Introduction
Cervical carcinoma (CC) is one of the leading causes
of cancer-related death in females (1), with an increasing incidence in
relatively young females (2). It is
of great importance to investigate the mechanisms of tumourigenesis
and development of CC to facilitate its therapy.
As a type of small non-coding RNA, microRNAs
(miRNAs) can post-transcriptionally modify target gene expression
by binding to 3′-untranslated regions (3′-UTRs), followed by
translational inhibition or degradation of mRNAs (3). miRNAs can either be cancer-promoting,
by inhibiting anti-oncogenes, or cancer-suppressing, by inhibiting
oncogenes (3). Accumulating
evidence has proven that miRNAs can impact tumour metastasis in a
broad spectrum of human malignancies. For instance, excessive
maturation of miR-25-3p via m6A modification, which can be induced
by cigarette smoking, promotes the development of pancreatic cancer
(4), and miR-375 has been described
as a crucial regulator of phagocyte infiltration (5). Moreover, miRNAs have been reported to
modulate tumour proliferation by regulating target genes.
MiRNA-145, for instance, was reported to inhibit the proliferation
of non-small cell lung cancer cells by targeting c-Myc (6). However, the roles of miRNAs,
especially proliferation-relevant miRNAs, in CC remain to be
clarified.
miR-302c-3p and miR-520a-3p are known as tumour
suppressors in several types of cancer, including glioma (7), renal cell carcinoma (8), and hepatocellular carcinoma (9). However, the biological functions of
miR-302c-3p and miR-520a-3p in CC, particularly in CC
proliferation, are still unclear. Moreover, given that miR-302 and
miR-520 can regulate the natural killer group 2 D ligands major
histocompatibility complex class I chain-related proteins A and B
and unique long 16-binding protein 2 to counter the resistance of
Kasumi-1 cells to natural killer cells (10), it remains unclear whether
miR-302c-3p and miR-520a-3p can act co-operatively to regulate CC
metastasis and proliferation.
C-X-C motif ligand (CXCL)8 (CXCL8), also known as
interleukin-8 (IL-8), is a pro-inflammatory CXC chemokine that is
associated with the promotion of neutrophil chemotaxis and
degranulation (11). Under normal
circumstances, the expression of CXCL8 in tissues is low or
undetectable; however, increased expression of CXCL8 is detected in
some human solid tumours, such as cervical cancer (12), ovarian cancer (13,14),
and non-small cell lung cancer (15). CXCL8 produced by cancer cells in
response to epithelial-mesenchymal transition (EMT) is involved in
various biological cell phenotypes, such as cell proliferation,
migration and metastasis (16).
More importantly, in microsatellite-unstable colorectal cancer,
CXCL8 production and cell proliferation is enhanced by the loss of
miR-484 (17), indicative of a
regulatory role for CXCL8 and miRNAs. Although CXCL8 is not a novel
target in CC, as it has previously been reported to be upregulated
in CC (18), the present study
focused on investigating the relationship between miR-302c-3p,
miR-520a-3p and CXCL8.
In the present study, differentially expressed
miRNAs between CC tissues and adjacent normal tissues were screened
by literature review and based on the author's unpublished data
(data not shown). Of the 25 selected miRNAs, the effect of
miR-302c-3p and miR-520a-3p on CC cell proliferation was
investigated. The aim of the present study was to explore the
cooperative effects of miR-302c-3p and miR-520a-3p on CC cells to
gain a deeper understanding of CC proliferation and provide a
potential target for the treatment of CC.
Materials and methods
Patient samples
In total, 20 patients with recently diagnosed (and
previously untreated) CC at the Department of Obstetrics and
Gynaecology of The First Affiliated Hospital of Soochow University
from July 2016 to August 2017 were selected in a single-centre
study and had a median age of 51 years (range, 39–75). The CC
tissues and tumour-adjacent tissues were collected from the
selected patients following diagnostic evaluation. Written informed
consent was obtained from each patient prior to the experiment. The
protocols were also approved by the local ethics committee of The
First Affiliated Hospital of Soochow University. All the clinical
samples collected in the present study are squamous cell
carcinoma.
Cell culture
Human CC cell lines (HeLa-S3 and C-33A),
immortalized cervical epithelial cell line (H8) and human embryonic
kidney cells (293T) were obtained from the Cell Resource Center of
the Chinese Academy of Sciences (Shanghai, China). Cells were
cultured in DMEM (HyClone, Cytiva) containing 10% foetal bovine
serum (Gibco, Thermo Fisher Scientific) and 100 U/ml
penicillin/streptomycin (HyClone, Cytiva) in a 37°C constant
incubator with 95% air and 5% CO2.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of tissues and cell lines was extracted
using the RNeasy Plus Universal Tissue kit (Sangon Biotech Co.)
based on the manufacturer's instructions. miRNAs from CC cell lines
were extracted using the miRNeasy Mini Kit (Sangon Biotech Co). PCR
primers for 25 potentially dysregulated miRNAs and U6 were
synthesized by Guangzhou RiboBio.
Primers for CXCL8 and GAPDH were synthesized by
Takara (Sangon Biotech Co.) (Table
I). cDNA was constructed using the PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd.). Real-time PCR was performed using
SYBR premix Ex Taq II (Takara Biotechnology Co., Ltd.), and
fluorescence intensity was detected in a Light Cycler 480 system
(Roche Applied Science) using the following thermocycling
conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C
for 1 min. GAPDH was used as the internal control. The comparative
Cq (ΔΔCq) method (19)
was used to calculate the fold change for each miRNA/mRNA. U6 and
GAPDH were treated as internal controls. The following formulas
were used to calculate the expression fold changes (X):
ΔCq=Cq (target miRNA/mRNA)-Cq (U6/GAPDH), ΔΔCq=ΔCq (target
miRNAs/mRNA)-ΔCq (target miRNAs/mRNA),
X=2−ΔΔCq.
 | Table I.Primers for RT-qPCR. |
Table I.
Primers for RT-qPCR.
| Gene name |
| Primer sequence
(5′→3′) |
|---|
| miR-302c-3p | RT primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCACTG |
|
| Forward |
GCGTGCTTCCATGTTTCAGTGG |
|
| Reverse |
CAGTGCAGGGTCCGAGGTAT |
| miR-520a-3p | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGTCCAAA |
|
| Forward |
ACACTCCAGCTGGGAAAGTGCTTCCC |
|
| Reverse |
CTCAACTGGTGTCGTGGA |
| CXCL8 | Forward |
CAGAGCAACGTGCTCCAAAGTC |
|
| Reverse |
GAAGCGTTGCTGTCGGTTCA |
| U6 | Forward |
CTCGCTTCGGCAGCACA |
|
| Reverse |
AACGCTTCACGAATTTGCGT |
| GAPDH | Forward |
CTGGGCTACACTGAGCACC |
|
| Reverse |
AAGTGGTCGTTGAGGGCAATG |
miRNAs, siRNAs and plasmids
Human miR-302c-3p and miR-520a-3p mimics as well as
scrambled miR-302c-3p mimics, miR-520a-3p mimics and small
interfering RNAs were obtained from Guangzhou RiboBio. A CXCL8
eukaryotic expression vector (CXCL8/pcDNA3.1) was generated by
inserting the open reading frame of CXCL8 into pcDNA3.1, using the
empty vector as a negative control. For luciferase reporters, the
3′-UTR of CXCL8 was amplified from human genomic DNA before the
miR-302c-3p and miR-520a-3p binding sites within the CXCL8 3′-UTR
were mutated to remove complementarity. The mutant or wild-type
3′-UTR of CXCL8 were cloned into the psiCHECK-2 luciferase vector
(cat. no. 78260; Promega Corporation). All plasmids were confirmed
by sequencing.
