Introduction
Ovarian cancer is one of the leading causes of
cancer-related deaths worldwide (1). Few patients present marked clinical
symptoms of ovarian cancer at initial stages, resulting in late
diagnoses. Although most patients with ovarian cancer are initially
responsive to cisplatin (DDP) -based chemotherapy regimens, most
patients with recurring ovarian cancer acquire DDP resistance
(2). Exploring novel agents and
targets aiming at curbing resistance to DDP-based chemotherapy is a
matter of utmost urgency.
PTEN, located at 10q23, exerts a tumor-suppressive
effect on numerous types of cancer by interfering in multiple
pathogenic events (3). As a
phosphatase, PTEN dephosphorylates phosphatidylinositol (3,4,5)-trisphosphate,
thus inhibiting PI3K/AKT signaling (4). PTEN deactivation in tumorigenesis
subsequently increases the activity of PI3K/AKT signaling,
therefore amplifying the ability of cancer cells to proliferate,
migrate and invade, while inhibiting the apoptosis of these cells
(5,6). Notably, PTEN downregulation is closely
associated with the DDP-resistance of ovarian cancer (7,8).
Concerning the PTEN downregulation mechanism, it has been reported
that the demethylation agent fails to restore the expression of
PTEN protein, indicating that PTEN is highly post-transcriptionally
modulated (9). Notably, PTEN is
identified as a direct downstream target of multiple microRNAs
(miRs), a series of short non-coding RNAs, including miR-214
(7), miR-205 (10), miR-221/miR-222 (11), miR-93 (12) and miR-106a (13–16).
Among them, miR-106a has been widely regarded as an oncogenic miRNA
in types of cancer such as stomach cancer, liver cancer, non-small
cell lung cancer, prostate cancer and ovarian cancers (13–17).
However, its specific effect on ovarian cancer cell resistance to
DDP remains to be elucidated. It is, therefore, hypothesized that
the miR-106a/PTEN axis might appear in ovarian cancer and have an
effect on the DDP-resistance of ovarian cancer.
Long non-coding RNAs (lncRNAs) are a type of RNA and
are defined as being transcripts with lengths exceeding 200
nucleotides that are not translated into proteins (18). lncRNAs can genetically,
epigenetically and post-transcriptionally modulate gene expression
(19,20), thus participating in the occurrence
and development of various types of cancer (21,22),
such as ovarian cancer (23,24).
By serving as competing endogenous RNAs (ceRNAs) for miRNAs,
lncRNAs act as natural miRNA decoys (25). Notably, miR-106a serves as a
critical component of several lncRNA-miRNA-mRNA networks affecting
PTEN expression. In gastric cancer, lncRNA-FER1L4 competes for
miR-106a-5p to regulate PTEN expression through its miRNA response
elements (26). In chronic myeloid
leukemia, lncRNA-BGL3 serves as a miR-106a ceRNA, inhibiting PTEN
expression from repressing Bcr-Abl-induced cell transformation
(27). It was therefore
hypothesized that lncRNAs could potentially be involved in ovarian
cancer cell resistance to DDP in a miR-106a/PTEN axis-related
manner.
To test the hypothesis, online data and microarray
expression files were procured from the Gene Expression Omnibus
(GEO) and The Cancer Genome Atlas (TCGA) databases. These data were
analyzed for lncRNAs that were positively associated with PTEN and
negatively associated with miR-106a in ovarian cancer. Among the
candidates, lncRNA HAND2-AS1 was selected as it was downregulated
in recurrent ovarian cancer and is hypothesized to be associated
with DDP-resistance. The specific effects of HAND2-AS1 on ovarian
cancer cells with resistance to DDP were subsequently investigated.
In further confirmation of the predicted lncRNA-miRNA-mRNA network,
the role of HAND2-AS1 in regulating PTEN/PI3K/AKT signaling and the
binding of miR-106a to HAND2-AS1 and PTEN 3′UTR was examined.
Finally, the dynamic effects of HAND2-AS1 and miR-106a on
PTEN/PI3K/AKT signaling pathway and ovarian cancer cells with
resistance to DDP were detected. In summary, a lncRNA-miRNA-mRNA
network modulating ovarian cancer cell resistance to DDP was
proposed.
Materials and methods
Clinical tissue samples
A total of 12 paired ovarian cancer tissues and
adjacent non-cancerous tissues (at least 0.5 cm in distance) were
harvested from patients (average age, 58.08±7.74 years old)
diagnosed with ovarian cancer and underwent surgical resection in
the Fourth Hospital of Changsha between January 2010 and May 2020.
Patients who had received chemotherapy before surgery were excluded
from this study. Ovarian cancer samples were histologically
examined and the diagnosis was established by clinical
pathologists. Clinical sampling was performed with the approval of
the Ethics Committee of Hunan Normal University [approval no.
2019(188); Changsha, China] Signed written informed consent was
obtained from all patients enrolled.
Cell line and cell transfection
SKOV3 cell line [American Type Culture Collection
(ATCC)® HTB-77™] was obtained from ATCC and cultured in
McCoy's 5a Medium Modified (ATCC) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.). DDP-resistant SKOV3/DDP (DDP) cell
line was purchased from the Chinese Academy of Medical Sciences and
Peking Union Medical College. Briefly, the SKOV3/DDP cell line was
established by exposing the original SKOV3 cell line to gradually
increasing DDP concentrations. SKOV3 cells were cultured in a
medium containing 0.02 mg/l DDP. The dose of DDP was steadily
increased until the cells reached a stable growth phase with the
medium containing 0.2 mg/l DDP. This induction culture lasted 6
months. All cells were cultured at 37°C in 5% CO2.
To generate lncRNA HAND2-AS1-overexpression or
-knockdown cells, 2×105 cells/ml target SKOV3 cells were
transfected with 2 µg lncRNA HAND2-AS1-overexpressing vector
(HAND2-AS1), HAND2-AS1 knockdown vector (sh-HAND2-AS1). The empty
vector (pLVX-puro) or pLVX-shRNA2-puro containing a scramble
sequence (sh-NC) were used as negative controls for overexpression
vector or knockdown vector transfection. For miR-106a-5p
overexpression or knockdown 2×105 cells/ml target SKOV3
cells were transfected with 50 pmol miR106a-5p mimics (miR-106a-5p)
or miR106a-5p inhibitor (anti-miR106a-5p). The mimics NC (miR-NC)
or inhibitor NC (anti-NC) were used as negative controls. All the
vectors or miRNAs were obtained from Shanghai GenePharma Co., Ltd.
The sequences are listed in Table
SI. Transfection was generated using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance
with the manufacturer's instructions. After incubation with the
DNA/RNA Lipofectamine 3000 complex for 6 h at 37°C in the cell
culture incubator, the SKOV3 cells were further cultured with fresh
complete medium for 48 h. Then, cells were harvested for further
experiments. The transfection efficiency was determined by reverse
transcription-quantitative PCR (RT-qPCR).
Bioinformatics analysis
The GEO dataset GSE15709 (expression profiling of
DDP-sensitive and resistant ovarian cancer cell lines, n=5
respectively) (28) and GSE14407
(expression profiling of ovarian surface epithelia and ovarian
cancer epithelia, n=12 respectively) (29) were downloaded using R language
GEOquery (version 3.12) (30)
package and the differential expression genes were analyzed by
Limma package (version 3.44.3) (31). The expression of lncRNA HAND2-AS1 in
primary ovarian carcinoma (n=354) and recurrent ovarian carcinoma
tissue samples (n=5) were obtained from TCGA-ovarian cancer (OV),
download from Xena data hub: https://ucscpublic.xenahubs.net.
