Introduction
As the global age increases, the morbidity and
mortality resulting from myocardial infarction (MI) also increases
each year (1). Although timely
reperfusion can effectively reduce mortality, the recovery of blood
flow through ischemic myocardium yields additional reperfusion
injury, including myocardial stunning, reperfusion arrhythmia and
myocardial necrosis (2). Myocardial
ischemia/reperfusion (I/R) injury is affected by a variety of
complex pathological mechanisms. For example, mitochondrial
Ca2+ overload, platelet activation and micro-thrombosis
formation, the disruption of mitochondrial membrane potential, free
radical or reactive oxygen species and inflammatory responses
(3). Among which, autophagy has
also been found to be involved in I/R (4). Studies have reported that ischemic
postconditioning (IPostC) can protect the heart from I/R injury by
transient intermittent I/R episodes prior to long-term ischemia or
hypoxia reperfusion (5). Wei et
al (6) found that IPostC could
improve autonomic function in acute ischemic stroke patients
through the enhancement of the total autonomic nerve activity and
vagus nerve activity. Another study also found that IPostC
attenuates the injury in I/R myocardium by upregulating microRNA
(miRNA/miR)-499 and inhibiting Toll-like receptor 2 activation
(5). miR-30a-5p, a member of the
miR-30 family, is regarded as a key miRNA in cardiovascular
pathophysiology (7). In addition,
our previous study demonstrated that hypoxia postconditioning
(HPostC) protected aged cardiomyocytes from hypoxia/reoxygenation
(H/R) injury via DNA methyltransferase 3B (DNMT3B)-regulated
miR-30a-5p, suggesting that miR-30a-5p may be a potential novel
target for ischemic MI (8).
However, the underlying mechanism involved in IPostC protection
against aging myocardial I/R injury has not yet been
elucidated.
Histone modification is a widely studied epigenetic
modification, which has been demonstrated to result in the
alteration of miRNA expression (9).
As a type of histone modification, histone acetylation is generally
correlated with transcriptional activation resulting from chromatin
decondensation and thereby allowing transcriptional machinery
access (10). This process is
catalyzed by histone acetyltransferases (HATs) and histone
deacetyltransferases (HDACs), which are responsible for adding and
removing acetyl groups from histone tails, respectively.
HDAC-mediated acetylation is associated with ischemic heart disease
(11). Application of trichostatin
A (TSA), a class I and class II HDAC inhibitor alters the response
to myocardial ischemic injury in the heart and limited areas of MI
(12). A study has shown that TSA
can reduce post-ischemic infarct size, prevent myocardial
remodeling after myocardial infarction, and protect myocardial
function after myocardial I/R by inhibiting HDAC activity (13). Hence, histone acetylation plays a
crucial role in ischemic heart disease, while its effect on aged
heart I/R injury remains unknown.
Histone acetylation is essential for transcriptional
regulation, which can change the affinity of transcription factors
to DNA binding sites (14). As an
important transcriptional factor, c-Myc regulates ~15% of genes in
the genome, and can drive several biological processes, including
cell apoptosis, proliferation, growth and differentiation (15). c-Myc widely participates in the
development of disease via target genes (16). c-Myc can upregulate a series of
transcriptional programs to directly regulate gene transcription
(17). Additionally, c-Myc alters
miRNA and long non-coding RNA expression patterns, which indirectly
influences target gene expression in various diseases (18). For example, it has been reported
that c-Myc functions as a modulator of polycystin-1 expression,
likely via a feed-forward regulatory loop mechanism, in autosomal
dominant polycystic kidney disease (19). Chen et al (20) found that vitamin D receptor
signaling could regulate the c-Myc/Mad-1 network to inhibit the
expression of the long non-coding RNA H19. Therefore, it is
necessary to elucidate the relationship between histone acetylation
and c-Myc responsible for miR-30a-5p transcription in senescent
cardiomyocytes.
The present study revealed the role and epigenetic
or transcriptional regulation mechanism of miR-30a-5p on the
effects of HPostC in senescent cardiomyocytes. It was found that
hyperacetylation of H3K14 promoted c-Myc binding to the miR-30a-5p
gene promoter, which led to the increased transcription of
miR-30a-5p and attenuated senescent cardiomyocyte I/R injury.
Materials and methods
Cell culture and treatment
H9C2 rat cardiomyocytes (The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin streptomycin in 5%
CO2 at 37°C in a humified atmosphere. Senescent
cardiomyocytes were established by 8 mg/ml D-galactose (Shanghai
Bio-Tech Co., Ltd.) treatment of H9C2 cells for 9 days, which was
based on findings from our previous study where increased
senescence-associated β-galactosidase (SA-β-gal)-positive cells
were identified using a Senescence β-Galactosidase staining kit
(Beijing Solarbio Science & Technology Co., Ltd.) (8). Furthermore, H/R and HPostC models were
established as previously reported (8). Senescent cardiomyocytes were treated
with 200 nmol/l Trichostatin A (TSA; cat. no. HY-15144;
MedChemExpress) or 1 µmol/l Romidepsin (cat. no. HY-15149;
MedChemExpress) treatment for 24 h, and then H/R or HPostC
treatment.
Cell adenovirus transduction
Adenovirus vectors ADV2 (U6/CMV-RFP) encoding Rattus
HDAC2 and c-Myc-specific short hairpin RNA (shRNA) were obtained
from Shanghai GenePharma Co., Ltd. The following sequences were
included in the present study: Adenovirus sh-HDAC2 (Ad-shHDAC2),
5′-GGTATAGATGACGAGTCATAT-3′; Ad-shc-Myc,
5′-GAATTTCTATCACCAGCAACA-3′; and Ad-sh negative control (NC),
5′-ACTACCGTTGTTATAGGTG-3′. The cytolysate products were centrifuged
in a table centrifuge at 1,006.2 × g for 15 min at room temperature
to harvest the viral supernatant. The AdEasy™ system (Shanghai
GenePharma Co., Ltd.) was used for adenoviral vector construction.
The pAdEasy (Stratagene; Agilent Technologies, Inc.) was used to
package the plasmid, and the ratio of the ADV shuttle vector and
packaging plasmid was 3~4:1. Subsequently, production and cell
transduction of the adenovirus in 293T cells (The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) was
performed, and cells were flow-cytometrically sorted to maintain a
GFP positivity rate >95%. Next, H9C2 cells cultured in a
25-cm2 culture flask were incubated with Ad-shHDAC2,
Ad-shc-Myc or Ad-shNC (the titer of the virus was 1×109
PFU/ml) at a multiplicity of infection of 100. DMEM without FBS was
used to dilute adenovirus. After incubation for 6 h, the medium was
changed to serum-containing medium for 24–48 h, followed by
treatments.
Cell viability assay
The cell viability of senescent cardiomyocytes was
determined using a Cell Viability Imaging kit (cat. no.
06432379001; Merck KGaA) according to the manufacturer's protocols.
Viable and dead cells are displayed as green and red fluorescence,
respectively, under a confocal microscope (magnification, ×10;
Olympus FV1000; Olympus Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
Rat cardiomyocyte RNA was extracted using RNAsimple
Total RNA Kit (cat. no. DP419; Tiangen Biotech Co., Ltd.) and RNA
was reverse transcribed into cDNA using a PrimeScript®
RT reagent kit (Takara Bio, Inc.), according to the manufacturer's
protocol. RT-qPCR was performed using SYBR Premix ExTaq™ (Takara
Bio, Inc.) on the FTC 3,000 RT-qPCR System (Funglyn Biotech, Inc.).
c-Myc, HDAC2 and β-actin primer sequences are listed in Table I, miR-30a-5p and U6 primers were
purchased from Guangzhou RiboBio Co., Ltd. β-actin and U6 were used
as internal reference genes. The 20 µl volume reaction system was
comprised as follows: 10 µl SYBR Premix ExTaq™, 0.8 µl primers (10
µM), 2 µl cDNA and 6.4 µl ddH2O. The following
thermocycling conditions were used for qPCR: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec.
