Introduction
GC is the fifth commonest malignancy and the third
leading cause of cancer-related mortality in the world (1). According to the World Health
Organization, GC is classified into several subtypes regarding to
the molecular pathogenesis and clinicopathological profiles,
including papillary, tubular and mucinous adenocarcinomas, mixed
carcinomas and poorly cohesive carcinomas, with or without a
component of ring cells (2). H.
pylori infection, which causes chronic inflammation in gastric
and subsequent cancer development, is the major risk factor for GC
(3). Due to vaccination or drugs
developed to treat H. pylori infection, the incidence of
H. pylori-related GC has decreased, whereas genetic
variation-induced GC patients has increased (4). Accumulating evidence has identified
that TP53, PIK3CA and APC are the most frequent mutated genes in GC
patients (5–7). Additionally, there are potentially
amplifications of receptor tyrosine kinases, including Erb-B2
Receptor Tyrosine Kinase 2 (ERBB2) and EGFR, also common in solid
tumors (8). However, there is a
lack of effective drugs to treat the patients carrying these
alterations. Novel disease avenues need to be explored for clinical
diagnosis and treatment.
microRNAs (miRNAs), which contain 21–24 nucleotides,
are types of non-coding RNA. They control different cellular
functions by targeting the abundance of the mRNA of key genes in
cells (9). Dysregulation of miRNAs
stimulates or suppresses cancer growth, proliferation and
metastasis by downregulating tumor suppressors or oncogenes
(10). Well-known oncogenic miRNAs
are miR-17 and miR-21. For example, cancer-associated fibroblasts
secrete miR-17, which enhances the aggressiveness of colon cancer
by regulating the Runt-related transcription factor
(RUNX)3/Myc/TGF-β1 cascade (11).
miR-17-5p is overexpressed in GC patients (12). Upregulation of miR-17-5p contributes
to GC growth and metastasis by downregulating RUNX3 (13). In addition, the oncogenic function
of miR-17 has been reported in thyroid cancer, laryngeal squamous
cell carcinoma, breast cancer and prostate cancer (14–16).
miR-21 is highly expressed in a wide range of types of cancer, such
as lung cancer, GC, colon cancer and prostate cancer (17–20).
miR-21 in serum is considered as a marker of colorectal cancer
(CRC) and its high expression is associated with shorter
progression free survival of CRC patients. As well as miR-21,
various miRNAs, including miR-20a-5p, miR-103a-3p, miR-106a-5p and
miR-143-5p, have been demonstrated to act as novel biomarkers for
CRC recurrence (21). miR-21
promotes the growth and metastasis of various types of cancer by
targeting different tumor suppressors, including SMAD7, PTEN and
HMG-box transcription factor 1 (22). As a previous study indicated, PTEN
deficiency, which might be associated with gene mutations, loss of
heterozygosity and promoter hypermethylation, serves as an
indicator of the pathological state of GC. When considered with
ERBB2 expression, PTEN deficiency was related to a more aggressive
proliferative capacity of tumors (23).
miR-486-5p was initially identified as a tumor
suppressive microRNA in lung cancer; Pang et al (24) found that among 26 miRNAs, miR-486-5p
is one of the most significantly decreased miRNAs in non-small-cell
lung cancer. A subsequent study showed that miR-486-5p is
downregulated in breast cancer patients (25). miR-486-5p also suppresses the growth
and proliferation of papillary thyroid carcinoma cells by
negatively regulating fibrillin-1 (26).
The present study investigated the expression of
miR-486-5p in GC tissues by analyzing The Cancer Genome Atlas
(TCGA) database (http://tcga-data.nci.nih.gov/tcga/) and reverse
transcription-quantitative (RT-q) PCR in samples of patients.
Overexpression by mimics and knockdown by inhibitors were used to
study the role of miR-486-5p in GC cell function based on CCK8,
colony formation, cell cycle analysis, apoptosis measurement and
Transwell analysis of migration. The findings suggested that
miR-486-5p is a tumor suppressor in GC.