Cell proliferation assay
Cell counting kit (CCK-8, Merck KGaA) and
ethynyl-2-deoxyuridine (EdU, Guangzhou RiboBio) staining were both
utilised to measure the proliferation ability of each cell line
under different conditions. For the CCK-8 assay, 5×103
cells in 100 µl were transferred to each well of a 96-well plate.
Cells were cultured at 37°C before CCK-8 solution (10 µl) was added
to each well 24, 48, 72, and 96 h later. Subsequently, the cells
were incubated at 37°C for an additional 4 h before OD values (450
nm) were measured using a microplate reader (Bio-Rad Laboratories,
Inc.). For the EdU assay, the cells were transfected with miR-302
and miR-520 mimic or inhibitor (100 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and incubated with 100 µl of 50 µM EdU per well
for 2 h at 37°C. Cells were visualized by fluorescence microscopy
and the ratio of EdU/DAPI was calculated to analyse the EdU assay.
Experiments were repeated at least three times.
Flow cytometry assay
Cell apoptosis was assessed with the Annexin V-FITC
apoptosis detection kit (Elabscience). Flow cytometry was performed
with FACSCalibur (BD Biosciences) and all experiments were
performed at least three times.
Luciferase assay
The potential direct target genes of miR-302c-3p and
miR-520a-3p were predicted by miRTarBase
(mirtarbase.mbc.nctu.edu.tw/php/index.php). For the luciferase
reporter assays, 293T cells were seeded onto 24-well plates before
miR-302c-3p or miR-520a-3p mimics (100 nM) and wild-type or
mut-type CXCL8 3′-UTR (1.5 µg) were co-transfected into 293T cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). After transfection for 48 h, cells were collected and lysed
for luciferase assays. The intensity of luciferase activity was
measured using the Dual-Luciferase Reporter Assay System (Promega
Corporation). Renilla luciferase was used for normalization. Each
experiment was performed at least three times.
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer
(Beyotime Institute of Biotechnology). Extracts were mixed with
SDS-PAGE loading buffer and denatured by boiling for 5 min. Equal
amounts of protein (~30 µg) were separated on 10% SDS-PAGE gels and
transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad
Laboratories, Inc.). Blots were blocked using 5% bovine serum
albumin (BSA, Gibco; Thermo Fisher Scientific, Inc.) for 2 h and
incubated overnight at 4°C with primary antibodies against CXCL8
(1:1,000; cat. no. ab154390; Abcam) and β-actin (1:5,000; cat. no.
4970S; Cell Signaling Technology, Inc.). PVDF membranes were washed
with Tris-buffered saline (TBS) supplemented with 0.05% Tween-20
before incubation with HRP-conjugated secondary antibody (1:2,000;
cat. no. 7074S; Cell Signaling Technology, Inc.) for 2 h at room
temperature. Signals were visualized using enhanced
chemiluminescence reagents (Thermo Fisher Scientific, Inc.).
Densitometric analysis of the blot bands was performed using ImageJ
1.48 software, and the intensity values were normalized against the
values of the β-actin loading control. All experiments were
performed at least three times.
Statistical analysis
Data were analysed using SPSS software (version 19;
IBM Corp.). Continuous data sets are expressed as the mean ± SEM.
Comparisons between two groups were analysed using t-tests. One-way
ANOVA was used for comparison of three or more groups, with
additional post-hoc Tukey's tests to account for multiple
comparisons. Statistical analysis of categorical variables was
performed using the χ2 test. Correlations between
miR-302c-3p and miR-520a-3p expression levels and CC profile and
between miR-302c-3p, miR-520a-3p and CXCL8 expression levels were
determined by Pearson's linear correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-302c-3p and miR-520a-3p are
downregulated in CC cell lines
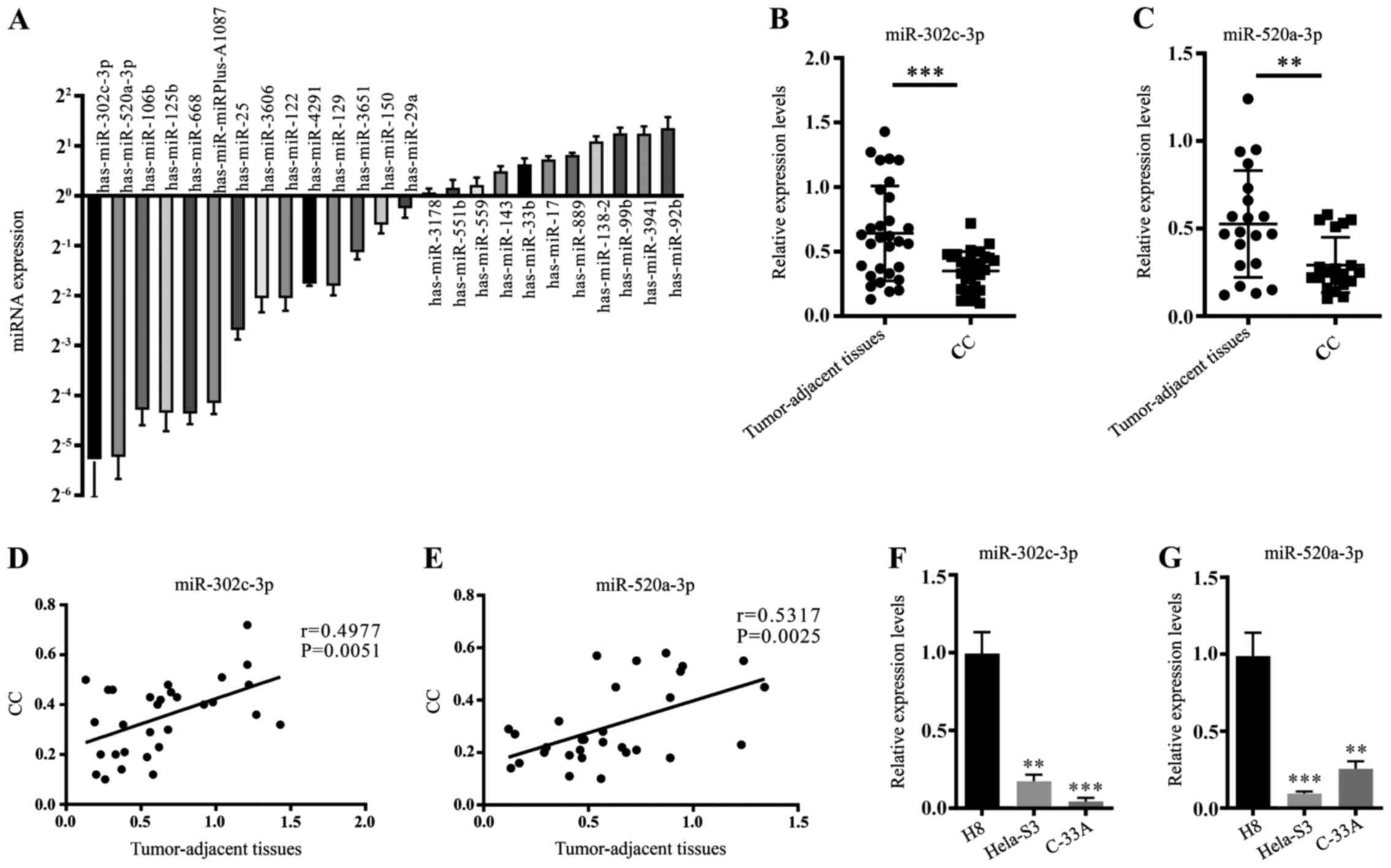
To investigate the potentially dysregulated miRNAs
between clinical CC tissues and tumour-adjacent tissues, RT-qPCR
was performed to measure the expression of 25 mature miRNA
sequences in both CC tissues and tumour-adjacent tissues (Fig. 1A). Two highly downregulated miRNAs,
miR-302c-3p and miR-520a-3p, were notable due to their
tumour-suppressing roles in other cancer cells (7,18).