Relative mRNA expression was calculated using the 2−ΔΔCq
method (21).
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | GenBank | Primer sequences
(5′→3′) |
|---|
| β-actin | NM_031144.3 | F:
TGTCACCAACTGGGACGATA |
|
|
| R:
GGGGTGTTGAAGGTCTCAAA |
| c-Myc | NM_012603.2 | F:
GCCTTTTCGTTGTTTTCCAA |
|
|
| R:
CACAGCAAACCTCCACACAG |
| HDAC2 | NM_053447.1 | F:
GGGCTGCTTCAACCTAACTG |
|
|
| R:
TTCACAATCAAGGGCAACTG |
Western blotting
Cells were lysed in NP-40 buffer (cat. no. P0013F;
Beyotime Institute of Biotechnology) containing 50 mM Tris (pH
7.4), 150 mM NaCl, 1% NP-40 and proteinase inhibitors. Protein
concentration was determined using a BCA kit (cat. no. KGP902;
Jiangsu KGI Biotechnology Co., Ltd.), according to the
manufacturer's instructions. Total protein (30 µg) was separated
via SDS-PAGE on 10% gel, and then separated proteins were
transferred onto PVDF membranes (EMD Millipore). Following blocking
with 5% non-fat milk for 2 h at room temperature, membranes were
incubated at 4°C overnight with LC3B-II/I (cat. no. ab192890;
1:1,000; Abcam), p62 (cat. no. ab56416; 1:1,000; Abcam), beclin-1
(BECN1; cat. no. ab210498; 1:1,000; Abcam), HDAC2 (cat. no.
ab32117; 1:1,000; Abcam), c-Myc (cat. no. ab32072; 1:1,000; Abcam)
and β-actin (cat. no. AC028; 1:10,000; ABclonal Biotech Co., Ltd.)
antibodies. The membranes were then incubated with secondary
antibodies (cat. no. M21002S; 1:5,000; Abmart Pharmaceutical
Technology Co., Ltd.) for 1 h at 37°C. Protein signals were
visualized with enhanced chemiluminescence HRP substrate (cat. no.
KF001; Affinity Biosciences) and semi-quantified using ImageJ
software version 5.1 (National Institutes of Health).
Chromatin immunoprecipitation
(ChIP)-PCR
An EZ-Magna ChIP™ kit (cat. no. 17-371;
Sigma-Aldrich; Merck KGaA) was used for ChIP. Chromatin was
immunoprecipitated for 24 h at 4°C using anti-HDAC2 (cat. no.
ab124974; 1 µg; Abcam), anti-H3K14ac (cat. no. ab203952; 1 µg;
Abcam) and anti-c-Myc (cat. no. 18583S; 1 µg; Cell Signaling
Technology, Inc.) antibody. Rabbit IgG immunoprecipitation was used
as a negative control. 1/100 of total cell lysate was used as an
internal control. qPCR was used to analyze precipitated DNA using
specific primers (Table II) and it
was performed as described above. The signals were calculated as
the percentage of input.
 | Table II.Primer sequences used for chromatin
immunoprecipitation-PCR assay. |
Table II.
Primer sequences used for chromatin
immunoprecipitation-PCR assay.
| Gene | Primer
sequence | Length (bp) |
|---|
| miR- | F:
ATGTTGTAGTCCTAGTAAGTCACCT | 25 |
| 30a-5p | R:
TCTGTAAACTGTAAAGCCTCGT | 22 |
Construction of deletion and
site-directed mutagenesis of the miR-30a gene promoter
miR-30a-5p gene promoter sequence analysis, reporter
gene construction and promoter activity study miR-30a-5p gene
promoter sequences (2,200 bp) were downloaded from the University
of California Santa Cruz Genome Browser Database (http://genome.ucsc.edu/). The c-Myc binding sites in
the promotor of the miR-30a-5p gene were analyzed using the JASPAR
database (http://jaspar.genereg.net/). The
truncated vectors PGL3-rno-miR-30a-5p-F1-mut1
(5′-GTGACGACCAGTGTGGACCT-3′), PGL3-rno-miR-30a-5p-F1-mut2
(5′-GTGTGGACCTTTGTACATGG-3′) and PGL3-rno-miR-30a-5p-F1-mut3
(5′-GTGACGACCAGTGTGGACCTTTGTACATGG-3′) were chemically synthesized,
and NheI/XhoI restriction sites were added to both
ends of the target fragment during synthesis. The synthesized gene
fragments were cloned into the PUC57 vector (Shanghai YingBiotech
Company) and fused to the firefly luciferase reporter vector
pGL3-basic (Shanghai YingBiotech Company).
Dual-luciferase assay
The core promoter sequence of miR-30a-5p was cloned
into the PUC57 vector to construct a luciferase reporter plasmid,
which had a c-Myc binding site at 700 bp upstream of the
transcription start site of miR-30a-5p. Then, the full-length
sequence of c-Myc was co-transfected with the full-length sequence
of miR-30a-5p gene promoter region (wild-type, −700/+1, pGL3-P1)
and the three full-length sequences of miR-30a-5p gene promoter
region with c-Myc binding site mutation (mutant-type, pGL3-MT1,
pGL3-MT2, pGL3-MT3) to detect the effect of c-Myc on the
transcriptional activity of miR-30a-5p gene promoter region. 293T
cells were seeded at 8×104 cells per well in 24-well
plates, and 24 h after plating, the cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. In
each well, 100 ng wild-type or mutant-type plasmid RNA vector and
Renilla luciferase were co-transfected. The former culture
medium was discarded 48 h after transfection, and the cells were
then rinsed twice with PBS. Then, 100 µl passive lysis buffer was
added in each well with cells and slightly shaken at room
temperature for 15 min, followed by collection of the cell lysate.
The program was set for 2 sec for pre-reading and 10 sec for value
reading, with 100 µl Stop & Glo® Reagent (Promega
Corporation) added for each sampling session. Then, prepared Stop
& Glo Reagent and the luminescent plate or tube containing the
cell lysate (20 µl/per sample) were placed into a bioluminescence
detector. Finally, the program was operated and data were recorded
after fluorescence reading. The Renilla luciferase signal
was normalized to the firefly luciferase signal. This process was
performed in triplicate for each target vector.
Database search
HDAC2 was found to regulate histone deacetylation
under HPostC treatment, as predicted by Uniprot (http://www.uniprot.org/). First, HDAC2 (accession no.
F7ENH8_RAT) was searched on the Uniprot website in the UniProtKB
field to query the protein function. Bioinformatics analysis of
HDAC2 in the ‘Biological process’ category of Gene Ontology (GO)
showed the association of HDAC2 with ‘histone H3 deacetylation’
(red underline), especially for ‘histone deacetylase activity
(H3-K14 specific)’ (red underline GO ID:0031078). The Uniprot
database was searched in order to identify the known GO terms
associated with HDAC. However, this was not the same as performing
bioinformatics analysis (GO enrichment analysis) (22).
