Introduction
Esophageal cancer (EC) is one of the most common
types of gastrointestinal cancer worldwide, which is accompanied by
a poor prognosis and unsatisfactory therapeutic outcomes (1,2).
According to 2015 statistics, ~245,651 new cases of EC were
diagnosed in China; due to the high incidence and mortality rates
of 17.87/100,000 and 13.68/100,000 individuals, respectively
(3), EC poses a serious health and
socioeconomic burden in China. In total, >95% of EC cases are
defined as squamous cell carcinomas or adenocarcinomas (3). EC is often asymptomatic during the
early stages and, therefore, the majority of cases are diagnosed at
an advanced stage (4). To date,
there has been a lack of specific prevention strategies for EC,
particularly in patients with distant metastases (5,6). Thus,
it is urgent to identify novel molecular biomarkers and targets for
the early screening and treatment of this malignancy.
Glioblastoma-amplified sequence (GBAS), also known
as nipsnap homolog 2 (NIPSNAP), belongs to the NIPSNAP family, and
is located in the vicinity of marker D7S499 on chromosome 7p12
(7). GBAS encodes a mitochondrial
protein that contains tyrosine phosphorylation sites, transmembrane
motifs and a mitochondrial targeting sequence (8), which is present on the surface of
mitochondria (9). Emerging evidence
has indicated the functional roles of GBAS in cellular activities.
For example, Abudu et al (10) reported that NIPSNAP family members,
including NIPSNAP1 and NIPSNAP2, serve crucial roles in mitophagy
by recruiting selective autophagy-related proteins (10). Multiple previous studies have also
suggested that GBAS may be involved in the initiation, development
and progression of several types of solid tumors alongside the
co-amplification with other cellular genes, such as EGFR, which is
an important oncogene in several cancer types (8). For example, a previous study revealed
that GBAS was co-amplified with EGFR, where the EGFR intronic
variant was associated with GBAS expression in early-stage
non-small cell lung cancer (NSCLC) (11). Another study demonstrated that GBAS
expression levels were downregulated in oral squamous cell
carcinoma (OSCC) and regulated the viability and apoptosis of
cancer cells through activating the p53 signaling pathway (12). Therefore, GBAS may serve as a
molecular biomarker of tumorigenesis; however, the mechanisms
underlying the role of GBAS in the pathogenesis of EC remain poorly
understood.
The present study aimed to perform a comprehensive
analysis of the function of GBAS as a potential important oncogene
in the development of EC. The effects of GBAS knockdown on the
viability, colony formation ability, cell cycle progression and
apoptosis of EC cells were investigated in vitro, in order
to determine whether GBAS may serve as a novel oncogene in the
development of EC.
Materials and methods
Patient studies
A total of 15 esophageal squamous cell carcinoma (5
female patients and 10 male patients) and matched adjacent tissues
(5 cm away from cancer tissue) were obtained at Sichuan Cancer
Hospital (Chengdu, China), between May 2019 and April 2020, from
the patients before any clinical interventions. The samples were
frozen and stored in liquid nitrogen until further use. All the
patients included had achieved R0 resection and provided complete
clinicopathological data. A total of 12 patients had tumors located
in the middle part of the thoracic esophagus. All patients were
diagnosed with EC by two pathologists. The protocols of the present
study were approved by the Ethics Committee of Sichuan Cancer
Hospital (approval no. SCCHEC-02-2017-043) and written informed
consent was obtained from all patients prior to participation.
Cell lines and culture
Human esophageal epithelial cells (HEEC) and human
EC cell lines (TE-1, KYSE-150, EC9706 and ECA-109) were obtained
from the American Type Culture Collection. 293T cells were
purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The HEEC and EC cell lines were
cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific,
Inc.), while the 293T cell was cultured in DMEM medium (Invitrogen;
Thermo Fisher Scientific, Inc.), supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 g/ml
streptomycin, and maintained in a humidified atmosphere with 5%
CO2 at 37°C.
Lentiviral-mediated GBAS
knockdown
The package of lentivirus was used 3rd generation
system. The GV115 (pGCSIL-GFP; Shanghai GeneChem Co., Ltd.)
lentivirus was used for constructing GBAS-knockdown vectors. The
short hairpin RNA (shRNA/sh) sequence targeting GBAS used was as
follows: Forward,
5′-CCGGCCATGTGAAAGTCCTGTTGAACTCGAGTTCAACAGGACTTTCACATGGTTTTTG-3′
and reverse,
5′-AATTCAAAAACCATGTGAAAGTCCTGTTGAACTCGAGTTCAACAGGACTTTCACATGG-3′.
After cloning the shGBAS or shCtrl sequence into the GV115 vectors,
the lentivirus construction system, including the GV115-GBAS vector
(20 µg), and Helper 1.0 (15 µg; Shanghai GeneChem Co., Ltd.) and
Helper 2.0 (10 µg; Shanghai GeneChem Co., Ltd.) packaging vectors,
were co-transfected into 293T cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following 72 h
of transfection, the freshly harvested virus supernatants were
collected by ultracentrifugation (25,000 × g at 4°C for 2 h) and
subsequently stored at −80°C. The multiplicity of infection for EC
cell transfection was 20. After 72 h, the transfection efficiency
of GBAS knockdown (shGBAS) was determined using reverse
transcription-quantitative PCR (RT-qPCR) and western blotting.