Furthermore, the expression of miR-302c-3p and miR-520a-3p was
examined in 20 pairs of clinical CC tissues and tumour-adjacent
normal tissues. The data showed that miR-302c-3p and miR-520a-3p
were downregulated in CC tissues compared with adjacent tissues
(Fig. 1B and C; ***P<0.001,
**P<0.01). Pearson's linear correlation analysis revealed that
miR-302c-3p and miR-520a-3p expression levels in adjacent normal
tissues were positively correlated with those in normal tissues
(Fig. 1D and E; P=0.0051,
P=0.0025). The expression levels of miR-302c-3p and miR-520a-3p
were measured in two CC cell lines (HeLa-S3 and C-33A), as well as
in the immortalized cervical epithelial cell line H8. It was found
that miR-302c-3p and miR-520a-3p were both downregulated in HeLa-S3
and C-33A cells (Fig. 1F-G;
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001). These
results demonstrate that the expressions of miR-302c-3p and
miR-520a-3p are downregulated in both CC tissues and cell
lines.
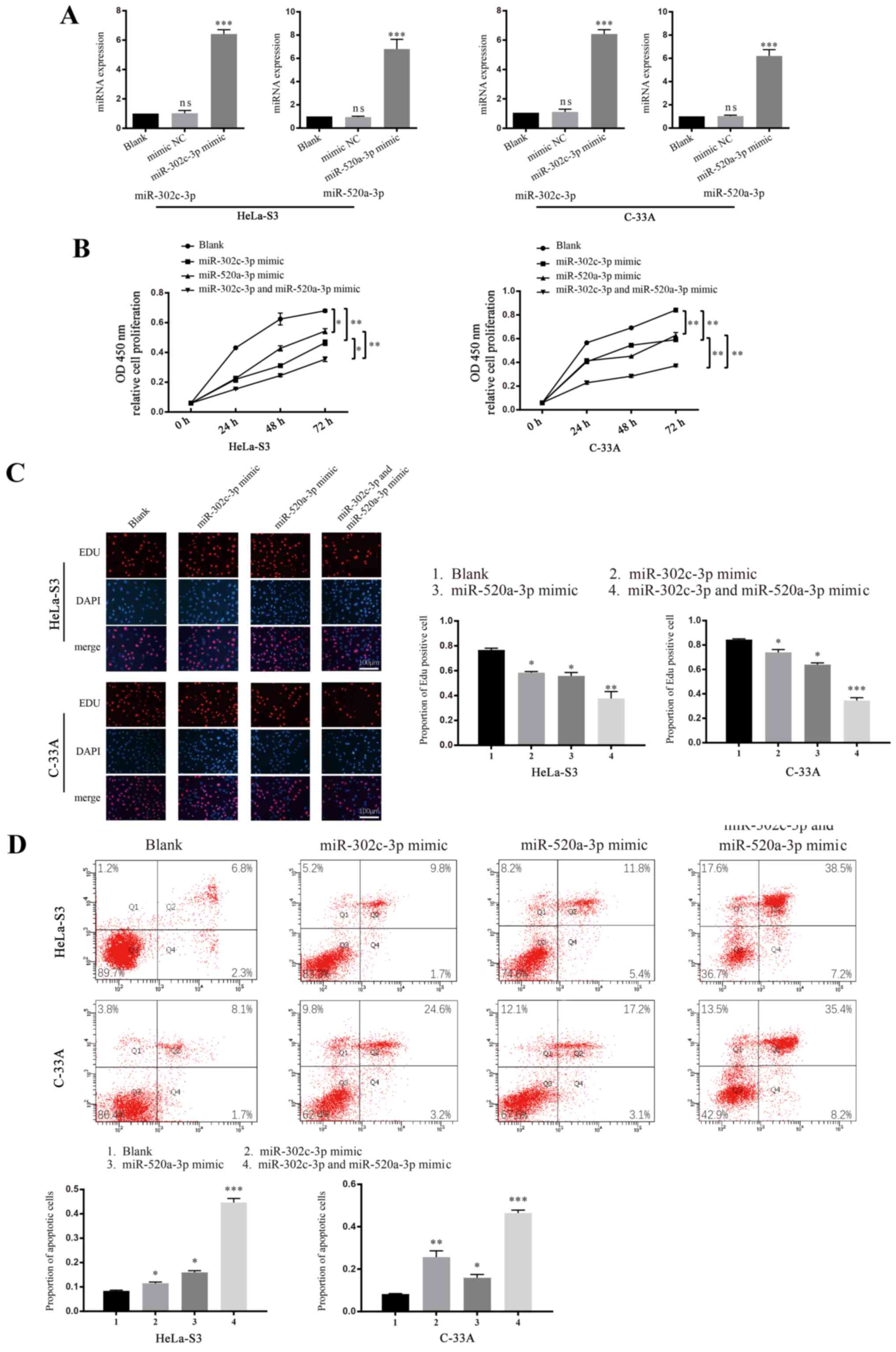
miR-302c-3p and miR-520a-3p inhibit
the proliferation of CC
miR-302c-3p is reported to inhibit the growth of
hepatocellular carcinoma (20),
while miR-520a-3p inhibits the proliferation of non-small cell lung
cancer cells (18). To investigate
whether miR-302c-3p or miR-520a-3p is involved in the proliferation
of CC, CCK-8 and EdU assays were performed using cells transfected
with mimics or inhibitors of miR-302c-3p and miR-520a-3p. The
transfection efficiency of both overexpression and knockdown of
miR-302c-3p and miR-520a-3p were verified using RT-qPCR (Figs. 2A and S1A; ****P<0.0001). A significant
decrease in cell proliferation was observed in both HeLa-S3 and
C-33A cells transfected with miR-302c-3p/miR-520a-3p mimics
(Fig. 2B; *P<0.05, **P<0.01),
whereas the cells treated with miR-302c-3p and miR-520a-3p
inhibitors had increased proliferation (Fig. S1B; **P<0.01) compared with the
control groups (blank cells were treated with an equivalent volume
of PBS). Co-transfection of miR-302c-3p and miR-520a-3p mimics led
to cooperative suppression of cell proliferation, which was greater
than that of cells transfected with miR-302c-3p or miR-520a-3p
alone. Results consistent with these observations were also
obtained from the EdU staining assay (Figs. 2C and S1C; **P<0.01, ****P<0.0001,
*P<0.05). Tumour tissue growth requires the rate of cell
proliferation to exceed that of cell death; thus, the
apoptosis-associated functions of miR-302c-3p and miR-520a-3p were
detected using flow cytometry. As expected, both HeLa-S3 and C-33A
cells (Fig. 2D; **P<0.01,
****P<0.0001) transfected with miR-302c-3p/miR-520a-3p mimics
showed significant increases in cell apoptosis, whereas the cells
treated with miR-302c-3p/miR-520a-3p inhibitor showed decreases in
cell apoptosis (Fig. S1D;
**P<0.01, ****P<0.0001 vs. control). These results
demonstrated that miR-302c-3p and miR-520a-3p can suppress the
growth and promote the apoptosis of CC cells in an independent or
cooperative manner.
