Introduction
Oesophageal cancer (EC) is one of the commonest
types of cancer worldwide with high morbidity, especially in
Eastern Asia (1–3). It is also the sixth leading cause of
cancer-related death globally, with 5.3% of all cancer deaths
(age-standardized rates, 5.5 per 100,000) (2). EC has two main histological subtypes:
Oesophageal adenocarcinoma (EA) and oesophageal squamous cell
carcinoma (ESCC) (4). ESCC is one
of the most aggressive squamous cell carcinomas and has a high
prevalence in Asia, especially in China (5–7).
Patients with advanced oesophageal squamous cell carcinoma who
cannot tolerate or refuse to undergo surgery can choose
radiotherapy (RT) (4,7,8). As a
number of patients may suffer radioresistance, the outcomes of
clinical treatment are unsatisfactory (9). Hence, it is necessary to improve the
clinical outcomes to explore the related molecular mechanisms of
the proliferation and radioresistance of ESCC.
Cell division cycle-associated protein 2 (CDCA2;
also known as RepoMan), regulates the phosphatase of the core
substrates throughout the cell cycle (10). It has been reported that CDCA2 can
promote cell proliferation in colorectal cancer by activating the
AKT/cyclin D1 pathway (11). CDCA2,
which is highly expressed in oral squamous cell carcinoma and lung
adenocarcinoma, respectively, can promote the growth of certain
types of tumour (12,13). Previous studies have also reported
that CDCA2 can modulate chromatin remodelling and DNA damage
checkpoint activation (14,15).
At present, the biological functions of CDCA2 in
ESCC have rarely been reported and hence the purpose of the present
study was to examine the expression and biological behaviours of
CDCA2 and evaluate its role in the process of tumour growth and
radioresistance in ESCC. Targeting CDCA2 may be a novel therapeutic
option to heighten the radiosensitivity of ESCC.
Materials and methods
The Cancer Genome Atlas (TCGA)
database
The expression data of all genes in ESCC were
downloaded from TCGA, comprising 93 samples (11 normal tissues and
82 tumour tissues).
Cell culture
A total of five types of human ESCC cell lines
(ECA109, KYSE150, KYSE450, TE10 and TE13) and an oesophageal
epithelial cell line (SHEE) were maintained at the Jiangsu Province
Hospital Core Facility Center. All cell lines involved in the
present study were purchased from the Shanghai Institute of
Biochemistry and Cell Biology. The cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and
100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All
cells were cultured in a humidified chamber with 5% CO2
at 37°C.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
In accordance with the manufacturer's instructions,
total RNA was extracted from cells using TRIzol® (Thermo
Fisher Scientific, Inc.). A NanoDrop spectrophotometer (ND-100,
Thermo Fisher Scientific, Inc.) was used to detect the quality and
concentration of RNA. RNA reverse transcription was conducted with
a New Poly (A) Tailing kit (Thermo Fisher Scientific, Inc.) and a
PrimeScript RT Master Mix kit (cat. no. RR036A; Takara Bio, Inc.).
The temperature and duration of RT were as follows: 37°C for 15
min, followed by 85°C for 5 sec and 4°C for 10 min. RT-PCR was
performed utilizing Universal SYBR Green Master Mix (cat. no.
4913914001; Roche Diagnostics) with a 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: 95°C for 10 min, followed by 40
cycles at 95°C for 5 sec, 55°C for 30 sec and 72°C for 30 sec.
Expression levels were calculated using the 2−ΔΔCq
method (16). The relative mRNA
expression was normalized to β-actin. All-in-one™ qPCR primers for
CDCA2 were purchased from GeneCopoeia, Inc. All experiments were
repeated three times. Primer sequences for CDCA2 and β-actin are as
follows: CDCA2 forward, 5′-TGCCGAATTACCTCCTAATCCT-3′ and reverse,
5′-TGCTCTACGGTTACTGTGGAAA-3′; and β-actin forward,
5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′.
Lentivirus transfection
Human CDCA2-targeting short hairpin (sh)RNA
sequences
(CCGGCTGTGGCAAGAGGGAAAGTAACTCGAGTTACTTTCCCTCTTGCCACAGTTTTTG) were
cloned into hU6-MCS-CMW-puromycin (Shanghai GenePharma Co., Ltd.).
The titre of lentivirus was 2.37×108 TU/ml, and the best
transduction efficiency in the current study was at multiplicity of
infection=5. Lentivirus transduction was conducted according to the
manufacturer's instructions. Lentivirus was added into cells at
room temperature. Cells were placed in an incubator at 37°C for 12
h, and then the medium was changed. As a negative control (NC), a
shRNA with a scrambled sequence was used. The transfected cells
were selected by puromycin until stably transfected cells were
obtained. The concentration of puromycin in medium was 3 µg/ml.
Transfection efficiency was assessed by western blot analysis and
RT-qPCR. Subsequent experiments were performed at least 72 h after
transfection.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded in triplicate into a 96-well plate
at a density of 3,000 cells per well. Following cell adherence, 100
µl mixed solution (CCK-8 solution: RPMI-1640 medium=1:10) was added
to each well according to the manufacturer's instructions at the
indicated time points (days 1, 2, 3 and 4). Following a 2-h
incubation period at 37°C, absorbance was determined with a
microplate reader (cat. no. ELx800; BioTek Instruments, Inc.).
5-Ethynyl-2′-deoxyuridine (EdU)
incorporation assay
An EdU assay kit (Guangzhou RiboBio Co., Ltd.) was
used to assess cellular proliferation. Cells were cultured in
RPMI-1640 medium containing 10% FBS at 37°C in 24-well plates in
triplicate at a density of 5×104 cells per well
overnight. Cells were then maintained for 2 h at 37°C in medium
containing 50 µM EdU and treated according to the manufacturer's
instructions. Typical images were captured under a fluorescence
microscope (magnification, ×40; Nikon Corporation). The proportion
of EdU-positive cells among the cells from three random fields was
analysed using ImageJ software (Java 1.6.0_20; National Institutes
of Health).
Clonogenic survival assays
Unequal numbers of exponentially growing cells (0
Gy: 300 cells, 2 Gy: 600 cells, 4 Gy: 1,200 cells, 6 Gy: 3,000
cells and 8 Gy: 6,000 cells) were plated into 6-well plates.
Following cell adherence, they were subjected to X-ray radiation.
