Introduction
Colorectal cancer (CRC) is the third most common
malignant tumour in China, and its mortality ranks fourth among all
malignant tumours (1,2). At present, the first choice of
treatment for CRC is surgical resection. However, the optimal time
window for surgery is often missed when the early symptoms of CRC
are not detected; thus, early screening for the diagnosis of CRC is
key to improving treatment outcomes (3). Identifying a marker that is specific
to CRC and easy to detect would provide new insight for the early
diagnosis, treatment and monitoring of CRC.
F-box and WD repeat domain-containing 7 (FBXW7) is a
novel ubiquitin ligase encoded by the tumour suppressor gene
FBXW7. Its active targets consist of a variety of
cancer-related factors, such as cyclin E, c-Myc, Mcl-1, c-Jun and
mTOR (4,5). Abnormal expression of these targets is
associated with CRC, gastric cancer and ovarian cancer (6). FBXW7 plays a role in physiological and
pathological processes, such as tumour cell proliferation, early
apoptosis and signal transduction (7,8).
Previous studies have indicated that the expression of FBXW7
serves an indispensable role in the occurrence, development and
metastasis of CRC (9–11).
MicroRNAs (miRNAs/miRs) are involved in a wide range
of cellular functions and normal biological processes, including
cell proliferation, differentiation, apoptosis and biotic stress
resistance (12,13). Notably, some miRNAs are considered
to have oncogenic activities, while others act as tumour
suppressors (14,15). Oncogenic miRNAs are upregulated in
cancer and promote cancer development by targeting tumour
suppressor genes through different mechanisms. For instance,
previous studies have identified a negative correlation between
miR-223 expression and FBXW7 expression in breast cancer,
hepatocellular carcinoma, non-small cell lung cancer and CRC
(16–19). Monitoring miRNAs that bind to the
FBXW7 gene, and regulate CRC development and progression,
could be an important approach for the prognosis of clinical
treatment effects. However, the precise regulatory mechanisms
underlying the effects of miR-223 on CRC progression remain
unclear.
Notch signalling is an evolutionarily conserved
pathway that is involved in various processes, including cell
proliferation, differentiation and apoptosis (20). Notch signalling is also considered
to be associated with tumorigenesis (21). Increasing evidence has revealed that
Notch1 is relevant to other signalling pathways, such as Akt/mTOR
and NF-κB signalling (22,23). In addition, miRNAs have been
reported to regulate genes within the Notch and mTOR signalling
pathways (24,25); therefore, the miR-223/FBXW7 axis may
be associated with Notch/mTOR signaling.
The aim of the present study was to examine the
regulatory relationship between miRNAs, FBXW7 and associated
signalling pathways in order to improve current understanding of
the mechanisms underlying CRC progression. The present study
demonstrated that miR-223 was upregulated in CRC tissue and could
bind to the 3′untranslated region (UTR) of the FBXW7 gene.
miR-223 also promoted HCT116 cell proliferation, whilst inhibiting
apoptosis.
Materials and methods
Clinical samples
A total of 20CRC primary tumour tissue samples and
matched adjacent non-tumour tissues were collected at the
Affiliated Renmin Hospital of Hubei University of Medicine (Shiyan,
China) between October 2016 and December 2018. All patients were
diagnosed in histopathologically and clinically according to the
American Joint Committee on Cancer criteria, and performed surgical
operation. The adjacent tissue was 5 cm away from the edge of the
tumour, all the tissue samples were snap frozen using liquid
nitrogen, and then stored at −80°C until further analysis. This
research was approved by the Ethics Committee of Hubei University
of Medicine, and all patients provided written informed consent for
the use of samples.
Plasmid construction
The FBXW7 gene was cloned into pTriEx-1.1
vector. The 3′UTR of the FBXW7 gene was inserted into
pMIR-REPORT vector (Ambion) to construct the recombinant plasmids
for luciferase reporter gene assays. The primers used for PCR are
listed in Table SI. The pTriEx-1.1
(Novagen) and pMIR-REPORT vectors were separately digested with
HindIII/SacI or NcoI/BamHI,
respectively, then the digested fragments were ligated with T4 DNA
Ligase (New England Biolabs, Inc.) after purification. The
recombinant plasmids were transformed into Escherichia coli
and screened by PCR, and their sequences verified by
sequencing.
Bioinformatics analysis
The potential miRNAs targeting the FBXW7 gene
(including its UTR) were analysed by TargetScan (version 7.1;
www.targetscan.org/). Target miRNA
expression was predicted using miRanda (www.microrna.org/microrna/). The bioinformatics
analyses, including expression and prognosis, were performed by
Sangerbox (http://sangerbox.com/Tool) using the
pan-cancer monogenic fast comprehensive analysis tool. The
expression data of normal mucosa tissues and CRC tissues were
obtained from Genotype-Tissue Expression (GTEx, http://gtexportal.org/home/) and The Cancer Genome
Atlas (TCGA, http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Differential gene expression levels was calculated as the
log2 of its upper quartile Fragments Per Kilobase of
transcript per Million mapped reads value. The expression levels of
FBXW7 mRNA were calculated as the log2 of its TPM
(transcripts per million) value after normalization of gene length
and sequencing depth.
Immunohistochemistry staining
The procedures for immunohistochemistry staining
were performed as previously reported (26). Sections (3-µm thick) were fixed
using 4% paraformaldehyde for 12 h. The paraformaldehyde-fixed,
paraffin-embedded sections were deparaffinized and rehydrated, and
antigen retrieval was performed in boiling citrate buffer for 10
min, followed by blocking with 3% hydrogen peroxide for 10 min at
room temperature. After washing with PBS three times, the sections
were incubated with anti-FBXW7 antibody (1:200; Abcam; cat. no.
ab109617) overnight at 4°C. The secondary antibody (1:50; Beyotime
Institute of Biotechnology; cat. no. A0208) was incubated for 1 h
at room temperature, followed by incubation with 3-diaminobenzidine
(DAB) and re-staining with haematoxylin. The staining results were
photographed under a BX46 light microscope (Olympus Corporation)
and evaluated by two pathologists in a blinded manner.
Cell culture and transfection
The human HCT116 cell line was purchased from
American Type Culture Collection and cultured in high-glucose DMEM
(Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) in a 37°C incubator with 5%
CO2. Cells in the logarithmic growth phase (80%
confluence) were used for experiments. The 50 ng pMIR-REPORT
luciferase reporter plasmids (Ambion; 50 ng), miR-223 mimics
(5′-UGUCAGUUUGUCAAAUACCCCA-3′), miR mimic control (miCtr,
5′-UUCUCCGAACGUGUCACGUTT-3′), miR-223 inhibitor
(5′-UGGGGUAUUUGACAAACUGACA-3′) and miR-223 inhibitor control (Ctr;
5′-CAGUACUUUUGUGUAGUACAA-3′) were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Small interfering RNA (siRNA) targeting
FBXW7 (siFBXW7−1, 5′-ACAGGACAGUGUUUACAAA-3′;
siFBXW7−2, 5′-CCAUGCAAAGUCUCAGAAU-3′) and negative control
(si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′) were transfected using
Entranster™-R4000 (Engreen Biosystem, Ltd.), according to the
manufacturer's instructions. All small nucleic acids were purchased
from Shanghai GenePharma Co., Ltd. and were used at a final
concentration of 20 nM. The cells were treated or collected at the
indicated times after transfection.
