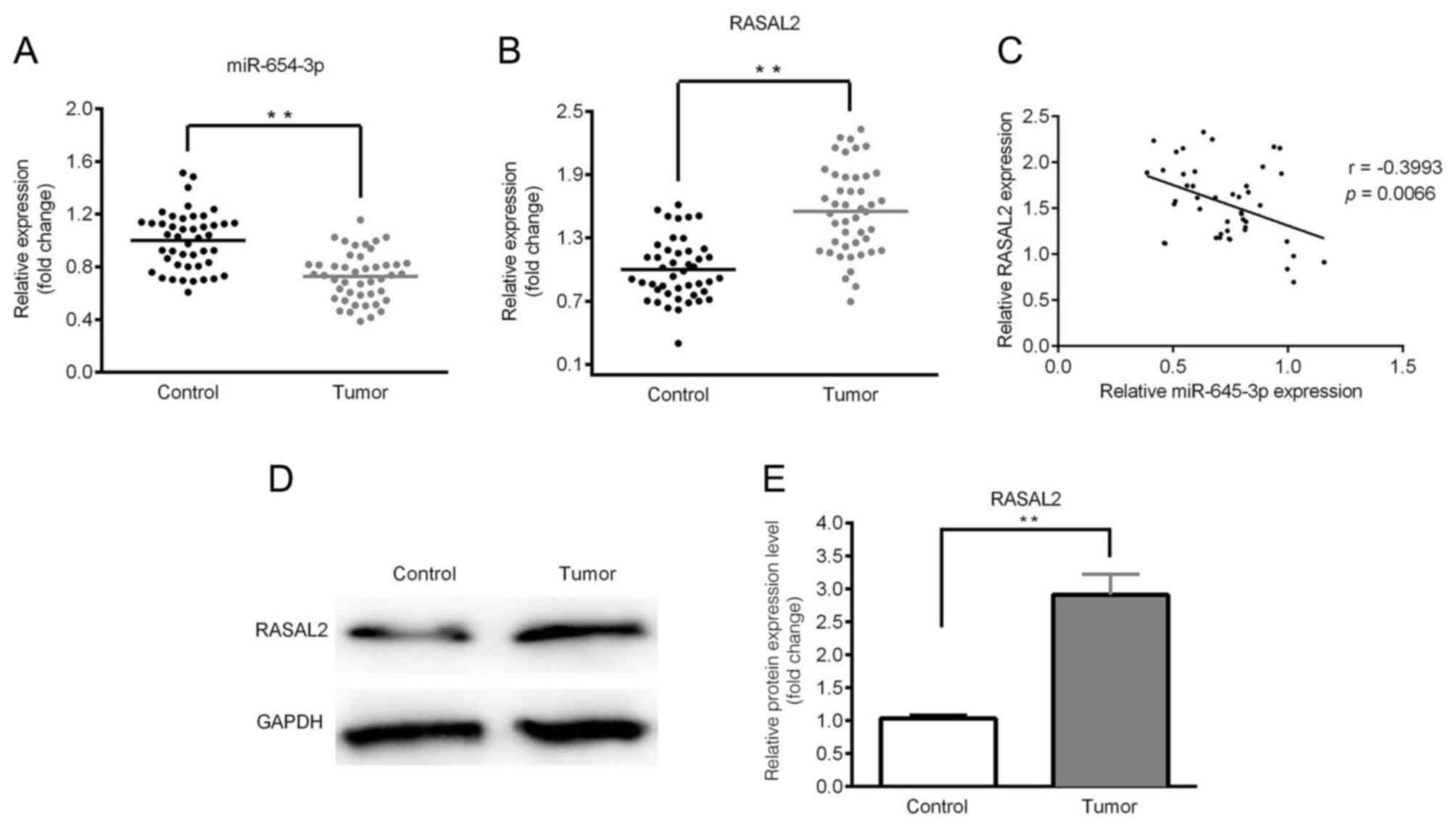
Downregulated expression of miR-654-3p
and upregulated expression of RASAL2 in lung cancer tissues
The expression levels of miR-654-3p and RASAL2 were
determined by RT-qPCR and western blotting. As presented in
Fig. 1A and B, significantly
downregulated expression of miR-654-3p and upregulated expression
of RASAL2 were observed in the tumor tissues compared with control.