For co-expression genes selection, GSE15709 was used
for analysis. Pearson's correlation coefficient analysis was
performed by R language Psych package (version 2.1.3) (32) to identify genes positively
correlated with HAND2-AS1 (|r|>0.95, P-value <0.01). Then,
these co-expressed genes were analyzed by Gene Set Enrichment
Analysis (GSEA) using R language ClusterProfiler package (version
3.16.1) (33) based on the tumor
marker pathway gene set (Hall mark genesets: h.aLL.v7.2.entrez.
GMT) from msigdb (https://www.gsea-msigdb.org).
For lncRNA and miRNA binding site prediction, online
tool LncTar (http://www.cuilab.cn/lnctar) was used.
MTT assay
Following treatment and transfection, target cells
were seeded in 96-well plates at a density of 2×104
cells/well. MTT (0.5 g/l; Sigma-Aldrich; Merck KGaA) was added to
each well. The cells were then incubated at 37°C for 0 and 48 h.
Following the removal of the medium, 50 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added and the cells were further
incubated at 37°C for 10 min. The absorbance of each sample was
subsequently measured at 450 nm using a plate reader.
Flow cytometry analysis
Following treatment and transfection, target cells
were seeded in 6-well plates at a density of 1×105
cells/ml medium for 24 h. An Annexin V-FITC Apoptosis Detection kit
(KeyGen Biotech Co., Ltd.) was then used to determine cell
apoptosis. Briefly, harvested cells were suspended in 500 µl
binding buffer and incubated with 5 µl Annexin V at 4°C for 10 min
in the dark. Then, cells were further incubated with 5 µl PI
solution at 4°C for 10 min in the dark and then subjected to flow
cytometry (BD FACSDiva Fusion; BD Biosciences). The early and late
apoptotic rate was statistically analyzed by FlowJo software
version 10.5 (FlowJo LLC).
RT-q)PCR
Total RNA from target cells with or without
treatment or transfection was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and converted into
cDNA using a PrimeScript® RT reagent kit (Takara Bio,
Inc.) following the manufacturer's instructions and a previous
method (34,35). qPCR was conducted using an SYBR
Premix Ex Taq II (Takara Bio, Inc.). The thermocycling conditions
were as follows: Initial denaturation at 95°C for 2 min, followed
by 40 cycles of denaturation at 95°C for 15 sec, and annealing and
extension at 60°C for 30 sec. U6 and GAPDH were used as the
internal references for miRNA and mRNA expression determination,
respectively. The 2−ΔΔCq method was used for the
calculation of relative expression (36). The primers are listed in Table SI.
Western blotting
Protein was extracted from target cells with or
without treatment or transfection using RIPA lysis buffer
containing PMSF (Beyotime Institute of Biotechnology). The protein
concentration was determined by Bradford Protein Assay Kit
(Beyotime Institute of Biotechnology). Protein was harvested,
diluted, denatured at 100°C for 5 min and then 30–50 µg protein
were separated via 10% SDS-PAGE gel, and subsequently transferred
to PVDF membranes (EMD Millipore). After blocking with 5% BSA
(Beyotime Institute of Biotechnology) at room temperature for 2 h,
the membranes were incubated overnight at 4°C with the following
primary antibodies (1:1,000) against cleaved-caspase-3 (cat. no.
ab2302), caspase-3 (cat. no. ab13847), poly-ADP ribose polymerase
(PARP; cat. no. ab74290), cleaved-PARP (cat. no. ab32064), Bax
(cat. no. ab32503), Bcl-2 (cat. no. ab32124), PTEN (cat. no.
ab32199), PI3K (cat. no. ab32089), phosphorylated (p-)PI3K (cat.
no. ab182651), AKT (cat. no. ab179463), p-AKT (cat. no. ab81283)
and GAPDH (cat. no. ab8245; all from Abcam). GAPDH was used as a
visual loading control. After having been incubated with the
HRP-conjugated secondary antibodies (1:5,000; cat. nos. SA00001-1
and SA00001-2; ProteinTech Group, Inc.) for 1 h at room
temperature, protein signals were detected using enhanced