Luciferase reporter gene assay
The 3′UTR sequences of FBXW7 were cloned into
the pMIR-REPORT vector to construct the FBXW7 luciferase
reporter plasmids. A total of 50 ng reporter plasmids were
co-transfected with 20 nM miR-223 mimics, miR-25 mimics, miR-223
inhibitor and mimics control (miCtr) into 293T cells. Luciferase
activity was measured 36 h later by fluorescence detection with the
Luciferase Assay System (Promega Corporation). The cells in the
plate were lysed with 5Xcell lysis reagent (Promega Corporation).
After centrifugation, the supernatant was collected and added to
100 µl luciferase assay reagent at room temperature, and luciferase
was measured using a Glomax 20/20 bioluminescence detector (Promega
Corporation). The luciferase activity was normalized to the
activity of Renilla luciferase.
Cellular viability assay
The viability of HCT116 cells was detected using a
Cell Counting Kit-8 (CCK-8) kit (Dojindo Molecular Technologies,
Inc.). CCK-8 reagent (10 µl/well) was added to 1×106
HCT116 cells/well at 0, 24, 48 and 72 h post-transfection, which
were then incubated at 37°C with 5% CO2 for 2 h. The
absorbance was measured in each well at a wavelength of 450 nm
using the xMark Microplate Absorbance Spectrophotometer (Bio-Rad
Laboratories, Inc.). Each group was set up in triplicate.
Apoptosis assay
HCT116 cells transfected with miR-223 mimics or
inhibitor were analysed using an Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Staining Kit
(cat. no. ab14085; Abcam). A total of 1×106 HCT116 cells
from each group were collected 48 h post-transfection. After
washing twice with PBS, the cells were resuspended in 500 µl
binding buffer, then mixed with 5 µl Annexin V-FITC followed by 5
µl PI and incubated in the dark for 10 min at room temperature.
Apoptosis in each group was detected by flow cytometry (Beckman
Coulter, Inc.; CytoFLEX), the frequency of apoptotic cells (Annexin
V+PI+) was obtained using software CytExpert
2.4 (Beckman Coulter, Inc.). The experiment was independently
repeated three times.
FBXW7 rescue assay
A total of 1×106 HCT116 cells/well were
transfected with miR-223 mimics or NC using Entranster™-R4000
(Engreen Biosystem, Ltd.), then transfected with 1 µg
pTriEx-FBXW7 overexpression plasmid or empty plasmid 12 h
later. The cells were collected 48 h later and lysed for protein
detection by western blotting. The cell viability was also detected
at the indicated times following miR-223 transfection and apoptosis
rates were detected by flow cytometry at 48 h
post-transfection.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from homogenised tissue
samples or from HCT116 cells harvested 36 h after transfection
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNA was obtained using a OneScript cDNA Synthesis kit
(Applied Biological Materials Inc., cat. no. G594) with an
oligo(dT) primer or miRNA-specific stem-loop primers, according to
manufacturer's protocol. The primers for RT and qPCR are listed in
Table SI. mRNA expression levels
were examined on an CFX96 Touch PCR system (Bio-Rad Laboratories,
Inc.) using SYBR Premix Ex Taq™ (Applied Biological Materials Inc.,
G892). The thermocycling conditions were as follows: 95°C for 30
sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec.
GAPDH or U6 were used as the internal reference for
genes or miRNAs, respectively. Each reaction was set up in
triplicate, and experiments were performed three times. The
relative expression of each target was analysed using the
2−ΔΔCq method (27).
Western blot analysis
Total proteins of each group were collected from
HCT116 cells 48 h post-transfection or from homogenised tissue and
lysed using RIPA reagent (Solarbio), then quantified using a BCA
kit (Beyotime Institute of Biotechnology). Proteins were separated
by SDS-PAGE on 10% gels and transferred to PVDF membranes (Bio-Rad
Laboratories, Inc.). The membranes were then blocked at 37°C for 1
h using 5% skimmed milk and incubated at 4°C overnight with mouse
or rabbit anti-human polyclonal antibodies specific for FBXW7
(1:2,000; ProteinTech Group, Inc.; cat. no. 28424-1-AP), Notch
intracellular receptor domain (NICD; 1:500; GeneTex, cat. no.
GTX28925), hes family bHLH transcription factor 1 (Hes-1; 1:1,000;
GeneTex; cat. no. GTX108356) and phosphorylated (p)-mTOR (1:1,000;
GeneTex; cat. no. GTX132803). Antibodies specific for Akt (cat. no.
10176-2-AP), p-Akt (cat. no. 66444-1-Ig), mTOR (cat. no.
66888-1-Ig, ProteinTech Group, Inc.) was used at 1:2,000 dilution,
and the mouse anti-human GAPDH monoclonal antibody was used at a
1:5,000 dilution (Beyotime Institute of Biotechnology; cat. no.
AF5009). After washing the membrane three times, goat anti-mouse
(cat. no. A0216) or anti-rabbit IgG (cat. no. A0208) (both at
1:5,000 dilution; Beyotime Institute of Biotechnology) was added,
and the mixture was incubated for 1 h at room temperature, and then
subjected to imaging using an ECL solution (Beyotime Institute of
Biotechnology). Densitometric analysis of the protein bands was
carried out using Image Lab 3.0 (Bio-Rad Laboratories, Inc.).
Protein expression levels are presented as the ratio of the
densitometric value of target proteins relative to the GAPDH
internal control.
Statistical analysis
The cell viability, apoptosis and RT-qPCR results
were statistically analysed using GraphPad Prism 5.0 software
(GraphPad Software, Inc.). The data are presented as the mean ± SD
of three independent experimental repeats. Comparisons between
multiple groups were analysed using one-way ANOVA followed by the
Tukey's HSD post hoc test. Student's t-test was used to compare two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Downregulation of FBXW7 expression in
CRC
First, the present study investigated the difference
in FBXW7 expression in datasets from TCGA and GTEx databases
representing 27 types of tumour tissue and adjacent normal tissue
samples (Fig. 1A). FBXW7 expression
was significantly downregulated in most tumour types, compared with
adjacent tissue. Moreover, the expression of FBXW7 was evaluated in
CRC tissue samples and adjacent normal tissue (n=20). The mRNA
expression levels of FBXW7 in CRC tissue were significantly
lower than in normal tissues (Fig.
1B). The protein expression of FBXW7 was also lower in CRC
tissues compared with in adjacent tissues (Fig. 1C and D). Immunohistochemical
staining also suggested reduced expression in the cells of CRC
tissue compared with in adjacent colon tissue (Fig. 1E). Disease-specific survival (DSS)
and overall survival (OS) time of patients with CRC did not
correlate with FBXW7 expression (Figs. S1 and S2).