A negative correlation between miR-654-3p and RASAL2 expression was
identified (Fig. 1C). Furthermore,
the expression of RASAL2 protein was notably upregulated in tumor
tissues compared with in the control (Fig. 1D and E). In addition, the expression
levels of miR-654-3p and RASAL2 were significantly associated with
the clinicopathological features of NSCLC, including tumor size and
lymph node metastasis (Table
I).
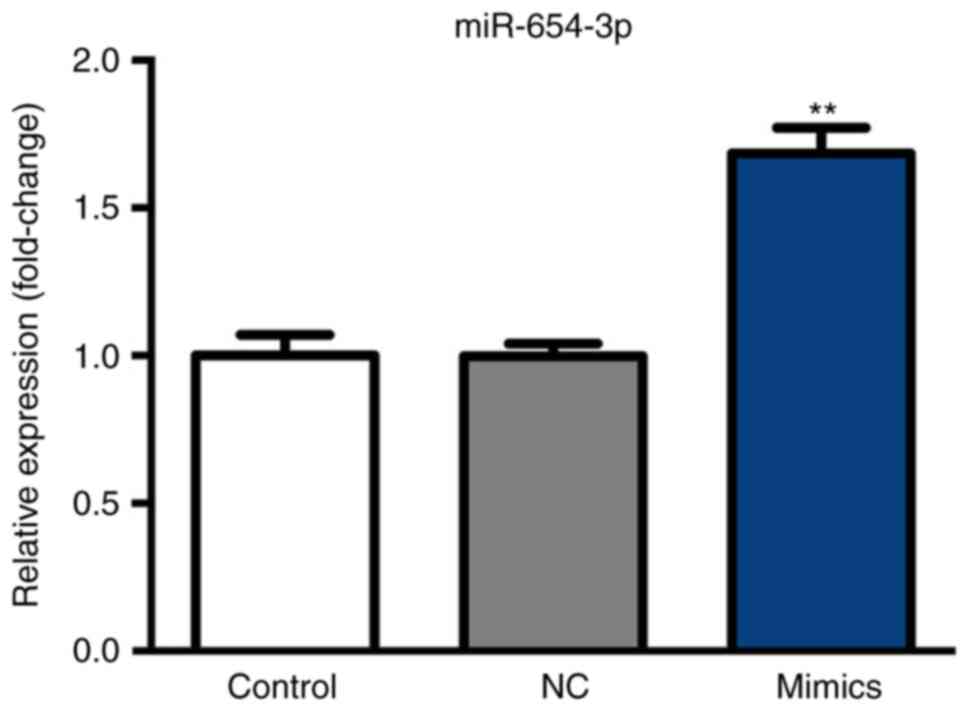
Expression of miR-654-3p following
transfection with miR-654-3p mimics
After 6 h following transfection with miR-654-3p
mimics and a 48-h incubation at 37°C, miR-654-3p expression was
analyzed by RT-qPCR. As presented in Fig. 2, miR-654-3p expression was
significantly upregulated in the mimics group compared with the
control groups.
