Introduction
Hepatocellular carcinoma (HCC) is the fourth major
cause of cancer mortality worldwide and the global morbidity of the
disease is increasing (1). Surgical
resection is the most effective treatment method of all therapy
methods; however, most patients are initially diagnosed as advanced
and untreatable by surgery (2).
High rates of postoperative recurrence and metastasis are also
reasons for the high mortality rate of HCC (3). Although great progress in the therapy
methods has been made in the last few decades, the survival rate of
patients with HCC remains poor (4).
Hence, the search for new effective therapeutic targets is of great
significance for HCC treatment.
In recent years, an increasing numbers of studies
have focused on the effect of lncRNAs on the occurrence and
development of HCC (5–7). Research has shown that aberrant
expression of long non-coding RNAs (lncRNAs) decelerates or
accelerates the progression of HCC. For instance, a study by Liu
et al (8) demonstrates that
lncRNA Ftx can inhibit proliferation and metastasis of HCC and that
higher lncRNA Ftx expression is associated with longer survival.
Furthermore, long intergenic non-protein coding (LINC)RNA-p21
overexpression reduces the levels of epithelial-mesenchymal
transition (EMT)-related proteins and decrease tumor metastasis in
HCC (7). The findings of Wang et
al (9) indicate that high
lncRNA ATB expression promotes proliferation of HCC and is
positively correlated with poorer survival of patients with HCC.
Gong et al (6) found that
LINC00238 is expressed at low levels in HCC samples. However, the
regulatory mechanism of LINC00238 in HCC remains to be
elucidated.
The transmembrane protein (TMEM) family is a class
of proteins containing one or more putative transmembrane segments
that span partially or completely through biological membranes
(10). As a member of the TMEM
family, TMEM106 is involved in several diseases, including cancer.
In a study on non-small cell lung carcinoma, Liu and Zhu (11) found that TMEM106A is an
anti-oncogene that inhibits the proliferation and invasion of lung
cancer cells. Lang et al (12) note that abnormal expression of
TMEM106B is associated with progression of frontotemporal lobar
degeneration. In addition, Assassi et al (13) found that TMEM106C is aberrantly
expressed in ankylosing spondylitis. Luo et al (14) have indicate that TMEM106C is
involved in HCC development. However, remains to be elucidated
whether lncRNAs are involved in the regulation of TMEM106C in HCC
progression.
The present study explored the regulatory role of
LINC00238 in HCC progression. The findings suggested that LINC00238
expression was decreased in HCC tissues and cell lines and the low
LINC00238 expression was associated with the poor prognosis of
patients with HCC. LINC00238 inhibited HCC progression by
regulating the TMEM106C-mediated apoptosis signaling pathway.
Furthermore, ATF3, the gene enriched in the apoptosis pathway,
could regulate LINC00238 expression, consequently preventing HCC
development.
Materials and methods
Bioinformatics analysis
The online platform Gene Expression Profiling
Interactive Analysis (GEPIA) (15)
was used to analyze the differentially expressed lncRNAs in HCC. A
total of 369 HCC cases and 160 normal were downloaded from The
Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The expression of
LINC00238, TMEM106C and ATF3 in HCC tumor tissues and normal
tissues were analyzed based on the downloaded data. The overall
survival information in this study was downloaded from TCGA
database. Survival analysis was performed with the R package
‘survival’ (16,17) by using the Kaplan-Meier curve method
and TANRIC platform. The binding sites between LINC00238 and
TMEM106C were predicted by LncTar (www.cuilab.cn/lnctar). Gene Set Enrichment Analysis
(GSEA) (18) was performed using
three gene set datasets [GO-BP (http://www.geneontology.org), KEGG (http://www.genome.jp/kegg/) and Hallmark gene sets
(19)] to analyze the impact of
TMEM106C on signaling pathways. The JASPAR (http:///jaspar.genereg.net/) and PROMO (alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)
databases were used to analyze the upstream transcription factor of
LINC00238.
Patients and samples
A total of 43 human HCC tissues and corresponding
normal tissues were collected from 22 male (age range, 32–64 years)
and 21 female (age range, 34–70 years) patients with HCC between
January 2018 and September 2019. None of the patients received any
treatment before surgical resection, including surgery,
radiotherapy and chemotherapy. The study was approved by the ethics
committee of Qingdao No. 6 People's Hospital [approval no. (2018)
26] and written informed consent was obtained from all
patients.
Cell lines and culture
The human normal hepatic cell line MIHA and HCC cell
lines Huh7, HLF, SNU-449, MHCC-97h and HCCLM3 were purchased from
Mlbio and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing antibiotics (Gibco; Thermo Fisher Scientific, Inc.) and
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), in a humidified
atmosphere containing 5% CO2 at 37°C.
Cell transfection
LINC00238-overexpressing plasmid pcDNA3.1
(pc-LINC00238), negative control plasmid (pc-NC), short interfering
(si)RNAs targeting LINC00238 (30 nM, si-LINC00238#1,
5′-GAGGATACTTAGGAGAGCTAT-3′; si-LINC00238#2,
5′-GGAGAGCTATGTATCCTATTT-3′; and si-LINC00238#3,
5′-GGCTGGAAGTGTTCAGTTAGT-3′) and negative control siRNA (30 nM,
si-NC, 5′-GAGGTATGGTATTCGGTCTGA-3′), TMEM106C-overexpressing
plasmid pcDNA3.1 (pc-TMEM106C) negative control plasmid (pc-NC),
siRNAs targeting ATF3 (30 nM, si-ATF3#1,
5′-GAGCAGTTCGGTGCATATGGT-3′; si-ATF3#2,
5′-GGGTTACTGGCAGGTTGAACT-3′; and si-ATF3#3,
5′-GGGAAACAGTTGAGAGGTTAT-3′) and negative control siRNA (30 nM,
si-NC, 5′-GCTTGGAAGGTCGGTCATGTA-3′), ATF3-overexpressing plasmid
pcDNA3.1 (pc-ATF3) negative control plasmid (pc-NC) were purchased
from Shanghai GenePharma Co., Ltd. The transfection was performed
by Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
cells were collected after transfection for 48 h.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from tissues and cells
(1×106) using TRIzol reagent (Thermo Fisher Scientific,
Inc.). The PrimeScript RT reagent kit (Takara Bio, Inc.) was used
to synthesize cDNA. qPCR was carried out using SYBR Premix Ex Taq
II kit (Takara Bio, Inc.) according to the manufacturer's protocol.