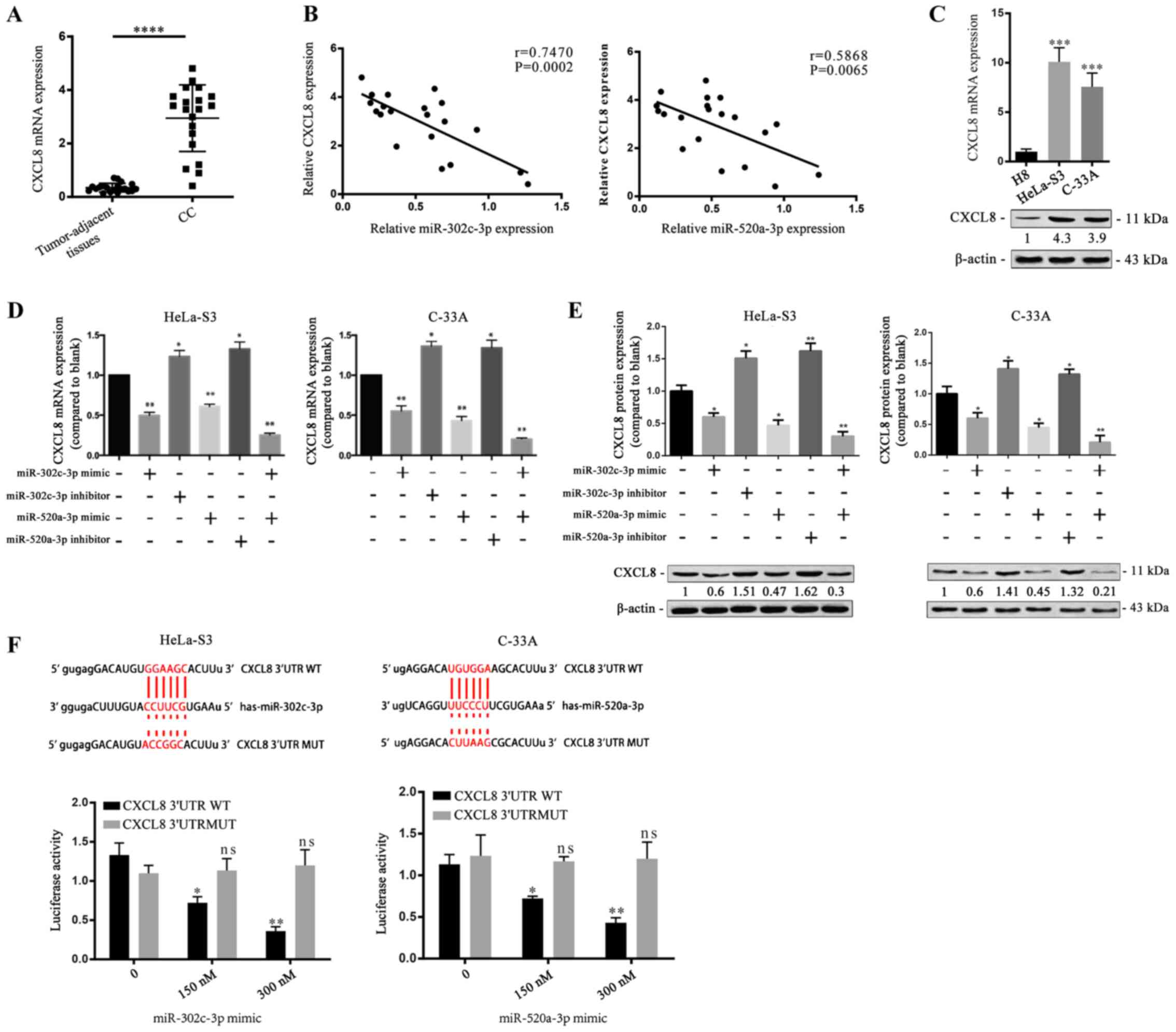
CXCL8 is a direct target of
miR-302c-3p and miR-520a-3p
To understand the proliferation-suppressive role of
miR-302c-3p and miR-520a-3p, the potential target genes of
miR-302c-3p and miR-520a-3p were predicted by using miRTarBase.
Among the candidates, CXCL8 has been reported to be upregulated in
various types of carcinomas (21,22)
and contributes to cancer cell proliferation (23,24).
Therefore, CXCL8 was selected for further evaluation and RT-qPCR
demonstrated that CC tissues had much higher CXCL8 expression than
tumour-adjacent tissues (Fig. 3A;
****P<0.0001). Pearson's linear correlation analysis found that
CXCL8 was negatively correlated with miR-302c-3p and miR-520a-3p
(Fig. 3B; P=0.0002, P=0.0065).
Moreover, RT-qPCR measured the relative expressions of CXCL8 and
miR-302c-3p/miR-520a-3p, which showed that high expression of
miR-302c-3p or miR-520a-3p corresponds to low expression of CXCL8
(Fig. S2A; **P<0.01). RT-qPCR
and western blot analysis revealed that the mRNA and protein
expression levels of CXCL8 were increased in HeLa-S3 and C-33A
cells compared with H8 cells (Fig.