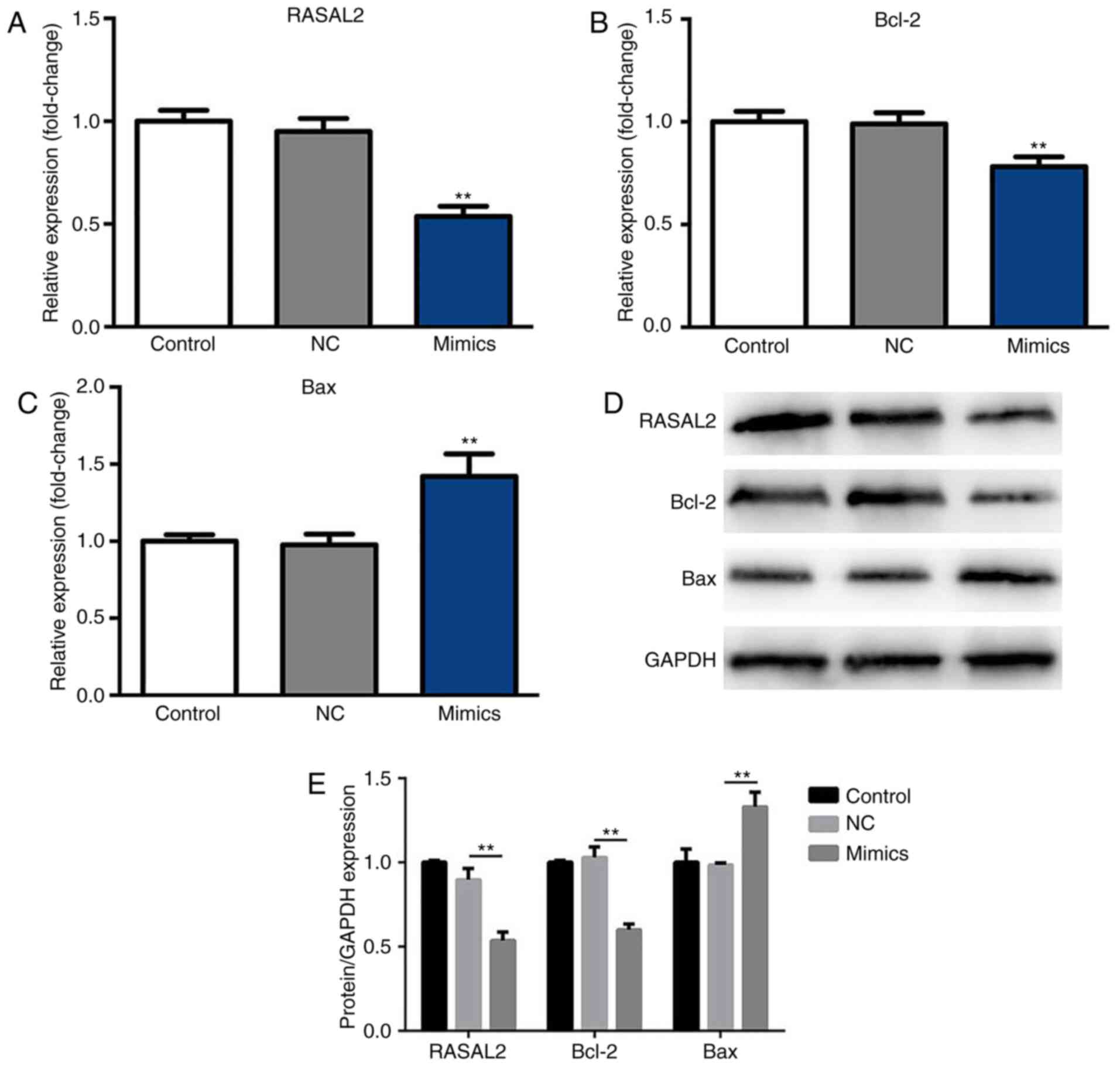
Suppressed RASAL2 and Bcl-2
expression, and upregulated Bax expression in A549 cells following
transfection with miR-654-3p mimics
After 48 h incubation at 37°C with 5%
CO2, the expression levels of RASAL2, Bax and Bcl-2 were
determined by RT-qPCR and western blotting. As presented in
Fig. 3A and B, the mRNA expression
levels of RASAL2 and Bcl-2 were significantly decreased in the
miR-654-3p mimics group compared with the control groups,
respectively. Furthermore, the mRNA expression levels of Bax were
significantly upregulated in the mimics group compared with the
control groups (Fig. 3C).
Additionally, compared with the NC group, significantly decreased
expression of RASAL2 and Bcl-2, and increased expression of Bax
protein were observed in response to miR-654-3p transfection
(Fig. 3D and E). Therefore,
miR-654-3p may inhibit the expression of RASAL2 and Bcl-2
expression, while inducing that of Bax.