RT-qPCR
HEEC, EC9760, ECA-109, KYSE-150 and TE-1 cells were
washed with PBS three times and total RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Total RNA was
reverse-transcribed into cDNA using a reverse-transcribed kit
according to the manufacturer's protocol (Promega Corporation).
qPCR was subsequently performed using SYBR Green Master mix (Takara
Bio, Inc.). The following thermocycling conditions were used for
qPCR: Initial denaturation at 95°C for 5 min; followed by 40 cycles
at 95°C for 15 sec, 60°C for 30 sec and 70°C for 10 sec. Relative
expression levels were calculated using the 2−ΔΔCq
method (13). The following primer
sequences were used for qPCR: GBAS forward,
5′-TTCGTAAGGCAAGAAGTGAC-3′ and reverse, 5′-GTCGGAGTTGGTAAGACCTG-3′;
and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH was used as the internal
reference gene.
Western blotting
KYSE-150 and TE-1 cells were centrifuged at 100 × g
for 5 min at room temperature and washed with cold PBS twice. The
collected cells were resuspended in RIPA lysis buffer (Beyotime
Institute of Biotechnology), supplemented with 1 mM PMSF
(Sigma-Aldrich; Merck KGaA), and maintained on ice for 30 min.
Following centrifugation at 12,000 × g for 15 min at 4°C, the
supernatant was transferred into new Eppendorf tubes and protein
quantification was performed using a BCA assay (Thermo Fisher
Scientific, Inc.). Subsequently, 25 µg protein/lane was separated
via SDS-PAGE on 12% gel, then separated proteins were transferred
onto PVDF membranes and blocked in 5% skimmed milk in TBS with 1%
Tween-20 (TBST) for 2 h at a room temperature. The membranes were
incubated with the following primary antibodies at 4°C overnight:
Anti-GBAS (1:2,000; cat. no. ab153833; Abcam), anti-caspase-3
(1:1,000; cat. no. 9662; CST) and anti-caspase-3 (1:1,000; cat. no.
9661; CST Biological Reagents Co., Ltd.) anti-GAPDH (1:2,000; cat.
no. sc-32233; Santa Cruz Biotechnology, Inc.). Following primary
antibody incubation, the membranes were incubated with anti-rabbit
(cat. no. 7074) and anti-mouse (cat. no. 7076) secondary antibodies
(1:2,000; Cell Signaling Technology, Inc.) at room temperature for
2 h. The membranes were washed three times with TBST and protein
bands were visualized using enhanced chemiluminescence reagent
(Cell Signaling Technology, Inc.).
Cell viability assay
TE-1 and KYSE-150 cells were seeded into 96-well
plates at a cell density of 2.0×103 cells/well, with
three replicates/group. The cells were subsequently cultured with
100 µl RPMI-1640 medium for a total of 5 days. Every 24 h, a Celigo
image cytometer system (Nexcelom Bioscience LLC) was used to
determine cell number, and the number of cells with green
fluorescence in each well was calculated using a Celigo cell count
(14).
Colony formation assay
TE-1 and KYSE-150 cells at the logarithmic growth
phase were treated with 0.5% trypsin (Thermo Fisher Scientific,
Inc.). After cell counting, 400 cells/well were seeded into a
six-well culture plate, with more than three replicates/group. The
transfected cells were cultured for 14 days, and the culture medium
was changed every 3 days. Following incubation, the cell colonies
were fixed with 4% paraformaldehyde for 20 min at a room
temperature and stained with Giemsa dye (Beyotime Institute of
Biotechnology) for 20 min at a room temperature.
Flow cytometric analysis of
apoptosis
Following the corresponding transfections, TE-1 and
KYSE-150 cells were digested with trypsin and collected. The cell
pellet was washed with ice-cold PBS and then resuspended in 200 µl
1X binding buffer. The cells were subsequently incubated with 10 µl
Annexin V-allophycocyanin (Beyotime Institute of Technology) at
room temperature for 15 min in the dark. All samples were detected
and analyzed using a BD Accuri™ C6 Plus flow cytometer (BD
Biosciences). The apoptosis (early + late apoptosis) were
quantified and analyzed using a BD Accuri C6 Software (BD
Biosciences).
Cell cycle analysis
Flow cytometry was performed to determine the cell
cycle distribution. Briefly, following transfection, TE-1 and
KYSE-150 cells were fixed with pre-cooled 75% ethanol and incubated
at 4°C for 1 h. The cells were subsequently washed once with cold
PBS and centrifuged at 100 × g for 5 min at 4°C. The cell pellets
were resuspended in propidium iodide (cat. no. P4170;
Sigma-Aldrich; Merck KGaA), RNase (cat. no. EN0531; Thermo Fisher
Scientific, Inc.), PBS and Triton X-100 (cat. no. 9002-93-1;
Sigma-Aldrich; Merck KGaA) at a ratio of 25:10:1,000:40. All
samples were detected and analyzed using a BD Accuri™ C6 Plus flow
cytometer (BD Biosciences). The distribution of EC cells in each
stage of the cell cycle was analyzed using FCS Express IVD software
(version 4; De Novo Software).
Statistical analysis
Statistical analysis was performed using SPSS 25.0
software (IBM Corp.) and GraphPad Prism version 8.0 (GraphPad
Software, Inc.). The data are presented as the mean ± SD of three
independent repeats. Statistical differences between two groups
were determined using an unpaired Student's t-test for unpaired
data, or a paired Student's t-test for paired data and statistical
differences among multiple groups were analyzed using one way-ANOVA
followed by a Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
GBAS expression knockdown by
lentivirus-mediated transfection
To analyze the expression levels of GBAS in patients
with EC, EC clinical tissues were obtained and the expression
levels of GBAS were determined using RT-qPCR. Baseline
clinicopathological characteristics of the esophageal squamous cell
carcinoma are shown in Table S1.