The reaction conditions were as follows: 95°C for 5 min, followed
by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. GAPDH was used
as the internal control for mRNA expression. Relative gene
expression levels were calculated using the 2−ΔΔCq
method (20). All experiments were
performed in triplicate. The primers used in the study were:
LINC00238: 5′-TTTGCAGAGTGGGTGCTAGG-3′ (forward) and
5′-TGCTTCATCTGGCAATGACCT-3′ (reverse); Transmembrane Protein 106C
(TMEM106C): 5′-GCCTGTCCAGCCAGATTCAG-3′ (forward) and
5′-TTCCTACAGCCCCCTACTCT-3′ (reverse); activating transcription
factor 3 (ATF3): 5′-AGGTTTGCCATCCAGAACAA-3′ (forward) and
5′-AGGCACTCCGTCTTCTCCTT-3′ (reverse); GAPDH:
5′-CTGGGCTACACTGAGCACC-3′ (forward) 5′-AGTGGTCGTTGAGGGCAATG-3′
(reverse).
Cell proliferation assay
HCC cells were trypsinized and seeded into 96-well
culture plates (4,000 cells/well). The cells were harvested at 24,
48, 72 and 96 h to detect proliferation using the
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) Cell Proliferation Colorimetric Assay kit (AmyJet Scientific,
Inc.) and the absorption was detected at a wavelength of 490
nm.
Cell apoptosis assay
The apoptosis of HCC cells was evaluated using
Annexin V/PI apoptosis-detection kit (Invitrogen; Thermo Fisher
Scientific, Inc.). HCC cells were washed with Annexin V-binding
buffer (Sigma-Aldrich; Merck KGaA), followed by incubation in
binding buffer containing Annexin V-FITC (25 µg/ml) and PI (25
µg/ml) in the dark for 10 min at room temperature. Apoptotic cells
(early + late apoptotic cells) were analyzed using a BD Accuri C6
Plus flow cytometer and FACSDiva software (version 6.13; BD
Biosciences).
Wound healing assay
HCC cells were seeded into 6-well plates and
incubated to achieve ≥90% confluence. The monolayer cells were
scratched with a 10-µl sterile micropipette tip and cellular debris
was removed by washing with PBS. Then, cells were incubated in
serum-free medium with proliferation inhibitor mitomycin C (10 µM;
MedChemExpress) at 37°C. Cell migration was observed at 0 and 48 h
under a light microscope (magnification, ×100, Olympus Corporation)
and analyzed with ImageJ version 1.49 software (NIH). All
experiments were performed in triplicate.
Transwell assay
The invasion of HCC cells was detected by Transwell
assay. The upper chambers (8 µm pore size) were coated with
Matrigel at 37°C for 30 min. Cells (1×104 cells/well)
were seeded into the upper chamber with serum-free DMEM. DMEM
containing 10% FBS was added to the lower chambers. The chambers
were incubated for 48 h at 37°C. Subsequently, the noninvasive
cells were removed with a cotton swab and the invading cells were
fixed with methanol. The cells were then stained with crystal
violet at room temperature for 15 min. The number of invasive cells
was observed under a light microscope (magnification, ×200; Olympus
Corporation).
Western blotting
HCC cells (1×106) were lysed with RIPA
buffer (Beyotime, Shanghai, China) to obtain total protein,
centrifuged for 10 min at 14,000 × g and the supernatant was
collected. The protein concentration was determined using a BCA kit
(Beijing Solarbio Science & Technology Co., Ltd.).
Subsequently, the proteins (20 µg) were separated by 10% SDS-PAGE
and transferred onto PVDF membranes. The membranes were incubated
with 5% non-fat dry milk for 1 h at room temperature and then
incubated with primary antibodies against TMEM106C (1:500; cat. no.
PA5-67816; Thermo Fisher Scientific, Inc.), ATF3 (1:1,000; cat. no.
18665S; Cell Signaling Technology, Inc.), caspase-7 (CASP7)
(1:1,000; cat. no. 12827S; Cell Signaling Technology, Inc.), tissue
inhibitor of metalloproteinase 2 (TIMP2) (1:1,000; cat. no. 5738S;
Cell Signaling Technology, Inc.), programmed cell death 4 (PDCD4)
(1:1,000; cat. no. 9535S; Cell Signaling Technology, Inc.) and
GAPDH (1:1,000; cat. no. 5174S; Cell Signaling Technology, Inc.)
overnight at 4°C, followed by incubation with the Goat anti-Rabbit
IgG Secondary Antibody (1:5,000; cat. no. 31466; Thermo Fisher
Scientific, Inc.) at room temperature. GAPDH served as the internal
reference. The bands were visualized using enhanced
chemiluminescence (ECL) detection reagents (Amersham Biosciences;
Cytiva). Blots were semi-quantitatively analyzed using ImageJ
software (version 2.0; National Institutes of Health).
RNA immunoprecipitation (RIP)
assay
To explore the binding relationship between
LINC00238 and TMEM106C, the RIP assay was performed using Magna RIP
kit (EMD Millipore) according the manufacturer's protocols. HCC
cells were lysed with RIPA lysis buffer (EMD Millipore) and then
incubated with anti-TMEM106C-coated Dynabeads Protein G (cat. no.
PA5-61466; Thermo Fisher Scientific, Inc.) beads at 4°C. IgG (cat.
no. ab182931; Abcam) was used as the control. The beads were washed
with PBS and incubated with TRIzol (Thermo Fisher Scientific, Inc.)
for 30 min at 55°C. The levels of LINC00238 and TMEM106C were
measured using RT-qPCR.
Statistical analysis
Data are shown as means ± SD and were analyzed using
SPSS 23.0 (IBM Corp.) and GraphPad Prism 7.0 (GraphPad Software,
Inc.). Comparisons between two groups were analyzed using unpaired
Student's t-test and the comparisons among multiple groups were
analyzed by one-way ANOVA followed by Tukey's post hoc test.
Pearson's correlation analysis was used to assess the correlation
among LINC00238, TMEM106C and ATF3 expression. All experiments were
performed in triplicate.