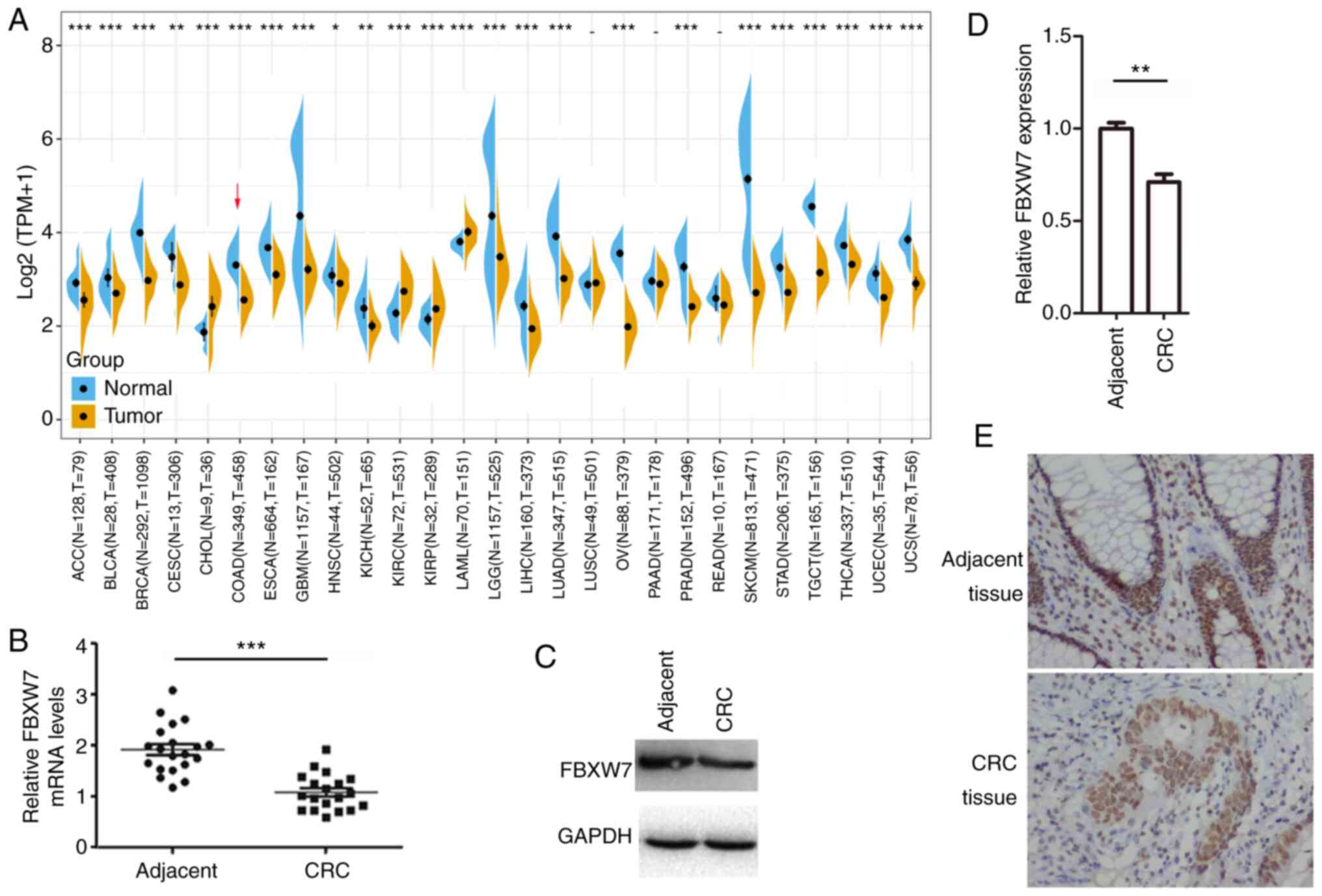 | Figure 1.FBXW7 mRNA and protein expression
levels in CRC tissues and adjacent normal tissues. (A) Differences
in FBXW7 expression in 27 types of tumour tissue and normal tissue
samples in The Cancer Genome Atlas and the Genotype Tissue
Expression database. The red arrow marks the expression in CRC
tissue, compared with healthy tissue. (B) FBXW7 mRNA
expression in CRC tissues and normal adjacent tissue. n=20 in each
group. (C) FBXW7 protein expression in CRC tissue and normal
adjacent tissue. (D) Western blotting results normalized to those
of GAPDH levels. (E) Immunohistochemical staining of FBXW7
expression in CRC and normal adjacent tissues. Magnification, ×200.
Data are presented as the mean ± SD of three replicates.
**P<0.01, ***P<0.001, vs. adjacent (paired Student's t-test).
CRC, colorectal cancer; N, normal; T, tumour; FBXW7, F-box and WD
repeat domain containing 7; log2(TPM+1), normalization
of gene expression; TPM, transcripts per million. |
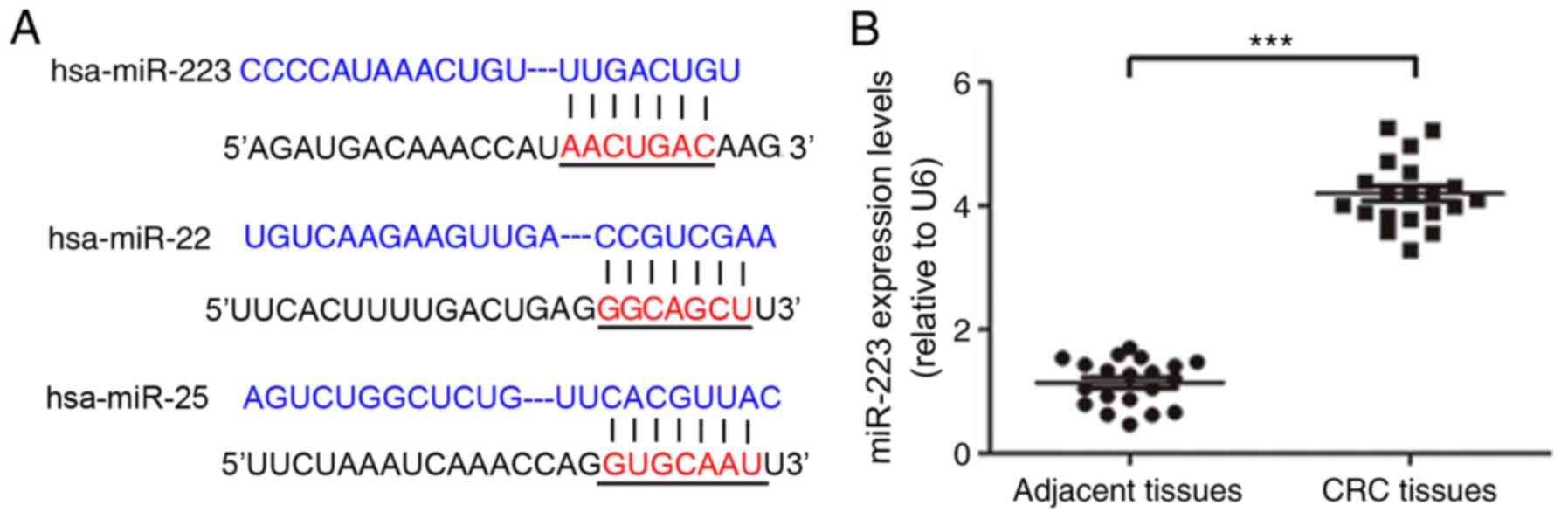
Upregulated miR-223 targets FBXW7 in
CRC cells
miRNA sequences complementary to the human
FBXW7 gene were predicted using the TargetScan and miRanda
(28) tools, which suggested
miR-223, miR-22 and miR-25 targeted the 3′UTR of FBXW7
(Fig. 2A). To determine the
expression levels of the predicted miRNAs in CRC cells, the levels
of miRNAs in HCT116 cells were measured using RT-qPCR. The relative
expression level of miR-223 was significantly increased in CRC
tissue compared with in adjacent tissues (Fig. 2B).