chemiluminescence (ECL, Amersham; Cytiva) and visualized with an
iBright™ CL1500 Imaging System (Invitrogen; Thermo Fisher
Scientific, Inc.). Densitometry was performed using ImageJ software
version 1.44 (National Institutes of Health).
Luciferase reporter assay
The wild type (wt) or mutant (mut) type of PTEN
3′UTR or HAND2-AS1 was inserted to the multiple cloning site of the
psiCHECK-2 vector (Promega Corporation). Mut reporter vector
contained a 6 or 8 bp mutation in the predicted miR-106a binding
site. 293T cells (ATCC) were co-transfected with
wt-PTEN-3UTR/mut-PTEN-3′UTR or wt-HAND2-AS1/mut-HAND2-AS1 and
miR-106a/anti-miR-106a. Transfection was performed using
Lipofectamine® 3000 in accordance with the
manufacturer's instructions. After 48 h, the Renilla
luciferase activity and firefly luciferase activity was ascertained
using a Dual-Luciferase Reporter Assay System (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Statistical analyses
All data were processed and analyzed using GraphPad
version 6.0 software (GraphPad Software, Inc.) and expressed as the
mean ± standard deviation. The experiments were repeated at least
three times. The differences between two groups were analyzed using
an unpaired Student's t-test. Whereas, differences between the
ovarian cancer tissues and paired normal ovarian tissues were
analyzed using a paired Student's t-test. Differences among more
than two groups were analyzed using a one-way analysis of variance,
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Selection of lncRNA associated with
ovarian cancer resistance to DDP
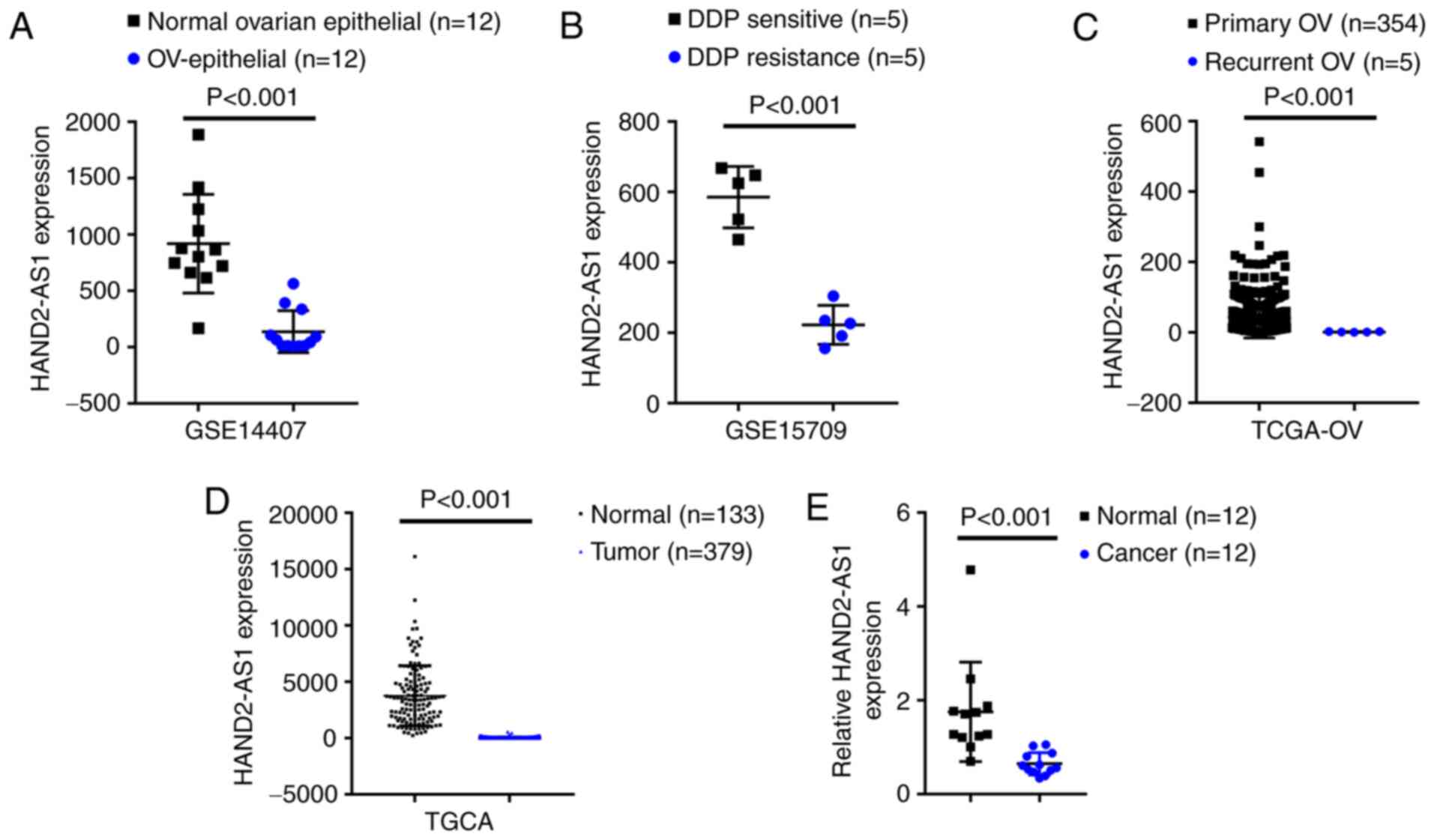
As confirmed by GSE14407, the expression of lncRNA
HAND2-AS1 was shown to be significantly downregulated within
epithelial tissue samples of ovarian cancer, compared with that
within non-cancerous ovarian epithelial tissue samples (Fig. 1A). According to GSE15709, the
expression of lncRNA HAND2-AS1 was markedly downregulated in
ovarian carcinoma cell line A2780 with resistance to DDP, compared
with that in A2780 with sensitivity to DDP (Fig. 1B). According to TCGA-OV database,
the expression of lncRNA HAND2-AS1 was markedly downregulated
within recurrent ovarian carcinoma tissue samples, compared with
that in primary ovarian cancer tissue samples (Fig. 1C). According to TCGA database, the
expression of lncRNA HAND2-AS1 was markedly downregulated within
ovarian carcinoma tissue samples, compared with that in
non-cancerous tissue samples (Fig.
1D). Additionally, lncRNA HAND2-AS1 expression in 12 paired
non-cancerous and ovarian carcinoma tissue samples was evaluated;
Fig. 1E showed that lncRNA
HAND2-AS1 expression was markedly downregulated within ovarian
carcinoma tissue samples, compared with that in normal tissue
samples.
In vitro effects of HAND2-AS1 on the
DDP-resistance of ovarian cancer cells
After confirming that HANDs-AS1 was downregulated
within ovarian carcinoma, particularly in DDP-resistant and
recurrent ovarian carcinoma, its specific effects on ovarian cancer
DDP-resistance were examined. Regular and DDP-resistant SKOV3/DDP
cells exposed to 0.125, 0.5, 2, 8 and 32 mg/l DDP were analyzed for
cell viability through MTT assays; the IC50 value for
SKOV3/DDP cells with resistance to DDP was much higher compared
with that for regular SKOV3 cells (IC50=2.963 and 15.80,
respectively; Fig. 2A). Consistent
with in vitro results, lncRNA HAND2-AS1 expression was shown
to have markedly decreased within SKOV3/DDP cells with resistance
to DDP compared with that in regular SKOV3 cells (Fig. 2B).
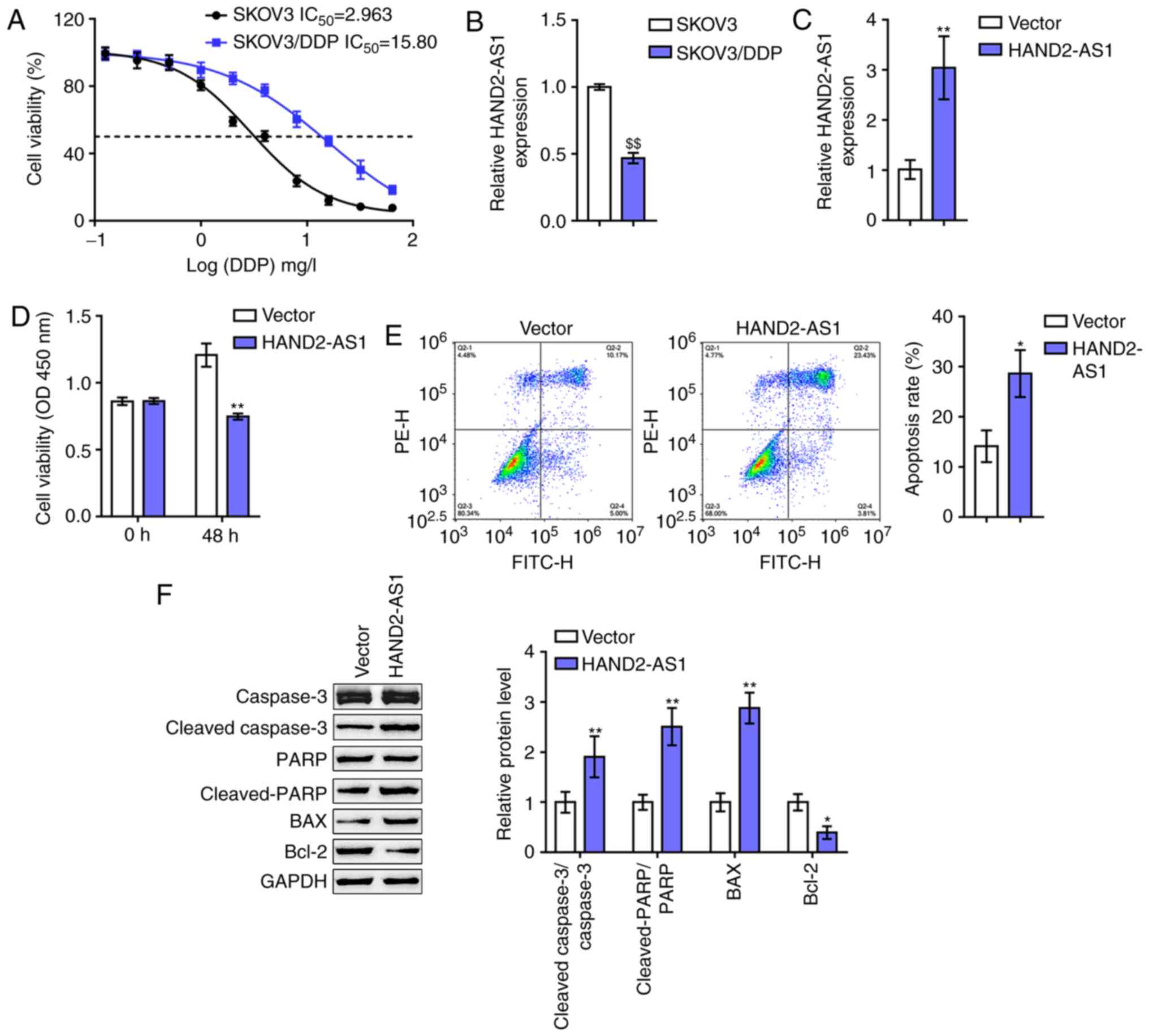 | Figure 2.In vitro effects of HAND2-AS1
on ovarian cancer cell resistance to DDP. (A) Regular SKOV3 cells
and DDP-resistant SKOV3/DDP cells were exposed to 0.125, 0.5, 2, 8
and 32 mg/l DDP and examined for cell viability via MTT assays.