Results
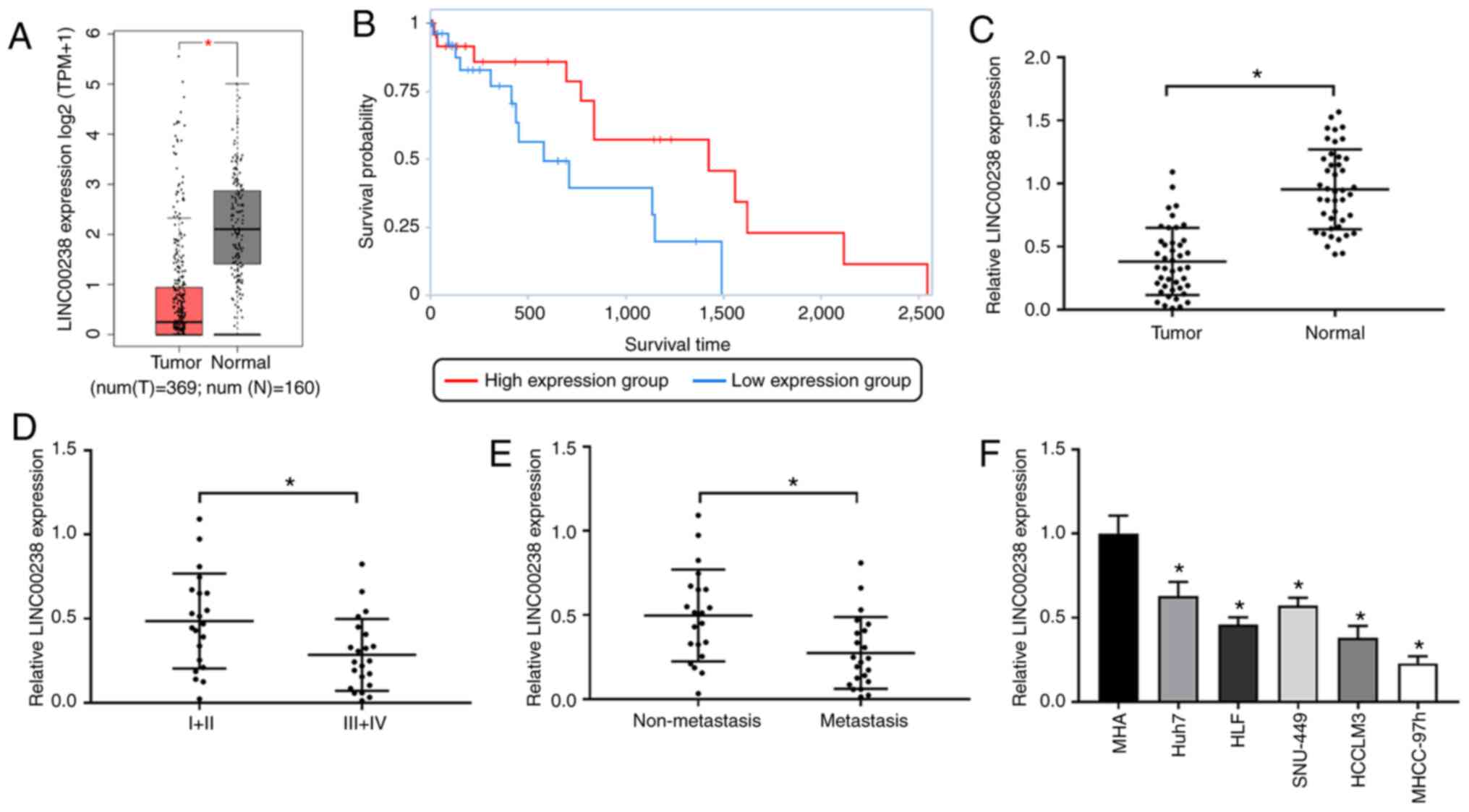
LINC00238 is downregulated in HCC
The online platform GEPIA was used to analyze the
differentially expressed lncRNAs in HCC and the results suggested
that LINC00238 was downregulated in HCC. The online analysis based
on TCGA suggested that LINC00238 levels in HCC tissues were
markedly lower compared with those in normal tissues (Fig. 1A) and that LINC00238 levels was
positively correlated with the overall survival of patients with
HCC (Fig. 1B). LINC00238 expression
was measured in HCC tissues and corresponding normal tissues using
RT-qPCR. The data showed that LINC00238 was significantly
downregulated in HCC tumor tissues (Fig. 1C). High levels of LINC00238
expression were associated with local TNM stage (I/II; Fig. 1D) and non-metastasis (Fig. 1E). LINC00238 expression in human
normal hepatic cell line MIHA and HCC cell lines (Huh7, HLF,
SNU-449, MHCC-97h and HCCLM3) was measured using RT-qPCR. The
results indicated that LINC00238 expression in HCC cell lines was
markedly decreased compared with that in the normal hepatic cell
line (Fig. 1F). HCCLM3 and MHCC-97h
cell lines were selected for further study because of the low
LINC00238 expression in these cells.
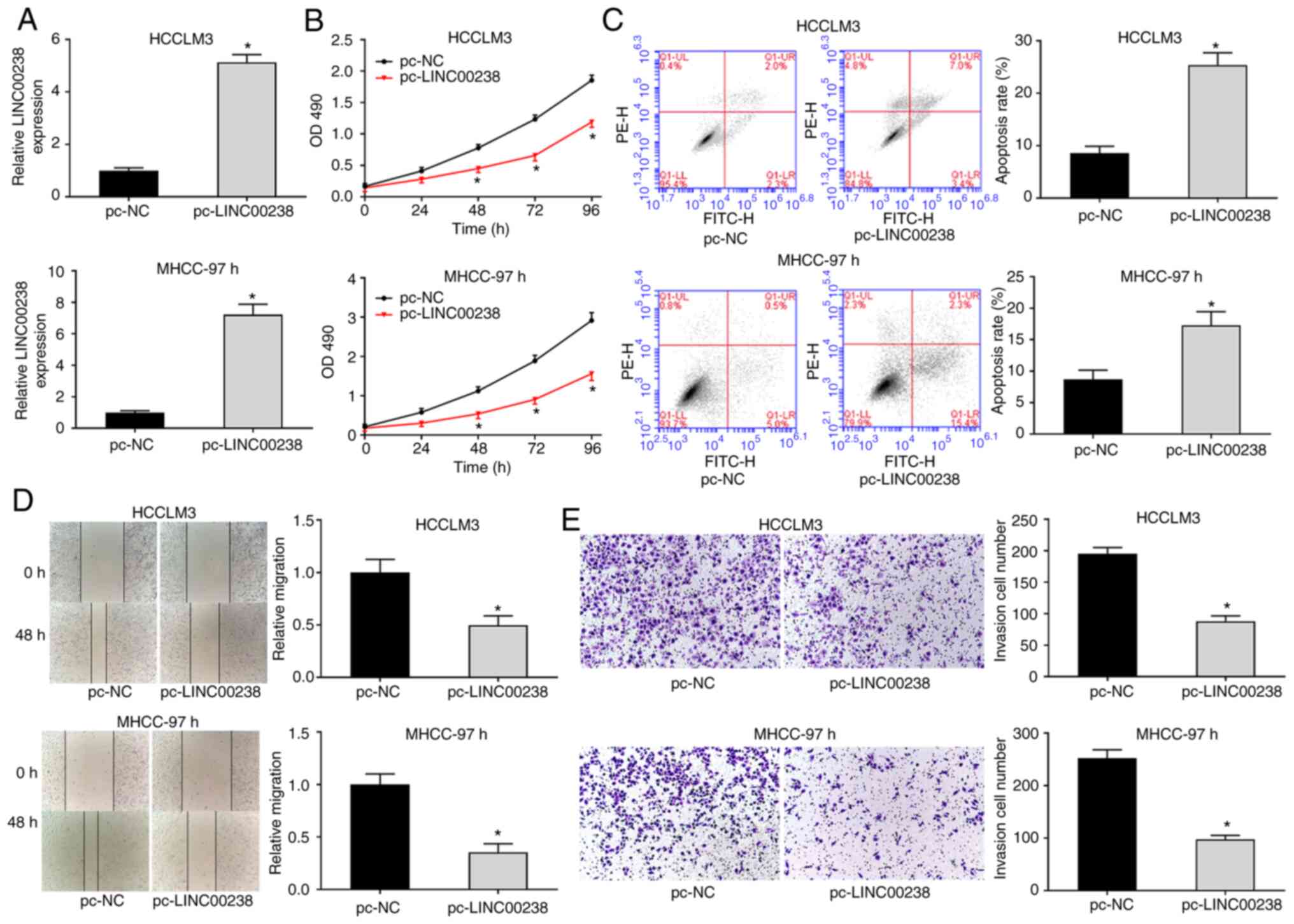
LINC00238 overexpression inhibits cell
proliferation, migration and invasion of HCC cells
The LINC00238 overexpressing plasmid pc-LINC00238
was transfected into HCCLM3 and MHCC-97h cells to detect the
influence of LINC00238 on the biological function of HCC cells. As
shown in Fig. 2A, LINC00238
expression was significantly increased in the pc-LINC00238 group
compared with that in pc-NC group. The proliferation of HCC cells
was measured using the MTS assay. The data suggested that LINC00238
overexpression suppressed the proliferation ability of HCCLM3 and
MHCC-97h cells (Fig. 2B). Flow
cytometry results revealed that the apoptotic rates of HCCLM3 and
MHCC-97h cells were significantly increased after LINC00238
overexpression (Fig. 2C). The
migration and invasion capacity of HCC cells was detected by wound
healing and Transwell assays, respectively. The results in Fig. 2D showed that LINC00238
overexpression markedly decreased the migration capacity of HCC
cells. Additionally, the number of invasive cells in the
pc-LINC00238 group was significantly reduced compared with that in
the pc-NC group (Fig. 2E).