The results revealed that there was no significant difference in
the expression of GBAS in EC tissues compared with those in normal
adjacent tissues (Fig. 1A). The
mRNA expression levels of GBAS were subsequently analyzed in normal
HEEC cells and four EC cell lines (TE-1, KYSE-150, ECA109 and
EC9706) using RT-qPCR. The results demonstrated that the expression
levels of GBAS were significantly upregulated in TE-1, KYSE-150,
ECA109 and EC9706 cells compared with those in HEEC cells (Fig. 1B). TE-1 and KYSE-150 cells were
selected for use in the following experiments due to the high
expression levels of GBAS. The lentiviral-mediated knockdown
efficiency of GBAS was verified at both the mRNA and protein
levels. RT-qPCR analysis demonstrated that the mRNA expression
levels of GBAS were significantly downregulated in both TE-1 and
KYSE-150 cells (Fig. 1C and D).
Consistent with these findings, the protein expression levels of
GBAS were also downregulated in shGBAS-transfected TE-1 and
shGBAS-transfected KYSE-150 cells (Fig.
1E). These results suggested that GBAS expression was
successfully knocked down in both TE-1 and KYSE-150 cells.
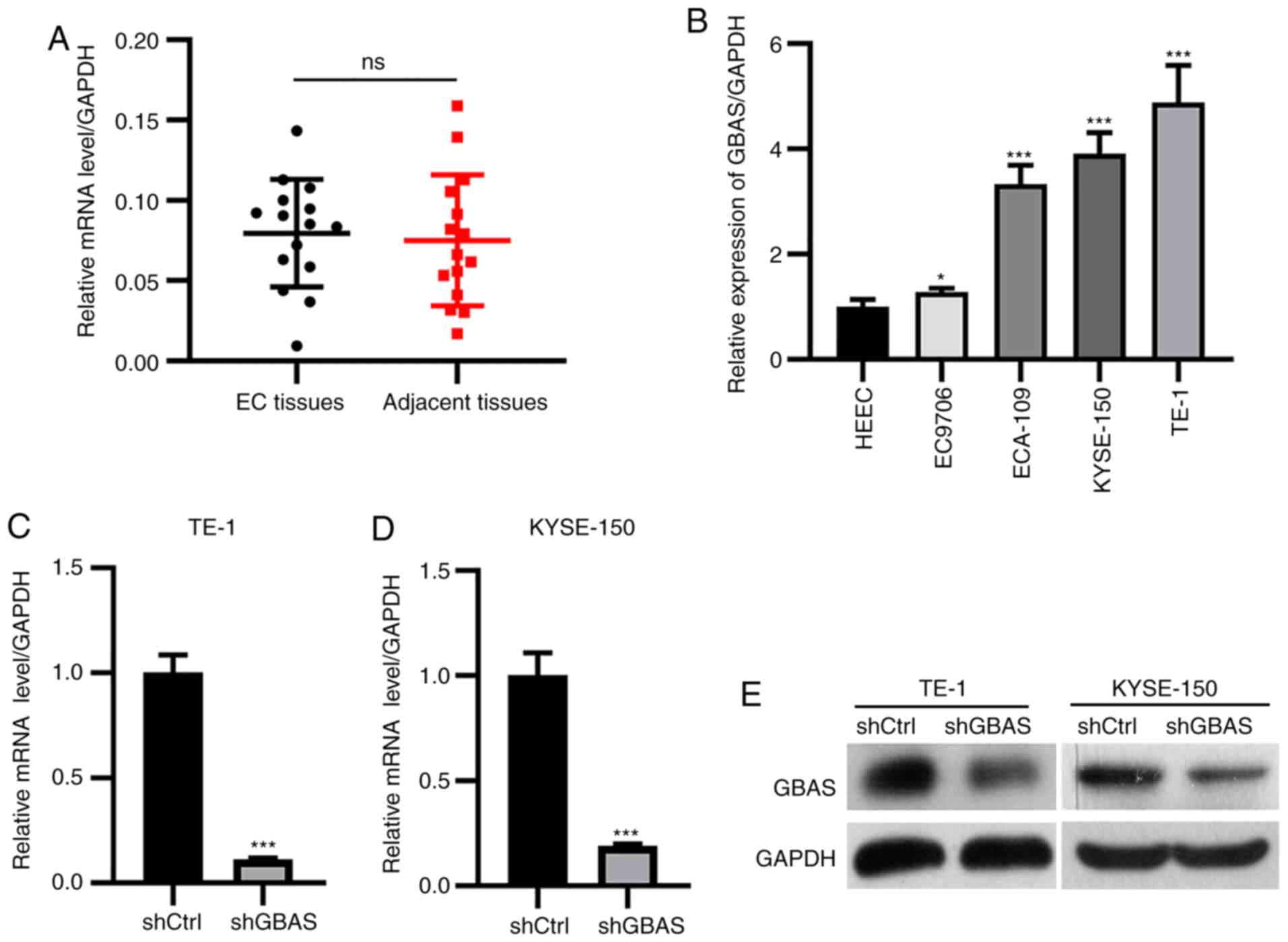 | Figure 1.Lentiviral-mediated knockdown of GBAS
expression. (A) The expression level of GBAS in patients with EC
and adjacent normal tissues detected by RT-qPCR. (B) RT-qPCR was
used to analyze mRNA expression levels in the four EC cell lines,
TE-1, KYSE-150, ECA-109 and EC9706. GAPDH served as the internal
reference control. *P<0.05, ***P<0.001 vs. HEEC cells.