3C; **P<0.01, ***P<0.001). To investigate whether
miR-302c-3p and miR-520a-3p regulate CXCL8 expression in CC cells,
HeLa-S3 and C-33A cells were transfected with miR-302c-3p and
miR-520a-3p mimics before measurement of CXCL8 expression. CXCL8
was downregulated in response to transfection of miR-302c-3p and
miR-520a-3p mimics into CC cells (Fig.
3D and E; *P<0.05, **P<0.01). Furthermore, a luciferase
reporter assay was performed to identify the binding of CXCL8 with
miR-302c-3p or miR-520a-3p. The results revealed that miR-302c-3p
mimics and miR-520a-3p mimics inhibited the luciferase reporter
activity of the wild-type CXCL8 3′-UTR construct (Fig. 3F; **P<0.01). Collectively, these
data indicated that CXCL8 is a direct common target of miR-302c-3p
and miR-520a-3p.
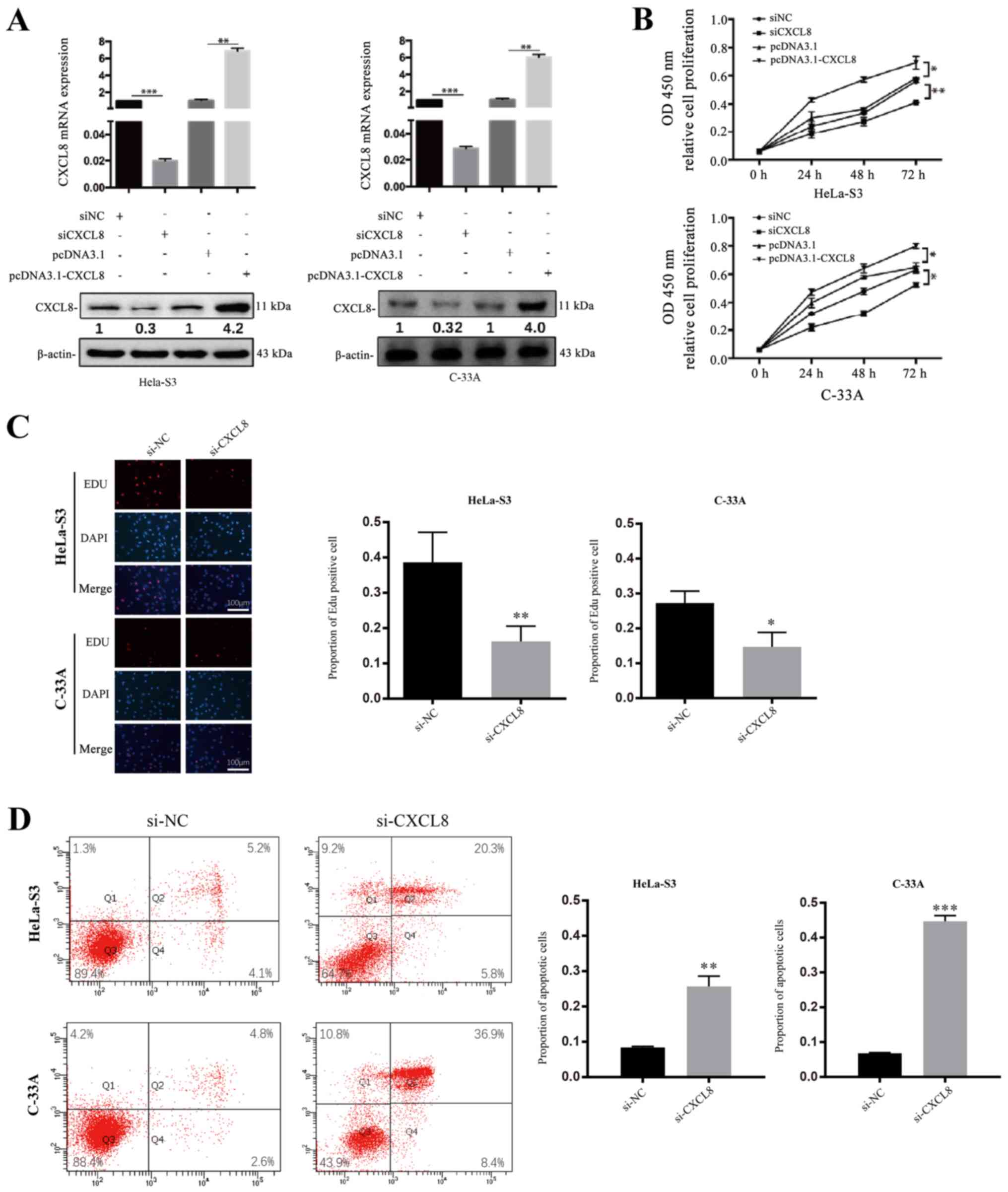
CXCL8 promotes the proliferation and
decreases the apoptosis of CC cells
As previously described, CXCL8 has been reported to
be involved in cell proliferation-promoting and
apoptosis-suppressive behaviour in cancerous cells (23,24).
To investigate whether CXCL8 is a tumour-related cytokine in
HeLa-S3 and C-33A cells, the effect of CXCL8 on CC cell
proliferation was detected by the CCK-8 assay. Briefly, the HeLa-S3
and C-33A cell lines were treated with CXCL8 siRNA or negative
control (NC) or with pcDNA3.1-CXCL8 or pcDNA3.1, and then both
CCK-8 and EdU staining assays were performed. Compared with the
levels in cells treated with si-NC, the mRNA and protein levels of
CXCL8 were decreased in cells transfected with si-CXCL8, whereas
cells treated with pcDNA3.1-CXCL8 (pcDNA3.1 used as control) showed
increased expression (Fig. 4A;
**P<0.01). The up- and downregulation of CXCL8 correspondingly
increased or decreased the proliferation ability of HeLa-S3 and
C-33A cells compared with the controls (Figs. 4B and C and S3A; *P<0.05, ****P<0.0001). To