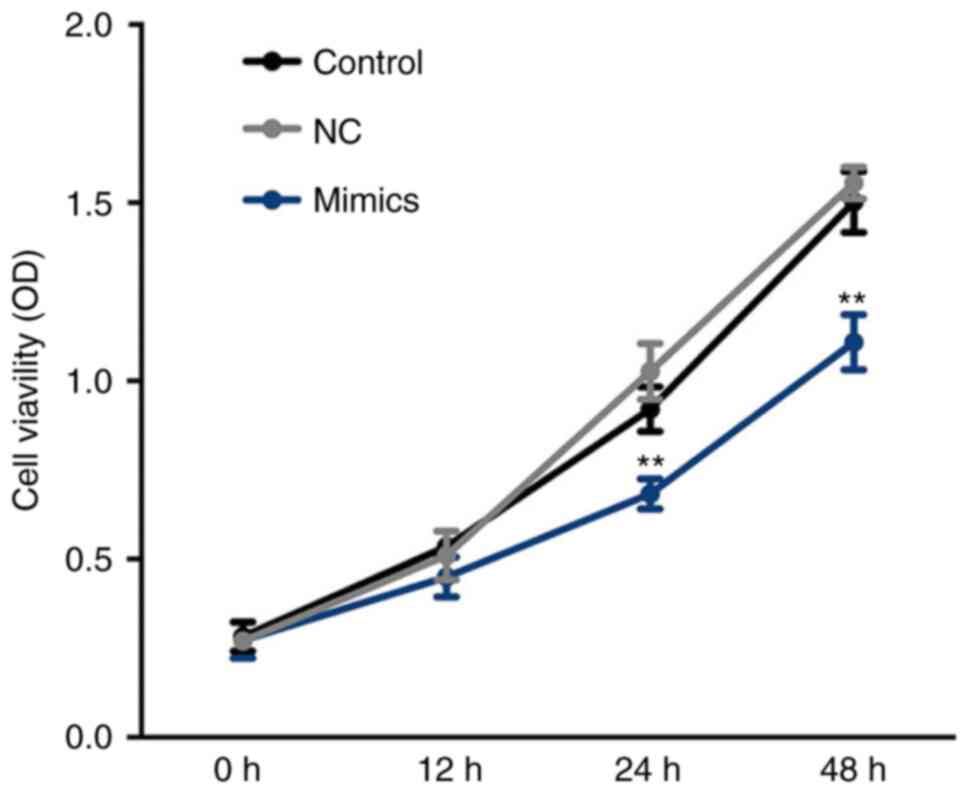
Suppressed viability of A549 cells
following transfection with miR-654-3p mimics
Cell viability was determined at 0, 12, 24 and 48 h.
As presented in Fig. 4, cell
viability was significantly inhibited in the miR-654-3p mimics
group compared with the control groups at 24 and 48 h. These
results suggest that cell viability is suppressed by miR-654-3p
upregulation.