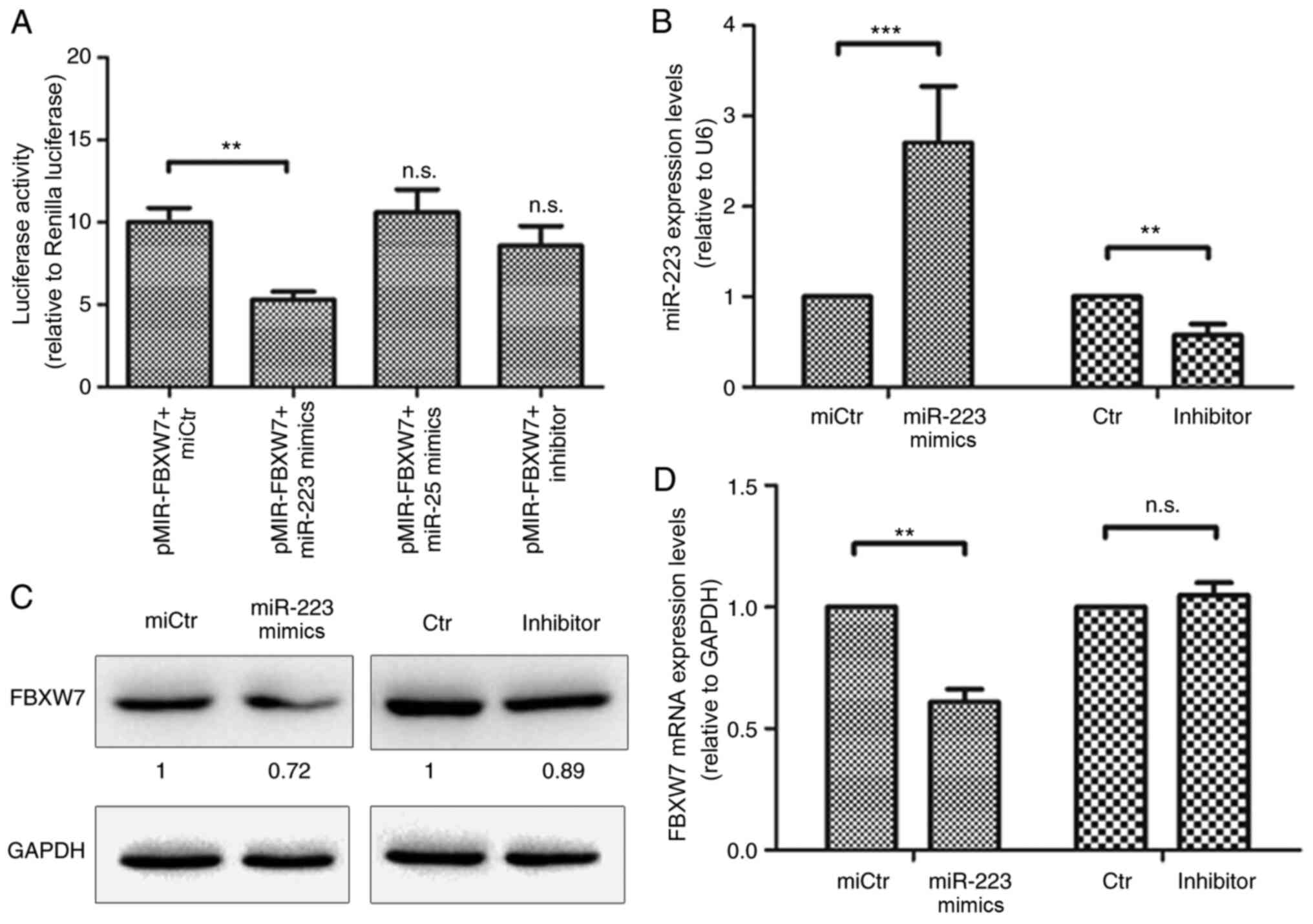
miR-223 binds to FBXW7 and inhibits
its expression in HCT116 cells
A FBXW7 luciferase vector was co-transfected
with miR-223 mimics, miR-25 mimics, miCtr or miR-223 inhibitor into
293T cells. Changes in luciferase activity in the transfected cells
were measured 36 h post-transfection. miR-223 significantly reduced
relative luciferase activity, while miR-25 did not influence
luciferase activity (Fig. 3A). This
indicates that miR-223 may interact with the 3′UTR of FBXW7.
The effect of miR-223 on the expression of FBXW7 was evaluated in
HCT116 cells. miR-223 mimics or inhibitor were used to transfect
HCT116 cells (Fig. 3B). FBXW7
protein expression levels were decreased after transfection with
miR-223 mimics compared with miCtr (Fig. 3C). mRNA expression levels were also
significantly downregulated (Fig.
3C). However, FBXW7 protein and mRNA expression levels were not
significantly altered after transfection with the inhibitor. This
indicated that miR-223 could directly inhibit FBXW7
expression.
Inhibition of FBXW7 by miR-223 affects
the proliferation and apoptosis of HCT116 cells
To investigate the function of miR-223 targeting
FBXW7 in CRC cells, HCT116 cells were transfected with
miR-223 mimics or inhibitor sequences, and cell viability was
evaluated at different time points. Transfection with miR-223
mimics increased cell proliferation after transfection for 48 h,
compared with miCtr (Fig. 4A).
However, transfection with the inhibitor had no effect. In
addition, apoptosis of HCT116 cells was detected by flow cytometry
(Fig. 4B). Transfection with
miR-223 mimics led to a decrease in the number of apoptotic cells,
compared with miCtr, whereas the miR-223 inhibitor promoted
apoptosis of HCT116 cells, compared with Ctr (Fig. 4C).
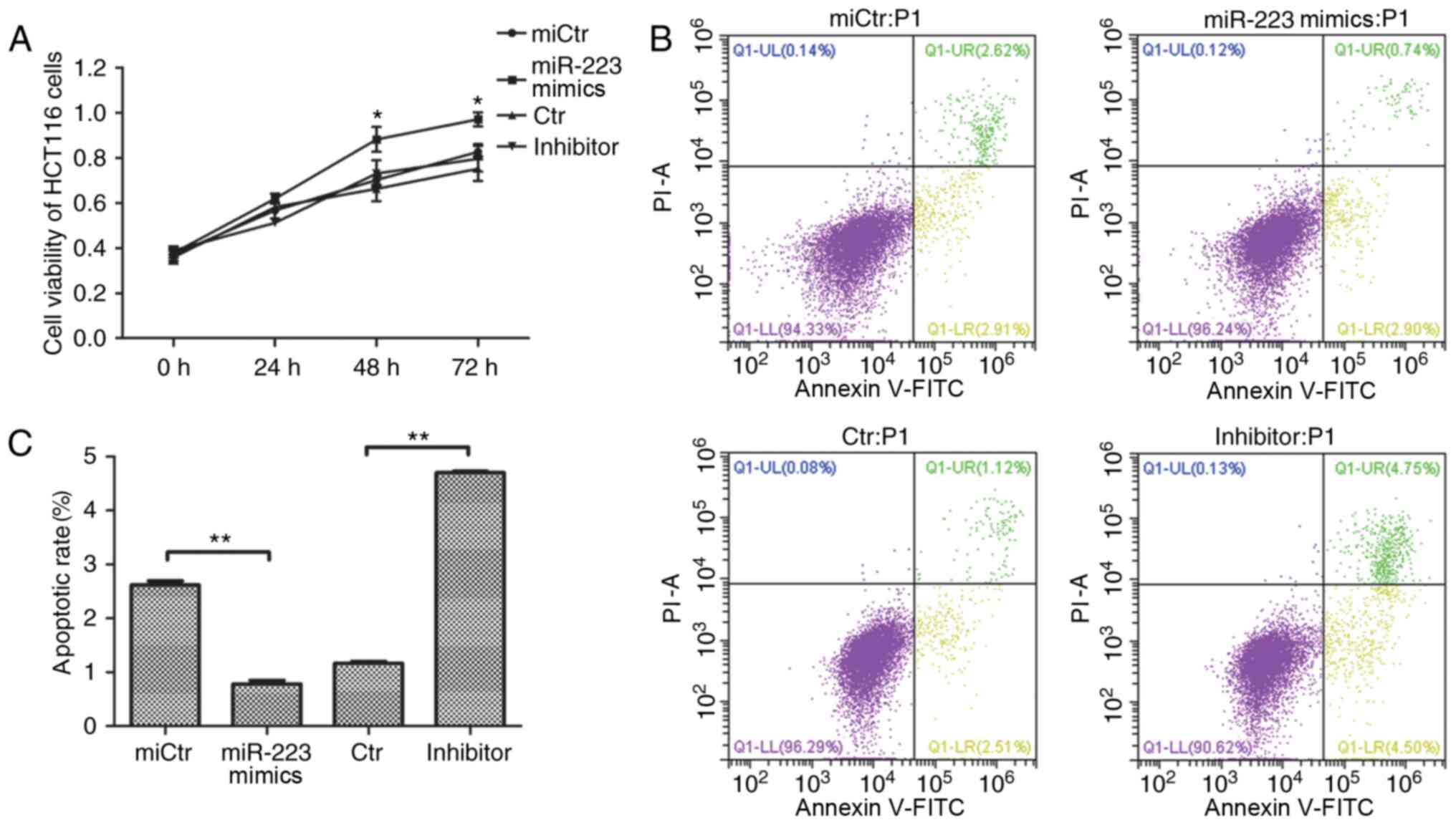 | Figure 4.Effect of miR-223 on cell viability
and apoptosis of HCT116 cells. (A) Effect of miR-223 mimics
transfection on the activity of HCT116 cells. Cells were detected
using the Cell Counting Kit-8 reagent after transfection with
miR-223 mimics or inhibitor from 0 to 72 h. *P<0.05 vs. miCtr
(one-way ANOVA and Tukey's post hoc test). (B) Effect of miR-223
mimics and inhibitor transfection on the early apoptosis of HCT116
cells. (C) Apoptosis rates were recorded following Annexin
V-FITC/PI double staining. Data are presented as the mean ± SD of
three replicates. **P<0.01 (one-way ANOVA and Tukey's post hoc