investigate whether CXCL8 can decrease CC cell apoptosis, flow
cytometry was also performed. A significant decrease in apoptosis
was observed in both HeLa-S3 and C-33A cells transfected with
pcDNA3.1-CXCL8, whereas the cells treated with si-CXCL8 showed the
opposite effects (Figs. 4D and
S3B; ***P<0.001,
****P<0.0001).
Downregulation of CXCL8 by miR-302c-3p
and miR-520a-3p inhibits CC cell proliferation
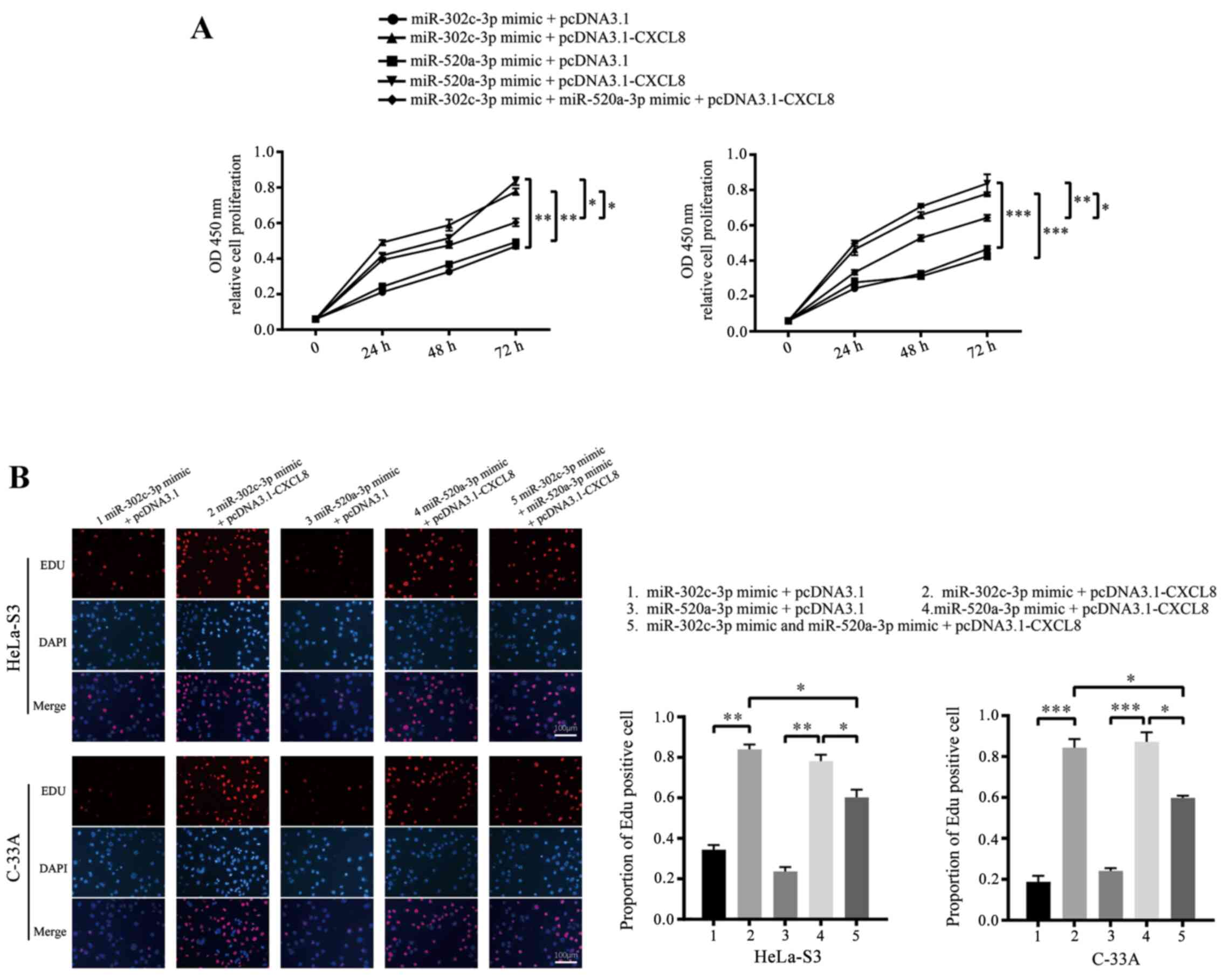
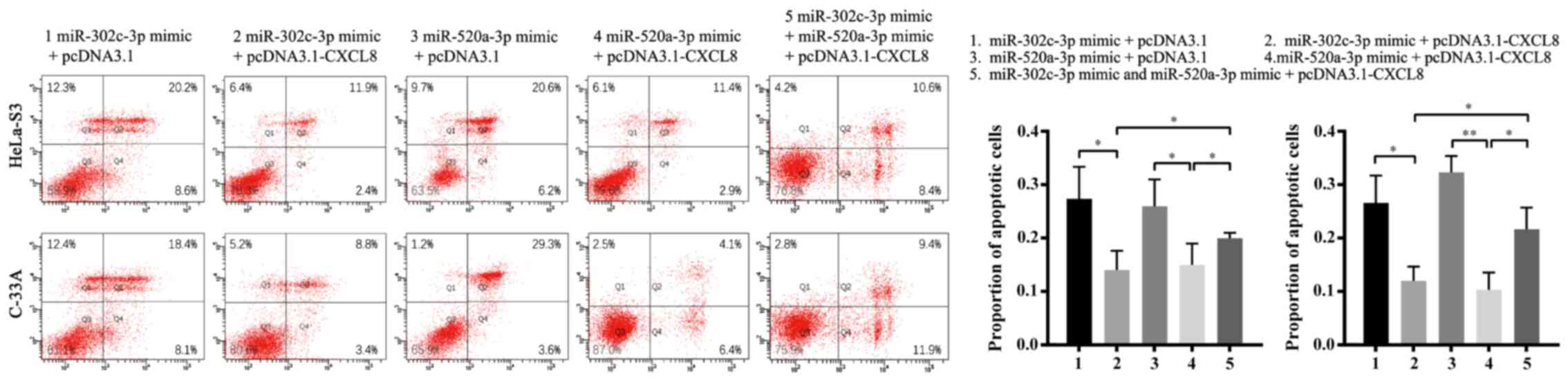
To confirm whether CXCL8 downregulation is the
mechanism by which miR-302c-3p and miR-520a-3p suppress
proliferation and promote apoptosis in CC cells, HeLa-S3 and C-33A
cells were co-transfected with pcDNA3.1-CXCL8 or pcDNA3.1 and
miR-302c-3p/miR-520a-3p mimics. As expected, restoration of CXCL8
partially blocked the miR-302c-3p and miR-520a-3p-mediated
suppression of CC cell proliferation and promotion of apoptosis
(Figs. 5A and B and 6; **P<0.01, ***P<0.001,
****P<0.0001, *P<0.05). In summary, these results indicated
that CXCL8 is a functional target of miR-302c-3p and miR-520a-3p to
suppress the proliferation of CC cells.
Discussion
To the author's best knowledge, the ability of
miR-302c-3p and miR-520a-3p to efficiently target CXCL8 and thus
modulate the proliferation of CC cells has not been reported
before. Although previous studies have revealed that miR-302c-3p
can suppress cell proliferation in glioma (7), renal cell carcinoma (8) and hepatocellular carcinoma (9), and miR-520a-3p can suppress cell
proliferation in non-small cell lung cancer (18), breast cancer (25) and osteosarcoma (26), the functions of miR-302c-3p and
miR-520a-3p in CC are still unknown. In the present study, the
expression of miR-302c-3p and miR-520a-3p was measured in CC
tissues and adjacent tissues, which revealed that both of these
miRs were downregulated in CC cells and tissues. Transfection of CC
cells demonstrated that miR-302c-3p and miR-520a-3p could inhibit
the proliferation and enhance the apoptosis of CC cells. These
results show that miR-302c-3p and miR-520a-3p are important
proliferation suppressors and apoptosis promoters in CC.