Immediately after radiation, the culture medium (RPMI-1640 medium
containing 10% FBS) was renewed and the cells were cultured at 37°C
for ≤12 days. The cells were fixed and then stained with crystal
violet at room temperature for 30 min. The number of colonies
(>50 cells/colony) were counted under a light microscope
(magnification, ×10). Plating efficiencies (PEs) were calculated as
the number of colonies divided by the number of cells seeded. The
surviving fraction (SF) of each radiation group was corrected by
the PE of the nonradiated control. Dose-response clonogenic
survival curves were plotted on a log-linear scale. Cell survival
curves based on the mean survival fractions of the cell line were
fitted to a multitarget single-hit model:
S=1-(1-e−D/D0)N (17). The experiment was repeated three
times.
Immunofluorescence
Immunofluorescence detection of γH2AX foci was
utilized to evaluate DNA double-strand breaks (DSBs) in ESCC cells.
The cells were seeded on a glass-bottomed dish and then irradiated
with a single 8 Gy dose. Then, 2, 6 and 24 h after radiation, the
cells were washed with PBS and fixed with 4% paraformaldehyde for
20 min at room temperature. Cells were stained with a rabbit
anti-γH2AX monoclonal antibody (1:200; cat. no. ab229914; Abcam)
overnight at 4°C and then with Alexa 555 Fluor secondary antibody
(1:500; cat. no. A0453; Beyotime Institute of Biotechnology) for
1.5 h at room temperature. The nuclei were counterstained with 2
µg/ml DAPI (cat. no. C1005; Beyotime Institute of Biotechnology)
for confocal microscopy(Zeiss AG; magnification, ×40) for 30 min at
room temperature.
Cell cycle detection
First, cells were seeded into a 6-well plate at a
density of 1×105 cells per well and treated with 6-MV
X-ray radiation at doses of 0 or 8 Gy. Then, the cells were
collected and stained with PI/RNase Staining Buffer (BD
Biosciences) for 20 min at room temperature, according to the
manufacturer's protocol. The cell cycle was detected using flow
cytometry (FACSCalibur; BD Biosciences) and interpreted using
FlowJo software (V10; FlowJo LLC). The experiment was repeated
three times.
Western blotting
Total protein was separated from cell lysates on ice
using RIPA buffer (Beyotime Institute of Biotechnology). An equal
amount of protein (40 µg), whose concentration was quantified by a
BCA kit (Beyotime Institute of Biotechnology), was separated on a
10% SDS-polyacrylamide gel and then transferred to PVDF membranes.
Afterwards, the membranes were blocked with 5% skimmed milk powder
for 2 h at room temperature, followed by incubation with primary
antibodies in dilution buffer at 4°C overnight. The next day,
HRP-linked anti-rabbit secondary antibodies (1:3,000; cat. no.
7074; Cell Signaling Technology, Inc.) were incubated at room
temperature for 2 h. Western blotting analysis was conducted using
a rabbit anti-CDCA2 monoclonal antibody (1:1,000; cat. no. 14976;
Cell Signaling Technology, Inc.). An anti-GAPDH polyclonal antibody
(1:5,000; cat. no. 10494-1-AP; ProteinTech Group, Inc.) was used as
a loading control. The signals were visualized via an enhanced
chemiluminescence detection kit (Thermo Fisher Scientific, Inc.)
and a chemiluminescence detection system.
Gene Set Enrichment Analysis
(GSEA)
GSEA was performed using the Kyoto Encyclopaedia of
Genes and Genomes (KEGG) pathway gene sets in the Molecular
Signatures Database against two probe-level expression matrices via
the GSEA v3.0 software (www.broadinstitute.org/gsea).
Xenograft tumours in nude mice
The present study was approved by the Institutional
Animal Care and Use Committee of Nanjing Medical University
(approval no. 2103063). In total, 24 male BALB/c nude mice (17-20
g; 4-6 weeks old) were obtained from the Nanjing Medical University
Animal Center and raised in a specific pathogen-free environment
under a 12-h light-dark cycle at 23±1°C and 50±5% humidity
atmosphere. The mice were divided into four groups (n=6): i) shNC;
ii) shCDCA2; iii) shNC and irradiation; and iv) shCDCA2 and
irradiation. A total of 2×106 KYSE450 cells were
subcutaneously implanted into the flanks of mice. At ~20 days post
injection, mice were exposed to irradiation (8 Gy) once in an RS
2000 Pro XRay Bioirradiator (Radsource) and then sacrificed at day
35. The lead shields were used to protect nontumor tissue from
radiation damage. The mice were sacrificed with an intraperitoneal
injection of 1% pentobarbital sodium at 100 mg/kg. The criteria for
death were sustained non-spontaneous breathing for 2-3 min and no
blink reflex.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Differences between groups were analysed using unpaired
Student's t-test or one-way and mixed design ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using GraphPad Prism software (version 5.0; GraphPad
Software, Inc.). All experiments were repeated at least three
times.