Materials and methods
Cell lines and regents
Gastric normal cells GES-1 and GC cells MKN-45, AGS,
HGC27 and MKN74 were obtained from the American Type Culture
Collection. miR-486-5p mimics, inhibitors and their controls were
purchased from Guangzhou RiboBio Co., Ltd. The CCK8 kit and cell
cycle assay kit were from Beyotime Institute of Biotechnology. The
apoptosis assay kit was from Invitrogen (Thermo Fisher Scientific,
Inc.).
Clinical analysis of miR-486-5p
The expression of miR-486-5p was analyzed in GC and
normal tissues based on The Cancer Genome Atlas (http://cancergenome.nih.gov) website and the samples
collected from First Affiliated Hospital of Jiamusi University
hospital between April 2019 and October 2019. A total of 372 cancer
and 32 normal tissues from TCGA database were used to analyze
miR-486-5p. A total of 23 cancer and 23 normal tissues (5 cm distal
to the cancer tissues) were collected from GC patients by surgery
(male:female, 10:13; age range: 30–65 years old). The tissues were
harvested before any drug interventions. The expression of
miR-486-5p between 23 cancer and 23 normal tissues was compared
using paired Student's t-test. A written informed consent was
obtained from each patient. The experiments were carried out
according to World Medical Association Declaration of Helsinki and
were supported by the Ethics Committee of First Affiliated Hospital
of Jiamusi University (Ethic approval no. 202067).
Cell culture
GES-1, MKN-45, AGS, HGC27 and MKN74 cells were grown
in Dulbecco modified Eagle's medium (DMEM) or RPMI-1640 (Corning,
Inc.), which was supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
antibiotics (Gibco; Thermo Fisher Scientific, Inc.). Cells were
cultured at 37°C containing 5% CO2.
miR-486-5p downregulation and
overexpression
To knock down and to overexpress miR-486-5p,
miR-486-5p inhibitors (40 nM), mimics (40 nM) and their controls
(40 nM) were used to transfect AGS and HGC27 cells (80% confluence)
by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h at 37°C, the cells were used for
CCK8, colony formation, cell cycle, apoptosis, migration, RT-qPCR
and immunoblotting analysis. The sequence of miR-486-5p mimics and
inhibitor were: miR-486-5p mimics: sense: UCCUGUACUGAGCUGCCCCGAG,
antisense: CUCGGGGCAGCUCAGUACAGGA; miR-486-5p inhibitor:
CUCGGGGCAGCUCAGUACAGGA; miR-486-5p mimics control: sense:
UUUGUACUACACAAAAGUACUG, antisense: CAGUACUUUUGUGUAGUACAAA;
miR-486-5p inhibitor control: CAGUACUUUUGUGUAGUACAAA. The mimics,
inhibitor and controls were purchased from Huzhou Hippo
Biotechnology Co., Ltd.
Ectopic expression of FGF9 by
lentivirus
The coding sequence of FGF9 (cat. no. NM_002010) was
synthesized by Tianyi Huiyuan Biotech Co., Ltd. and then cloned
into overexpressing GV115 lentivirus vectors (Shanghai GeneChem).
The GV115 (20 µg), pHelper1.0 (15 µg) and pHelper2.0 (10 µg) were
cotransfected into 293T cells at 37°C for 6 h using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The virus were concentrated and used to infect
AGS and HGC27 cells when cell density reached 70% confluence. The
subsequent transfection with the lentiviruses was for 72 h (MOI=10)
at 37°C.
RNA isolation and RT-qPCR
Human GC, normal tissues and cells were collected
and homogenized in TRIzol® (Thermo Fisher Scientific,
Inc.). Total RNA of 100 mg tissues or 1×106 cells was
extracted from the tissues following the manufacturer's protocols.