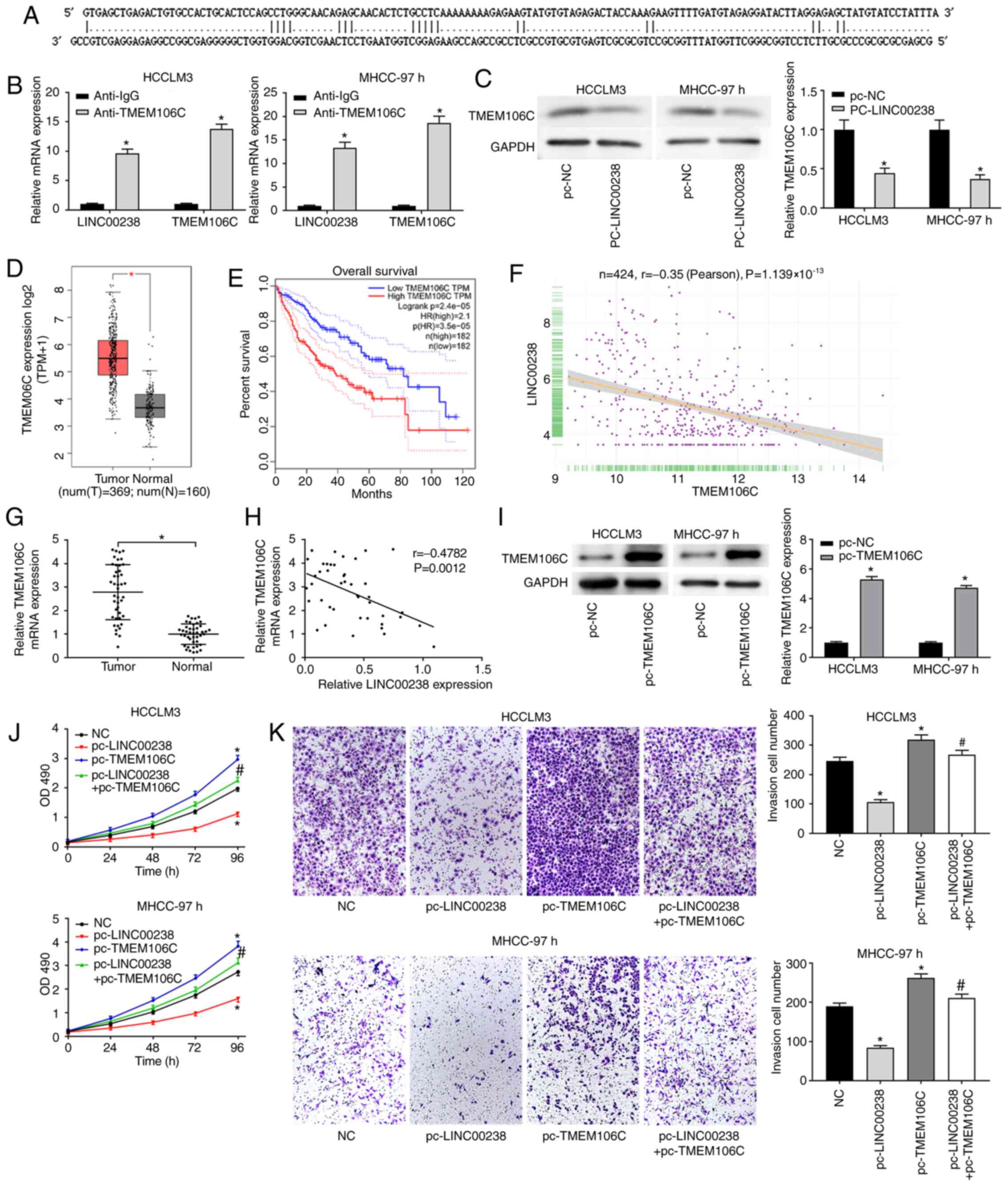
LINC00238 negatively regulates the
expression of TMEM106C in HCC
A recent study showed that TMEM106C is highly
expressed in HCC and serves an important role in HCC progression
(14). LncTar was performed to
determine whether there is a targeted relationship between
LINC00238 and TMEM106C (21) and
the results (Fig. 3A) suggested
that TMEM106C was the target gene of LINC00238. The target
relationship between LINC00238 and TMEM106C was verified using RIP
assay. The data suggested that LINC00238 expression was
significantly increased in TMEM106C immunoprecipitates in HCCLM3
and MHCC-97h cells compared with that the Anti-IgG group,
suggesting that TMEN106C could couple with LINC00238 directly
(Fig. 3B). The protein levels of
TMEM106 in HCC cells after LINC00238 overexpression was evaluated
using western blotting. The data showed that LINC00238
overexpression significantly reduced TMEM106 expression (Fig. 3C). The online data based on TCGA
suggested that TMEM106C is more highly expressed in HCC tumor
tissues than in normal tissues (Fig.
3D) and a high level of TMEM106C expression is associated with
poor overall survival (Fig. 3E).