Results
CDCA2 is upregulated in human ESCC
tissues and cell lines
First, ESCC-related gene expression profiles of
tumours and normal tissues were analysed using information
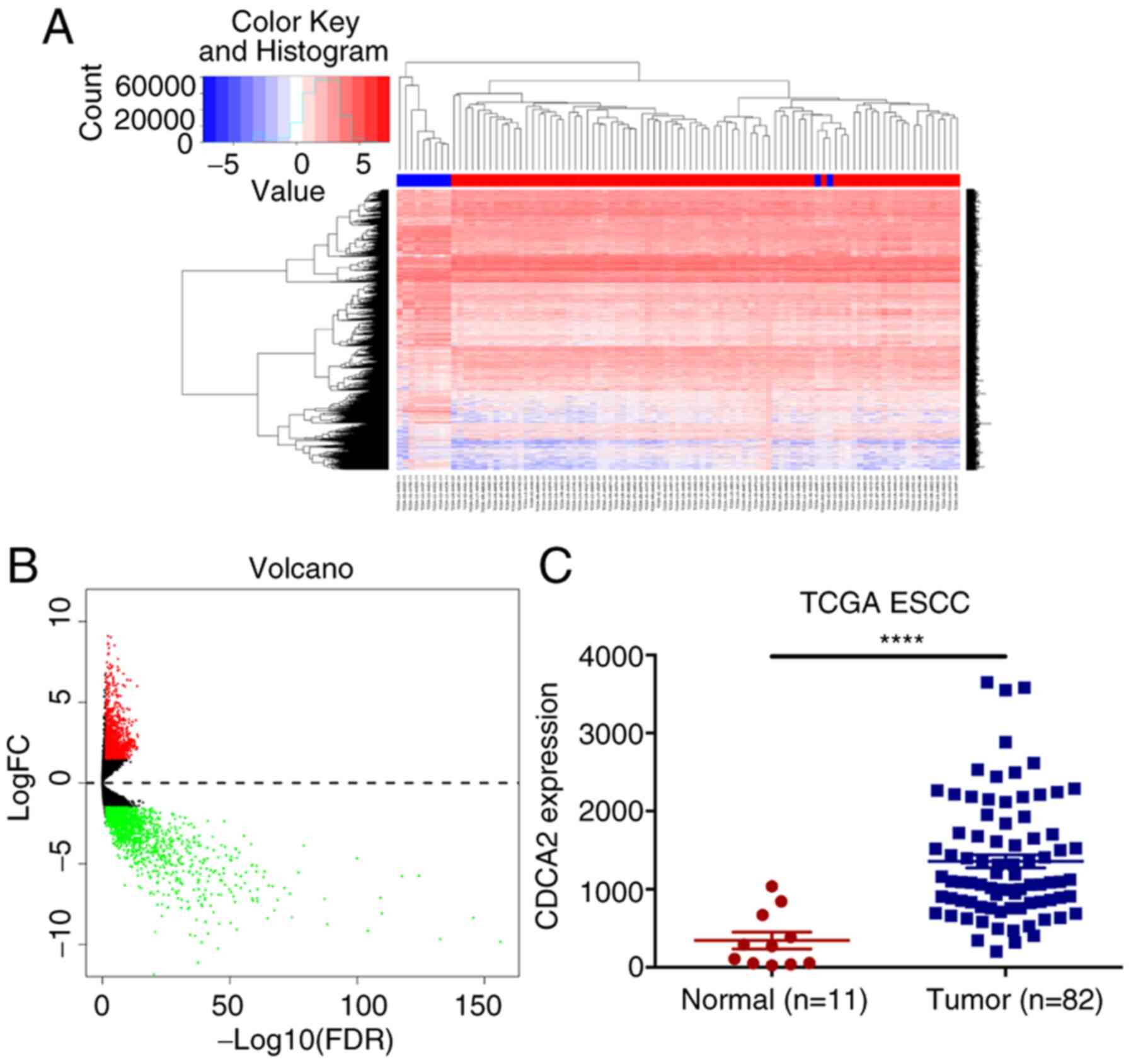
extracted from TCGA database (Fig. 1A
and B). The results showed that, compared with the expression
in normal tissues, the expression of CDCA2 in tumour samples was
significantly upregulated (Fig.
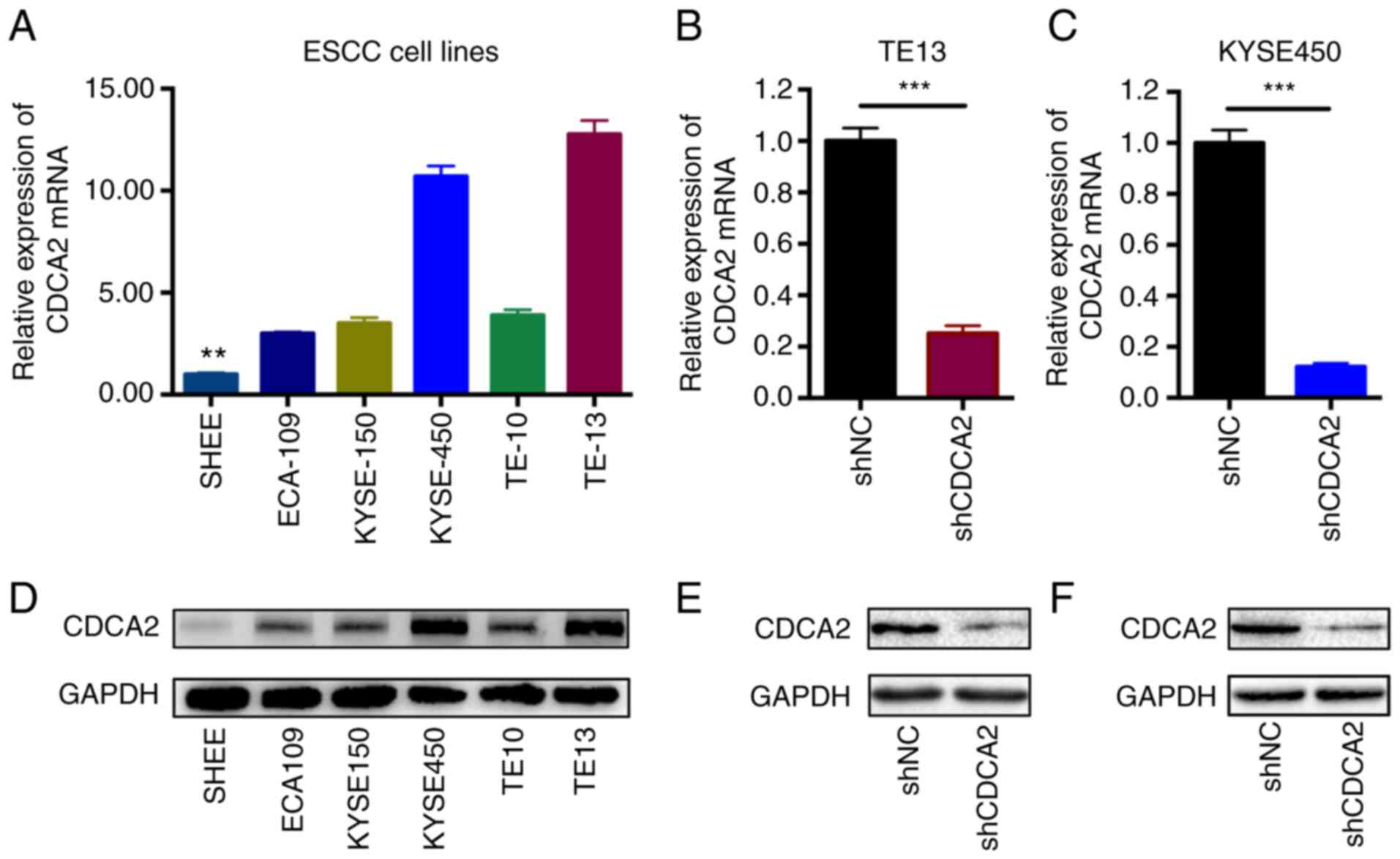
1C). Afterwards, the CDCA2 expression levels were detected by
RT-qPCR and western blot analysis in ESCC cell lines (Fig. 2A and D). As the results showed that
CDCA2 was highly expressed in ESCC cells, especially in KYSE450 and
TE13 cell lines, KYSE450 and TE13 cells were ultimately selected
for further experiments. According to these results, it was
hypothesized that CDCA2 might act as a regulator of tumour
biological behaviour in ESCC. Since the function and mechanism of
CDCA2 in the proliferation and radioresistance of ESCC cells remain
unclear, it was intended to conduct related experiments.