Data are shown as IC50 values. (B) lncRNA HAND2-AS1
expression in regular SKOV3 cells and DDP-resistant SKOV3/DDP cells
determined by RT-qPCR. (C) HAND2-AS1 overexpression generated in
SKOV3/DDP cells by transfection of HAND2-AS1-overexpressing vector,
as confirmed by RT-qPCR. Next, SKOV3/DDP cells were transfected
with HAND2-AS1 upon DDP treatment and examined for (D) cell
viability by MTT assay and (E) cell apoptosis by flow cytometry.
(F) The protein levels of cleaved-caspase-3, caspase-3,
cleaved-PARP, PARP, Bax and Bcl-2 were determined via western
blotting. $$P<0.01 vs. SKOV3 group; *P<0.05,
**P<0.01 vs. vector. DDP, cisplatin; lncRNA, long non-coding
RNA; RT-qPCR, reverse transcription-quantitative PCR; PARP,
poly-ADP ribose polymerase. |
To evaluate the effect of HAND2-AS1 on ovarian
cancer cell resistance to DDP, HAND2-AS1 overexpression was
generated in SKOV3/DDP cells by transfection of
HAND2-AS1-overexpressing vector, and confirmed by RT-qPCR (Fig. 2C). Next, the changes in the
viability and apoptosis of cells and Bcl-2/caspase-3 apoptotic
signaling were examined in transfected SKOV3/DDP cells upon DDP
treatment. As shown in Fig. 2D and
E, the overexpression of HAND2-AS1 downregulated SKOV3/DDP cell
viability, while upregulating SKOV3/DDP cell apoptosis. As for the
signaling pathway involved, HAND2-AS1 overexpression led to
significant increases in cleaved-caspase-3/caspase-3 ratios,
cleaved-PARP/PARP ratios and Bax proteins, while reducing Bcl-2
protein levels (Fig. 2F). These
data indicated that HAND2-AS1 overexpression could potentially
re-sensitize DDP-resistant SKOV3 cells to DDP.
PTEN expression is positively
correlated with HAND2-AS1 in ovarian cancer
To further investigate the underlying mechanism,
differentially-expressed genes that were positively associated
withHAND2-AS1 were further analyzed. According to GSE15709, 124
genes were positively correlated and 78 were negatively correlated
with HAND2-AS1 (r >0.95; Fig.
S1A). Gene Set Enrichment Analysis analyzed these co-expressed
genes and showed that signaling of
‘hallmark_PI3K_AKT_MTOR_signaling’ was found to be negatively
associated with HAND2-AS1 (Fig. S1B
and C). As the natural inhibitor of the PI3K/AKT pathway, PTEN
has been widely reported to modulate cell chemosensitivity of
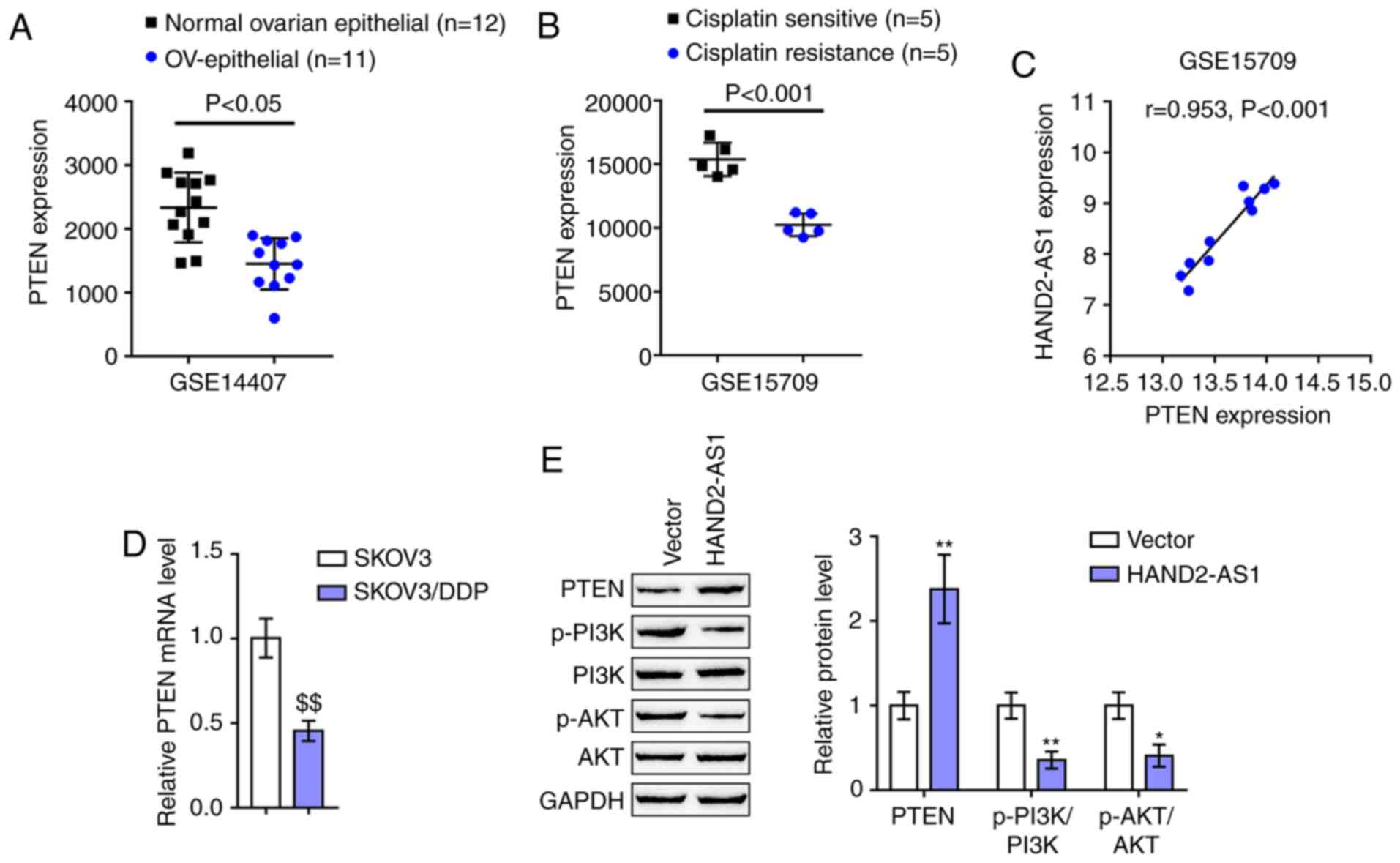
several cancer types (37,38), including ovarian cancer (8). According to GSE14407, the expression
of PTEN was downregulated within epithelial tissue samples of
ovarian carcinoma, compared with that within non-cancerous ovarian
epithelial tissue samples (Fig.
3A). According to GSE15709, the expression of PTEN was
distinctly downregulated within ovarian carcinoma cell line A2780
with resistance to DDP, compared with that in A2780 with
sensitivity to DDP (Fig. 3B).