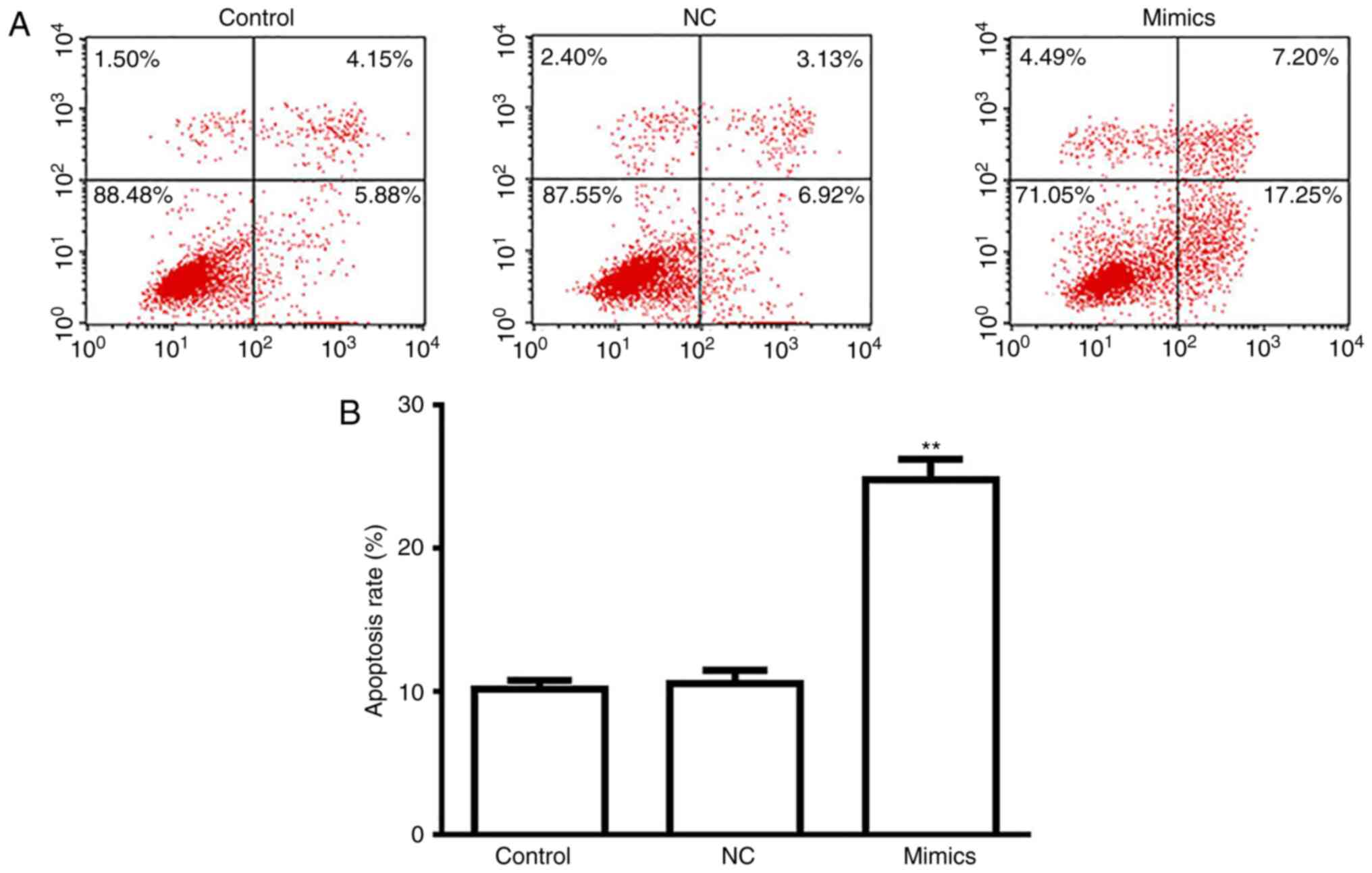
A549 cell apoptosis is induced
following transfection with miR-654-3p mimics
As presented in Fig.
5, a significantly increased rate of apoptosis was observed in
the miR-654-3p mimics group compared with the control groups
(Fig. 5A and B). Thus,
overexpression of miR-654-3p may induce apoptosis by targeting
RASAL2.