RT-qPCR was used to confirm knockdown efficency of GBAS in (C) TE-1
and (D) KYSE-150 cells. (E) Western blotting was used to confirm
knockdown efficency of GBAS in TE-1 and KYSE-150 cells. GAPDH
served as the internal loading control. ***P<0.001 vs. shCtrl.
Ns, no significance; GBAS, glioblastoma-amplified sequence;
RT-qPCR, reverse transcription-quantitative PCR; EC, esophageal
cancer; sh, short hairpin RNA; Ctrl, control. |
Knockdown of GBAS expression
suppresses the viability and colony formation of EC cells
To determine the functional roles of GBAS in
regulating the development of EC, cell viability was determined
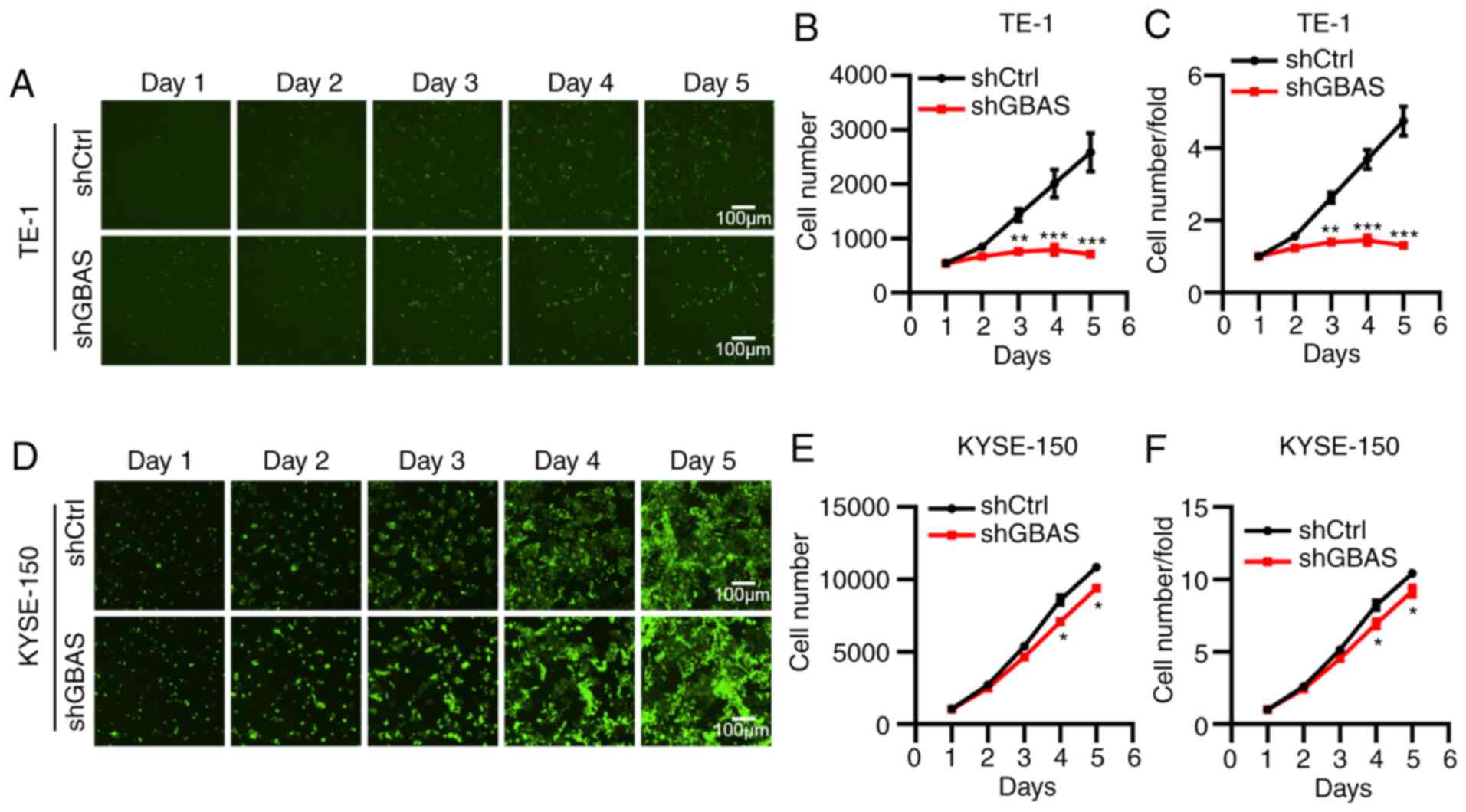
using a Celigo image cytometer system. As shown in Fig. 2, knockdown of GBAS significantly
inhibited cell viability following culture for 5 days in both TE-1
(Fig. 2A-C) and KYSE-150 (Fig. 2D-F) cells. In addition, compared
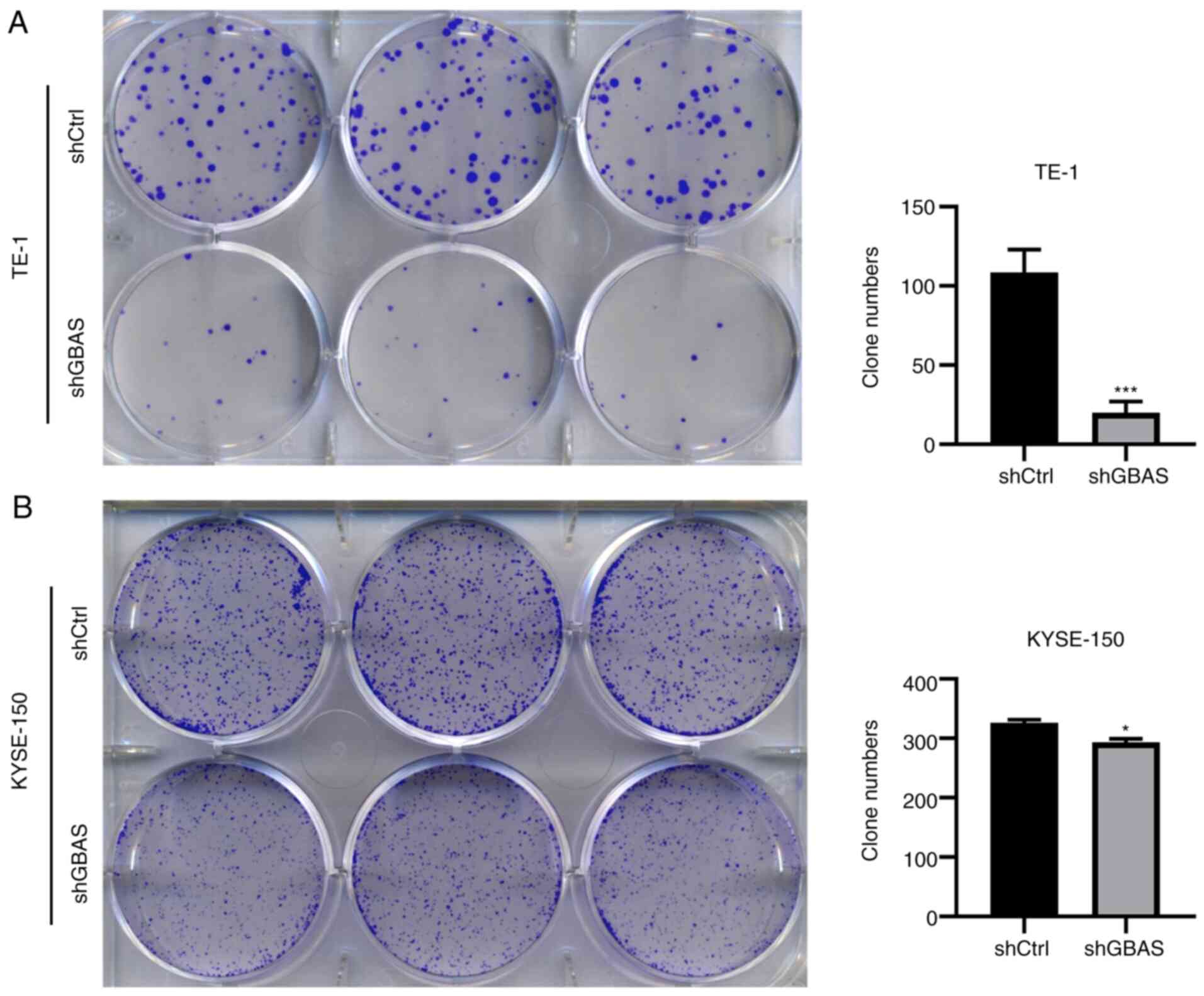
with TE-1 cells transfected with shCtrl, colony formation was