Notably, HAND2-AS1 and PTEN expression was strongly positively
correlated in GSE15709 (Fig. 3C).
As with HAND2-AS1, PTEN expression was markedly downregulated in
SKOV3/DDP cells, compared with that in SKOV3 cells (Fig. 3D). SKOV3/DDP cells were then
transfected with HAND2-AS1 and examined for protein levels of
PTEN/PI3K/AKT signaling factors, such as PTEN, p-PI3K, PI3K, p-AKT
and AKT. As shown in Fig. 3E,
HAND2-AS1 overexpression in SKOV3/DDP cells significantly increased
PTEN protein levels, while reducing p-PI3K/PI3K ratios and
p-AKT/AKT ratios. In summary, PTEN could potentially participate in
HAND2-AS1 effects on ovarian cancer DDP-resistance.
miR-106a directly binds to PTEN 3′UTR
and HAND2-AS1
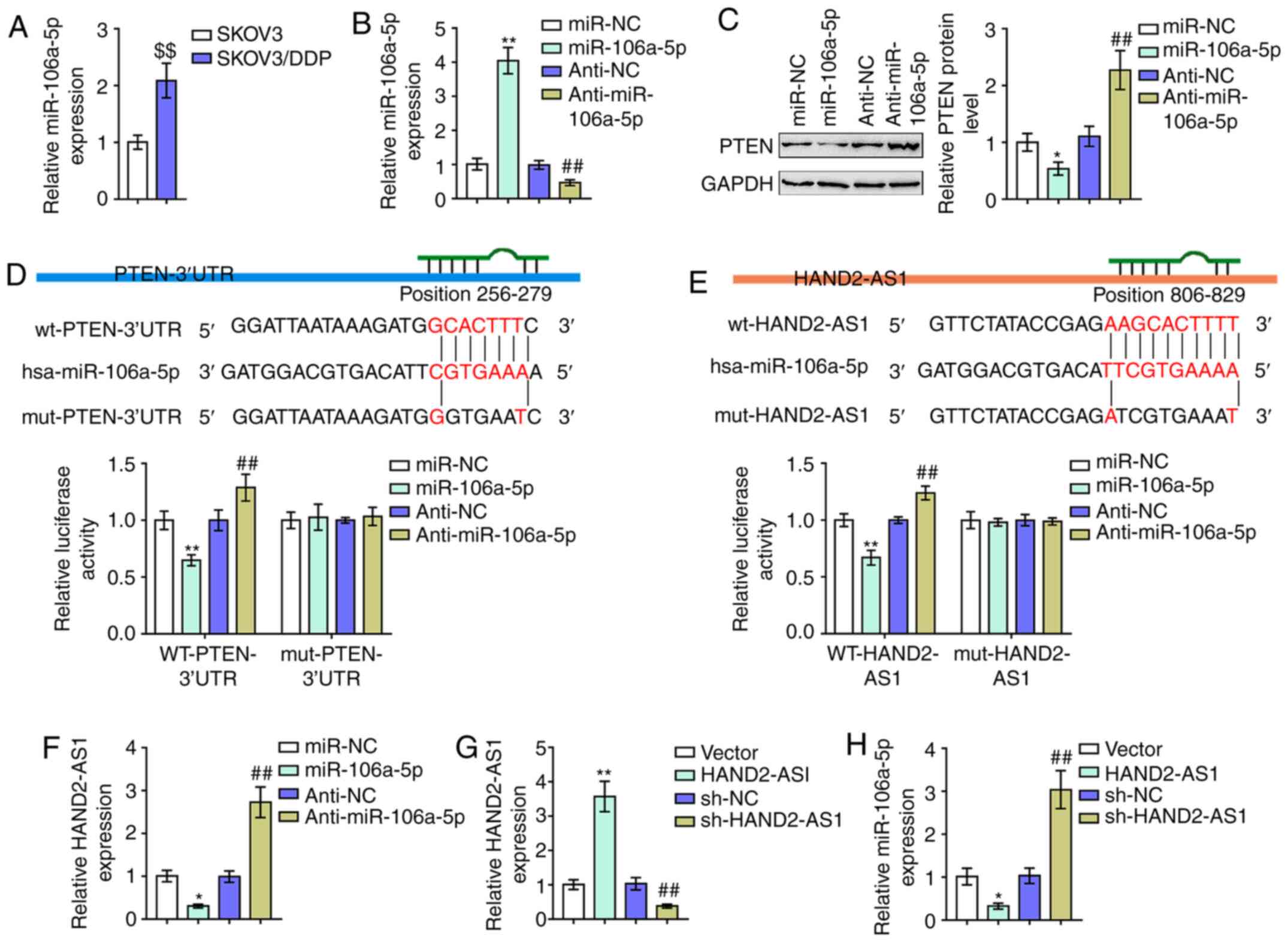
As aforementioned, miR-106a serves as an oncogenic
miRNA by targeting PTEN (15,17,39).
Notably, online tools LncTar predicted that miR-106a could directly
bind to HAND2-AS1 (Fig. 4E). The
potential of the competitive binding of HAND2-AS1 to miR-106a to
counteract miR-106a-mediated suppression on PTEN was then
investigated. Consistent with previous studies on the association
between miR-106a expression and cancer DDP-resistance (14), miR-106a expression was demonstrated
to be markedly upregulated in SKOV3/DDP cells with resistance to
DDP, compared with that in regular SKOV3 cells (Fig. 4A). miR-106a/anti-miR-106a was
transfected to generate miR-106a expression in SKOV3/DDP cells with
resistance to DDP; RT-qPCR was performed to verify the transfection
efficiency (Fig. 4B). SKOV3/DDP
cells with resistance to DDP were subsequently transfected with
miR-106a/anti-miR-106a and the protein levels of PTEN were
examined. As presented in Fig. 4C,
it was demonstrated that the overexpression of miR-106a
significantly decreased, while the inhibition of miR-106a
increased, PTEN proteins.
To verify the reported and predicted bindings of
miR-106a and PTEN 3′UTR and HAND2-AS1, luciferase reporter assays
were performed. As described above, two different types of PTEN
3′UTR and HAND2-AS1 luciferase reporter vectors, wt and mut, were
constructed (Fig. 4D and E). These
vectors were co-transfected with miR-106a/anti-miR-106a in 293T
cells and the luciferase activity was determined and showed that
miR-106a was significantly suppressed. At the same time, miR-106a
inhibition enhanced luciferase activity in both the wt PTEN 3′UTR
and HAND2-AS1 vectors. Mutating the putative miR-106a binding site
could eliminate the luciferase activity alterations (Fig. 4D and E). In SKOV3/DDP cells,
miR-106a-5p decreased the levels of HAND2-AS1 and anti-miR-106a-5p
increased levels of HAND2-AS1 (Fig.
4F), whereas HAND2-AS1 overexpression or knockdown successfully
increased or reduced the levels of HAND2-AS1 (Fig. 4G) and also negatively regulated
miR-106a-5p expression (Fig. 4H).
In brief, miR-106a directly targeted PTEN 3′UTR and HAND2-AS1.
Dynamic effects of HAND2-AS1 and
miR-106a on ovarian cancer cells
After confirming the binding of miR-106a to PTEN
3′UTR and HAND2-AS1, the dynamic effects of these factors on
SKOV3/DDP cell resistance to DDP were subsequently investigated.
SKOV3/DDP cells were co-transfected with HAND2-AS1 and miR-106a and
were first examined for the mRNA expression of PTEN. PTEN mRNA
expression was significantly upregulated by HAND2-AS1
overexpression, but downregulated by miR-106a overexpression. The
effects of HAND2-AS1 overexpression were partially reversed by
miR-106a overexpression (Fig.