CXCL8 is upregulated in a variety of solid tumours,
including gastric cancer (21),
ovarian cancer (27), non-small
cell lung cancer (28) and breast
cancer (29) and is reported to be
relevant to a broad spectrum of biological behaviours of cancer
cells, such as increased proliferation (30), invasion (31) and metastasis (32). In addition, CXCL8 has also been
reported to be associated with the severity of cervical cancer
malignancy (33,34). It has been reported that inhibition
of CXCL8 is associated with inhibition of angiogenesis in CC
(35). Overexpression of CXCL8 can
increase the proliferation and survival of cancer cells through
autocrine signalling pathways: MAPK (36,37),
focal adhesion kinase and sarcoma signalling (38). Queries about the function of CXCL8
in CC, together with the evidence of the proliferation-promoting
activity of CXCL8 in other carcinomas, prompted further
investigation of this predicted target gene of miR-302c-3p and
miR-520a-3p. In the present study, it was found that CXCL8 is
upregulated both in CC cells and clinical CC tissues. Knockdown of
CXCL8 attenuated the proliferation and increased the apoptosis of
HeLa-S3 and C-33A cells. Importantly, CXCL8 is directly targeted by
miR-302c-3p and miR-520a-3p, and the inhibition of cell
proliferation induced by miR-302c-3p and miR-520a-3p can be
abrogated by CXCL8 overexpression in CC cell lines.
The findings of the present study provide direct
evidence that the downregulation of CXCL8 induced by miR-302c-3p
and miR-520a-3p results in decreased proliferation and increased
apoptosis activity in CC cells. Specifically, it was found that
miR-302c-3p and miR-520a-3p synergistically downregulated CXCL8.
Members of the primate-specific miRNA family, miR-302c-3p and
miR-520-3p share similar seed sequences. Accumulating evidence has
indicated that miRNAs may target multiple genes in a coordinated
way. Therefore, only a subset of collaborating miRNAs can be
identified on the basis of miRNA expression analysis. To explore
the full interaction of miRNAs, a more efficient screening method
should be performed; however, methods for such studies are
currently unavailable. The present study was limited in several
aspects, as relevant data concerning migration, invasion and
angiogenesis in CC were missing. However, this can be a direction
for future work to explore the possible role of both miR-302c-3p
and miR-520a-3p in CC.
There are several limitations that are noteworthy.
Firstly, in the present study, expression of CXCL8 was lower in
C33A cells compared with that in Hela-S3 cells. However, TCGA data
show that HPV-positive cases, e.g., Hela cells derived from
HPV-positive adenocarcinoma cells, have decreased expression of
CXCL8 compared to HPV-negative cases, e.g., C-33A cells derived
from HPV-negative squamous cell carcinoma cells. This discrepancy
may possibly arise due to the varieties and differences between
tumor samples and cell lines in TCGA data. Considering the
discrepancy, further measurements and analysis on the expression of
CXCL8 among CC cells can form a future study. Secondly, it has been
reported that inhibition of CXCL8 is associated with inhibition of
angiogenesis (37). In the present
study, the effect of miR-302c-3p and miR-520a-3p on proliferation
and apoptosis of CC cells was the main focus. Although no related
data concerning angiogenesis was mentioned, this could serve as the
future direction for study. Moreover, although no relevant data
concerning the migration and invasion of CC cells is presented it
is a promising direction for future research.
The present study demonstrated that miR-302c-3p and
miR-520a-3p were downregulated in CC cell lines and clinical CC
tissues. Furthermore, the overexpression of miR-302c-3p and
miR-520a-3p suppressed CC cell proliferation and promoted CC cell
apoptosis in vitro by targeting CXCL8. The
miR-302c/520a-CXCL8 axis deepens the understanding of the
mechanisms underlying the proliferation of tumour cells and may
provide a potential therapeutic strategy for the targeted treatment
of CC. Furthermore, as many studies have documented that CXCL8 is
overexpressed in a large set of diseases in advanced stages,
suppressing the effects of CXCL8 may have important implications
for the systemic treatment of aggressive and metastatic
diseases.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81672560).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HMD, HZ and YGC conceptualized the study and
designed the experiments. JW and JHZ performed the experiments.
FRS, RNJ and JYS analyzed the data. HMD, HZ and JYS provided
critical materials. HMD, HZ and JYS wrote the manuscript. YGC
supervised the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients prior to the experiment. The protocols were approved by
the local ethics committee of The First Affiliated Hospital of
Soochow University (approval no. SH20181203).
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing
interests.
References
|
1
|
DiPaolo JA and Alvarez-Salas LM: Advances
in the development of therapeutic nucleic acids against cervical
cancer. Expert Opin Biol Ther. 4:1251–1264. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6-methyladenosine stimulated by cigarette smoke promotes
pancreatic cancer progression. Nat Commun. 10:18582019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frank AC, Ebersberger S, Fink AF, Lampe S,
Weigert A, Schmid T, Ebersberger I, Syed SN and Brüne B: Apoptotic
tumor cell-derived microRNA-375 uses CD36 to alter the
tumor-associated macrophage phenotype. Nat Commun. 10:11352019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng
A and Hu J: miRNA-145 inhibits non-small cell lung cancer cell
proliferation by targeting c-Myc. J Exp Clin Cancer Res.
29:1512010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Wei Y, Tong H, Chen L, Fan Y, Ji
Y, Jia W, Liu D and Wang G: miR-302c-3p suppresses invasion and
proliferation of glioma cells via down-regulating metadherin (MTDH)
expression. Cancer Biol Ther. 16:1308–1315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gu DH, Mao JH, Pan XD, Zhu H, Chen X,
Zheng B and Shan Y: microRNA-302c-3p inhibits renal cell carcinoma
cell proliferation by targeting Grb2-associated binding 2 (Gab2).
Oncotarget. 8:26334–26343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang L, Guo Y, Liu X, Wang T, Tong X, Lei
K, Wang J, Huang D and Xu Q: The tumor suppressive miR-302c-3p
inhibits migration and invasion of hepatocellular carcinoma cells
by targeting TRAF4. J Cancer. 9:2693–2701. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Min D, Lv XB, Wang X, Zhang B, Meng W, Yu
F and Hu H: Downregulation of miR-302c and miR-520c by 1,25(OH)2D3
treatment enhances the susceptibility of tumour cells to natural
killer cell-mediated cytotoxicity. Br J Cancer. 109:723–730. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia L, Li F, Shao M, Zhang W, Zhang C,
Zhao X, Luan H, Qi Y, Zhang P, Liang L, et al: IL-8 is upregulated
in cervical cancer tissues and is associated with the proliferation
and migration of HeLa cervical cancer cells. Oncol Lett.