There was a negative correlation between LINC00238 and TMEM106C
expression in HCC (Fig. 3F).
TMEM106C expression was detected with RT-qPCR and the correlation
between LINC00238 and TMEM106C expression analyzed. The results
indicated that TMEM106C expression was significantly upregulated in
tumor tissues (Fig. 3G) and
negatively correlated with LINC00238 expression (Fig. 3H).
To investigate whether LINC00238 served its role
through TMEM106C, pc-LINC00238 and pc-TMEM106C were co-transfected
into HCCLM3 and MHCC-97h cells to detect the influence of LINC00238
and TMEM106C on HCC progression. As shown in Fig. 3I, the TMEM106C expression was
significantly increased by TMEM106C overexpression. The
proliferation ability of HCC cells was markedly enhanced by
TMEM106C overexpression (Fig. 3J).
Additionally, the capacity of invasion (Fig. 3K) was significantly higher in the
p-TMEM106C group than that in the pc-NC group. In addition, the
inhibitory effect of LINC00238 overexpression on the proliferation
and invasion of HCC cells was eliminated with TMEM106C
overexpression.
TMEM106C regulates the apoptosis
pathway and LINC00238 is transcriptionally regulated by ATF3 in
HCC
The present study performed GESA analysis of the
gene sequences of TMEM106C in HCC cells (18). The results showed that TMEM106C
suppressed the apoptosis signaling pathway (Fig. 4A). The protein expression of genes
enriched in the apoptosis signaling pathway was then detected.
Western blot analysis showed that the protein expression of
capsase-7 (CASP7), TIMP2, PDCD4 and ATF3 was significantly
increased by LINC00238 overexpression and decreased by TMEM106C
overexpression (Fig. 4B). The
inhibitory effect of TMEM106C on protein expression of ASP7, TIMP2,
PDCD4 and ATF3 in HCC cells was eliminated by LINC00238
overexpression (Fig. 4B).
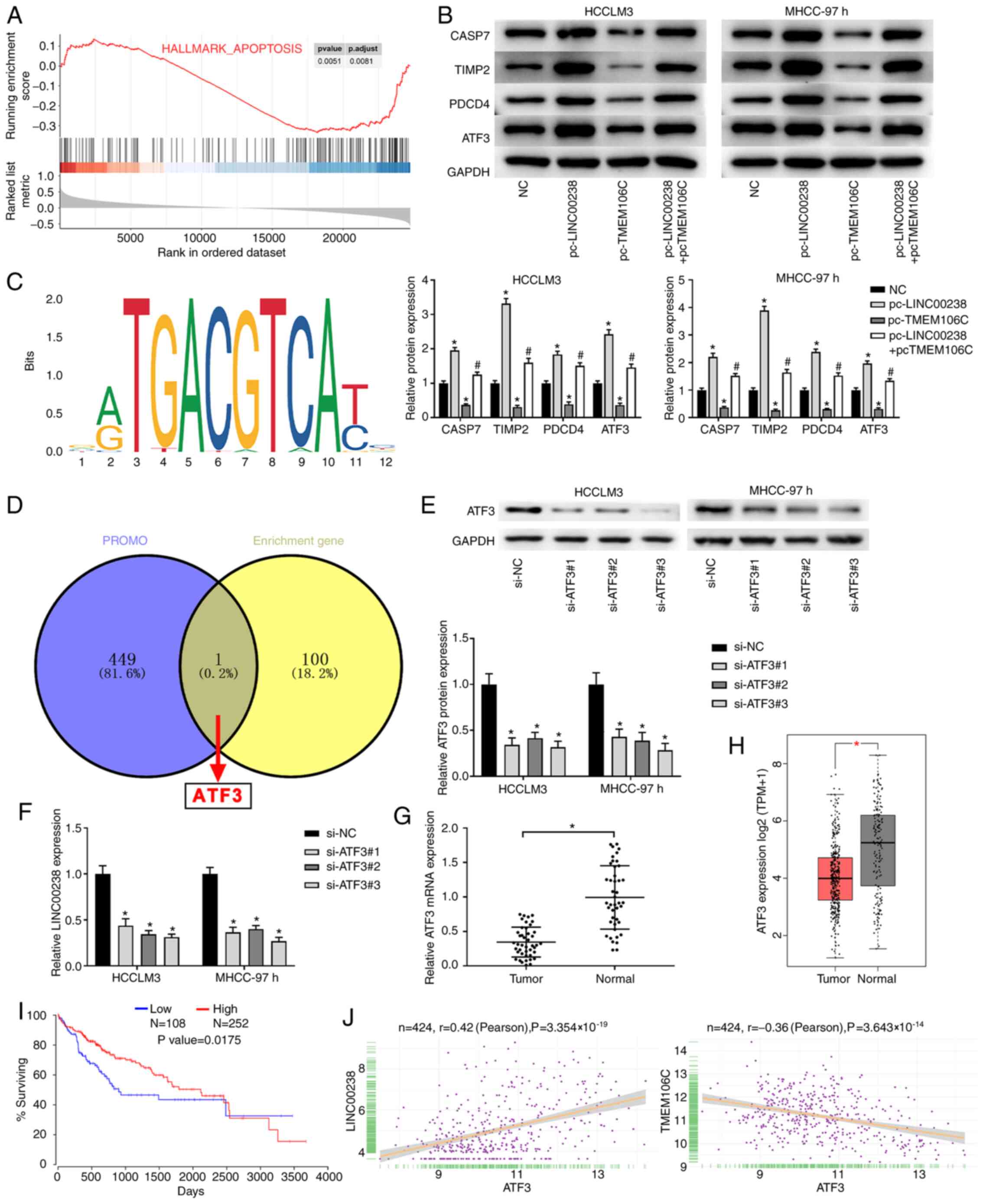 | Figure 4.TMEM106C regulates apoptosis pathway
and LINC00238 is transcriptionally regulated by ATF3 in HCC. (A)
The Enriched KEGG pathway analysis showed that TMEM106C suppressed
the apoptosis signaling pathway. (B) The results of western
blotting showed that protein expression of ASP7, TIMP2, PDCD4 and
ATF3 in HCCLM3 and MHCC-97h cells was increased by LINC00238
overexpression while decreased by TMEM1106C overexpression.