significantly suppressed in TE-1 cells transfected with shGBAS
(Fig. 3A). GBAS knockdown also
inhibited colony formation in KYSE-150 cells (Fig. 3B). Collectively, these results
indicated that the knockdown of GBAS expression may suppress cell
viability and colony formation in EC cell lines.
Knockdown of GBAS promotes apoptosis
of EC cells
Accumulating evidence has indicated that cell
apoptosis predetermines the onset and progression of malignant
tumors due to its regulatory role in cell viability and role in
maintaining a constant number of cells (15,16).
Thus, the present study aimed to investigate the effect of GBAS
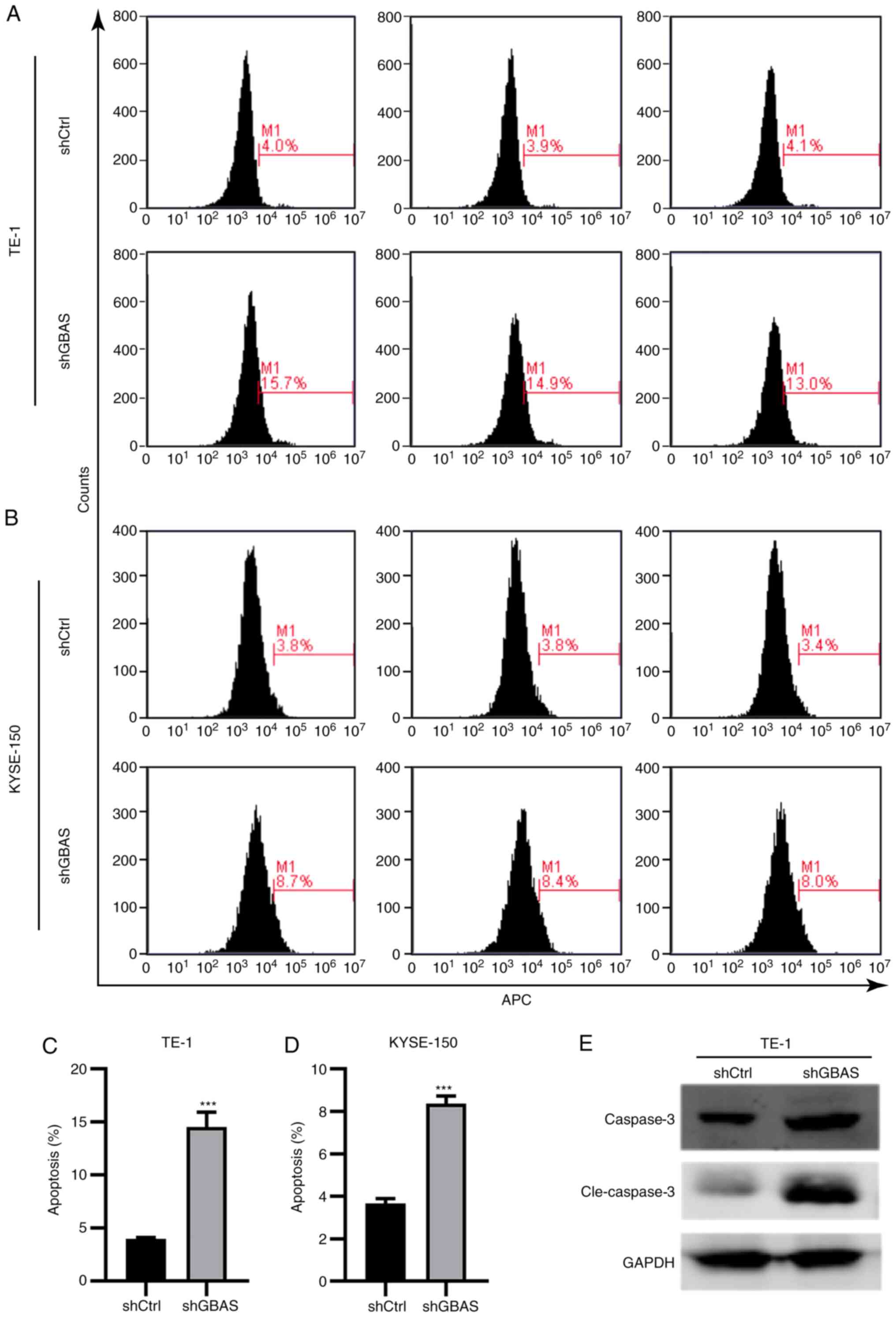
knockdown on the apoptosis of TE-1 and KYSE-150 cells by Annexin V
staining. The knockdown of GBAS significantly increased the
percentage of apoptotic TE-1 (Fig. 4A
and C) and KYSE-150 (Fig. 4B and
D) cells. Accordingly, the expression levels of apoptotic
markers, including caspase-3 and cleaved caspase-3, in TE-1 cells