To determine the important roles of CDCA2 in
contributing to ESCC radioresistance, stable CDCA2 knockdown cell
lines of both KYSE450 and TE13 cells were successfully generated by
utilizing a specific shRNA against CDCA2. Then, RT-qPCR was
performed to check the downregulation of CDCA2 in the two cell
lines (Fig. 2B and C). Western blot
analysis was also conducted to confirm the results (Fig. 2E and F).
CDCA2 promotes the proliferation of
ESCC cells
CCK-8 assays were conducted to explore the influence
of CDCA2 knockdown on ESCC proliferation. The proliferation rates
of the cells in which the expression of CDCA2 was downregulated
were significantly lower than those of the control groups (Fig. 3A and B). The results indicated that
CDCA2 could regulate tumour growth and it was hypothesized that it
might induce radioresistance in ESCC. The role of CDCA2 in cell
proliferation was also verified via a more sensitive and specific
EdU assay. As demonstrated in Fig. 3C
and D, CDCA2 depletion in KYSE450 cells reduced the number of
EdU-positive cells. Similar results were observed in TE13 cells
(Fig. 3E and F). According to these
results, the downregulation of CDCA2 could effectively inhibit ESCC
cell proliferation.
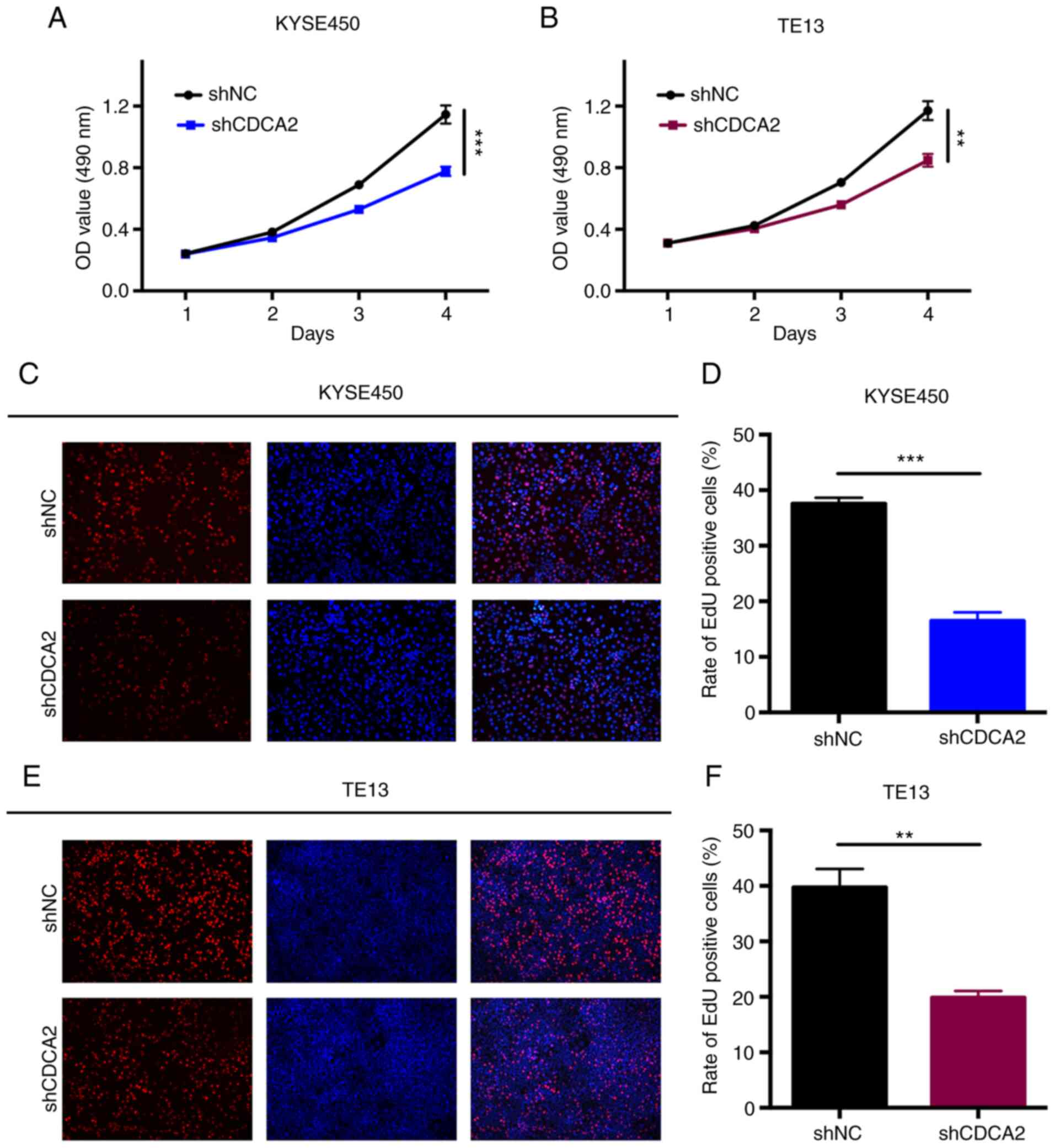 | Figure 3.The knockdown of CDCA2 suppresses
cell proliferation. CCK-8 assays were conducted to measure the rate
of ESCC cell proliferation in (A) KYSE450/shCDCA2 and (B)
TE13/shCDCA2 cells compared to that of the control groups. Mean ±
standard deviation, n=3, **P<0.01, ***P<0.001. Typical images
of the EdU incorporation assays and mean percentage of EdU positive
cells in (C and D) KYSE450 cells and (E and F) TE13 cells with
CDCA2 knockdown compared with the controls (blue fluorescence, cell
nuclei; red fluorescence, EdU-positive cells) (magnification, ×40).