RNA to cDNA conversion was performed using miRNA First Strand cDNA
Synthesis kit (Vazyme Biotech Co., Ltd.), according to the
manufacturer's protocols. Quantification of miRNA was determined by
using SYBR Master Mixture (TransGen Biotech Co., Ltd.) on the
Bio-Rad qPCR system (Bio-Rad Laboratories, Inc.), according to the
manufacturer's protocols. The following thermocycling conditions
were used for qPCR: Initial denaturation at 95°C for 30 sec;
followed by 40 cycles at 95°C for 10 sec, 60°C for 30 sec; lastly
95°C for 25 sec, 60°C for 60 sec, 95°C for 15 sec. Relative
expression levels were calculated using the 2−ΔΔCq
method (27). The experiments were
performed three times. U6 was used as an internal control. Primer
sequences were: miR-486-5p forward: 5′-GCCGTCCTGTCATGAGCTGC-3′ and
reverse: 5′-GTGCAGGGTCCGAGGT-3′; U6 forward:
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse:
5′-ACGCTTCACGAATTTGCGTGTC-3′.
Western blotting
Total proteins were extracted from the cells using
the RIPA lysis buffer (Beyotime Institute of Biotechnology). The
concentration of total protein was detected using a BCA assay kit
(Thermo Fisher Scientific, Inc.) Equal amount of the proteins (30
µg) were separated on 10–12% SDS-PAGE gels and transferred onto
PVDF membranes. The membranes were blocked by 5% non-fat milk
dissolved in PBST for 1 h at room temperature and incubated with
indicated primary antibodies at 4°C overnight, including antibodies
against E-cadherin (1:1,000; cat. no. 14472), N-cadherin (1:1,000;
cat. no. 13116), Vimentin (1:1,000; cat. no. 5741) and cleaved
caspase-3 (1:1,000; cat. no. 9661) were from Cell Signaling
Technology, Inc. Antibody against FGF9 (1:1,000; cat. no. ab206408)
was from Abcam. GAPDH (1:1,000; cat. no. 60004-1-Ig), β-actin
(1:1,000; cat. no. 66009-1-Ig). And the secondary antibodies,
including (HRP-conjugated Affinipure Goat Anti-Mouse IgG (H + L),
1:10,000; cat. no. SA00001-1; HRP-conjugated Affinipure Goat
Anti-Mouse IgG (H+L), 1:10,000; cat. no. SA00001-1; HRP-conjugated
Affinipure Goat Anti-Rabbit IgG (H+L), 1:10,000; cat. no.
SA00001-2), obtained from ProteinTech Group, Inc., were incubated
for 2 h at 4°C. The protein was visualized using Pierce ECL Western
Blotting substrate (Thermo Fisher Scientific, Inc.), and analyzed
using Image Lab 6.1 software (Bio-Rad Laboratories, Inc.).
CCK8 assay
After transfecting GC cells with miR-486-5p,
miR-486-5p inhibitors, mimics and their controls for 48 h, the
cells were counted and a total of 2,000 cells were seeded in
triplicate into 96-well plates and cultured at 37°C and 5%
CO2. At indicated time points, 10% of CCK8 regent was
incubated with culture medium for 3 h at 37°C. Then, the plates
were vibrated for 30 sec and absorbance at 450 nm was detected on a
microplate reader.
Colony formation assay
After transfecting GC cells with miR-486-5p,
miR-486-5p inhibitors, mimics and their controls for 48 h, the
cells were counted and a total of 2,000 cells were seeded into
6-well plates in triplicate and cultured at 37°C and 5%
CO2. Colonies were formed 10 days later and culture
medium was removed. Then, the colonies were washed by PBS for three
times and fixed by methanol for 30 min at room temperature.
Following staining with 0.2% crystal violet (Beijing Solarbio
Science & Technology Co., Ltd.) for 30 min at room temperature,
the cell colonies (>50 cells) were washed and images captured
using a camera.
Cell cycle
Cell cycle was analyzed by using propidium iodide
(PI) staining. After transfecting GC cells with miR-486-5p,
miR-486-5p inhibitors, mimics and their controls for 48 h, the
cells were harvested by trypsin and were washed with iced PBS for
three times. The cells were fixed in 70% ethanol overnight at
−20°C. Then the cells were stained with PI at 37°C for 30 min and
cell cycle was analyzed on a flow cytometry system (cytoFLEX,
Beckman Coulter, Inc.). The data were analyzed by CytExpert
(Version 2.4.0.28; Beckman Coulter, Inc.).