test). miR, microRNA; FBXW7, F-box and WD repeat domain containing
7; miCtr, mimics control; Ctr, inhibitor control; FITC, fluorescein
isothiocyanate; PI, propidium iodide; UL, upper left; UR, upper
right; LL, lower left; LR, lower right. |
siRNA silencing of FBXW7 regulates
HCT116 cell proliferation and apoptosis
siRNA targeting FBXW7 expression was used to further
verify the effect of FBXW7 on HCT116 cell proliferation and
apoptosis. Compared with si-NC, the relative mRNA and protein
expression levels of FBXW7 were reduced after transfection with
siFBXW7-1 and siFBXW7-2 (Fig. 5A and
B). The cell viability assay indicated that siFBXW7-2 increased
HCT116 cell proliferation, compared with si-NC (Fig. 5C). Moreover, compared with the
control, both siRNA transfections reduced the apoptotic cell rate
(Fig. 5E). In addition, rescuing
FBXW7 through overexpression after transfection with miR-223 mimics
inhibited cell viability and increased apoptosis compared with
miR-223 mimics alone (Fig. 5D and
G). FBXW7 protein levels were upregulated following
co-transfection with FBXW7 overexpression vector and miR-223
mimics, compared with mimics alone (Fig. 5F). Altogether, these results
indicated that miR-223 may promote the proliferation and suppress
the apoptosis of HCT116 cells by targeting FBXW7.
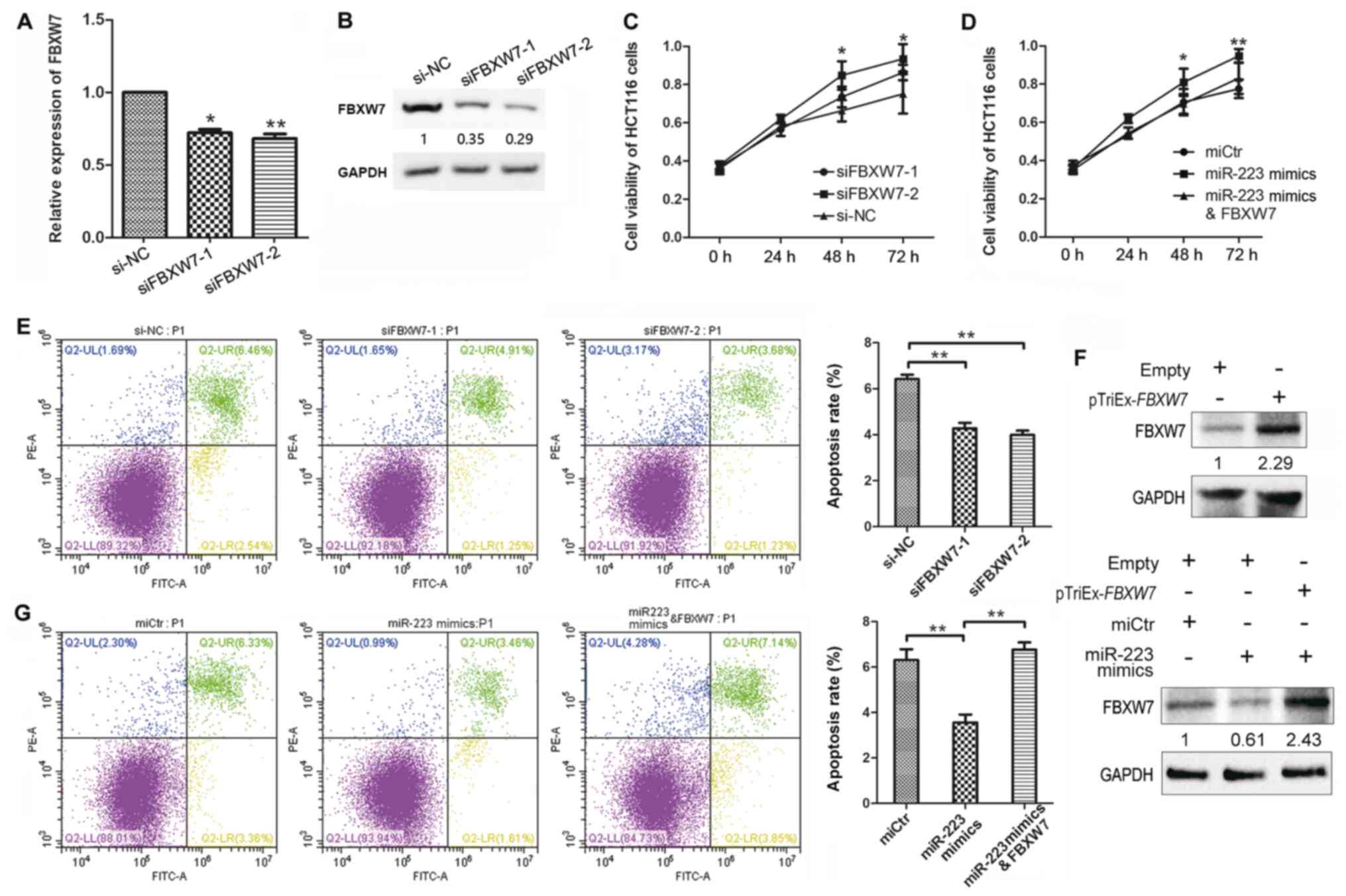 | Figure 5.Effect of FBXW7 knockdown on
the viability and apoptosis of HCT116 cells. (A) FBXW7 mRNA
expression levels in HCT116 cells transfected with siRNA targeting
FBXW7 or si-NC. (B) FBXW7 protein expression levels were
detected by western blotting. (C) HCT116 cell viability following
transfection with siFBXW7-1 and −2, or si-NC. (D) Cell viability
following transfection with miR-223 mimics, miCtr or
co-transfection with miR-223 mimics and FBXW7 overexpression
plasmid. (E) HCT116 cell apoptosis following transfection with
siFBXW7. Apoptotic rates are calculated as the frequency of Annexin
V+PI+ cells relative to total cells. (F)
FBXW7 protein expression levels in HCT116 cells transfected with
miR-223 mimics and FBXW7 overexpression plasmid. The upper panel
shows overexpression of FBXW7 induced by pTriEx-FBXW7 alone,
the lower panel shows expression of FBXW7 in cells transfected with
pTriEx-FBXW7 and miR-223 mimics. (G) Apoptosis of HCT116
cells following transfection with miR-223 mimics, miCtr or
co-transfection with miR-223 mimics and FBXW7 overexpression
plasmid. Data are presented as the mean ± SD of three replicates.