Statistical analysis
All data were collected and analyzed with GraphPad
Prism 6.0 (GraphPad Software, Inc.). Data are expressed as the mean
± SD. All experiments were conducted three times. Differences
between two groups were evaluated using an unpaired Student's
t-test. Differences between no more than three groups were
compared using one-way ANOVA followed by Student-Newman-Keul's
test. Differences between four or more groups were analyzed using
ANOVA followed by Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
HDAC2-mediated H3K14 hyperacetylation
in senescent cardiomyocytes under HPostC treatment
Histone acetylation plays a vital role in the
regulation of gene expression (23). To investigate whether miR-30a-5p
upregulation was affected by histone modification in senescent
cardiomyocytes under HPostC treatment, TSA, a HDAC inhibitor, was
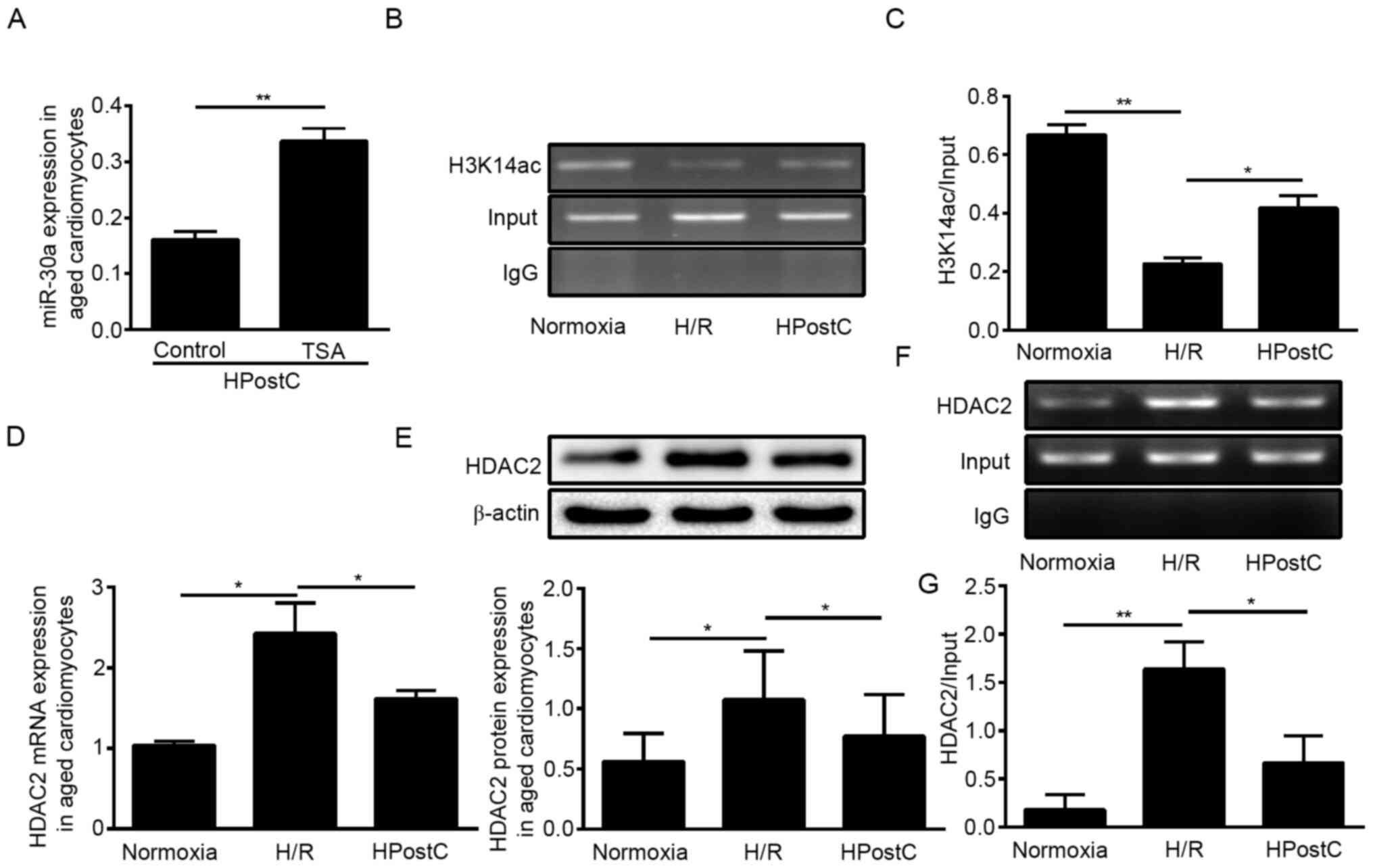
applied to suppress HDAC enzyme activity. As shown in Fig. 1A, TSA significantly enhanced
miR-30a-5p expression, which implied that histone acetylation
participated in miR-30a-5p transcription under HPostC treatment. In
addition, ChIP-PCR was used to detect H3K14 acetylation levels of
the miR-30a-5p promoter in senescent cardiomyocytes. The results
showed that, compared with the normoxia conditions, H/R
significantly decreased the levels of H3K14ac at the miR-30a-5p
gene promoter in senescent cardiomyocytes, whereas HPostC led to
increased H3K14ac levels in senescent cardiomyocytes compared with
H/R (Fig. 1B and C).
To determine the cause of the hyperacetylation at
the miR-30a-5p gene promoter under HPostC treatment, the functions
of HDAC2 was predicted by searching Uniprot. The Uniprot website
showed that under the GO ‘Biological process’ category HDAC2 was
found to be associated with ‘histone H3 deacetylation’ (underlined
in red), specifically ‘histone deacetylase activity (H3-K14
specific)’ (underlined in red, GO ID:0031078) (Fig. S1). RT-qPCR and western blotting
results showed a significant decrease in HDAC2 expression in
senescent cardiomyocytes exposed to HPostC compared with the H/R
group (Fig. 1D and E). Next,
ChIP-PCR was performed to determine whether HDAC2 directly binds to
the coding sequence region of miR-30a-5p. Cross-linked chromatin
samples were extracted from the senescent cardiomyocytes and
precipitated with an anti-HDAC2 antibody. As presented in Fig. 1F and G, HPostC inhibited the binding
of HDAC2 to the miR-30a-5p gene promoter compared with the H/R
group. These results confirmed that decreased HDAC2 binding to the
miR-30a-5p promoter facilitated H3K14 hyperacetylation in senescent
cardiomyocytes under HPostC treatment.
HDAC2-mediated H3K14 hyperacetylation
inhibits senescent cardiomyocyte autophagy under HPostC
To determine whether HDAC2 was involved in the
regulation of miR-30a-5p transcription in senescent cardiomyocytes,
HDAC2 was knocked down in senescent cardiomyocytes. As presented in
Fig. 2A and B, HDAC2 knockdown in
senescent cardiomyocytes enhanced the enrichment of H3K14
acetylation levels at the miR-30a-5p gene promoter, which resulted
in miR-30a-5p upregulation (Fig.
2C). Additionally, the expression of autophagy-related proteins
LC3II/I, BECN1 and p62 were detected by western blotting to
investigate the effects of HDAC2 on senescent cardiomyocyte
autophagy. The results showed that HDAC2 knockdown significantly
repressed LC3B-II/I and BECN1 expression, while increasing p62
expression under HPostC treatment (Fig.
2D). Subsequently, cell viability staining was performed, which
exhibited that the viability of senescent cardiomyocytes exposed to
H/R or HPostC significantly increased following romidepsin
treatment, which suggested that H3K14 hyperacetylation facilitated
the protection exerted by HPostC against H/R injury (Fig. 2E). These results demonstrated that
HDAC2-mediated H3K14 hyperacetylation under HPostC treatment could
facilitate miR-30a-5p expression to inhibit senescent cardiomyocyte
autophagy.