Apoptosis
Cell apoptosis was analyzed by using PI/Annexin V
staining. After transfecting GC cells with miR-486-5p, miR-486-5p
inhibitors, mimics and their controls for 48 h, the cells were
harvested by using EDTA-free trypsin. Then, the cells were washed
by PBS and stained with PI and Annexin V for 15 min at room
temperature. Immediately, cell apoptosis was analyzed on the flow
cytometry system (cytoFLEX, Beckman Coulter, Inc.). CytExpert
software (Version 2.4.0.28, Beckman Coulter, Inc.) was used for
data analysis. The apoptosis rate was calculated as the percentage
of early and late apoptotic cells.
Luciferase reporter assay
The wild-type or mutant 3′UTR of FGF9 was inserted
into psi-CHECK vectors (Promega Corporation). The vectors were
transfected into 293T cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). 48 h later,
luciferase activity was detected by using Dual-Luciferase Reporter
Assay System (Promega Corporation). Renilla luciferase
activity indicated transfection efficiency.
Statistical analysis
Statistical analysis was performed using SPSS 26
software (IBM Corp.). Students' t-test was performed to compare
difference between two groups and one-way analysis of variance
(ANOVA) followed by Tukey's post hoc test was applied for
comparison between ≥3 groups. Linear regression was performed to
analysis correlation between FGF9 and miR-486-5p using the public
database http://gepia.cancer-pku.cn/.
P<0.05 was considered to indicate a statistically significant
difference.
Results
The expression of miR-486-5p is
detected in GC tissues and cells
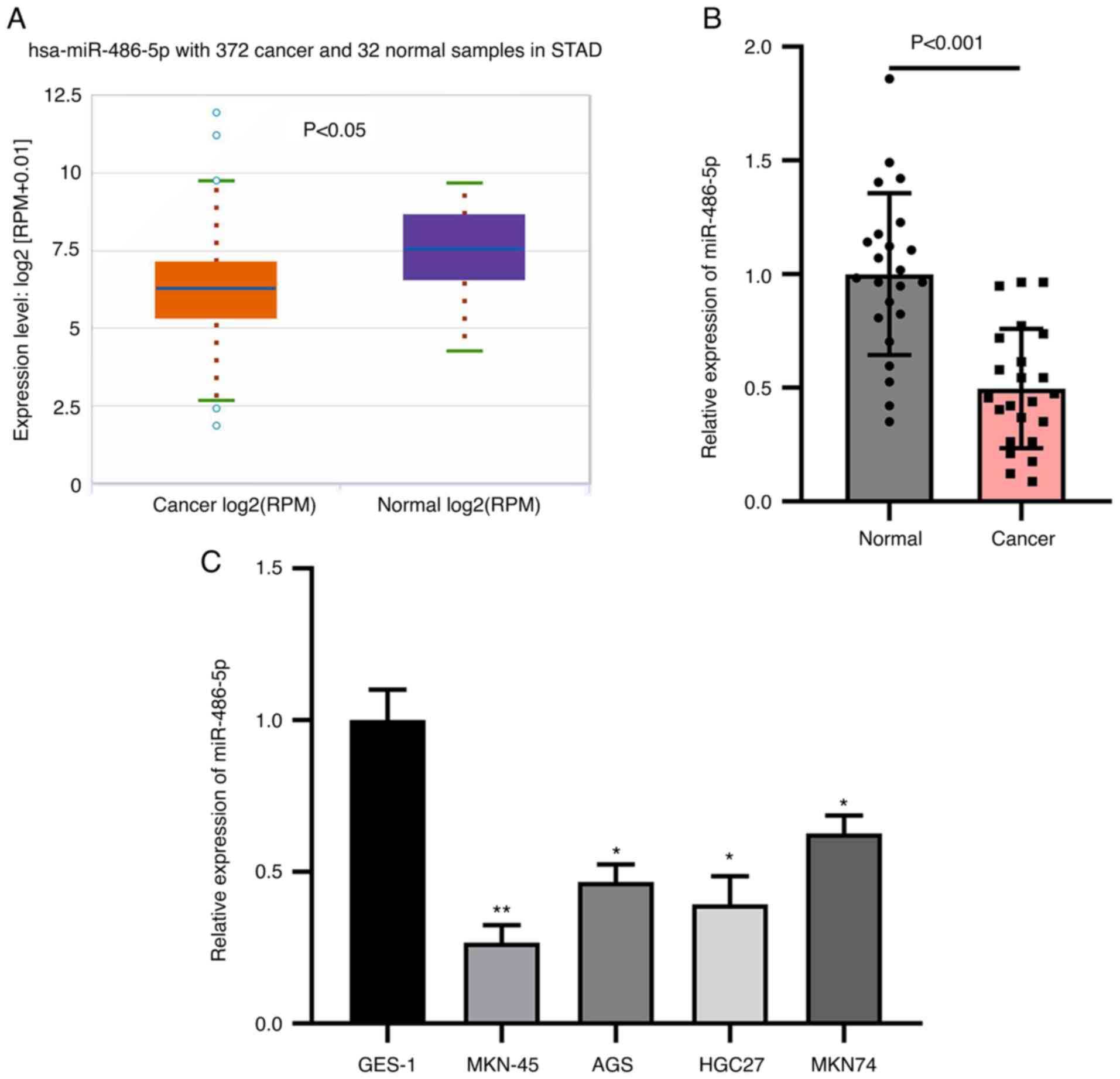
The expression of miR-486-5p in GC and normal
tissues was examined by analyzing the data from TCGA database. The
abundance of miR-486-5p was analyzed in a total of 372 stomach
adenocarcinoma (STAD) and 32 normal tissues. It was found that
miR-486-5p was downregulated in STAD tissues compared with normal
tissues (Fig. 1A). Then, GC tissues
and adjacent tissues were collected from the patients before any
interventions. RT-qPCR analysis of miR-486-5p results found that
miR-486-5p level was reduced in GC tissues comparing to normal
tissues (Fig. 1B), which was
consistent with the data analyzed from TCGA database. miR-486-5p
level was decreased in GC cells, such as MKN-45, AGS, HGC27 and
MKN74, comparing with gastric normal cells GES-1 (Fig. 1C). These results indicate that
miR-486-5p is downregulated in GC tissues and cells.