were detected using western blotting. As shown in Fig. 4E, the expression levels of caspase-3
and cleaved caspase-3 were notably upregulated following GBAS
knockdown in TE-1 cells. These results suggested that GBAS
knockdown may promote the apoptosis of EC cell lines.
Knockdown of GBAS blocks cell cycle
progression in EC cells
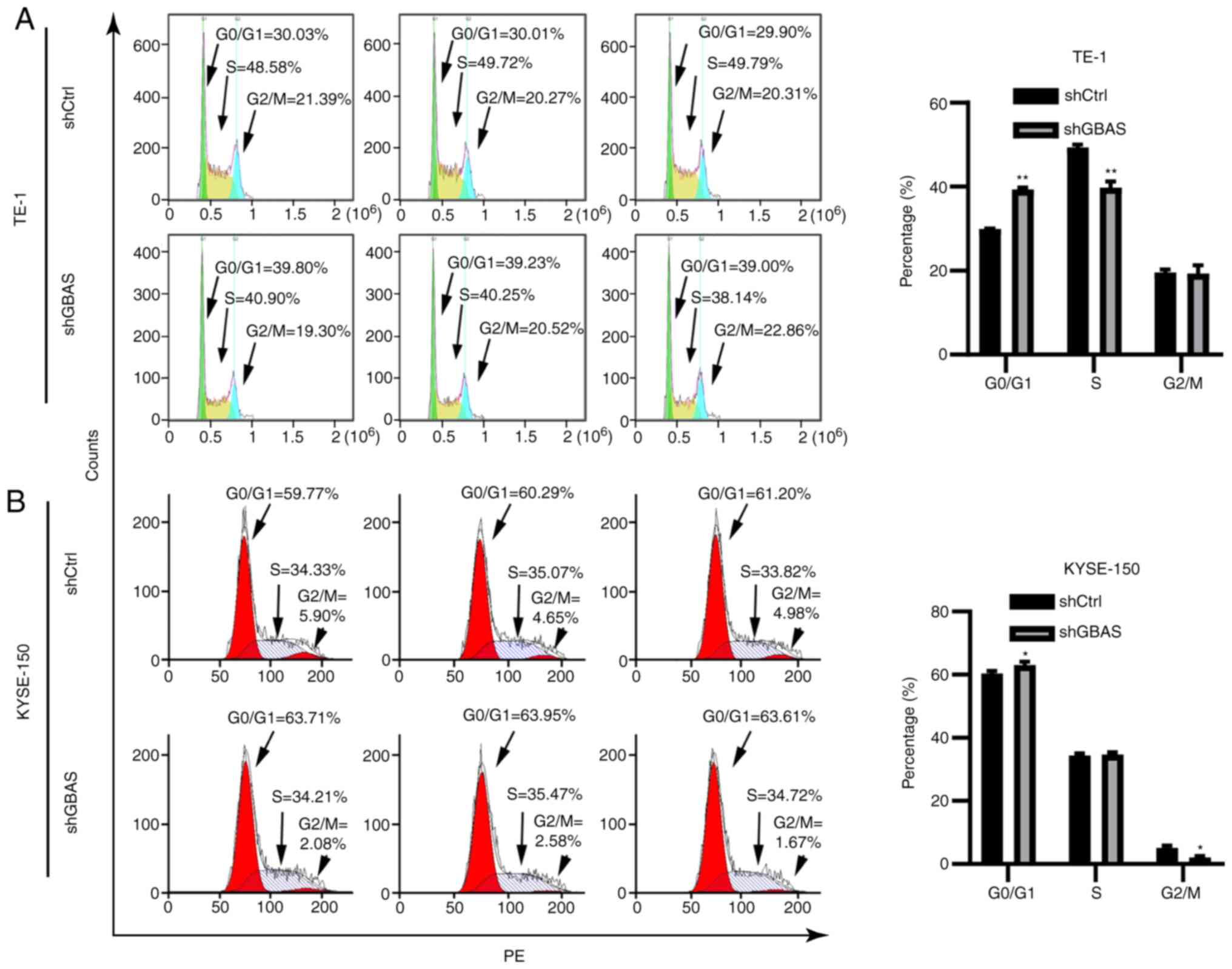
Whether GBAS was involved in cell cycle regulation
was determined using flow cytometry. Indeed, the knockdown of GBAS
in both TE-1 and KYSE-150 cells contributed to cell cycle
redistribution. Specifically, following GBAS knockdown, the number
of TE-1 and KYSE-150 cells in the G1 phase was
significantly increased. The number of cells in the S phases was
significantly decreased in TE-1 cells, while the number of cells in
the G2/M phases was significantly decreased in KYSE-150
cells (Fig. 5A and B). Taken
together, these results suggested that the knockdown of GBAS may
block cell cycle progression in EC cell lines.