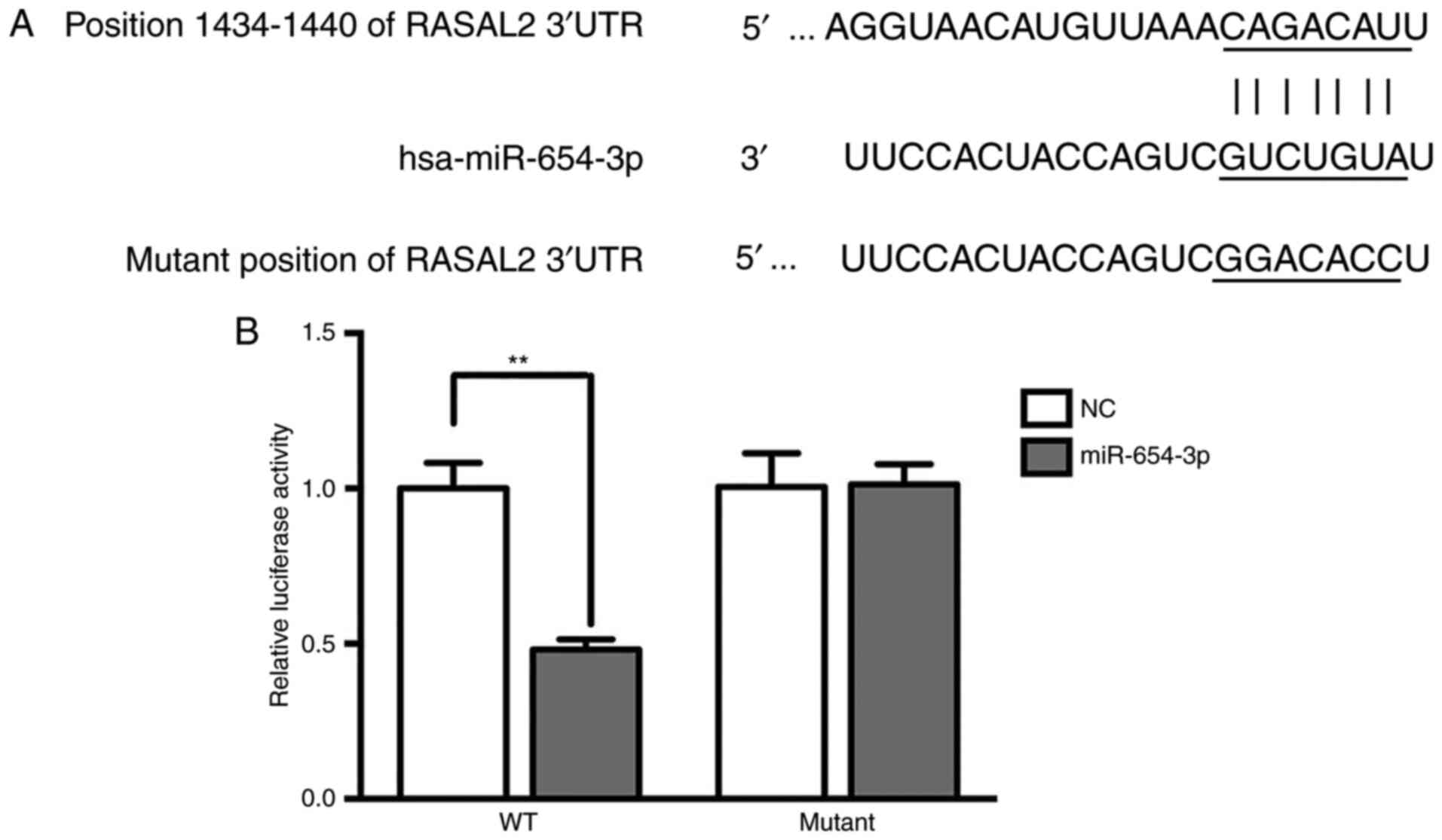
Target verification by luciferase
reporter assay
Bioinformatics analysis using TargetScan suggested
that RASAL2 was a target of miR-654-3p (Fig. 6A). Additionally, the luciferase
activity was significantly decreased in the RASAL2-3′UTR-WT group
treated with miR-654-3p mimics compared with the control (Fig. 6B), which indicated that RASAL2 was a
target of miR-654-3p.
Discussion
In the present study, it was suggested that RASAL2
was a target of miR-654-3p in NSCLC as predicted by TargetScan
analysis and demonstrated via a luciferase activity assay (17). Overexpression of miR-654-3p may
inhibit the expression of RASAL2 and Bcl-2, while inducing that of
Bax.
The present study aimed to investigate the effects
of miR-645-3p on the viability and apoptosis of NSCLC cells. The
results indicated that miR-654-3p expression was downregulated in
tumor tissues and NSCLC cells, which was also reported by Xu et
al (9). Furthermore,
bioinformatics and luciferase reporter analyses revealed RASAL2 as
a target of miR-654-3p in the present study. It was also suggested
that the upregulated expression of RASAL2 in A549 cells was
reversed by overexpression of miR-654-3p, which resulted in reduced
RASAL2 and Bcl-2 expression, and increased Bax expression, and
further resulted in the suppression of cell viability and the
induction of apoptosis of A549 cells. These results indicated that
miR-654-3p was associated with RASAL2 expression, and the viability
and apoptosis of NSCLC cells.