*P<0.05, **P<0.01 (one-way ANOVA and Tukey's post hoc test).
miR, microRNA; FBXW7, F-box and WD repeat domain containing 7;
miCtr, mimics control; si, small interfering RNA; NC, negative
control; FITC, fluorescein isothiocyanate; PI, propidium iodide;
UL, upper left; UR, upper right; LL, lower left; LR, lower
right. |
FBXW7 functions through the Notch and
Akt/mTOR pathways
Multiple studies have demonstrated that FBXW7 plays
a key role in cancer by regulating the Notch and mTOR pathways
(29,30). Thus, the possible association
between miR-223/FBXW7 axis and Notch-mTOR signalling
pathways was examined. Following transfection with miR-223 mimics
or siFBXW7−2, the mRNA expression levels of Notch and
its target gene Hes-1 were increased, compared with miCtr +
si-NC group (Fig. 6A and B), and
the expression levels of NICD and Hes-1 were increased after
inhibition of FBXW7 (Fig. 6C). The
protein expression levels of total mTOR and p-mTOR, as well as the
upstream components of the mTOR pathway, Akt and p-Akt were also
examined (Fig. 6D). Transfection
with miR-223 mimics and siFXBW7−2 both increased the ratio
of p-mTOR to total mTOR and the ratio of p-Akt to total Akt
(Fig. 6F). In addition, the
expression level of Bcl-2 was also upregulated following FBXW7
downregulation either through miR-223 mimics or siFXBW7−2,
while the levels of Bax were downregulated (Fig. 6E), and the ratio of Bax to Bcl-2 was
decreased (Fig. 6F). These results
demonstrated that FBXW7 functioned through the Notch and
Akt/mTOR signalling pathways.
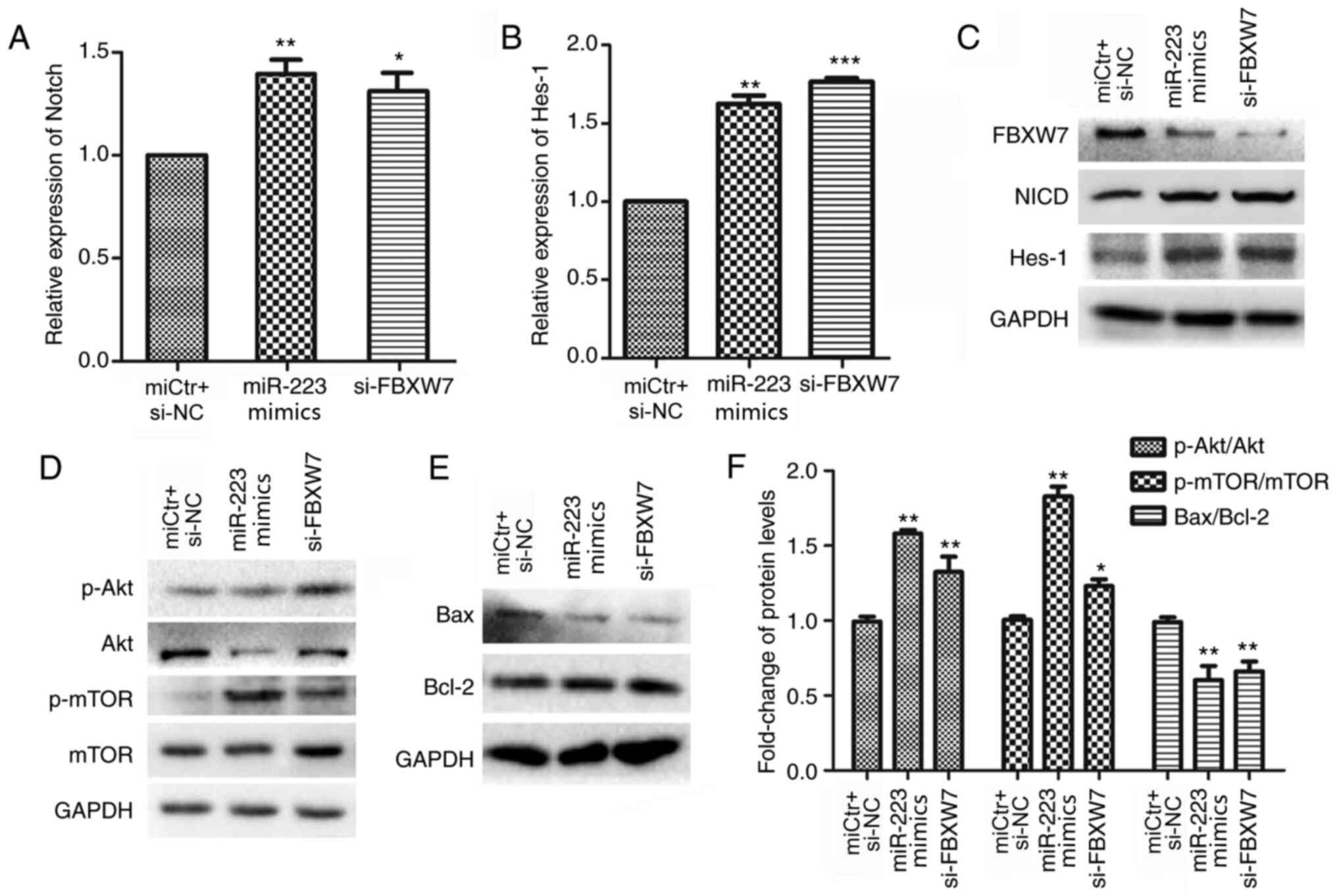 | Figure 6.FBXW7 regulates the progression of
CRC through the Notch and AKT/mTOR signalling pathways. Relative
mRNA expression levels of Notch and Hes-1 in HCT116
cells following (A) miR-223 mimics or (B) siFBXW7−2
transfection. miCtr + si-NC co-transfection was used as a control.
(C) Protein expression levels of NICD and Hes-1 in HCT116 cells
following miR-223 mimic or siFBXW7−2 transfection. (D)
Protein expression levels of total Akt, p-Akt, mTOR and p-mTOR in
HCT116 cells following miR-223 mimic or siFBXW7−2
transfection. (E) Protein expression levels of Bax and Bcl-2 after
transfection with miR-223 mimics or siFBXW7−2 transfection.
(F) Ratios of p-Akt/Akt, p-mTOR/mTOR and Bax/Bcl-2 calculated using
the densitometric value of the protein bands, after normalization
to GAPDH. Data are presented as the mean ± SD of three replicates.
*P<0.05, **P<0.01, ***P<0.001, vs. control (one-way ANOVA
and Tukey's post hoc test). miR, microRNA; FBXW7, F-box and WD
repeat domain containing 7; si, small interfering RNA; Hes-1; hes
family bHLH transcription factor 1; p, phosphorylated; NICD, Notch
intracellular receptor domain. |
Discussion
The tumour suppressor gene FBXW7 has been
reported to inhibit tumour progression by suppressing cell
proliferation and inducing apoptosis (31,32).