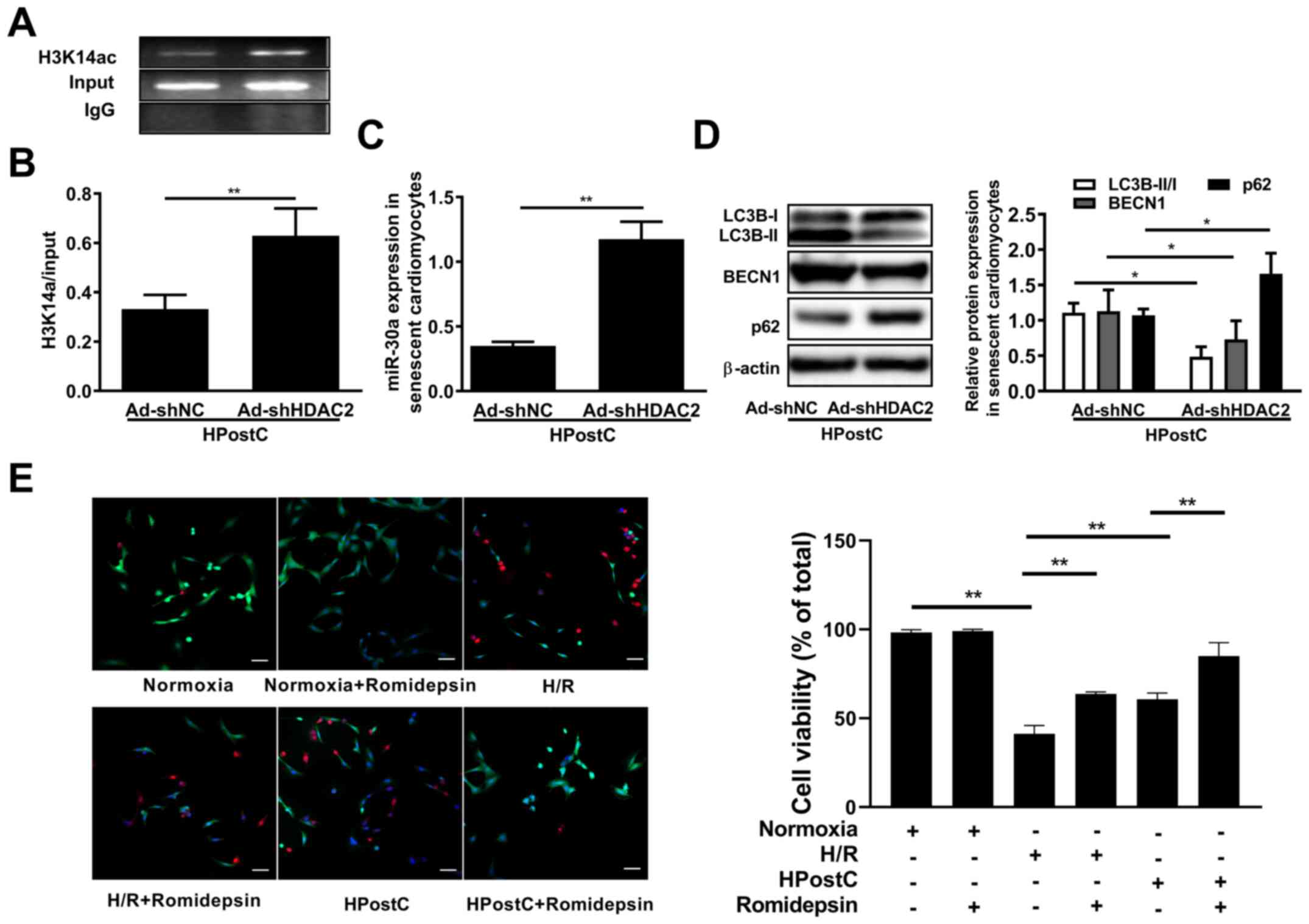 | Figure 2.miR-30a-5p negatively regulated by
HDAC2 is involved in HPostC-induced autophagy inhibition. (A and B)
The enrichment of H3K14 acetylation at the miR-30a-5p gene promoter
was identified by chromatin immunoprecipitation-PCR analysis in
cells infected with Ad-shNC and Ad-shHDAC2 for 48 h. (C) miR-30a-5p
mRNA expression in senescent cardiomyocytes treated as
aforementioned. (D) The relative protein expression levels of
LC3B-II/I, BECN1 and p62 after knockdown of HDAC2. (E) The cell
viability staining of romidepsin-treated aged H9C2 cell under
normoxia, H/R or HPostC conditions (scale bar, 50 µm). Data are
presented as the mean ± SD from three independent experiments.
*P<0.05, **P<0.01. HPostC, hypoxia postconditioning; miR,
microRNA; HDAC2, histone deacetylase 2; H/R, hypoxia/reoxygenation;
Ad-, adenovirus; sh, short hairpin RNA; NC, negative control; LC3B,
light chain 3β; BECN1, beclin-1. |
c-Myc binds to the miR-30a-5p gene
promoter to positively regulate miR-30a-5p transcriptional
activity
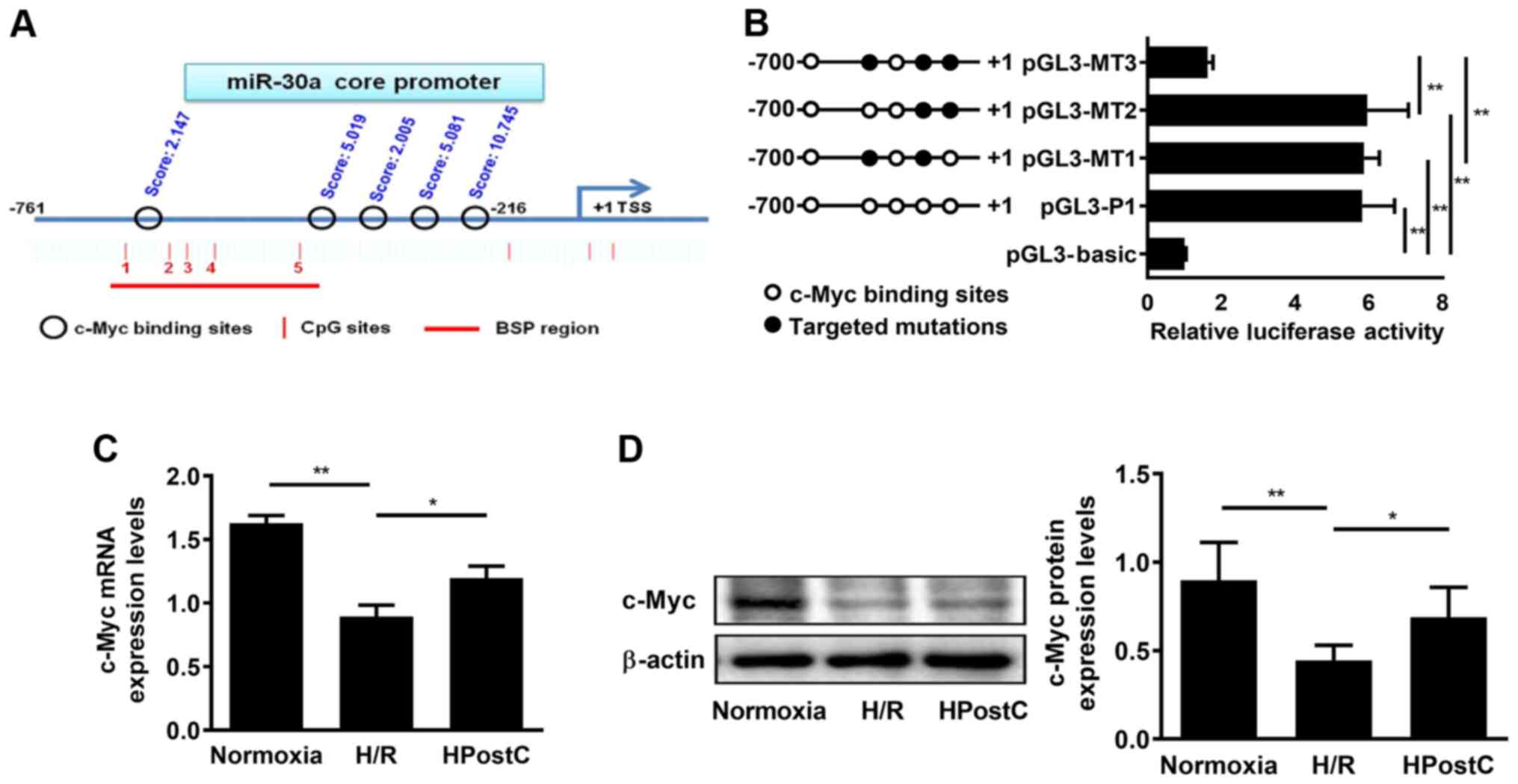
To investigate whether c-Myc participated in the
transcriptional regulation of miR-30a-5p, the possible c-Myc
binding sites at the miR-30a-5p gene promoter were analyzed using
the JASPAR database. Five putative transcription factor c-Myc
binding sites was found at the miR-30a-5p gene core promoter (−761
to −216): i) −664/-673; ii) −326/-335; iii) −294/-303; iv)
−263/-272; and v) −218/-227. Three binding sites with high scores
were selected for subsequent experiments (Figs. 3A and S2). Subsequently, substitution mutations
of the three identified c-Myc binding sites (−326/-335, −263/-272
and −218/-227) were generated and a luciferase reporter assay was
performed to confirm which binding site was functionally required
for c-Myc to regulate miR-30a-5p transcriptional activity. pGL3-P1,
which contained all c-Myc-binding sites, presented maximum promoter
activity (Fig. 3B). Mutation of the
region containing the −326/-335 and −263/-272 site (pGL3-MT1) and
−263/-272 and −218/-227 (pGL3-MT2) showed similar promoter activity
as pGL3-P1 (Fig. 3B). In addition,
a significant reduction in miR-30a-5p gene promoter activity was
observed when three identified binding sites were all mutated
(pGL3-MT3) compared with pGL3-MT1 and pGL3-MT2 (Fig. 3B). These results demonstrated that
both −218/-227 and −326/-335 regions were essential for c-Myc to
regulate miR-30a-5p gene promoter activity. Meanwhile, c-Myc
expression was measured using RT-qPCR and western blotting, and the