*P<0.05 vs. NC group; #P<0.05 vs. pc-LINC00238
group. (C) Recognition motif of ATF3 from the JASPAR database. (D)
Venn diagram showing ATF3 is upstream transcription factor of
LINC00238 of LINC00958 predicted by PROMO and KEGG. (E) The results
of western blotting showed that ATF3 protein expression in HCCLM3
and MHCC-97h cells was decreased by ATF3 siRNAs. (F) The results of
RT-qPCR showed that LINC00238 expression in HCCLM3 and MHCC-97h
cells was decreased by ATF3 siRNAs. (G) The results of RT-qPCR
showed that ATF3 expression was significantly lower in HCC tissues
than that in corresponding normal tissues. (H) Scatter plots
comparing ATF3 expression in HCC samples (n=396) and normal tissue
samples (n=160) showed that ATF3 was downregulated in HCC tumor
samples compared with normal samples. (I) Kaplan-Meier analysis
indicated an association between high TMEM106 and poor overall
survival in patients with HCC. (J) The correlation analysis among
ATF3, LINC00238 and TMEM106C showed that the ATF3 expression was
positively correlated with LINC00238 and negatively correlated with
TMEM106C expression. *P<0.05. TMEM106C, transmembrane protein
106C; LINC, long intergenic non-protein coding RNA; HCC,
hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and
Genomes; ATF3, activating transcription factor 3; CASP7, caspase-7;
TIMP2, tissue inhibitor of metalloproteinase 2; PDCD4, programmed
cell death 4; pc-, plasmid; si, short interfering; RT-qPCR, reverse
transcription-quantitative PCR. |
To investigate the potential mechanism of LINC00238
in HCC, the JASPAR and PROMO databases were analyzed and ATF3 was
identified as the upstream transcription factor of LINC00238
(Fig. 4C and D). The regulatory
effect of ATF3 on LINC00238 expression was detected after ATF3
siRNAs (si-ATF3#1, si-ATF3#2 and si-ATF3#3) were transfected into
HCC cells. As shown in Fig. 4E,
ATF3 levels were significantly decreased in the si-ATF3 groups. The
expression of LINC00238 was markedly suppressed in the si-ATF3
groups compared with that in the si-NC group (Fig. 4F). ATF3 expression in HCC tissues
and corresponding normal tissues was measured using RT-qPCR. The
results indicated that ATF3 expression in tumor tissues was lower
compared with that in normal tissues (Fig. 4G). Online data based on TCGA
suggested that ATF3 was expressed at low levels in HCC tumor
tissues (Fig. 4H) and low levels of
ATF3 expression were associated with poor overall survival
(Fig. 4I). Furthermore, there was a
positive correlation between ATF3 and LINC00238 and a negative
correlation between ATF3 and TMEM106C in HCC (Fig. 4J).
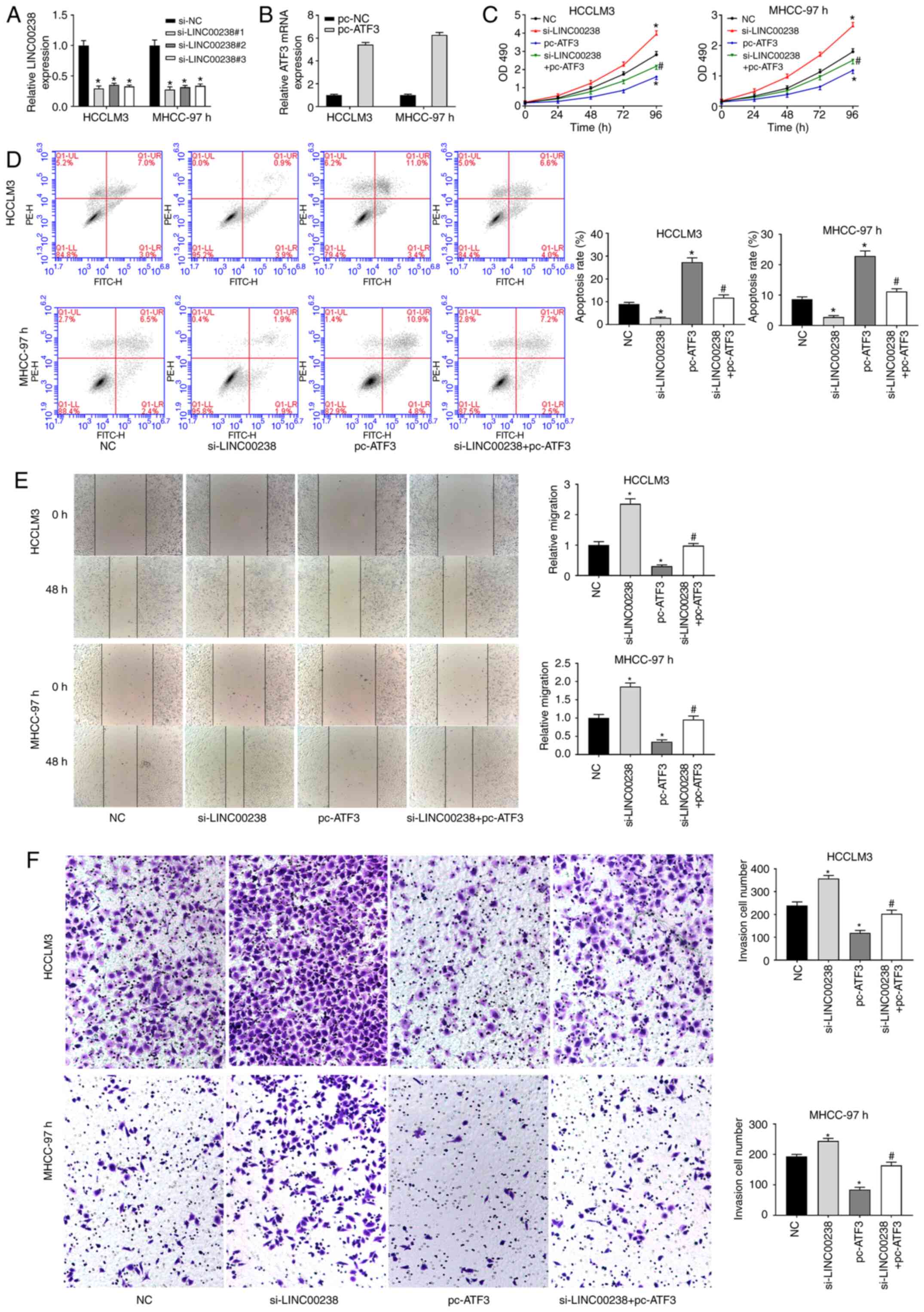
ATF3 overexpression reverses the
promoting effect of LINC00238 silencing on HCC progression
The si-LINC00238 and ATF3-overexpressing plasmid
were co-transfected into HCC cells to explore the regulatory effect
of ATF3 on LINC00238 in HCC progression. As shown in Fig. 5A, LINC00238 expression was
significantly decreased by LINC00238 silencing. The ATF3 expression
was markedly increased by ATF3 overexpression (Fig. 5B). The MTS data showed that
LINC00238 silencing enhanced cell proliferation, whereas ATF3
overexpression reduced the proliferation of HCC cells (Fig. 5C). Compared with the si-LINC00238
group, the proliferation ability of the si-LINC00238+pc-ATF3 group
was significantly decreased (Fig.
5C). Apoptosis of HCC cells was measured using flow cytometry.