5A).
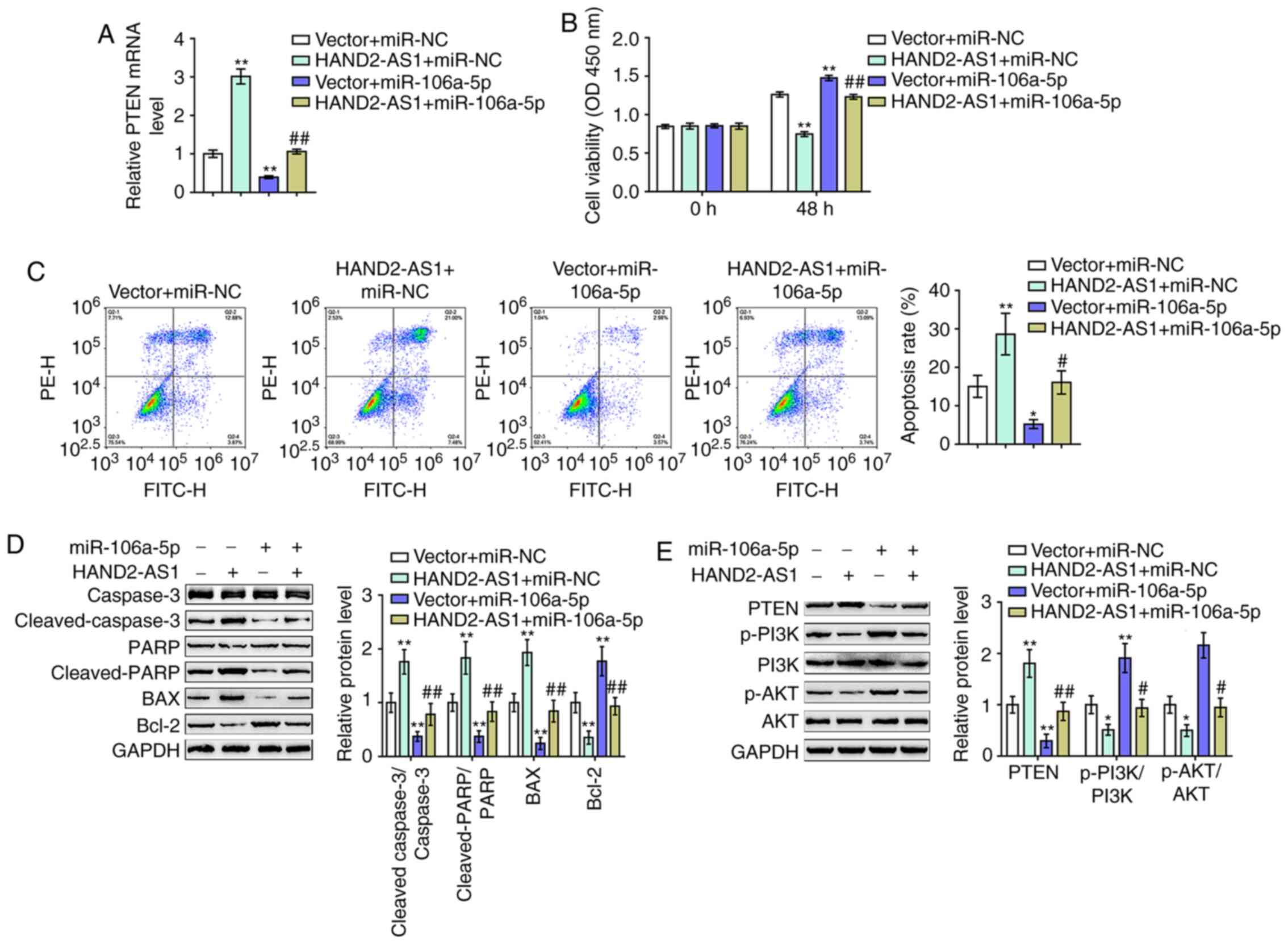 | Figure 5.Dynamic effects of HAND2-AS1 and
miR-106a on ovarian cancer cells. DDP-resistant SKOV3/DDP cells
were co-transfected with HAND2-AS1 and miR-106a and examined for
(A) mRNA expression of PTEN by reverse transcription-quantitative
PCR. (B) Cell viability was assessed using an MTT assay. (C) Cell
apoptosis was determined by flow cytometry. (D) The protein levels
of caspase-3, cleaved-caspase-3, PARP, cleaved-PARP, Bax and Bcl-2
were measured by western blotting. (E) Protein levels of PTEN,
p-PI3K, PI3K, p-AKT and AKT were determined by western blotting.
*P<0.05, **P<0.01 vs. vector + miR-NC group;
#P<0.05, ##P<0.01 vs. vector + miR-106a
group. miR, microRNA; DDP, cisplatin; p-, phosphorylated; PARP,
poly-ADP ribose polymerase; NC, negative control. |
As for the dynamic effects of HAND2-AS1 and miR-106a
on SKOV3/DDP cells with resistance to DDP, cleaved-caspase-3,
caspase-3, cleaved-PARP, PARP, Bax, Bcl-2, PTEN, p-PI3K, PI3K,
p-AKT and AKT cell viability, cell apoptosis and protein levels
were then examined. HAND2-AS1 overexpression significantly reduced
cell viability, while it increased cell apoptosis, whereas miR-106a
exerted the opposite effects. Notably, the effects of HAND2-AS1
overexpression were markedly reversed by miR-106a overexpression
(Fig. 5B and C).
Regarding the Bcl-2/caspase-3 apoptotic signaling
pathway, HAND2-AS1 overexpression significantly increased the
cleaved-caspase-3/caspase-3 ratios, the cleaved PARP/PARP ratios
and Bax proteins, while it decreased Bcl-2 protein levels (Fig. 5D). miR-106a overexpression exerted
opposite effects on these factors (Fig.
5D). The overexpression of miR-106a could significantly
attenuate the effects of HAND2-AS1 overexpression (Fig. 5D). The HAND2-AS1/miR-106a axis
modulated Bcl-2/caspase-3 apoptotic signaling to affect the
viability and apoptosis of SKOV3/DDP cells.
Regarding the PTEN/PI3K/AKT signaling pathway,
HAND2-AS1 overexpression significantly increased PTEN protein
levels, while it decreased p-PI3K/PI3K ratios and p-AKT/AKT ratios
(Fig. 5E). miR-106a overexpression
exerted opposite effects on these factors (Fig. 5E). Similarly, the overexpression of
miR-106a could significantly attenuate the effects of HAND2-AS1
overexpression on PTEN/PI3K/AKT signaling (Fig. 5E). In summary, HAND2-AS1
counteracted miR-106a-mediated suppression on PTEN through its role
as a ceRNA.
Discussion
The present study observed that lncRNA HAND2-AS1
expression was significantly downregulated within ovarian
carcinoma, especially within recurrent and DDP-resistant ovarian
carcinoma. The expression of HAND2-AS1 was also markedly inhibited
in SKOV3/DDP cells with resistance to DDP. In SKOV3/DDP cells,
HAND2-AS1 overexpression inhibited cell viability and promoted cell
apoptosis upon DDP treatment through Bcl-2/caspase-3 apoptotic
signaling. In agreement with the hypothesis of the present study,
PTEN mRNA expression was also markedly inhibited in SKOV3/DDP
ovarian cancer cells, while HAND2-AS1 overexpression rescued PTEN
proteins and blocked PI3K/AKT signaling activation. In addition,
miR-106a was directly bound to PTEN 3′UTR and HAND2-AS1. Upon DDP
treatment, miR-106a overexpression in SKOV3/DDP cells promoted cell
viability, inhibited cell apoptosis through Bcl-2/caspase-3
apoptotic signaling, downregulated the protein levels of PTEN and
upregulated PI3K/AKT signaling activity. Notably, miR-106a
overexpression partially reversed the effect of HAND2-AS1
overexpression upon PTEN proteins and SKOV3/DDP cell proliferation
upon DDP treatment.