results showed that HPostC significantly increased c-Myc expression
compared with H/R (Fig. 3C and D).
These results suggested that c-Myc may positively regulate
miR-30a-5p gene promoter transcriptional activity in senescent
cardiomyocytes under HPostC treatment.
H3K14 hyperacetylation facilitates
c-Myc binding to the miR-30a-5p gene promoter to inhibit
autophagy
To further verify the effect of c-Myc on the
regulation of miR-30a-5p, senescent cardiomyocytes were infected
with Ad-shRNA to knockdown c-Myc expression (Fig. S3). As expected, knockdown of c-Myc
significantly inhibited the expression of miR-30a-5p (Fig. 4A), which was accompanied by
increased LC3B-II/I and BECN1 expression and downregulation of p62
expression under HPostC treatment (Fig.
4B). This suggested that c-Myc may be involved in the
inhibition of autophagy in senescent cardiomyocytes under HPostC
treatment. In addition, cell viability staining showed that the
viability of senescent cardiomyocytes were attenuated by c-Myc
knockdown under H/R or HPostC conditions, which suggested that
silencing of c-Myc promoted the protection exerted by HPostC
against H/R injury (Fig. 4C). To
determine the association between H3K14 hyperacetylation and c-Myc
in miR-30a-5p transcription, senescent cardiomyocytes were treated
with romidepsin, and c-Myc binding at the miR-30a-5p gene promoter,
as well as miR-30a-5p expression, was examined by ChIP and RT-qPCR.
As shown in Fig. 4D and E,
romidepsin treatment significantly enhanced c-Myc binding to the
miR-30a-5p gene promoter, which led to the increased expression of
miR-30a-5p. Silencing c-Myc expression under romidepsin treatment
suppressed c-Myc binding to the miR-30a-5p gene, and decreased the
expression of miR-30a-5p. Collectively, these data suggested that
H3K14 hyperacetylation promoted c-Myc binding to the miR-30a-5p
gene promoter, which increased miR-30a-5p transcription in
senescent cardiomyocytes.
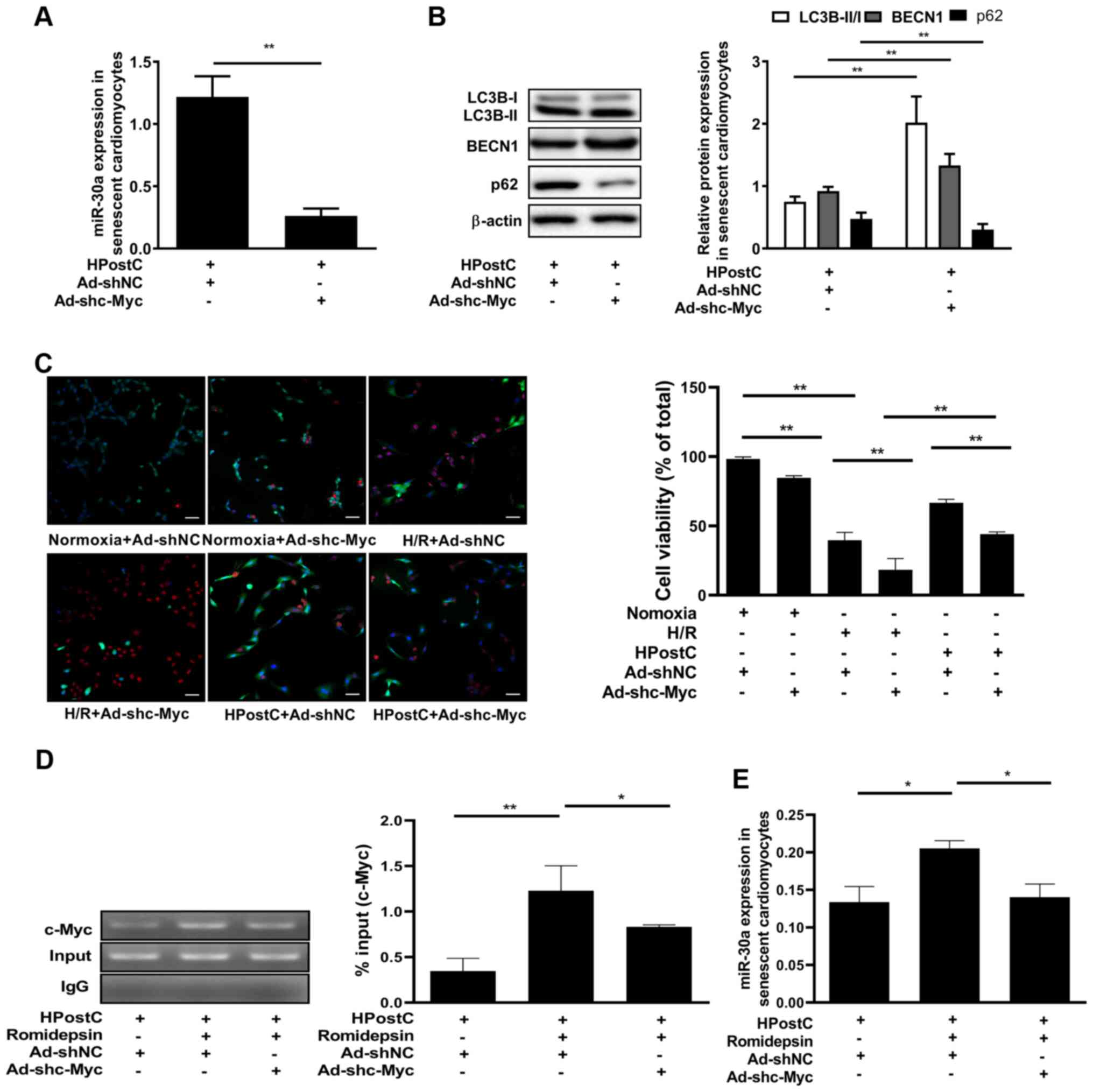 | Figure 4.Acetylation of H3K14 promotes c-Myc
binding to the miR-30a-5p gene promoter to inhibit autophagy of
senescent cardiomyocytes. (A) miR-30a-5p mRNA expression was
detected after knockdown of c-Myc expression under HPostC
treatment. (B) Relative protein detection in the senescent
cardiomyocytes treated as aforementioned. (C) Live/dead cell
imaging in senescent cardiomyocytes treated as aforementioned
(scale bar, 50 µm). (D) Chromatin immunoprecipitation-PCR assay of
c-Myc binding at the miR-30a-5p gene promoter. (E) miR-30a-5p mRNA
expression was examined by reverse transcription-quantitative PCR.