15:1350–1356. 2018.PubMed/NCBI
|
|
13
|
Sanguinete MMM, Oliveira PH, Martins-Filho
A, Micheli DC, Tavares-Murta BM, Murta EFC and Nomelini RS: Serum
IL-6 and IL-8 correlate with prognostic factors in ovarian cancer.
Immunol Invest. 46:677–688. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yung MM, Tang HW, Cai PC, Leung TH, Ngu
SF, Chan KK, Xu D, Yang H, Ngan HY and Chan DW: GRO-α and IL-8
enhance ovarian cancer metastatic potential via the CXCR2-mediated
TAK1/NFκB signaling cascade. Theranostics. 8:1270–1285. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu J, Cheng T, Liu L, Heng J, Liu X, Sun
Z, Wang W, Li K and Yang N: Mast cells induce
epithelial-to-mesenchymal transition and migration in non-small
cell lung cancer through IL-8/Wnt/beta-catenin pathway. J Cancer.
10:55672019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang CP, Zhou HJ, Qin J, Luo Y and Zhang
T: MicroRNA-520c-3p negatively regulates EMT by targeting IL-8 to
suppress the invasion and migration of breast cancer. Oncol Rep.
38:3144–3152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mei Q, Xue G, Li X, Wu Z, Li X, Yan H, Guo
M, Sun S and Han W: Methylation-induced loss of miR-484 in
microsatellite-unstable colorectal cancer promotes both viability
and IL-8 production via CD137L. J Pathol. 236:165–174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu J, Tan Q, Deng B, Fang C, Qi D and Wang
R: The microRNA-520a-3p inhibits proliferation, apoptosis and
metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J
Cancer Res. 5:802–811. 2015.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu K, Pan Q, Jia LQ, Dai Z, Ke AW, Zeng
HY, Tang ZY, Fan J and Zhou J: miR-302c inhibits tumor growth of
hepatocellular carcinoma by suppressing the endothelial-mesenchymal
transition of endothelial cells. Sci Rep. 4:55242014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin C, He H, Liu H, Li R, Chen Y, Qi Y,
Jiang Q, Chen L, Zhang P, Zhang H, et al: Tumour-associated
macrophages-derived CXCL8 determines immune evasion through
autonomous PD-L1 expression in gastric cancer. Gut. 68:1764–1773.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jayatilaka H, Tyle P, Chen JJ, Kwak M, Ju
J, Kim HJ, Lee JSH, Wu PH, Gilkes DM, Fan R and Wirtz D:
Synergistic IL-6 and IL-8 paracrine signalling pathway infers a
strategy to inhibit tumour cell migration. Nat Commun. 8:155842017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li MQ, Luo XZ, Meng YH, Mei J, Zhu XY, Jin
LP and Li DJ: CXCL8 enhances proliferation and growth and reduces
apoptosis in endometrial stromal cells in an autocrine manner via a
CXCR1-triggered PTEN/AKT signal pathway. Hum Reprod. 27:2107–2116.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu YM, Webster SJ, Flower D and Woll PJ:
Interleukin-8/CXCL8 is a growth factor for human lung cancer cells.
Br J Cancer. 91:1970–1976. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Wei J, Mei Z, Yin Y, Li Y, Lu M and
Jin S: Suppressing role of miR-520a-3p in breast cancer through
CCND1 and CD44. Am J Transl Res. 9:146–154. 2017.PubMed/NCBI
|
|
26
|
Wang X, Xu Y, Chen X and Xiao J: [ARTICLE
WITHDRAWN] Dexmedetomidine inhibits osteosarcoma cell proliferation
and migration, and promotes apoptosis by regulating miR-520a-3p.
Oncol Res. 26:495–502. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shathasivam P, Kollara A, Spybey T, Park
S, Clarke B, Ringuette MJ and Brown TJ: VEPH1 expression decreases
vascularisation in ovarian cancer xenografts and inhibits VEGFA and
IL8 expression through inhibition of AKT activation. Br J Cancer.
116:1065–1076. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Campbell J, Stenmark MH, Zhao J,
Stanton P, Matuszak MM, Ten Haken RK and Kong FS: Plasma levels of
IL-8 and TGF-β1 predict radiation-induced lung toxicity in
non-small cell lung cancer: A validation study. Int J Radiat Oncol
Biol Phys. 98:615–621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Liu J, Jiang Q, Deng J, Xu F, Chen
X, Cheng F, Zhang Y, Yao Y, Xia Z, et al: Human adipose-derived
mesenchymal stem cell-secreted CXCL1 and CXCL8 facilitate breast
tumor growth by promoting angiogenesis. Stem Cells. 35:2060–2070.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng J, Li Y, Liu S, Jiang Y, Ma J, Wan
L, Li Q and Pang T: CXCL8 derived from mesenchymal stromal cells
supports survival and proliferation of acute myeloid leukemia cells
through the PI3K/AKT pathway. FASEB J. 33:4755–4764. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Wang Y, Wei P, Shi D, Wen S, Wu F,
Liu L, Ye N and Zhou H: Bisphenol A affects trophoblast invasion by
inhibiting CXCL8 expression in decidual stromal cells. Mol Cell
Endocrinol. 470:38–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tong H, Ke JQ, Jiang FZ, Wang XJ, Wang FY,
Li YR, Lu W and Wan XP: Tumor-associated macrophage-derived CXCL8
could induce ERα suppression via HOXB13 in endometrial cancer.
Cancer Lett. 376:127–136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Osiagwu DD, Azenabor AE, Osijirin AA,
Awopetu PI and Oyegbami FR: Evaluation of interleukin 8 and
interleukin 10 cytokines in liquid based cervical cytology samples.
Pan Afr Med J. 32:1482019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan R, Shuai H, Luo X, Wang X and Guan B:
The clinical and prognostic value of CXCL8 in cervical carcinoma
patients: Immunohistochemical analysis. Biosci Rep.
37:BSR201710212017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang W, Wu Q, Wang C, Yang L, Liu P and
Ma C: AKIP1 promotes angiogenesis and tumor growth by upregulating
CXC-chemokines in cervical cancer cells. Mol Cell Biochem.
448:311–320. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Glynn PC, Henney E and Hall IP: The
selective CXCR2 antagonist SB272844 blocks interleukin-8 and
growth-related oncogene-alpha-mediated inhibition of spontaneous
neutrophil apoptosis. Pulm Pharmacol Ther. 15:103–110. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li A, Dubey S, Varney ML, Dave BJ and
Singh RK: IL-8 directly enhanced endothelial cell survival,
proliferation, and matrix metalloproteinases production and
regulated angiogenesis. J Immunol. 170:3369–3376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Eitel J, Heise T, Thiesen U and Dersch P:
Cell invasion and IL-8 production pathways initiated by YadA of
Yersinia pseudotuberculosis require common signalling molecules
(FAK, c-Src, Ras) and distinct cell factors. Cell Microbiol.
7:63–77. 2005. View Article : Google Scholar : PubMed/NCBI
|