HAND2-AS1 is a lncRNA transcribed antisense adjacent
to HAND2 in chromosome 4q33-34. HAND2-AS1 is considered to be
tumor-suppressive on a variety of types of cancer (40–42).
In endometrioid endometrial carcinoma, lncRNA HAND2-AS1 inactivates
neuromedin U to suppress tumor invasion and metastasis (42). Through interacting with TGFβ1,
lncRNA HAND2-AS1 can suppress the invasion and metastasis of
non-small cell lung cancer and maintain stem cell activity
(43). Serving as a sponge for
micRNA-1275, lncRNA HAND2-AS1 suppresses cancer cell proliferation
and enhances cell apoptosis in chronic myeloid leukemia cells
(44). lncRNA HAND2-AS1
overexpression inhibits the capacity of esophagus squamous cell
carcinoma cells to proliferate, migrate and invade (45). According to GSE data (28,29),
lncRNA HAND2-AS1 is significantly inhibited within ovarian cancer,
particularly in recurrent and DDP-resistant ovarian cancer. Based
on the experimental results of the present study, lncRNA HAND2-AS1
expression was markedly reduced in SKOV3/DDP cells with resistance
to DDP, compared with regular SKOV3 cells, suggesting its
underlying effect on ovarian cancer cell resistance to DDP. As
hypothesized, lncRNA HAND2-AS1 overexpression re-sensitized
SKOV3/DDP cells to DDP treatment by inhibiting the viability and
the enhancement of cell-apoptosis through Bcl-2/caspase-3 apoptotic
signaling.
PTEN downregulation and consequent increase in
PI3K/AKT signaling activity are critical events within ovarian
cancer DDP-resistance (10–12,46).
Consistent with these previous studies, the expression of PTEN is
markedly lower in ovarian carcinoma with resistance to DDP
according to online GSE data (28)
and notably reduced within SKOV3/DDP cells, compared with regular
SKOV3 cells according to experimental results from the present
study. lncRNA HAND2-AS1 overexpression in SKOV3/DDP cells
significantly rescued PTEN proteins and suppressed PI3K/AKT
activity, suggesting that PTEN could be involved in lncRNA
HAND2-AS1 regulation of ovarian cancer DDP-resistance. Considering
the common mechanisms of lncRNAs exerting their roles in types of
cancer, namely serving as ceRNAs for miRNAs to counteract
miRNA-mediated suppression on target mRNAs (26,47,48),
the involvement of miRNAs is also hypothesized.
Based on analysis of the GSE and TCGA-OV data and
online tool LncTar prediction, attention was drawn to an oncogenic
miRNA, miR-106a. Although there have been numerous reports that
miR-106a exerts cancer-promoting effects on a variety of malignant
tumors, only a few studies focus on the mechanisms of miR-106a
enhancing ovarian cancer cell resistance to medication (49,50).
It was reported by Li et al (49) that miR-106a reduces ovarian cancer
cell sensitivity to DDP via binding to PDCD4. Huh et al
(50) revealed that the increase in
miR-106a is associated with the ovarian cancer cell and human tumor
sample resistance to paclitaxel. Consistent with the observation by
Li et al (49) that miR-106a
expression is increased in ovarian cancer OVCAR3/CIS cells with
resistance to DDP, compared with parental OVCAR3 cells, the present
study also found that the expression of miR-106a is markedly
upregulated in SKOV3/DDP cells with resistance to DDP, compared
with regular SKOV3 cells. lncRNAs serve as ceRNAs to sponge miRNAs,
thereby regulating gene expression (51). miRNAs have been shown to bind and
regulate lncRNA stability and induce miRNA-mediated decay (52). In the present study, miR-106a formed
a lncRNA-miRNA-mRNA network with lncRNA HAND2-AS1 and PTEN to
regulate PTEN protein levels. miR-106a overexpression reduced
HAND2-AS1 levels and further increased the SKOV3/DDP cells
resistance to DDP upon DDP treatment; in addition, the
overexpression of miR-106a markedly reversed the effects of
HAND2-AS1 overexpression on SKOV3/DDP cells upon DDP treatment,
indicating that miR-106a served as an oncogenic miRNA through
enhancement of ovarian cancer DDP-resistance. Notably, the
mechanism was shown to be similar to a previous study (49).
In conclusion, a lncRNA HAND2-AS1/miR-106a/PTEN axis
re-sensitized DDP-resistant SKOV3/DDP cells to DDP treatment.
Notably, one study reported that miR-106a inhibits ovarian cancer
A2780 cell resistance to DDP via binding to myeloid cell leukemia-1
(53), which differs from the
present findings. This might be attributed to the different targets
of miR-106a. Since miRNAs target multiple downstream targets, the
present findings require further in vivo and clinical
investigations to extend their application scope.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LijL and LiL were responsible for the acquisition
and analysis of data. LH and TL performed the bioinformatics and
data analyses. DX and XL were responsible for collecting the
clinical samples and analysis of data. LijL wrote the manuscript.
LiL supervised the present study. LH and TL confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Clinical sampling was performed with the approval of
the Ethics Committee of Hunan Normal University (Changsha, China).
Signed written informed consent was obtained from all patients
enrolled.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Ali AY, Farrand L, Kim JY, Byun S, Suh
J-Y, Lee HJ and Tsang BK: Molecular determinants of ovarian cancer
chemoresistance: New insights into an old conundrum. Ann N Y Acad
Sci. 1271:58–67. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li F, Guo Y, Han L, Duan Y, Fang F, Niu S,
Ba Q, Zhu H, Kong F, Lin C, et al: In vitro and in vivo growth
inhibition of drug-resistant ovarian carcinoma cells using a
combination of cisplatin and a TRAIL-encoding retrovirus. Oncol
Lett. 4:1254–1258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Milella M, Falcone I, Conciatori F, Cesta
Incani U, Del Curatolo A, Inzerilli N, Nuzzo CMA, Vaccaro V, Vari
S, Cognetti F, et al: PTEN: Multiple functions in human malignant
tumors. Front Oncol. 5:242015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cantley LC and Neel BG: New insights into
tumor suppression: PTEN suppresses tumor formation by restraining
the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA.
96:4240–4245. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bermúdez Brito M, Goulielmaki E and
Papakonstanti EA: Focus on PTEN regulation. Front Oncol. 5:1662015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou
H and Jia L: MicroRNA-130b targets PTEN to mediate drug resistance
and proliferation of breast cancer cells via the PI3K/Akt signaling
pathway. Sci Rep. 7:419422017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang H, Kong W, He L, Zhao J-J, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA
amplification contributes to cisplatin resistance in an ovarian
cancer cell line. Gynecol Oncol. 97:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schöndorf T, Ebert MP, Hoffmann J, Becker
M, Moser N, Pur S, Göhring UJ and Weisshaar MP: Hypermethylation of
the PTEN gene in ovarian cancer cell lines. Cancer Lett.