Data are presented as the mean ± SD from three independent
experiments. *P<0.05, **P<0.01. HPostC, hypoxia
postconditioning; miR, microRNA; H/R, hypoxia/reoxygenation; Ad-,
adenovirus; sh, short hairpin RNA; NC, negative control; LC3B,
light chain 3β; BECN1, beclin-1. |
Discussion
Ischemic heart disease has resulted in increased
morbidity and mortality worldwide (24). Numerous preclinical reports have
indicated that aging increases the vulnerability of the heart to
I/R injury, and studies have aimed to find ways to reduce cardiac
myocyte death following I/R (25).
For example, it has been shown that I/R damage is likely to be the
consequence of enhanced oxidative stress with ageing (26). Griecsová et al (27) demonstrated that the loss of
preconditioning protection was associated with an age-dependent
reduction of Akt phosphorylation and endothelial nitric oxide
synthase and protein kinase C ε levels in the hearts of mature rats
compared with the younger rats. Furthermore, it has been
demonstrated that aged myocytes accumulate more diastolic
Ca2+ in ischemia and early reperfusion than younger
hearts, cells may account for the increased sensitivity to ischemia
and reperfusion injury in the aging heart (28). Myocardial IPostC is an endogenous
cardioprotective phenomenon that can increase the tolerance of the
heart to reperfusion injury when exposed to short-term I/R
(29). Although the protection of
IPostC on myocardial I/R injury has been confirmed, the underlying
mechanisms in senescent myocardium are still unclear.
Autophagy is a vital physiological process in cells,
which is considered the key to maintaining the normal structure and
function of the heart (30). It has
been reported that autophagy plays a dual role in myocardial I/R,
low levels of autophagy can relieve energy depletion, maintain
protein homeostasis, remove damaged cells and play a protective
role in cell survival during ischemia (31), but long-term upregulation of
autophagy can lead to excessive cell degradation and death
(32). First, autophagy can
effectively remove inflammasome and inhibit the activity of
inflammatory transcription factors, such as NF-κB (33). However, excessive autophagy may lead
to the release of inflammatory factors (34). On the other hand, using the
autophagy inhibitor 3-MA or knocking down BECN1 expression can
reduce cell death during myocardial I/R (35), and autophagy-related proteins Atg5
and BECN1 may turn into pro-apoptotic proteins in the case of being
proteolyzed by proteases such as calpain (36). However, whether autophagy plays a
beneficial or harmful role in myocardial I/R injury are still
controversial. In the present study, autophagy was discovered to
perform a destructive role in I/R injury.
Clinical studies have revealed significant
prognostic benefits of IPostC in elderly patients with acute MI.
For example, it has been reported that the peak of creatine kinase
isoenzyme, postoperative myocardial troponin I and high sensitive C
reaction protein are significantly attenuated by post-conditioning
when compared with the control group (37). Similarly, it has also been found
that 43 patients with ST-segment elevation myocardial infarction
who underwent post-conditioning had a significant reduction in
infarct size (38), which was
similar to our previous study demonstrating that HPostC protected
senescent H9C2 cells from H/R injury via DNMT3B-dependent
miR-30a-5p activation (8). To
further investigate the underlying mechanisms of miR-30a-5p
upregulation in senescent cardiomyocytes under HPostC treatment,
the present study explored another epigenetic modification that
potentially involves miR-30a-5p transcriptional regulation. In this
study, it was found that HDAC2-mediated H3K14 hyperacetylation
contributed to miR-30a-5p upregulation in HPostC. In recent years,
accumulating evidence has demonstrated that miR-30a-5p participates
in cardiovascular pathophysiology (39–41).
Although several studies have indicated the importance of
miR-30a-5p in cardiovascular diseases, there are still conflicting
views. Previous studies indicated that miR-30a may be a
compensatory upregulation to protect the myocardium of patients
with heart failure (42). Li et
al (43) found that miR-30a was
significantly decreased in I/R conditions, and knockdown of miR-30a
could reverse the anti-autophagy effects of salvianolic acid B
against I/R injury. By contrast, Shen et al (44) indicated that miR-30 was upregulated
in a murine MI model and a cardiomyocyte hypoxic model. Another
study also reported that the expression of circulating miR-30a in
patients with MI was significantly elevated, which was also
demonstrated to be a potential predictor of acute MI (45).
In addition to DNA methylation, the present study
confirmed that the hyperacetylation of H3K14 at the miR-30a-5p gene
promoter was responsible for the abnormal transcription of
miR-30a-5p in senescent cardiomyocytes subjected to HPostC. Histone
acetylation is associated with an ‘open’ chromatin conformation
that promotes transcription (46).
Acetylation weakens the electrostatic interaction between DNA and
histones in nucleosome fibers, which reduces DNA affinity and
allows chromatin to adopt a more relaxed structure to recruit basic
transcription mechanisms (47). For
example, the acetylated histone markers H3K4ac, H3K39ac and H3K14ac
are correlated with transcriptional activation (48). In addition, histone acetylation has
been reported to be catalyzed via two primary mechanisms (49): HATs destroy interactions between the
DNA and histones, enabling transcription factors to enter the DNA
(50); and HDACs can coagulate
chromatin and inhibit transcription (51). In the present study, it was observed
that decreased HDAC2 binding at the miR-30a-5p gene promoter
resulted in H3K14 hyperacetylation in senescent cardiomyocytes
under HPostC treatment. These data supported the hypothesis that
histone hyperacetylation at the gene promoter contributed to
transcriptional activation.
Several studies have suggested that histone
acetylation facilitates chromatin opening and promotes the binding
of transcription factors to DNA (52,53).
Therefore, the present study further investigated whether
hyperacetylation of H3K14 in the miR-30a-5p gene promoter affected
the ability of transcription factors to bind to miR-30a-5p promoter
regions. The results showed that five c-Myc putative binding sites
were found, and only-218/-227 and −326/-335 sites were essential
for c-Myc to regulate miR-30a-5p gene promoter activity at the
miR-30a-5p gene promoter. c-Myc positively regulated miR-30a-5p
gene promoter transcriptional activity in senescent cardiomyocytes
under HPostC treatment. As a result, the present study indicated
that hyperacetylation of H3K14 promoted c-Myc binding to the
miR-30a-5p gene promoter, which led to the upregulation of
miR-30a-5p transcription in senescent cardiomyocytes under HPostC
treatment. Zhang et al (54)
reported that high acetylation of the glial cell-derived
neurotrophic factor (GDNF) promoter region promoted GDNF
transcription via increasing early growth response protein 1
binding to the GDNF promoter, which was similar to the findings of
the current study.