Mean ± standard deviation, n=3, **P<0.01, ***P<0.001. CDCA2,
cell division cycle-associated 2; ESCC, oesophageal squamous cell
carcinoma; sh, short hairpin; OD, optical density; sh, short
hairpin; NC, negative control; EdU, 5-Ethynyl-2′-deoxyuridine. |
CDCA2 regulates the radiosensitivity
of ESCC cells
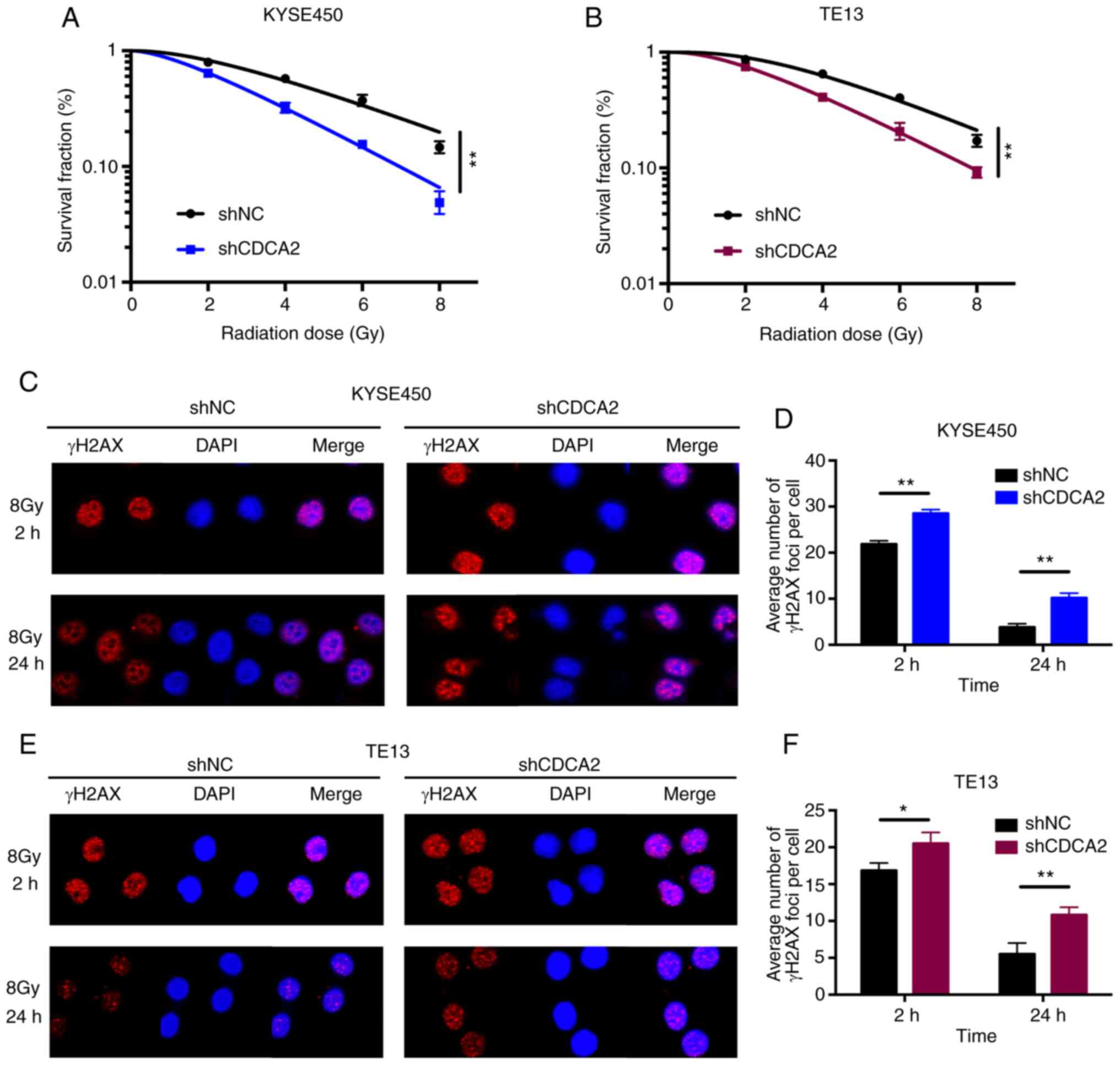
The role of CDCA2 in the radioresistance of ESCC was
further determine. First, clonogenic survival assays were performed
to assess the influence of CDCA2 on radiosensitivity. As
demonstrated in Fig. 4A, the
surviving cell fraction of KYSE450 cells with CDCA2 downregulation
was distinctly lower compared with the control group, which was
exposed to X-ray radiation. TE13 cells with reduced CDCA2
expression showed a similar outcome (Fig. 4B).
Immunofluorescence detection of γ-H2AX foci in
KYSE450 and TE13 cell lines following X-ray radiation was used to
explore whether CDCA2 was able to regulate radiation-induced DNA
DSBs. As demonstrated in Fig. 4C-F,
there were increased γ-H2AX signals at different time points (2 and
24 h) after irradiation in CDCA2-depleted cells compared with the
control group. This suggested that downregulation of CDCA2 in ESCC
cells could induce more DNA DSBs during radiation exposure.
Collectively, downregulation of CDCA2 expression in oesophageal
cancer cells enhanced cell radiation sensitivity.
CDCA2 influences the cell cycle
distribution of ESCC cells
Previous studies have demonstrated that
G2/M arrest is a pivotal mechanism for regulating
radioresistance in ESCC (18–21).
To explore whether CDCA2 could influence radiosensitivity in ESCC
cells by regulating cell cycle distribution, every phase percentage
with CDCA2 knockdown after radiation was quantified (Fig. 5A-D). When exposed to X-ray radiation
(8 Gy), the CDCA2-downregulated cells had a higher G2/M
phase percentage than the controls in KYSE450 and TE13 cell lines
(Fig. 5E and F).