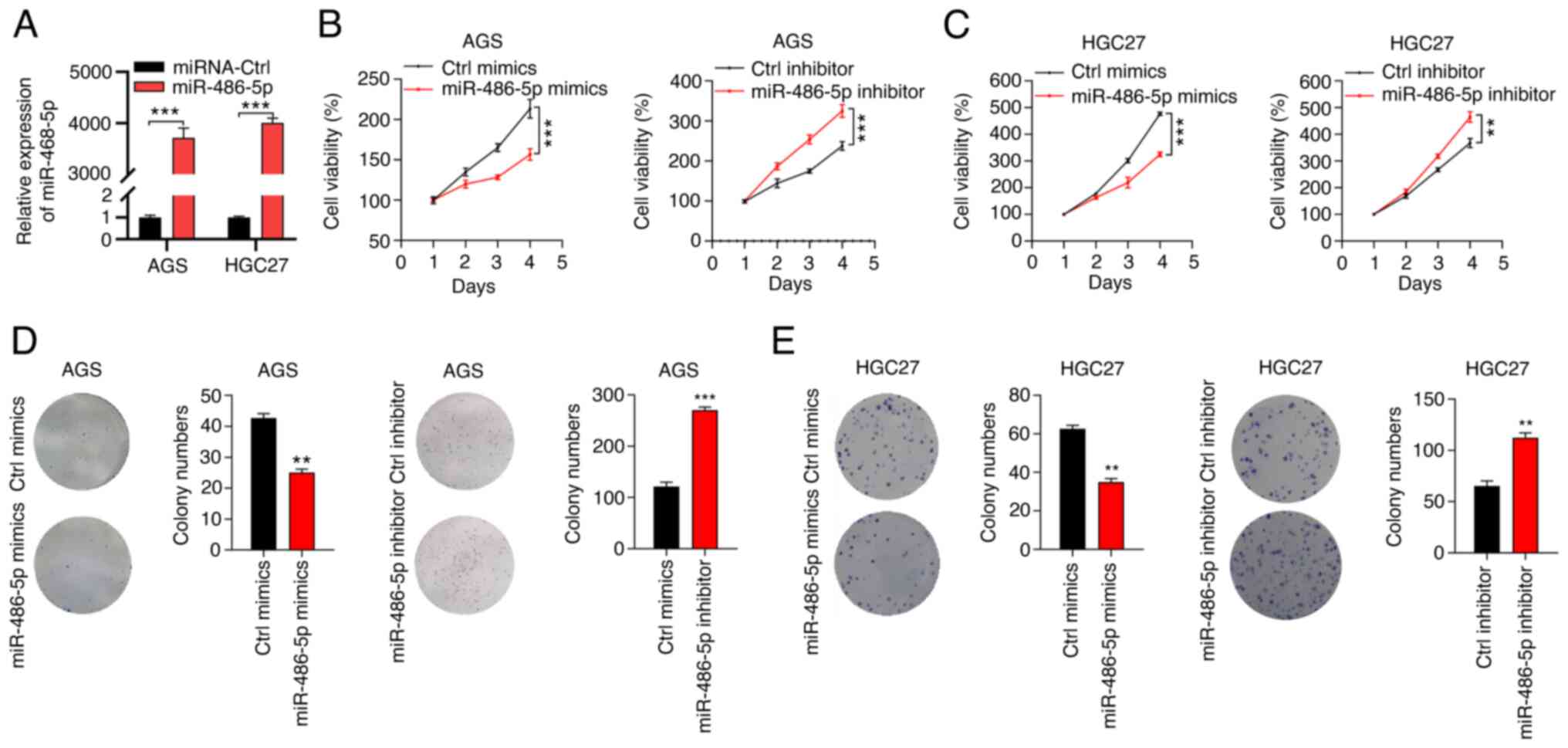
miR-486-5p retards the proliferation
ability of GC cells
To study the function of miR-486-5p in GC, mimics
and inhibitors were applied to overexpress and to downregulate
miR-486-5p in GC cells. Due to the difficulties in performing the
Transwell assay on MKN45 and MKN74 cells, AGS and HGC27 cells were
used to in the present study. As RT-qPCR results suggested, the
miR-486-5p mimics effectively overexpressed miR-486-5p expression
in both AGS and HGC27 cells (Fig.
2A), while there were no obvious changes in miR-486-5p
expression levels following transfection with miR-486-5p inhibitor
(Fig. S1). CCK8 assay was
performed to detect cell growth. The results showed that miR-486-5p
overexpression by mimics suppressed the cell growth of AGS cells.
Similar results were found in HGC27 cells after overexpressing
miR-486-5p. By contrast, miR-486-5p downregulation enhanced the
growth and proliferation capacity of AGS and HGC27 cells (Fig. 2B and C). miR-486-5p ectopic
expression suppressed the colony formation ability of AGS and HGC27
cells. By contrast, miR-486-5p knockdown resulted in accelerated
colony growth in AGS and HGC27 cells (Fig. 2D and E). Thus, miR-486-5p was a
tumor suppressor in GC.