In conclusion, the present study found that
HDAC2-mediated H3K14 hyperacetylation of the miR-30a-5p gene
promoter facilitated c-Myc binding to the miR-30a-5p gene promoter,
which contributed to the protective effects of HPostC against H/R
injury on senescent cardiomyocytes. These findings provided novel
insights into the mechanism underlying HPostC-mediated abnormal
transcription of miR-30a-5p in senescent cardiomyocytes.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Scientific Grants (grant nos. 81870225, 81870332 and
81860044), Ningxia Key Research and Development Projects (grant no.
2018BEG03026) and Natural Scientific Grants of Ningxia Province
(grant nos. 2019AAC03075, 2019AAC03076 and 2020AAC03380).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LBX and HPZ contributed to the conception of the
study, performed statistical analysis and wrote the manuscript.
LBX, HPZ, YHW, WG and LYG performed the experiments and acquired
data. ANY and SCM provided conceptual advice and interpreted the
data. YY and KW mainly helped with data collection and study
design. YDJ designed and supervised the study. LBX and HPZ confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Curran J, Burkhoff D and Kloner RA: Beyond
reperfusion: Acute ventricular unloading and cardioprotection
during myocardial infarction. J Cardiovasc Transl Res. 12:95–106.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akhmedov A, Montecucco F, Costantino S,
Vdovenko D, Schaub Clerigué A, Gaul DS, Burger F, Roth A, Carbone
F, Liberale L, et al: Cardiomyocyte-specific JunD overexpression
increases infarct size following ischemia/reperfusion cardiac
injury by downregulating Sirt3. Thromb Haemost. 120:168–180. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang YM, Zhang ZY and Wang RX: Protective
mechanisms of quercetin against myocardial ischemia reperfusion
injury. Front Physiol. 11:9562020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao R, Xie E, Yang X and Gong B: Alliin
alleviates myocardial ischemia-reperfusion injury by promoting
autophagy. Biochem Biophys Res Commun. 512:236–243. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang XY, Huang Z, Li QJ, Zhong GQ, Meng
JJ, Wang DX and Tu RH: Ischemic postconditioning attenuates the
inflammatory response in ischemia/reperfusion myocardium by
upregulating miR-499 and inhibiting TLR2 activation. Mol Med Rep.
22:209–218. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei L, Liang H, Mo M, Liu Z, Ye R, Ye H,
Ouyang W, Yu W, Zhao W and Zhang X: The effect of remote ischemic
postconditioning on autonomic function in patients with acute
ischemic stroke: A randomized controlled trail. Complement Ther
Med. 54:1025412020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang C, Liao P, Liang R, Zheng X and Jian
J: Epigallocatechin gallate prevents mitochondrial impairment and
cell apoptosis by regulating miR-30a/p53 axis. Phytomedicine.
61:1528452019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Hao Y, Zhang H, Xu L, Ding N, Wang
R, Zhu G, Ma S, Yang A, Yang Y, et al: DNA Hypomethylation of
miR-30a mediated the protection of hypoxia postconditioning against
aged cardiomyocytes hypoxia/reoxygenation injury through inhibiting
autophagy. Circ J. 84:616–625. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kao SH, Cheng WC, Wang YT, Wu HT, Yeh HY,
Chen YJ, Tsai MH and Wu KJ: Regulation of miRNA biogenesis and
histone modification by K63-polyubiquitinated DDX17 controls cancer
stem-like features. Cancer Res. 79:2549–2563. 2019.PubMed/NCBI
|
|
10
|
Rymen B, Kawamura A, Lambolez A, Inagaki
S, Takebayashi A, Iwase A, Sakamoto Y, Sako K, Favero DS, Ikeuchi
M, et al: Histone acetylation orchestrates wound-induced
transcriptional activation and cellular reprogramming in
Arabidopsis. Commun Biol. 2:4042019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei F, Tang D, Li Z, Kashif MH, Khan A, Lu
H, Jia R and Chen P: Molecular cloning and subcellular localization
of six HDACs and their roles in response to salt and drought stress
in kenaf (Hibiscus cannabinus L.). Biol Res. 52:202019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan H, Li H, Yu P, Fan Q, Zhang X, Huang
W, Shen J, Cui Y and Zhou W: Involvement of HDAC6 in ischaemia and
reperfusion-induced rat retinal injury. BMC Ophthalmol. 18:3002018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao TC, Cheng G, Zhang LX, Tseng YT and
Padbury JF: Inhibition of histone deacetylases triggers
pharmacologic preconditioning effects against myocardial ischemic
injury. Cardiovasc Res. 76:473–481. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Liu H, Liu X, Zhang X, Wu J, Yuan
L, Du X, Wang R, Ma Y, Chen X, et al: Histone acetylation plays an
important role in MC-LR-induced apoptosis and cycle disorder in SD
rat testicular cells. Chemosphere. 241:1250732020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parrot C, Kurbegovic A, Yao G, Couillard
M, Côté O and Trudel M: c-Myc is a regulator of the PKD1 gene and
PC1-induced pathogenesis. Hum Mol Genet. 28:751–763. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karagiannis P, Takahashi K, Saito M,
Yoshida Y, Okita K, Watanabe A, Inoue H, Yamashita JK, Todani M,
Nakagawa M, et al: Induced pluripotent stem cells and their use in
human models of disease and development. Physiol Rev. 99:79–114.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saravia J, Zeng H, Dhungana Y, Bastardo
Blanco D, Nguyen TM, Chapman NM, Wang Y, Kanneganti A, Liu S,
Raynor JL, et al: Homeostasis and transitional activation of
regulatory T cells require c-Myc. Sci Adv. 6:eaaw64432020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Fu Y and Guo H: c-Myc-induced
long non-coding RNA small nucleolar RNA host gene 7 regulates
glycolysis in breast cancer. J Breast Cancer. 22:533–547. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee EJ, Seo E, Kim JW, Nam SA, Lee JY, Jun
J, Oh S, Park M, Jho EH, Yoo KH, et al: TAZ/Wnt-β-catenin/c-MYC
axis regulates cystogenesis in polycystic kidney disease. Proc Natl
Acad Sci USA. 117:29001–29012. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Bu D, Ma Y, Zhu J, Chen G, Sun L,
Zuo S, Li T, Pan Y, Wang X, et al: H19 overexpression induces
resistance to 1,25(OH)2D3 by targeting VDR through miR-675-5p in
colon cancer cells. Neoplasia. 19:226–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo W, Zhang H, Yang A, Ma P, Sun L, Deng
M, Mao C, Xiong J, Sun J, Wang N, et al: Homocysteine accelerates
atherosclerosis by inhibiting scavenger receptor class B member1
via DNMT3b/SP1 pathway. J Mol Cell Cardiol. 138:34–48. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
UniProt C: UniProt: the universal protein
knowledgebase in 2021. Nucleic Acids Res. 49:D480–D489. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wen Z, Lian L, Ding H, Hu Y, Xiao Z, Xiong
K and Yang Q: LncRNA ANCR promotes hepatocellular carcinoma
metastasis through upregulating HNRNPA1 expression. RNA Biol.