FBXW7 may be an independent factor affecting the survival of
patients with CRC, and its expression has been reported to be
associated with the occurrence, development and prognosis of CRC
(33,34). Iwatsuki et al (35) detected the expression levels of
FBXW7 mRNA and protein in 93 CRC samples and found that the
expression of FBXW7 was lower in CRC tumour tissues than in
adjacent tissue samples. Notably, patients with low expression of
FBXW7 had a poor prognosis (35).
miR-223 is significantly associated with the tumour
size and TNM stage of gastric cancer, as well as the invasion and
distant metastasis of CRC (36,37).
Therefore, miR-223 may have a role in the development of CRC,
similar to miR-200a and miR-125b (38,39).
FBXW7 is one of the identified targets of various miRNAs
(40,41). Sano et al (42) demonstrated that miR-25 could
downregulate the expression of FBXW7, which may affect the
proliferation, invasion and apoptosis of gastric cancer cells. In
addition, a previous study confirmed that miR-25 and miR-223 could
reduce the mRNA expression levels of FBXW7, which affects
the activity of cyclin E and cell cycle progression (43). In the present study, inhibiting
miR-223 increased the number of apoptotic cells, possibly as a
result of endogenous FBXW7 reducing the activity of cyclin E
through an as yet unknown mechanism that ultimately promotes cell
apoptosis. Moreover, siRNA was used to directly interfere with the
expression of FBXW7, resulting in increased HCT116 cell viability
and inhibition of apoptosis. In addition, rescuing FBXW7 after
miR-223 transfection produced the opposite result. This indicates
that a direct relationship may exist between the proliferation and
apoptosis of HCT116 cells and the effect of FBXW7.
miR-223 was upregulated in CRC tissues compared with
normal tissues, whereas FBXW7 was downregulated in CRC
tissues. The important role of FBXW7 in human cancer suggests that
downregulation of FBXW7 through overexpression of miR-223 may
become an important indicator of cancer development (44). Dysregulation of miR-223 can
attenuate the tumour suppressor activity of FBXW7 during cell
transformation (45). Therefore,
further investigations of the function of FBXW7 and its downstream
pathways in CRC progression are necessary. Blocking the Notch and
mTOR pathways has been shown to affect the proliferation of various
cancer cells, including endometrial carcinoma, breast cancer and
retinoblastoma cells (46,47), indicating that these signalling
pathways are involved in the development of these types of cancer.
Previous studies have demonstrated that the Notch and NF-κB
pathways are involved in cancer cell progression after
transcriptionally regulating the expression of miR-223 (31,47).
NICD, which is degraded by FBXW7, acts as a transcriptional
activator that positively regulates target genes, including Hes-1.
Hes-1 directly promotes cell proliferation (48). A recent study concluded that the
active status of mTOR, p-mTOR, may be related to cell proliferation
(23). In addition, Senoo et
al (49) reported that Akt
phosphorylation directly mediated Rhonull cell migration
and apoptosis. Based on the aforementioned studies and the present
results, it may be concluded that inhibition of FBXW7 expression by
miR-223 promotes the proliferation and suppresses the apoptosis of
HCT116 cells through the Notch and Akt/mTOR pathways. miR-223 has a
positive impact on the Notch pathway and Akt/mTOR activation, and
whether this effect is mediated directly by miR-223 or indirectly
by FBXW7 after miR-223 binding needs to be further clarified.
In summary, the present study demonstrated that
upregulated miR-223 binds to the 3′UTR of the FBXW7 gene and
inhibits the expression of FBXW7, ultimately promoting the
proliferation and preventing the apoptosis of CRC cells through the
Notch and Akt/mTOR signalling pathways. Further examination of the
upstream molecular mechanism of miR-223 upregulation in CRC cells,
and how FBXW7 affects cell invasion and migration, will lead to a
better understanding of the occurrence and development of CRC and
provide new insight for early screening and diagnosis of CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by the Natural Science
Foundation of Hubei Province (grant nos. 2018CFB185, 2018CFB093 and
2017CFB238), the Natural Science Foundation of Hubei Provincial
Department of Education (grant nos. Q20192105 and Q20192104), the
Faculty Development Grants of Hubei University of Medicine (grant
nos. 2016QDJZR03 and 2017QDJZR08), and the Hubei Provincial
Training Program of Innovation and Entrepreneurship for
Undergraduates (grant nos. S201910929032 and S201913249006).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and ZL contributed to the design of experiments.
ZL, TM, JD and XL conducted the experiments. TM and XL provided
reagents. JD and ZL analysed the data. LL and ZL wrote the paper.
LL edited the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All procedures performed in the study involving
human participants were approved by the ethics committee of Hubei
University of Medicine, in accordance with the Declaration of
Helsinki. Written informed consent was obtained from all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang K, Song P, Gao J, Li G, Zhao X and
Zhang S: Perspectives on a combined test of multi serum biomarkers
in China: Towards screening for and diagnosing hepatocellular
carcinoma at an earlier stage. Drug Discov Ther. 8:102–109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Romano M, De Francesco F, Pirozzi G,
Gringeri E, Boetto R, Di Domenico M, Zavan B, Ferraro GA and Cillo
U: Expression of cancer stem cell biomarkers as a tool for a
correct therapeutic approach to hepatocellular carcinoma.
Oncoscience. 2:443–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for
ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeh CH, Bellon M and Nicot C: FBXW7: A
critical tumor suppressor of human cancers. Mol Cancer. 17:1152018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takeishi S and Nakayama KI: Role of Fbxw7
in the maintenance of normal stem cells and cancer-initiating
cells. Brit J Cancer. 111:1054–1059. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davis RJ, Welcker M and Clurman BE: Tumor
suppression by the Fbw7 ubiquitin ligase: Mechanisms and
opportunities. Cancer Cell. 26:455–464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kawashita Y, Morine Y, Ikemoto T, Saito Y,
Iwahashi S, Yamada S, Higashijima J, Imura S, Ogawa H, Yagi T and
Shimada M: Loss of Fbxw7 expression is a predictor of recurrence in
colorectal liver metastasis. J Hepatobiliary Pancreat Sci.
24:576–583. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Y, Wang M, Cui C, Zhang L, Liao F, Li
H and Wu X: Significance of combined tests of serum golgi
glycoprotein 73 and other biomarkers in diagnosis of small primary
hepatocellular carcinoma. Cancer Biomark. 15:677–683. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Zhang J, Zhou L, Sun W, Zheng ZG,
Lu P, Gao Y, Yang XS, Zhang ZC, Tao KS and Dou KF: Fbxw7 regulates
hepatocellular carcinoma migration and invasion via Notch1
signaling pathway. Int J Oncol. 47:231–243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li G, Luna C, Qiu J, Epstein DL and
Gonzalez P: Alterations in microRNA expression in stress-induced
cellular senescence. Mech Ageing Dev. 130:731–741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ramalinga M, Roy A, Srivastava A,
Bhattarai A, Harish V, Suy S, Collins S and Kumar D: MicroRNA-212
negatively regulates starvation induced autophagy in prostate
cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis
and cellular senescence. Oncotarget. 6:34446–34457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Welch C, Chen Y and Stallings RL:
MicroRNA-34a functions as a potential tumor suppressor by inducing
apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li R, Wu S, Chen X, Xu H, Teng P and Li W:
miR-223/FBW7 axis regulates doxorubicin sensitivity through
epithelial mesenchymal transition in non-small cell lung cancer. Am
J Transl Res. 8:2512–2524. 2016.PubMed/NCBI
|
|
17
|
Ibusuki M, Yamamoto Y, Shinriki S, Ando Y
and Iwase H: Reduced expression of ubiquitin ligase FBXW7 mRNA is
associated with poor prognosis in breast cancer patients. Cancer
Sci. 102:439–445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu J, Zhang W, Gao F, Liu YX, Chen ZY,
Cheng LY, Xie SF and Zheng SS: FBW7 increases chemosensitivity in
hepatocellular carcinoma cells through suppression of
epithelial-mesenchymal transition. Hepatobiliary Pancreat Dis Int.