207:215–220. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi X, Xiao L, Mao X, He J, Ding Y, Huang
J, Peng C and Xu Z: miR-205-5p mediated downregulation of PTEN
contributes to cisplatin resistance in C13K human ovarian cancer
cells. Front Genet. 9:5552018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Amini-Farsani Z, Sangtarash MH, Shamsara M
and Teimori H: MiR-221/222 promote chemoresistance to cisplatin in
ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway.
Cytotechnology. 70:203–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gong J, Xing C, Wang LY, Xie SS and Xiong
WD: L-Tetrahydropalmatine enhances the sensitivity of human ovarian
cancer cells to cisplatin via microRNA-93/PTEN/Akt cascade. J BUON.
24:701–708. 2019.PubMed/NCBI
|
|
13
|
Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu
FQ, Hu R and Wang MS: miR-106a promotes growth and metastasis of
non-small cell lung cancer by targeting PTEN. Int J Clin Exp
Pathol. 8:38272015.PubMed/NCBI
|
|
14
|
Fang Y, Shen H, Li H, Cao Y, Qin R, Long
L, Zhu X, Xie C and Xu W: miR-106a confers cisplatin resistance by
regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim
Biophys Sin (Shanghai). 45:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dhar S, Kumar A, Rimando AM, Zhang X and
Levenson AS: Resveratrol and pterostilbene epigenetically restore
PTEN expression by targeting oncomiRs of the miR-17 family in
prostate cancer. Oncotarget. 6:27214–27226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu B, Cai H, Zheng R, Yang S, Zhou Z and
Tu J: Long non-coding RNA 657 suppresses hepatocellular carcinoma
cell growth by acting as a molecular sponge of miR-106a-5p to
regulate PTEN expression. Int J Biochem Cell Biol. 92:34–42. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen L, Zhang F, Sheng XG, Zhang SQ, Chen
YT and Liu BW: MicroRNA-106a regulates phosphatase and tensin
homologue expression and promotes the proliferation and invasion of
ovarian cancer cells. Oncol Rep. 36:2135–2141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheetham S, Gruhl F, Mattick J and Dinger
M: Long noncoding RNAs and the genetics of cancer. Br J Cancer.
108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren C, Li X, Wang T, Wang G, Zhao C, Liang
T, Zhu Y, Li M, Yang C, Zhao Y, et al: Functions and mechanisms of
long noncoding RNAs in ovarian cancer. Int J Gynecol Cancer.
25:566–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sanchez-Mejias A and Tay Y: Competing
endogenous RNA networks: Tying the essential knots for cancer
biology and therapeutics. J Hematol Oncol. 8:302015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen
Y, Ouyang J, Zhang L, Tan H, Chen R, et al: A long noncoding RNA
critically regulates Bcr-Abl-mediated cellular transformation by
acting as a competitive endogenous RNA. Oncogene. 34:1768–1779.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bowen NJ, Walker LD, Matyunina LV, Logani
S, Totten KA, Benigno BB and McDonald JF: Gene expression profiling
supports the hypothesis that human ovarian surface epithelia are
multipotent and capable of serving as ovarian cancer initiating
cells. BMC Med Genomics. 2:712009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davis S and Meltzer PS: GEOquery: A bridge
between the Gene Expression Omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Revelle WR (Photographer), . psych:
Procedures for Personality and Psychological Research 2017.
|
|
33
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao J, Wu N, Liu X, Xia Y, Chen Y, Li S
and Deng Z: MicroRNA-142-3p inhibits cell proliferation and
chemoresistance in ovarian cancer via targeting sirtuin 1. Exp Ther
Med. 15:5205–5214. 2018.PubMed/NCBI
|
|
35
|
Zhang C, Wang M, Shi C, Shi F and Pei C:
Long non-coding RNA Linc00312 modulates the sensitivity of ovarian
cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Biosci Trends. 12:309–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Juric D, Castel P, Griffith M, Griffith
OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et
al: Convergent loss of PTEN leads to clinical resistance to a
PI(3)Kα inhibitor. Nature. 518:240–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luo B, Kang N, Chen Y, Liu L and Zhang Y:
Oncogene miR-106a promotes proliferation and metastasis of prostate
cancer cells by directly targeting PTEN in vivo and in vitro.
Minerva Med. 109:24–30. 2018.PubMed/NCBI
|
|
40
|
Kang Y, Zhu X, Xu Y, Tang Q, Huang Z, Zhao
Z, Lu J, Song G, Xu H, Deng C, et al: Energy stress-induced lncRNA
HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits
osteosarcoma progression. Am J Cancer Res. 8:5262018.PubMed/NCBI
|
|
41
|
Zhou J, Lin J, Zhang H, Zhu F and Xie R:
LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer
progression by upregulating KLF14. Biochem Biophys Res Commun.
503:1848–1853. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang X, Wang CC, Lee WY, Trovik J, Chung
TK and Kwong J: Long non-coding RNA HAND2-AS1 inhibits invasion and
metastasis in endometrioid endometrial carcinoma through
inactivating neuromedin U. Cancer Lett. 413:23–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Miao F, Chen J, Shi M, Song Y, Chen Z and
Pang L: LncRNA HAND2-AS1 inhibits non-small cell lung cancer
migration, invasion and maintains cell stemness through the
interactions with TGF-β1. Biosci Rep. Jan 11–2019.(Epub ahead of
print). doi: 10.1042/BSR20181525. View Article : Google Scholar
|
|
44
|
Yang J, Shi M and Zeng Y: LncRNA HAND2-AS1
inhibits proliferation and promotes apoptosis of chronic myeloid
leukemia cells by sponging with micRNA-1275. Eur Rev Med Pharmacol
Sci. 23:2103–2111. 2019.PubMed/NCBI
|
|
45
|
Yan Y, Li S, Wang S, Rubegni P, Tognetti
L, Zhang J and Yan L: Long noncoding RNA HAND2-AS1 inhibits cancer
cell proliferation, migration, and invasion in esophagus squamous
cell carcinoma by regulating microRNA-21. J Cell Biochem.
120:9564–9571. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fu X, Tian J, Zhang L, Chen Y and Hao Q:
Involvement of microRNA-93, a new regulator of PTEN/Akt signaling
pathway, in regulation of chemotherapeutic drug cisplatin
chemosensitivity in ovarian cancer cells. FEBS Lett. 586:1279–1286.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Abdollahzadeh R, Daraei A, Mansoori Y,
Sepahvand M, Amoli MM and Tavakkoly-Bazzaz J: Competing endogenous
RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A
new look at hallmarks of breast cancer. J Cell Physiol.
234:10080–10100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li H, Xu H, Shen H and Li H: microRNA 106a
modulates cisplatin sensitivity by targeting PDCD4 in human ovarian
cancer cells. Oncol Lett. 7:183–188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huh JH, Kim TH, Kim K, Song JA, Jung YJ,
Jeong JY, Lee MJ, Kim YK, Lee DH and An HJ: Dysregulation of
miR-106a and miR-591 confers paclitaxel resistance to ovarian
cancer. Br J Cancer. 109:452–461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Subramanian S: Competing endogenous RNAs
(ceRNAs): New entrants to the intricacies of gene regulation. Front
Genet. 5:82014.PubMed/NCBI
|
|
52
|
Ballantyne M, McDonald R and Baker A:
lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol
Ther. 99:494–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rao Y, Shi H, Ji M and Chen C: MiR-106a
targets Mcl-1 to suppress cisplatin resistance of ovarian cancer
A2780 cells. J Huazhong Univ Sci Technolog Med Sci. 33:567–572.
2013. View Article : Google Scholar : PubMed/NCBI
|