17:381–394. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen K, Miller EJ and Sadeghi MM:
PET-based imaging of ischemic heart disease. PET Clin. 14:211–221.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Mu N, Gu C, Liu M, Yang Z, Yin Y,
Chen M, Wang Y, Han Y, Yu L and Ma H: Metformin mediates
cardioprotection against aging-induced ischemic necroptosis. Aging
Cell. 19:e130962020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Webster I, Salie R, Marais E, Fan WJ,
Maarman G, Huisamen B and Lochner A: Myocardial susceptibility to
ischaemia/reperfusion in obesity: A re-evaluation of the effects of
age. BMC Physiol. 17:32017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Griecsová L, Farkašová V, Gáblovský I,
Khandelwal VK, Bernátová I, Tatarková Z, Kaplan P and Ravingerová
T: Effect of maturation on the resistance of rat hearts against
ischemia. Study of potential molecular mechanisms. Physiol Res. 64
(Suppl):S685–S696. 2015. View Article : Google Scholar
|
|
28
|
O'Brien JD, Ferguson JH and Howlett SE:
Effects of ischemia and reperfusion on isolated ventricular
myocytes from young adult and aged Fischer 344 rat hearts. Am J
Physiol Heart Circ Physiol. 294:H2174–H1783. 2008. View Article : Google Scholar
|
|
29
|
Bayrami G, Karimi P, Agha-Hosseini F,
Feyzizadeh S and Badalzadeh R: Effect of ischemic postconditioning
on myocardial function and infarct size following reperfusion
injury in diabetic rats pretreated with vildagliptin. J Cardiovasc
Pharmacol Ther. 23:174–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chávez MN, Morales RA, López-Crisosto C,
Roa JC, Allende ML and Lavandero S: Autophagy activation in
zebrafish heart regeneration. Sci Rep. 10:21912020. View Article : Google Scholar
|
|
31
|
Shi B, Ma M, Zheng Y, Pan Y and Lin X:
mTOR and Beclin1: Two key autophagy-related molecules and their
roles in myocardial ischemia/reperfusion injury. J Cell Physiol.
234:12562–12568. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Torres-Esquivel C, Montiel T,
Flores-Méndez M and Massieu L: Effect of β-hydroxybutyrate on
autophagy dynamics during severe hypoglycemia and the hypoglycemic
coma. Front Cell Neurosci. 14:5472152020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen C, Li J, Zhang W, Shah SWA and Ishfaq
M: Mycoplasma gallisepticum triggers immune damage in the chicken
thymus by activating the TLR-2/MyD88/NF-κB signaling pathway and
NLRP3 inflammasome. Vet Res. 51:522020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang Y, Liao Y, Zhang H and Li S: Lead
exposure induces cell autophagy via blocking the Akt/mTOR signaling
in rat astrocytes. J Toxicol Sci. 45:559–567. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Song A, Wu H, Sun Y and Dai M:
Paeonol inhibits apoptosis of vascular smooth muscle cells via
up-regulation of autophagy by activating class III PI3K/Beclin-1
signaling pathway. Life Sci. 264:1187142021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Campbell GR, Bruckman RS, Chu YL, Trout RN
and Spector SA: SMAC mimetics induce autophagy-dependent apoptosis
of HIV-1-infected resting memory CD4+ T cells. Cell Host Microbe.
24:689–702.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J, Zhang X, Cui Y, Ferdous M, Cui L
and Zhao P: Different postconditioning cycles affect prognosis of
aged patients undergoing primary percutaneous coronary
intervention. Cardiol J. 25:666–673. 2018.PubMed/NCBI
|
|
38
|
Slettom G, Jonassen AK, Dahle GO, Seifert
R, Larsen TH, Berge RK and Nordrehaug JE: Insulin postconditioning
reduces infarct size in the porcine heart in a dose-dependent
manner. J Cardiovasc Pharmacol Ther. 22:179–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maciejak A, Kostarska-Srokosz E, Gierlak
W, Dluzniewski M, Kuch M, Marchel M, Opolski G, Kiliszek M, Matlak
K, Dobrzycki S, et al: Circulating miR-30a-5p as a prognostic
biomarker of left ventricular dysfunction after acute myocardial
infarction. Sci Rep. 8:98832018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lv XB, Niu QH, Zhang M, Feng L and Feng J:
Critical functions of microRNA-30a-5p-E2F3 in cardiomyocyte
apoptosis induced by hypoxia/reoxygenation. Kaohsiung J Med Sci.
37:92–100. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding H, Wang Y, Hu L, Xue S, Wang Y, Zhang
L, Zhang Y, Qi H, Yu H, Aung LHH, et al: Combined detection of
miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217
for screening of early heart failure diseases. Biosci Rep.
40:BSR201916532020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao DS, Chen Y, Jiang H, Lu JP, Zhang G,
Geng J, Zhang Q, Shen JH, Zhou X, Zhu W and Shan QJ: Serum miR-210
and miR-30a expressions tend to revert to fetal levels in Chinese
adult patients with chronic heart failure. Cardiovasc Pathol.
22:444–450. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li D, Wang J, Hou J, Fu J, Liu J and Lin
R: Salvianolic acid B induced upregulation of miR-30a protects
cardiac myocytes from ischemia/reperfusion injury. BMC Complement
Altern Med. 16:3362016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shen Y, Shen Z, Miao L, Xin X, Lin S, Zhu
Y, Guo W and Zhu YZ: miRNA-30 family inhibition protects against
cardiac ischemic injury by regulating cystathionine-γ-lyase
expression. Antioxid Redox Signal. 22:224–240. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Long G, Wang F, Duan Q, Yang S, Chen F,
Gong W, Yang X, Wang Y, Chen C and Wang DW: Circulating miR-30a,
miR-195 and let-7b associated with acute myocardial infarction.
PLoS One. 7:e509262012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kubiak M, Jurek A, Kamińska K, Kowalewski
J, Huang S and Lewandowska MA: Chromosome conformation capture
reveals two elements that interact with the PTBP3 (ROD1)
transcription start site. Int J Mol Sci. 20:2422019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee J and Lee TH: How protein binding
sensitizes the nucleosome to histone H3K56 acetylation. ACS Chem
Biol. 14:506–515. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu Y, Lai Y, Chen X, Zhou DX and Zhao Y:
Distribution pattern of histone marks potentially determines their
roles in transcription and RNA processing in rice. J Plant Physiol.
249:1531672020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Narita T, Weinert BT and Choudhary C:
Functions and mechanisms of non-histone protein acetylation. Nature
reviews. Mol Cell Biol. 20:156–174. 2019.PubMed/NCBI
|
|
50
|
Okonkwo A, Mitra J, Johnson GS, Li L,
Dashwood WM, Hegde ML, Yue C, Dashwood RH and Rajendran P:
Heterocyclic analogs of sulforaphane trigger DNA damage and impede
DNA repair in colon cancer cells: Interplay of HATs and HDACs. Mol
Nutr Food Res. 62:e18002282018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Han KA, Shin WH, Jung S, Seol W, Seo H, Ko
C and Chung KC: Leucine-rich repeat kinase 2 exacerbates neuronal
cytotoxicity through phosphorylation of histone deacetylase 3 and
histone deacetylation. Hum Mol Genet. 26:1–18. 2017.PubMed/NCBI
|
|
52
|
Bascom GD and Schlick T: Chromatin fiber
folding directed by cooperative histone tail acetylation and linker
histone binding. Biophys J. 114:2376–2385. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Church M, Smith KC, Alhussain MM, Pennings
S and Fleming AB: Sas3 and Ada2(Gcn5)-dependent histone H3
acetylation is required for transcription elongation at the
de-repressed FLO1 gene. Nucleic Acids Res. 45:4413–4430.
2017.PubMed/NCBI
|
|
54
|
Zhang BL, Guo TW, Gao LL, Ji GQ, Gu XH,
Shao YQ, Yao RQ and Gao DS: Egr-1 and RNA POL II facilitate glioma
cell GDNF transcription induced by histone hyperacetylation in
promoter II. Oncotarget. 8:45105–45116. 2017. View Article : Google Scholar : PubMed/NCBI
|