Discussion
GBAS expression has been reported to be co-amplified
alongside EGFR in multiple types of cancer, which is associated
with the upregulated expression levels of the protein (8,17).
Previous research has shown that GBAS knockdown can regulate OSCC
viability and apoptosis through the p53 signaling pathway (12). However, whether GBAS may be involved
in the development and progression of solid tumor types,
particularly EC, remains unclear. Thus, further investigations are
required to elucidate the biological function and underlying
molecular mechanisms of GBAS in EC progression. The results of the
present study demonstrated that GBAS regulated cell viability,
colony formation and cell apoptosis, as well as the cell cycle
distribution of EC cells, which in turn may serve important
functional roles in the development of EC. To the best of our
knowledge, these findings have not been reported before in EC.
To further elucidate the clinical relevance of GBAS
expression in EC, the present study analyzed the expression levels
of GBAS in EC and adjacent normal tissues. The present study
revealed that the expression level of GBAS was not significantly
different in EC tissues compared with the adjacent normal tissues.
Unfortunately, owing to the limited availability of EC tissues,
only 15 paired samples were used in the present analysis. Thus, one
of the major limitations in the present study was the very small
sample size making it difficult to analyze the association between
EC tissues and GBAS expression; more clinical samples need to be
collected for further study. Nevertheless, previous studies suggest
that the expression of GBAS is dysregulated in several types of
malignancies and GBAS expression is co-amplified with EGFR in human
gliomas (8,11,18).
However, GBAS is downregulated in oral squamous cell carcinoma and
involved in controlling proliferation and apoptosis via the p53
signaling pathway (12). Therefore,
it was hypothesized that GBAS may be an oncogene associated with EC
development. However, the results suggested that GBAS may function
as a novel oncogene, although its expression levels were not
significantly different in EC tissues compared with normal
tissues.
The present study subsequently aimed to elucidate
the functional biological roles of GBAS in EC cells via
loss-of-function in vitro studies. The results revealed that
the knockdown of GBAS could effectively inhibit EC cell viability
and colony formation and induce apoptosis. Considering that the
dysregulation of cell viability and apoptosis are usually
considered as markers of tumorigenesis (19,20),
it was hypothesized that the dysregulated expression of GBAS in EC
may be associated with the overall survival of patients. Similar
findings have also been reported in other types of cancer,
including OSCC, bladder cancer and early-stage NSCLC (11). As reported by Hong et al
(11), overexpression of GBAS
promoted the centrosome amplification rate, as well as cell
migration and invasion in bladder cancer cells. In addition, other
assays in this previous study revealed that GBAS expression
co-amplified with a variant of EGFR, rs9642391C>G, was
associated with survival outcomes in patients with early-stage
NSCLC (11). Another previous study
suggested that GBAS expression was upregulated in OSCC, and
knockdown of GBAS could regulate OSCC cell growth and apoptosis via
activating the p53 signaling pathway (12). However, as the viability and
proliferative ability of EC cells following overexpression of GBAS
were not investigated in the present study, this will be further
investigated in future studies.
In addition to the determined functional roles of
GBAS in cell viability and apoptosis, the results of the flow
cytometric analysis revealed that the knockdown of GBAS altered the
cell cycle distribution of EC cells. Notably, GBAS knockdown
arrested the cell cycle at the G1 phase, which suggested
that GBAS may regulate the development of EC through arresting the
cell cycle. Of note, the role of GBAS in EC is consistent with the
previously reported roles of other members of the NIPSNAP family
(21–23). However, the mechanism underlying the
GBAS-induced regulation of the EC cell cycle still remains poorly
understood. The present found that following GBAS knockdown, the
results among two EC cell lines were not consistent, which might be
owing to the difference in the characteristics and culture
conditions of the two cell lines.
In conclusion, the findings of the present study
suggested that GBAS may promote the progression and development of
EC by regulating cell viability, apoptosis and cell cycle
progression, suggesting that GBAS may play a role in EC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JP and JTH performed the experiments and designed
the study. JP and KM conceptualized the study and performed the
experiments. HR, BX and JZ analyzed and interpreted the data. JP
and JTH drafted and revised the manuscript. JP and JTH confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study protocol was approved by the
ethics committee of Sichuan Cancer Hospital (Sichuan, China) and
written informed consent was obtained from all patients prior to
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hagymasi K and Tulassay Z: Risk factors
for esophageal cancer, and possible genetic background. Orv Hetil.