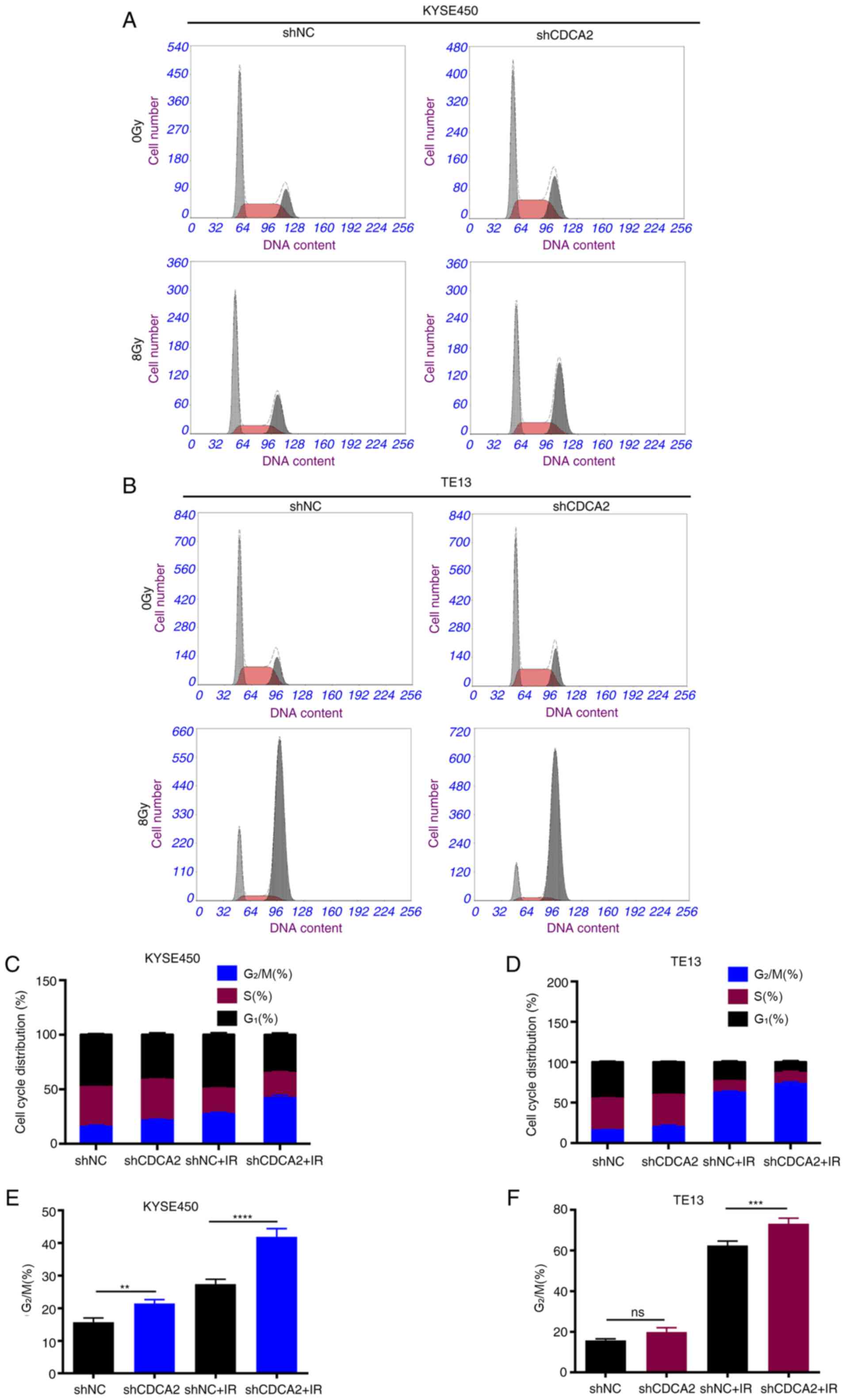 | Figure 5.Inhibition of CDCA2 changes the cell
cycle distribution in X ray-exposed ESCC cells. Cell cycle
distribution of (A) KYSE450/shCDCA2 and (B) TE13/shCDCA2 cells. (C
and D) Phase percentage with CDCA2 knockdown following radiation.
(E and F) Downregulation of CDCA2 had a higher G2/M
phase percentage. Mean ± standard deviation, n=3, **P<0.01,
***P<0.001, ****P<0.0001. CDCA2, cell division
cycle-associated 2; ESCC, oesophageal squamous cell carcinoma; sh,
short hairpin; NC, negative control; IR, ionising radiation; ns,
not significant. |
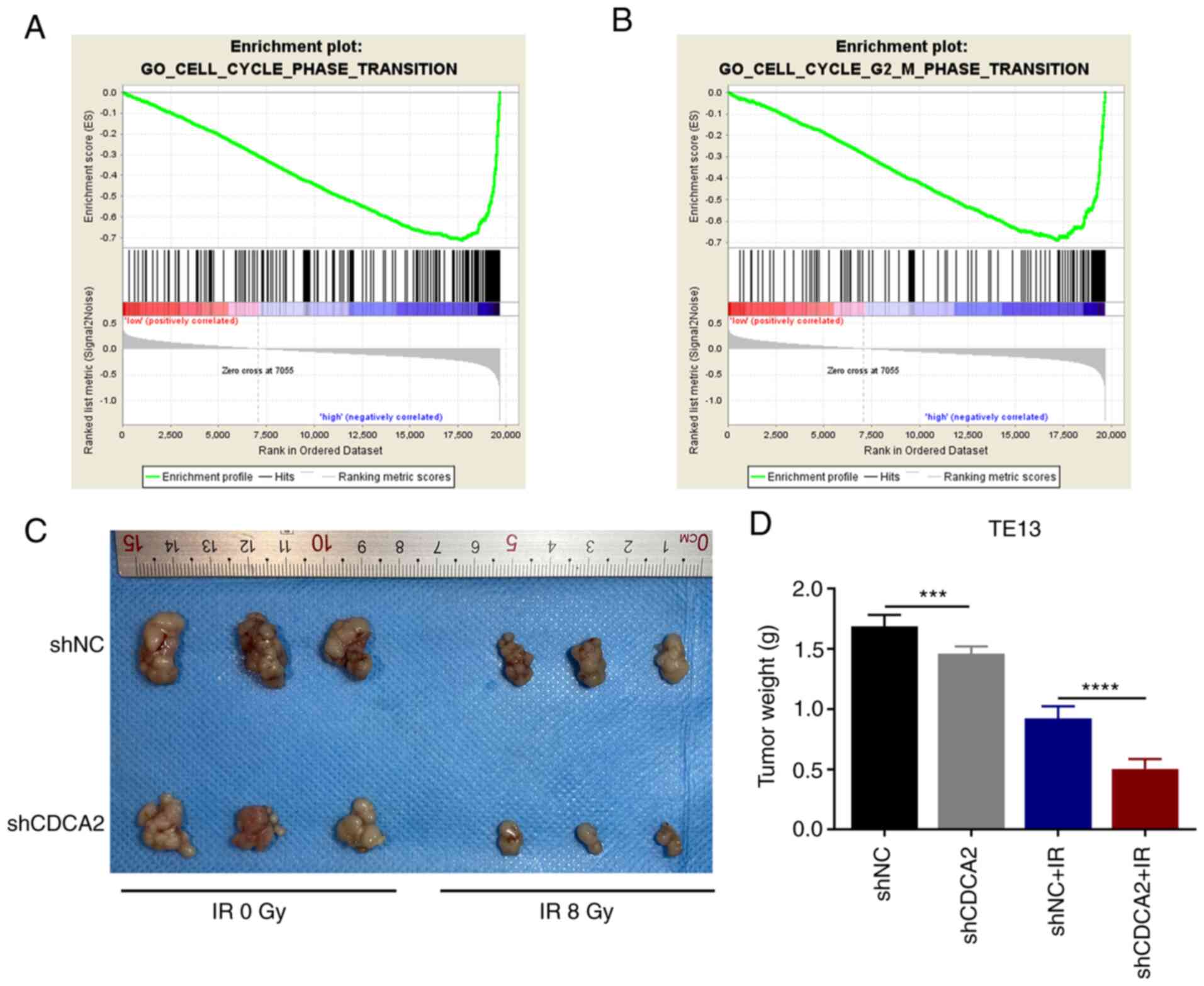
To study the possible link between CDCA2 and the
cell cycle phase transition pathway, GSEA was performed using the
KEGG pathway gene sets in the Molecular Signatures Database against
two probe-level expression matrices via the GSEA v3.0 software
(www.broadinstitute.org/gsea). It was
found that higher expression of CDCA2 was significantly associated
with the expression of related components of the cell cycle phase
transition and G2/M phase transition pathways (Fig. 6A and B). Collectively, the results
revealed that CDCA2 could regulate G2/M phase arrest to
further influence radioresistance in ESCC cells.