13:184–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ding J, Zhao Z, Song J, Luo B and Huang L:
miR-223 promotes the doxorubicin resistance of colorectal cancer
cells via regulating epithelial-mesenchymal transition by targeting
FBXW7. Acta Biochim Biophys Sin (Shanghai). 50:597–604. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu G and Wang J: Dynamic changes in
routine blood parameters of a severe COVID-19 case. Clin Chim Acta.
508:98–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gharaibeh L, Elmadany N, Alwosaibai K and
Alshaer W: Notch1 in cancer therapy: Possible clinical implications
and challenges. Mol Pharmacol. 98:559–576. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vacca A, Felli MP, Palermo R, Di Mario G,
Calce A, Di Giovine M, Frati L, Gulino A and Screpanti I: Notch3
and pre-TCR interaction unveils distinct NF-kappaB pathways in
T-cell development and leukemia. EMBO J. 25:1000–1008. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu J, Lu AD, Zhang LP, Zuo YX and Jia YP:
Study of clinical outcome and prognosis in pediatric core binding
factor-acute myeloid leukemia. Zhonghua Xue Ye Xue Za Zhi.
40:52–57. 2019.(In Chinese). PubMed/NCBI
|
|
24
|
Li Y, Guessous F, Zhang Y, Dipierro C,
Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen
TD, et al: MicroRNA-34a inhibits glioblastoma growth by targeting
multiple oncogenes. Cancer Res. 69:7569–7576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji Q, Hao X, Zhang M, Tang W, Yang M, Li
L, Xiang D, Desano JT, Bommer GT, Fan D, et al: MicroRNA miR-34
inhibits human pancreatic cancer tumor-initiating cells. PLoS One.
4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao G, Li Y, Wang M, Li X, Qin S, Sun X,
Liang R, Zhang B, Du N, Xu C, et al: FBXW7 suppresses
epithelial-mesenchymal transition and chemo-resistance of
non-small-cell lung cancer cells by targeting snai1 for
ubiquitin-dependent degradation. Cell Prolif. 51:e124732018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam J and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife,. 4:e050052015. View Article : Google Scholar
|
|
29
|
Bilinska K, Jakubowska P, Von Bartheld CS
and Butowt R: Expression of the SARS-CoV-2 entry proteins, ACE2 and
TMPRSS2, in cells of the olfactory epithelium: Identification of
cell types and trends with age. ACS Chem Neurosci. 11:1555–1562.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thompson WW, Shay DK, Weintraub E, Brammer
L, Cox N, Anderson LJ and Fukuda K: Mortality associated with
influenza and respiratory syncytial virus in the United States.
JAMA. 289:179–186. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Chen F, He Y, Yi L, Ge C, Shi X,
Tang C, Wang D, Wu Y and Nian W: Sensitivity of non-small cell lung
cancer to erlotinib is regulated by the Notch/miR-223/FBXW7
pathway. Biosci Rep. 37:BSR201604782017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun XF, Sun JP, Hou HT, Li K, Liu X and Ge
QX: MicroRNA-27b exerts an oncogenic function by targeting Fbxw7 in
human hepatocellular carcinoma. Tumour Biol. 37:15325–15332. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K and
Baba H: Overexpression of microRNA-223 regulates the ubiquitin
ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer.
106:182–188. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Liu Y, Lu J, Zhang P, Wang Y, Xu
Y, Wang Z, Mao JH and Wei G: Rapamycin inhibits FBXW7 loss-induced
epithelial-mesenchymal transition and cancer stem cell-like
characteristics in colorectal cancer cells. Biochem Biophys Res
Commun. 434:352–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iwatsuki M, Mimori K, Ishii H, Yokobori T,
Takatsuno Y, Sato T, Toh H, Onoyama I, Nakayama KI, Baba H and Mori
M: Loss of FBXW7, a cell cycle regulating gene, in colorectal
cancer: Clinical significance. Int J Cancer. 126:1828–1837. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li ZW, Yang YM, Du LT, Dong Z, Wang LL,
Zhang X, Zhou XJ, Zheng GX, Qu AL and Wang CX: Overexpression of
miR-223 correlates with tumor metastasis and poor prognosis in
patients with colorectal cancer. Med Oncol. 31:2562014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pichler M, Ress AL, Winter E, Stiegelbauer
V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn SW,
Kiesslich T, et al: miR-200a regulates epithelial to mesenchymal
transition-related gene expression and determines prognosis in
colorectal cancer patients. Br J Cancer. 110:1614–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
40
|
He D, Huang C, Zhou Q, Liu D, Xiong L,
Xiang H, Ma G and Zhang Z: HnRNPK/miR-223/FBXW7 feedback cascade
promotes pancreatic cancer cell growth and invasion. Oncotarget.
8:20165–20178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Z, Liu X, Liu S and Cao Q: Cholesterol
promotes the migration and invasion of renal carcinoma cells by
regulating the KLF5/miR-27a/FBXW7 pathway. Biochem Biophys Res
Commun. 502:69–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sano H, Kawahito Y, Wilder RL, Hashiramoto
A, Mukai S, Asai K, Kimura S, Kato H, Kondo M and Hla T: Expression
of cyclooxygenase-1 and −2 in human colorectal cancer. Cancer Res.
55:3785–3789. 1995.PubMed/NCBI
|
|
43
|
Xu Y, Sengupta T, Kukreja L and Minella
AC: MicroRNA-223 regulates cyclin E activity by modulating
expression of F-box and WD-40 domain protein 7. J Biol Chem.
285:34439–34446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yeh CH, Bellon M, Pancewicz-Wojtkiewicz J
and Nicot C: Oncogenic mutations in the FBXW7 gene of adult T-cell
leukemia patients. Proc Natl Acad Sci USA. 113:6731–6736. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hua J, Ding T and Yang L: Dysfunction of
microRNA-32 regulates ubiquitin ligase FBXW7 in multiple myeloma
disease. Onco Targets Ther. 9:6573–6579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Thomas MR, Marston L, Rafferty GF, Calvert
S, Marlow N, Peacock JL and Greenough A: Respiratory function of
very prematurely born infants at follow up: Influence of sex. Arch
Dis Child Fetal Neonatal Ed. 91:F197–F201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kumar V, Palermo R, Talora C, Campese AF,
Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S,
et al: Notch and NF-kB signaling pathways regulate miR-223/FBXW7
axis in T-cell acute lymphoblastic leukemia. Leukemia.
28:2324–2335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Espinosa L, Cathelin S, D'Altri T,
Trimarchi T, Statnikov A, Guiu J, Rodilla V, Inglés-Esteve J,
Nomdedeu J, Bellosillo B, et al: The Notch/Hes1 pathway sustains
NF-κB activation through CYLD repression in T cell leukemia. Cancer
Cell. 18:268–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Senoo H, Kamimura Y, Kimura R, Nakajima A,
Sawai S, Sesaki H and Iijima M: Phosphorylated Rho-GDP directly
activates mTORC2 kinase towards AKT through dimerization with
Ras-GTP to regulate cell migration. Nat Cell Biol. 21:867–878.
2019. View Article : Google Scholar : PubMed/NCBI
|