The data suggested that apoptosis of HCC cells was markedly
decreased by LINC00238 silencing and increased by ATF3
overexpression (Fig. 5D). The
inhibition of LINC00238 silencing on HCC cell apoptosis was
eliminated by ATF3 overexpression (Fig.
5D). Additionally, the migration and invasion capacity of HCC
cells following co-transfection with si-LINC00238 and pc-ATF3 were
measured. The data suggested that the capacity of migration and
invasion of HCC cells was significantly enhanced by LINC00238
silencing and reduced by ATF3 overexpression (Fig. 5E and F). Besides, ATF3
overexpression reversed the accelerating effect of LINC00238
silencing on the migration and invasion of HCC cells (Fig. 5E and F).
Discussion
HCC is one of the commonest cancers with high
mortality rates worldwide. High rates of metastasis and
postoperative recurrence lead to high mortality rate in patients
with HCC (3). The present study
found that LINC00238 was downregulated in HCC. Overexpression of
LINC00238 suppressed proliferation, migration and invasion of HCC
cells by activating the TMEM106C-mediated apoptosis signaling
pathway. Furthermore, it was found that ATF3, a gene enriched in
the apoptosis signaling pathway, was the upstream promoter of
LINC00238.
Previous studies have suggested that lncRNAs serve a
vital role in the initiation and progression of HCC. Mo et
al (22) revealed that
LINC01287 expression is increased in HCC cells and knockdown of
LINC01287 decreases the proliferation and invasion ability of HCC
cells. The lncRNA CTC-297N7.9 acts as a tumor suppressor in HCC and
low expression of lncRNA CTC-297N7.9 is associated with poor
prognosis in patients with HCC (23). In the present study, the expression
of LINC00238 was significantly decreased in both HCC tissues and
cell lines. High levels of LINC00238 were associated with a good
prognosis. Overexpression of LINC00238 inhibited the proliferation,
migration and reduced invasion capacity of HCC cells. These data
indicated that LINC00238 acted as a tumor suppressor in HCC and
LINC00238 overexpression inhibited HCC progression.
TMEM106C is a member of the TMEM106 family and is
expressed at low levels in patients with ankylosing spondylitis
(13). Duan et al (24) found that TMEM106C expression is
positively related to CENPM and negatively related to DLC-1 in HCC
tumor tissues. Luo et al (14) showed that FOXO1 and FOXO3 may be the
key target genes of TMEM106C in HCC. In the present study,
LINC00238 expression was higher in HCC tissues compared with that
in normal tissues. The predicted result of LncTar suggested that
TMEM106C was the target of LINC00238. LINC00238 overexpression
reduced the protein expression of TMRM106C. TCGA analysis showed
that high TMEM106C expression was associated with poor prognosis of
HCC and negatively correlated with LINC00238 expression. In
addition, TMEM106C overexpression reversed the inhibitory effect of
LINC00238 overexpression on the proliferation, migration and
invasion of HCC cells. These data suggested that LINC00238
inhibited HCC progression by suppressing TMEM106C expression.
The expression of the ATF3, a member of the ATF
family of transcription factors, can be induced by a variety of
stress signals (25). Previous
studies have demonstrated that ATF3 acts as a tumor inhibitor in
cancer. ATF3 dysfunction allows normal cells to be easily
transformed by oncogenes (26). For
instance, Li et al (27)
found that overexpression of ATF3 inhibits the proliferation and
migration in esophageal squamous cell carcinoma. The findings of
Hackl et al (28) indicated
that downregulation of ATF3 promotes the migration capacity of
colon cancer cells in vitro and facilitates tumor growth
in vivo. Lv et al (29) observed that ATF3 is significantly
downregulated in human intrahepatic cholangiocarcinoma with tumor
metastasis. The results of the present study were consistent with
those of previous studies. The expression of ATF3 was significantly
lower in HCC tumor tissues than that in normal tissues. Low ATF3
expression was associated with poor prognosis in patients with HCC.
ATF3 overexpression inhibited proliferation, migration and invasion
of HCC cells. In addition, it was found that ATF3 was an upstream
transcription factor of LINC00238. ATF3 expression was positively
correlated to LINC00238 and LINC00238 expression could be reduced
by ATF3 silencing. Overexpression of ATF3 reversed the promoting
effect of LINC00238 silencing on proliferation, migration and
invasion of HCC cells. These results indicated that ATF3 could
suppress HCC progression and strengthen the inhibitory effect of
LINC00238 on HCC.
Avoidance of apoptosis a major cause of cancer
development and progression (30).
CASP7 is a member of the caspases family that participates in
cervical cancer progression (31).
Palmerini et al (32)
indicated that CASP7 is downregulated in colon cancer. Proteins
encoded by TIMPs are natural inhibitors of matrix
metalloproteinases. Peeney et al (33) demonstrated that TIMP2 inhibits the
proliferation and EMT progression of triple-negative breast cancer.
PDCD4 is a tumor suppressor gene for various cancers and can
inhibit cell proliferation, migration and invasion, as well as
promote tumor cell apoptosis (34).
Hwang et al (35) indicated
that overexpression of PDCD4 induces anti-proliferation and
apoptosis-induced effects on human lung cancer. In the present
study, western blotting data showed that LINC00238 overexpression
increased the protein expression of CASP7, TIMP2 and PDCD4.
However, the promoting effect of LINC00238 on CASP7, TIMP2 and
PDCD4 was eliminated by TMEM106C overexpression. These results
suggested that LINC00238 inhibited the proliferation, migration and
invasion of HCC cells by upregulating these genes that were
enriched in the apoptosis pathway.
In conclusion, LINC00238 was downregulated in HCC
tissues and cell lines and served a vital role in restraining HCC
progression. Overexpression of LINC00238 decreased TMEM106C
expression and increased the expression of CASP7, TIMP2, PDCD4 and
ATF3, which are apoptotic pathway genes and thus inhibited the
proliferation, migration and invasion of HCC cells. Notably, ATF3
positively regulated the expression of LINC00238, thereby
participating in the inhibitory regulation of HCC by LINC00238.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CJ and FL designed the experiments, MY, JD and SF
collected data. JL and SS analyzed the data and wrote the
manuscript. FL and CJ confirm the authenticity of all the raw data.