CDCA2 influences ESCC cell
proliferation and radiosensitivity in vivo
To confirm the effects of CDCA2 on ESCC cell
proliferation in vivo, BALB/c nude mice were injected with
TE13 cells transfected with shNC and shCDCA2. The tumour weight of
shCDCA2 group was significantly lower compared with control group.
This effect was evident after exposure to radiation (Fig. 6C and D) and markedly evident
following exposure to radiation. The in vivo results were
consistent with the in vitro results. This further confirmed
that downregulation of CDCA in ESCC cells can inhibit cell
proliferation and improve tumour radiation sensitivity.
Discussion
Numerous studies have demonstrated that the
differences in the expression of various genes between cancer and
adjacent normal tissues led to innovations in diagnostic techniques
and chemotherapy treatment strategies for cancer (22,23).
However, RT is regarded as the standard treatment modality in
patients with ESCC who cannot be radically resected or those who
refuse surgery (i.e., not fit for surgery) (4,7,8). RT
serves a vital role in the local control of ESCC4 (1). However, distant metastasis and local
recurrence frequently occur, thus causing RT resistance (1,9).
Therefore, it is necessary to carry out genetic studies of
oesophageal cancer. The expression levels of all known genes in
oesophageal cancer and adjacent tissues were analysed in TCGA
database. The present study confirmed that the expression of CDCA2
was higher in ESCC tissues compared with normal tissues. Several
studies have demonstrated that CDCA2, as a cell division
cycle-associated protein, is associated with tumour occurrence,
progression and proliferation in several types of cancer, including
melanoma, colorectal cancer, neuroblastoma, squamous cell carcinoma
and others (11–13,24,25).
Ionizing radiation affects cell proliferation through the cell
cycle and cells at different stages of the cycle exhibit different
radiation sensitivities (26,27).
Hence, the present study explored the association between CDCA2 and
radioresistance. To date, no reports, to the best of the authors'
knowledge have examined the role of CDCA2 in ESCC radioresistance
to induce radiosensitivity.
The in vitro experiments validated the
abovementioned hypothesis. CDCA2 knockdown decreased ESCC cell
proliferation. Dose-dependent clone formation was used to explore
whether CDCA2 can affect radiosensitivity and the downregulation of
CDCA2 inhibited the formation of cloning. These results revealed
the action of CDCA2 in the regulation of ESCC radiosensitivity. DNA
is the main target of ionizing radiation (28). The efficacy of RT depends on its
ability to induce DNA damage in cancer cells (29). The ability of a cell to repair DNA
damage caused by radiation will finally affect whether it could
succumb to cell death (30). The
proficiency of DNA damage repair and DNA repair processes is
associated with tumour resistance to ionizing radiation (29). Inhibition of DNA damage repair can
cause cell cycle arrest or programmed cell death (31). Histone H2AX is phosphorylated near
DNA double-strand breaks and γH2AX can be used as a marker for DNA
DSBs in chromatin and can be used to assess radiosensitivity
(32). γH2AX phosphorylation foci
were significantly higher in shCDCA2 cells at 2 and 24 h after
radiation compared with the control groups. The persistence of
γH2AX lesions has previously been demonstrated to be associated
with radiosensitivity and the loss of lesions depends on effective
DNA repair. As DNA is the main target of ionizing radiation and
that DNA DSBs are a key lesion that causes cell death, CDCA2
knockdown can increase ESCC sensitivity to X-ray radiation
(31,33).
Generally, cells are sensitive to radiation-induced
DNA damage during G2/M and G1/S, while cells
in the late S phase are most resistant to ionizing radiation
(34). G2/M phase arrest
is the most sensitive stage of cell damage. After cells were
exposed to X-ray irradiation, CDCA2 knockdown increased
radiation-induced G2/M phase arrest, as demonstrated by
flow cytometry analysis.
Previous reports have demonstrated that
downregulation of CDCA2 can induce G1 arrest of lung
adenocarcinoma cells and oral squamous cell carcinoma cells
(12,13). In the present study, CDCA2 showed a
regulatory effect on the cycle distribution of oesophageal squamous
cell carcinoma cells, however, it was the G2/M phase
rather than the G1 phase. Radiation can increase the
distribution in the G2/M phase and the present study
showed that radiation combined with downregulation of CDCA2 can
aggravate this effect. The reason may be that CDCA2 did not serve
the G1-arresting function in these two cell lines, or
the sample size was too small. The results of the present study did
not contradict previous studies. To explore the relationship
between CDCA2 and the cell cycle, GSEA was also conducted using the
data of the ESCC cohorts from the TCGA database. Please provide a
reference for this statement, remembering to update the reference
list and in-text citations accordingly. The results of the present
study revealed that CDCA2 could regulate the cell cycle
distribution to further influence radioresistance in ESCC cells.
However, the findings and mechanisms in the present study should be
further tested in animal models and patient samples.
In conclusion, the present study found that
silencing CDCA2 could suppress the growth and enhance the
radiosensitivity of ESCC cells. CDCA2 is a potential molecular
target of radiosensitization.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China [grant no. 81672983
(BA16)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BX, HuC, HoC and XS conceived the study, BX and HuC
participated in data collection, analysis and interpretation, and
drafted the manuscript. ZX and XY contributed to collecting samples
and materials, and analyzing data. BX, HoC, ZX and XY participated
in the analysis and interpretation of the results. All authors read
and approved the final manuscript, and consented to publish this
manuscript. XS and HYC confirmed the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care and Use Committee of Nanjing Medical University
(approval no. 2103063) and was in accordance with China's National
Code of the Animal Care for Scientific Experimentation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RJ, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Pineros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Le Bras GF, Farooq MH, Falk GW and Andl
CD: Esophageal cancer: The latest on chemoprevention and state of
the art therapies. Pharmacol Res. 113A:A236–A244. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao J, He YT, Zheng RS, Zhang SW and Chen
WQ: Analysis of esophageal cancer time trends in China, 1989-2008.