150:407–413. 2009.(In Hungarian). PubMed/NCBI
|
|
2
|
Tsuyama S, Kohsaka S, Hayashi T, Suehara
Y, Hashimoto T, Kajiyama Y, Tsurumaru M, Ueno T, Mano H, Yao T and
Saito T: Comprehensive clinicopathological and molecular analysis
of primary malignant melanoma of the oesophagus. Histopathology.
78:240–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen R, Zheng RS, Zhang SW, Zeng HM, Wang
SM, Sun KX, Gu XY, Wei WW and He J: Analysis of incidence and
mortality of esophageal cancer in China, 2015. Zhonghua Yu Fang Yi
Xue Za Zhi. 53:1094–1097. 2019.(In Chinese). PubMed/NCBI
|
|
4
|
Cools-Lartigue J, Spicer J and Ferri LE:
Current status of management of malignant disease: Current
management of esophageal cancer. J Gastrointest Surg. 19:964–972.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Laarhoven HW: Is chemotherapy for
advanced or metastatic oesophageal squamous cell carcinoma no
longer needed? Lancet Oncol. 21:743–745. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatehi Hassanabad A, Chehade R, Breadner D
and Raphael J: Esophageal carcinoma: Towards targeted therapies.
Cell Oncol (Dordr). 43:195–209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seroussi E, Pan HQ, Kedra D, Roe BA and
Dumanski JP: Characterization of the human NIPSNAP1 gene from
22q12: A member of a novel gene family. Gene. 212:13–20. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang XY, Smith DI, Liu W and James CD:
GBAS, a novel gene encoding a protein with tyrosine phosphorylation
sites and a transmembrane domain, is co-amplified with EGFR.
Genomics. 49:448–451. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abudu YP, Pankiv S, Mathai BJ, Lamark T,
Johansen T and Simonsen A: NIPSNAP1 and NIPSNAP2 act as ‘eat me’
signals to allow sustained recruitment of autophagy receptors
during mitophagy. Autophagy. 15:1845–1847. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abudu YP, Pankiv S, Mathai BJ, Håkon
Lystad A, Bindesbøll C, Brenne HB, Yoke Wui Ng M, Thiede B,
Yamamoto A, Mutugi Nthiga T, et al: NIPSNAP1 and NIPSNAP2 Act as
‘Eat Me’ signals for mitophagy. Dev Cell. 49:509–525.e12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong MJ, Lee SY, Choi JE, Kang HG, Do SK,
Lee JH, Yoo SS, Lee EB, Seok Y, Cho S, et al: Intronic variant of
EGFR is associated with GBAS expression and survival outcome of
early-stage non-small cell lung cancer. Thorac Cancer. 9:916–923.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X, Bai Y, Han Y, Meng J and Liu H:
Downregulation of GBAS regulates oral squamous cell carcinoma
proliferation and apoptosis via the p53 signaling pathway. Onco
Targets Ther. 12:3729–3742. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L and Ouyang L: Effects of EIF3B gene
downregulation on apoptosis and proliferation of human ovarian
cancer SKOV3 and HO-8910 cells. Biomed Pharmacother. 109:831–837.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Green DR: Cancer and apoptosis: Who is
built to last? Cancer Cell. 31:2–4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaczanowski S: Apoptosis: Its origin,
history, maintenance and the medical implications for cancer and
aging. Phys Biol. 13:0310012016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng LS, Davis RC, Raffel LJ, Xiang AH,
Wang N, Quiñones M, Wen PZ, Toscano E, Diaz J, Pressman S, et al:
Coincident linkage of fasting plasma insulin and blood pressure to
chromosome 7q in hypertensive hispanic families. Circulation.
104:1255–1260. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Li S and Liu H: Co-delivery of
chitosan nanoparticles of 5-aminolevulinic acid and shGBAS for
improving photodynamic therapy efficacy in oral squamous cell
carcinomas. Photodiagnosis Photodyn Ther. 34:1022182021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gregory CD, Ford CA and Voss JJ:
Microenvironmental effects of cell death in malignant disease. Adv
Exp Med Biol. 930:51–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pachl J, Duska F, Waldauf P, Fric M, Fanta
J and Zdarsky E: Apoptosis as an early event in the development of
multiple organ failure? Physiol Res. 54:697–699. 2005.PubMed/NCBI
|
|
21
|
Malhotra A, Shibata Y, Hall IM and Dutta
A: Chromosomal structural variations during progression of a
prostate epithelial cell line to a malignant metastatic state
inactivate the NF2, NIPSNAP1, UGT2B17, and LPIN2 genes. Cancer Biol
Ther. 14:840–852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schoeber JP, Topala CN, Lee KP, Lambers
TT, Ricard G, van der Kemp AW, Huynen MA, Hoenderop JG and Bindels
RJ: Identification of Nipsnap1 as a novel auxiliary protein
inhibiting TRPV6 activity. Pflugers Arch. 457:91–101. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okuda-Ashitaka E, Minami T, Tsubouchi S,
Kiyonari H, Iwamatsu A, Noda T, Handa H and Ito S: Identification
of NIPSNAP1 as a nocistatin-interacting protein involving pain
transmission. J Biol Chem. 287:10403–10413. 2012. View Article : Google Scholar : PubMed/NCBI
|