RASAL2 has been studied in breast cancer,
hepatocellular carcinoma and colorectal cancer, among others;
however, its role in NSCLC requires further investigation (18–20).
Yan et al (21) reported
that miR-136 acts as a tumor suppressor by targeting RASAL2, thus
inhibiting cell invasion and metastasis in triple-negative breast
cancer. miR-136 was also demonstrated to be an oncogene in human
NSCLC (21). Therefore, the
combined effect of numerous miRNAs on the regulation of RASAL2
remains to be elucidated. Furthermore, RASAL2 is a member of the
RAS GTPase-activating proteins, which catalyzes the
dephosphorylation of GTP into GDP, inactivating Ras (14,22).
RASAL2 was proposed to affect epithelial-mesenchymal transition via
the mitogen-activated protein kinase/SRY-box 2 pathway, which
contributes to alterations in the migration and invasion of breast
and lung cancer cells (13,23). Therefore, investigations into the
effects of miR-654-3p on cell migration and invasion are required
to fully characterize the roles of miR-654-3p of A549 in NSCLC.
In addition, Geraldo et al (11) revealed that 14q32-encoded miRNAs,
including miR-495-3p, miR-654-3p, miR-376a-3p and miR-487b-3p,
could function as tumor suppressor genes when their expression was
downregulated. Therefore, future studies should investigate the
potential of these miRNAs in the treatment of NSCLC. In the present
study, it was demonstrated that RASAL2 was a target of miR-654-3p
in NSCLC; however, the specific mechanism underlying the effects of
miR-654-3p in the cell cycle, proliferation, migration, invasion
and apoptosis should be investigated beyond the RASAL2 pathway.
Additionally, although the number of patients involved in the
present study was small, future investigations can be conducted
using greater patient cohorts.
In conclusion, the present study reported that
miR-139-5p is downregulated and RASAL2 is upregulated in NSCLC.
Overexpression of miR-139-5p could suppress the viability and
promote the apoptosis of NSCLC cells by targeting RASAL2.
Therefore, miR-139-5p may act as a potential therapeutic target for
the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX drafted the manuscript. JX,SX, ZD, YL and PL
collected and analyzed the data. JX, SX, ZD, YL, PL, LN and QX
performed the experiments. PY designed the study. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Linyi Central Hospital. Written informed consent was
obtained from all patients prior to enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviation
Abbreviations:
|
NSCLC
|
non-small-cell lung cancer
|
References
|
1
|
Huang G, Sun X, Liu D, Zhang Y, Zhang B,
Xiao G, Li X, Gao X, Hu C, Wang M, et al: The efficacy and safety
of anti-PD-1/PD-L1 antibody therapy versus docetaxel for pretreated
advanced NSCLC: A meta-analysis. Oncotarget. 9:4239–4248. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quintanal-Villalonga Á, Ojeda-Márquez L,
Marrugal Á, Yagüe P, Ponce-Aix S, Salinas A, Carnero A, Ferrer I,
Molina-Pinelo S and Paz-Ares L: The FGFR4-388arg variant promotes
lung cancer progression by N-cadherin induction. Sci Rep.
8:23942018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gouvinhas C, De Mello RA, Oliveira D,
Castro-Lopes JM, Castelo-Branco P, Dos Santos RS, Hespanhol V and
Pozza DH: Lung cancer: A brief review of epidemiology and
screening. Future Oncol. 14:567–575. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rios J, Gosain R, Goulart BH, Huang B,
Oechsli MN, McDowell JK, Chen Q, Tucker T and Kloecker GH:
Treatment and outcomes of non-small-cell lung cancer patients with
high comorbidity. Cancer Manag Res. 10:167–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song YJ, Gao XH, Hong YQ and Wang LX:
Direct bilirubin levels are prognostic in non-small cell lung
cancer. Oncotarget. 9:892–900. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Schooneveld E, Wildiers H, Vergote I,
Vermeulen PB, Dirix LY and Van Laere SJ: Dysregulation of microRNAs
in breast cancer and their potential role as prognostic and
predictive biomarkers in patient management. Breast Cancer Res.