All authors reviewed and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the ethics committee of
Qingdao No. 6 People's Hospital [Approval no. (2018) 26] and
written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bellissimo F, Pinzone MR, Cacopardo B and
Nunnari G: Diagnostic and therapeutic management of hepatocellular
carcinoma. World J Gastroenterol. 21:12003–12021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng D, Deng J, Zhang B, He X, Meng Z, Li
G, Ye H, Zheng S, Wei L, Deng X, et al: LncRNA HOTAIR
epigenetically suppresses miR-122 expression in hepatocellular
carcinoma via DNA methylation. EBioMedicine. 36:159–170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lou W, Liu J, Gao Y, Zhong G, Ding B, Xu L
and Fan W: MicroRNA regulation of liver cancer stem cells. Am J
Cancer Res. 8:1126–1141. 2018.PubMed/NCBI
|
|
5
|
Huang Z, Zhou JK, Peng Y, He W and Huang
C: The role of long noncoding RNAs in hepatocellular carcinoma. Mol
Cancer. (19): 772020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gong D, Feng PC, Ke X-F, Kuang HL, Pan LL,
Ye Q and Wu JB: Silencing long non-coding RNA LINC01224 inhibits
hepatocellular carcinoma progression via MicroRNA-330-5p-induced
inhibition of CHEK1. Mol Ther Nucleic Acids. 19:482–497. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jia M, Jiang L, Wang YD, Huang JZ, Yu M
and Xue HZ: LincRNA-p21 inhibits invasion and metastasis of
hepatocellular carcinoma through Notch signaling induced
epithelial-mesenchymal transition. Hepatol Res. 46:1137–1144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu F, Yuan JH, Huang JF, Yang F, Wang TT,
Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al: Long noncoding RNA
FTX inhibits hepatocellular carcinoma proliferation and metastasis
by binding MCM2 and miR-374a. Oncogene. 35:5422–5434. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang CZ, Yan GX, Dong DS, Xin H and Liu
ZY: LncRNA-ATB promotes autophagy by activating Yes-associated
protein and inducing autophagy-related protein 5 expression in
hepatocellular carcinoma. World J Gastroenterol. 25:5310–5322.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marx S, Dal Maso T, Chen JW, Bury M,
Wouters J, Michiels C and Le Calvé B: Transmembrane (TMEM) protein
family members: Poorly characterized even if essential for the
metastatic process. Semin Cancer Biol. 60:96–106. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J and Zhu H: TMEM106A inhibits cell
proliferation, migration, and induces apoptosis of lung cancer
cells. J Cell Biochem. Nov 19–2018.(Epub ahead of print). doi:
10.1002/jcb.28057.
|
|
12
|
Lang CM, Fellerer K, Schwenk BM, Kuhn PH,
Kremmer E, Edbauer D, Capell A and Haass C: Membrane orientation
and subcellular localization of transmembrane protein 106B
(TMEM106B), a major risk factor for frontotemporal lobar
degeneration. J Biol Chem. 287:19355–19365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Assassi S, Reveille JD, Arnett FC, Weisman
MH, Ward MM, Agarwal SK, Gourh P, Bhula J, Sharif R, Sampat K, et
al: Whole-blood gene expression profiling in ankylosing spondylitis
shows upregulation of toll-like receptor 4 and 5. J Rheumatol.
38:87–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo X, Han G, Lu R, Guan S, Wang Y, Ju L,
Chen L, Shao J and Bian Z: Transmembrane protein 106C promotes the
development of hepatocellular carcinoma. J Cell Biochem.
121:4484–4495. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Therneau TM: Survival Analysis [R package
survival version 2.39-5]. 46:111–112. 2015.
|
|
17
|
R Core Team: A Language And Environment
For Statistical Computing. R Foundation for Statistical Computing;
Vienna: 2012
|
|
18
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The Molecular Signatures
Database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Ma W, Zeng P, Wang J, Geng B, Yang J
and Cui Q: LncTar: A tool for predicting the RNA targets of long
noncoding RNAs. Brief Bioinform. 16:806–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mo Y, He L, Lai Z, Wan Z, Chen Q, Pan S,
Li L, Li D, Huang J, Xue F, et al: LINC01287/miR-298/STAT3 feedback
loop regulates growth and the epithelial-to-mesenchymal transition
phenotype in hepatocellular carcinoma cells. J Exp Clin Cancer Res.
37:149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu S, Huang X, Zhang K, Tan W, Lin Z, He
Q, Chen Y and Shang C: Low expression of long noncoding RNA
CTC-297N7.9 predicts poor prognosis in patients with hepatocellular
carcinoma. Cancer Med. 8:7679–7692. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Duan J, Qian Y, Fu X, Chen M, Liu K, Liu
H, Yang J, Liu C and Chang Y: TMEM106C contributes to the malignant
characteristics and poor prognosis of hepatocellular carcinoma.
Aging (Albany NY). 13:5585–5606. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hai T, Wolford CC and Chang YS: ATF3, a
hub of the cellular adaptive-response network, in the pathogenesis
of diseases: Is modulation of inflammation a unifying component?
Gene Expr. 15:1–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan C and Boyd DD: ATF3 regulates the
stability of p53: A link to cancer. Cell Cycle. 5:926–929. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Yang Z, Chen Z, Bao Y, Zhang H, Fang
X and Yang W: ATF3 suppresses ESCC via downregulation of ID1. Oncol
Lett. 12:1642–1648. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hackl C, Lang SA, Moser C, Mori A,
Fichtner-Feigl S, Hellerbrand C, Dietmeier W, Schlitt HJ, Geissler
EK and Stoeltzing O: Activating transcription factor-3 (ATF3)
functions as a tumor suppressor in colon cancer and is up-regulated
upon heat-shock protein 90 (Hsp90) inhibition. BMC Cancer.
10:6682010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv L, Wei M, Lin P, Chen Z, Gong P, Quan Z
and Tang Z: Integrated mRNA and lncRNA expression profiling for
exploring metastatic biomarkers of human intrahepatic
cholangiocarcinoma. Am J Cancer Res. 7:688–699. 2017.PubMed/NCBI
|
|
30
|
Reed JC: Mechanisms of apoptosis avoidance
in cancer. Curr Opin Oncol. 11:68–75. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi TY, He J, Wang MY, Zhu ML, Yu KD, Shao
ZM, Sun MH, Wu X, Cheng X and Wei Q: CASP7 variants modify
susceptibility to cervical cancer in Chinese women. Sci Rep.
5:9225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Palmerini F, Devilard E, Jarry A, Birg F
and Xerri L: Caspase 7 downregulation as an immunohistochemical
marker of colonic carcinoma. Hum Pathol. 32:461–467. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peeney D, Jensen SM, Castro NP, Kumar S,
Noonan S, Handler C, Kuznetsov A, Shih J, Tran AD, Salomon DS, et
al: TIMP-2 suppresses tumor growth and metastasis in murine model
of triple-negative breast cancer. Carcinogenesis. 41:313–325. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Afonja O, Juste D, Das S, Matsuhashi S and
Samuels HH: Induction of PDCD4 tumor suppressor gene expression by
RAR agonists, antiestrogen and HER-2/neu antagonist in breast
cancer cells. Evidence for a role in apoptosis. Oncogene.
23:8135–8145. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hwang SK, Jeong YJ and Chang Y-C: PDCD4
inhibits lung tumorigenesis by the suppressing p62-Nrf2 signaling
pathway and upregulating Keap1 expression. Am J Cancer Res.
10:424–439. 2020.PubMed/NCBI
|