Asian Pac J Cancer Prev. 13:4613–4617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang H, Fan JH and Qiao YL: Epidemiology,
etiology, and prevention of esophageal squamous cell carcinoma in
China. Cancer Biol Med. 14:33–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Rossum PSN, Mohammad NH, Vleggaar FP
and van Hillegersberg R: Treatment for unresectable or metastatic
oesophageal cancer: Current evidence and trends. Nat Rev
Gastroenterol Hepatol. 15:235–249. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dai T and Shah MA: Chemoradiation in
oesophageal cancer. Best Pract Res Clin Gastroenterol. 29:193–209.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prevost M, Chamousset D, Nasa I, Freele E,
Morrice N, Moorhead G and Trinkle-Mulcahy L: Quantitative
fragmentome mapping reveals novel, domain-specific partners for the
modular protein RepoMan (recruits PP1 onto mitotic chromatin at
anaphase). Mol Cell Proteomics. 12:1468–1486. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng Y, Qian W, Zhang Y, Peng W, Li J, Gu
Q, Ji D, Zhang Z, Wang Q, Zhang D and Sun Y: CDCA2 promotes the
proliferation of colorectal cancer cells by activating the
AKT/CCND1 pathway in vitro and in vivo. BMC Cancer. 19:5762019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uchida F, Uzawa K, Kasamatsu A, Takatori
H, Sakamoto Y, Ogawara K, Shiiba M, Bukawa H and Tanzawa H:
Overexpression of CDCA2 in human squamous cell carcinoma:
Correlation with prevention of G1 phase arrest and apoptosis. PLoS
One. 8:e563812013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi R, Zhang C, Wu Y, Wang X, Sun Q, Sun
J, Xia W, Dong G, Wang A, Jiang F and Xu L: CDCA2 promotes lung
adenocarcinoma cell proliferation and predicts poor survival in
lung adenocarcinoma patients. Oncotarget. 8:19768–19779. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng A, Lewellyn AL, Schiemann WP and
Maller JL: Repo-man controls a protein phosphatase 1-dependent
threshold for DNA damage checkpoint activation. Curr Biol.
20:387–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vagnarelli P: Repo-man at the intersection
of chromatin remodelling, DNA repair, nuclear envelope
organization, and cancer progression. Adv Exp Med Biol.
773:401–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Yi J, Tao L, Huang G, Chu X, Song
H and Chen L: Wnt signaling induces radioresistance through
upregulating HMGB1 in esophageal squamous cell carcinoma. Cell
Death Dis. 9:4332018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dillon MT, Good JS and Harrington KJ:
Selective targeting of the G2/M cell cycle checkpoint to improve
the therapeutic index of radiotherapy. Clin Oncol (R Coll Radiol).
26:257–265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sinclair WK and Morton RA: X-ray
sensitivity during the cell generation cycle of cultured Chinese
hamster cells. Radiat Res. 29:450–474. 1966. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Terasima T and Tolmach LJ: Variations in
several responses of HeLa cells to x-irradiation during the
division cycle. Biophys J. 3:11–33. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng N, Zhang S, Wu W, Zhang N and Wang
J: Regulatory mechanisms and therapeutic targeting of vasculogenic
mimicry in hepatocellular carcinoma. Pharmacol Res. 166:1055072021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Petersen EV, Chudakova DA, Skorova EY,
Anikin V, Reshetov IV and Mynbaev OA: The extracellular
matrix-derived biomarkers for diagnosis, prognosis, and
personalized therapy of malignant tumors. Front Oncol.
10:5755692020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krasnoselsky AL, Whiteford CC, Wei JS,
Bilke S, Westermann F, Chen QR and Khan J: Altered expression of
cell cycle genes distinguishes aggressive neuroblastoma. Oncogene.
24:1533–1541. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao B, Chen L, Ke Y, Hang J, Cao L, Zhang
R, Zhang W, Liao Y, Gao Y, Chen J, et al: Identification of
methylation sites and signature genes with prognostic value for
luminal breast cancer. BMC Cancer. 18:4052018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang L, Graham PH, Hao J, Ni J, Bucci J,
Cozzi PJ, Kearsley JH and Li Y: PI3K/Akt/mTOR pathway inhibitors
enhance radiosensitivity in radioresistant prostate cancer cells
through inducing apoptosis, reducing autophagy, suppressing NHEJ
and HR repair pathways. Cell Death Dis. 5:e14372014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He G, Di X, Yan J, Zhu C, Sun X and Zhang
S: Silencing human epidermal growth factor receptor-3
radiosensitizes human luminal A breast cancer cells. Cancer Sci.
109:3774–3782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buckley AM, Lynam-Lennon N, O'Neill H and
O'Sullivan J: Targeting hallmarks of cancer to enhance
radiosensitivity in gastrointestinal cancers. Nat Rev Gastroenterol
Hepatol. 17:298–313. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sadoughi F, Mirsafaei L, Dana PM,
Hallajzadeh J, Asemi Z, Mansournia MA, Montazer M, Hosseinpour M
and Yousefi B: The role of DNA damage response in chemo- and
radio-resistance of cancer cells: Can DDR inhibitors sole the
problem? DNA Repair (Amst). 101:1030742021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lynam-Lennon N, Reynolds JV, Pidgeon GP,
Lysaght J, Marignol L and Maher SG: Alterations in DNA repair
efficiency are involved in the radioresistance of esophageal
adenocarcinoma. Radiat Res. 174:703–711. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu C, Gross N, Li Y, Li G, Wang Z, Zhong
S, Li Y and Hu G: PARP inhibitor Olaparib increases the
sensitization to radiotherapy in FaDu cells. J Cell Mol Med.
24:2444–2450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sak A and Stuschke M: Use of gammaH2AX and
other biomarkers of double-strand breaks during radiotherapy. Semin
Radiat Oncol. 20:223–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bouzinab K, Summers HS, Stevens MFG, Moody
CJ, Thomas NR, Gershkovich P, Weston N, Ashford MB, Bradshaw TD and
Turyanska L: Delivery of temozolomide and N3-propargyl analog to
brain tumors using an apoferritin nanocage. ACS Appl Mater
Interfaces. 12:12609–12617. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maity A, McKenna WG and Muschel RJ: The
molecular basis for cell cycle delays following ionizing radiation:
A review. Radiother Oncol. 31:1–13. 1994. View Article : Google Scholar : PubMed/NCBI
|