17:212015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang XY, Liu DJ, Yuan RB, Zhang DH, Li
SR, Zhang SH and Zhang LY: Low expression of miR-597 is correlated
with tumor stage and poor outcome in breast cancer. Eur Rev Med
Pharmacol Sci. 22:456–460. 2018.PubMed/NCBI
|
|
9
|
Xu C, Zheng Y, Lian D, Ye S, Yang J and
Zeng Z: Analysis of microRNA expression profile identifies novel
biomarkers for non-small cell lung cancer. Tumori. 101:104–110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi GL, Chen Y, Sun Y, Yin YJ and Song CX:
Significance of serum microRNAs in the auxiliary diagnosis of
non-small cell lung cancer. Clin Lab. 63:133–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Geraldo MV, Nakaya HI and Kimura ET:
Down-regulation of 14q32-encoded miRNAs and tumor suppressor role
for miR-654-3p in papillary thyroid cancer. Oncotarget.
8:9597–9607. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Teferedegne B, Macauley J, Foseh G,
Dragunsky E, Chumakov K, Murata H, Peden K and Lewis AM Jr:
MicroRNAs as potential biomarkers for VERO cell tumorigenicity.
Vaccine. 32:4799–4805. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hui K, Gao Y, Huang J, Xu S, Wang B, Zeng
J, Fan J, Wang X, Yue Y, Wu S, et al: RASAL2, a RAS
GTPase-activating protein, inhibits stemness and
epithelial-mesenchymal transition via MAPK/SOX2 pathway in bladder
cancer. Cell Death Dis. 8:e26002017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maertens O and Cichowski K: An expanding
role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv
Biol Regul. 55:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Wang J, Su Y and Zeng Z: RASAL2
inhibited the proliferation and metastasis capability of
nasopharyngeal carcinoma. Int J Clin Exp Med. 8:18765–18771.
2015.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang JF, Zhao HP, Wang ZF and Zheng SS:
Upregulation of RASAL2 promotes proliferation and metastasis, and
is targeted by miR-203 in hepatocellular carcinoma. Mol Med Rep.
15:2720–2726. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olsen SN, Wronski A, Castaño Z, Dake B,
Malone C, De Raedt T, Enos M, DeRose YS, Zhou W, Guerra S, et al:
Loss of RasGAP tumor suppressors underlies the aggressive nature of
luminal B breast cancers. Cancer Discov. 7:202–217. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia Z, Liu W, Gong L and Xiao Z:
Downregulation of RASAL2 promotes the proliferation,
epithelial-mesenchymal transition and metastasis of colorectal
cancer cells. Oncol Lett. 13:1379–1385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen H, Wu X, Zhang Y, Deng G, Ma J, Qu Y
and Zeng S: Expression of RASAL2 in hepatocellular carcinoma and
the clinical significance. Zhong Nan Da Xue Xue Bao Yi Xue Ban.
40:250–255. 2015.(In Chinese). PubMed/NCBI
|
|
21
|
Yan M, Li X, Tong D, Han C, Zhao R, He Y
and Jin X: miR-136 suppresses tumor invasion and metastasis by
targeting RASAL2 in triple-negative breast cancer. Oncol Rep.
36:65–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noto S, Maeda T, Hattori S, Inazawa J,
Imamura M, Asaka M and Hatakeyama M: A novel human RasGAP-like gene
that maps within the prostate cancer susceptibility locus at
chromosome 1q25. FEBS Lett. 441:127–131. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li N and Li S: RASAL2 promotes lung cancer
metastasis through epithelial-mesenchymal transition. Biochem
Biophys Res Commun. 455:358–362. 2014. View Article : Google Scholar : PubMed/NCBI
|