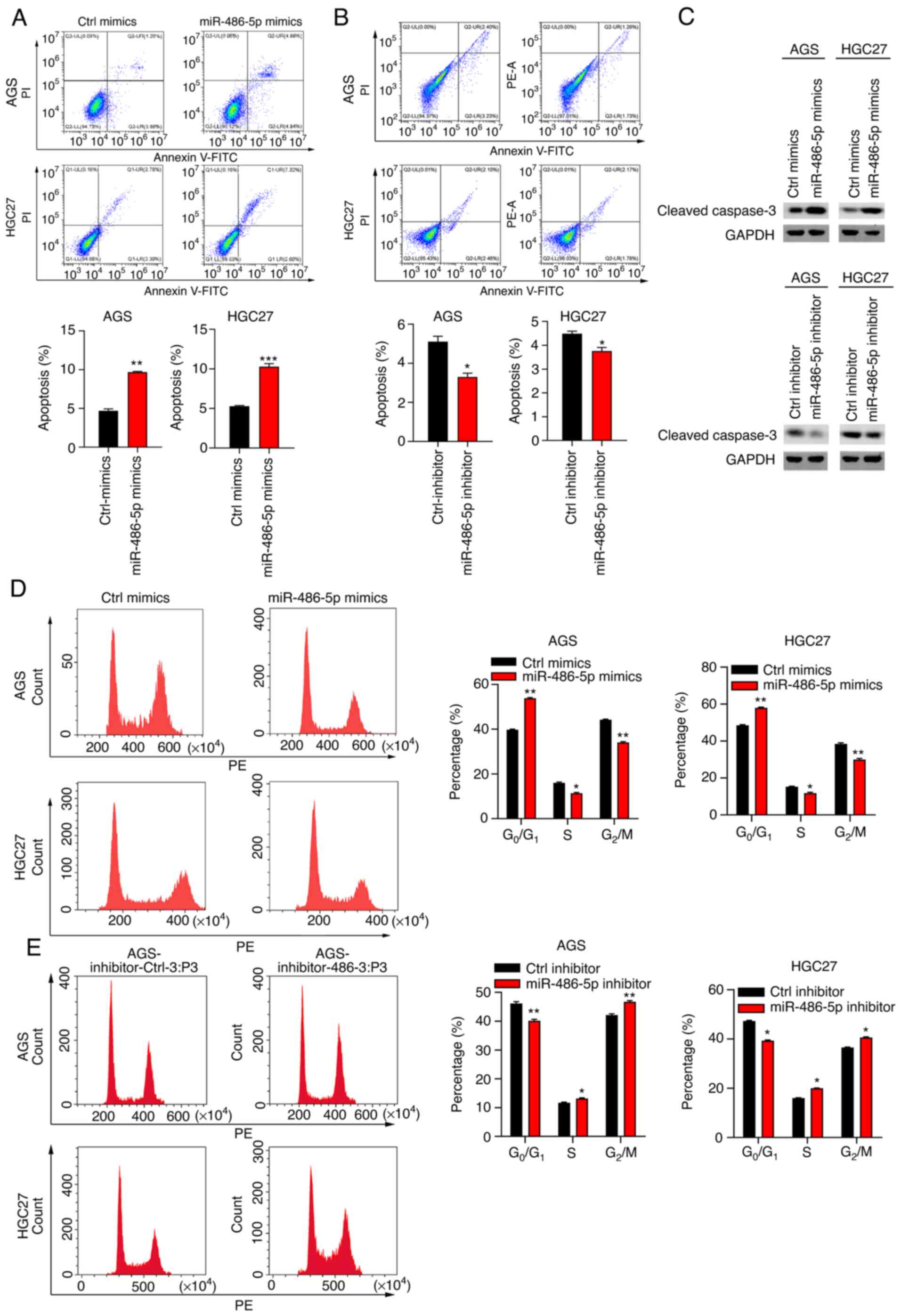
miR-486-5p induces cell apoptosis and
cell cycle arrest at G0/G1 phase
Suppressed cell apoptosis is a hallmark of cancer
(28). The present study next
explored the effect of miR-486-5p on cell apoptosis in GC cells by
staining the cells with PI/Annexin V-APC and flow cytometry
analysis. The results showed that miR-486-5p mimics enhanced cell
apoptosis in AGS and HGC27 cells, while opposite results were found
in the cells which were transfected with miR-486-5p inhibitors
(Fig. 3A and B). In addition, the
expression of cleaved-caspase 3 was effectively upregulated
following treatment with miR-486-5p mimics, while downregulated in
AGS and HGC27 cells transfected with miR-486-5p inhibitors
(Fig. 3C), which was consistent
with the apoptosis detection results. It has been shown that
miR-486-5p regulates cell cycle progress in lung cancer cells
(29). To study the role of
miR-486-5p in cell cycle in GC cells, the cells transfected with
Ctrl-mimics, miR-486-5p mimics and miR-486-5p inhibitors were
subjected to PI staining and flow cytometry analysis. It was found
that miR-486-5p overexpression resulted in increased
G0/G1 phase and decreased G2/M
phase in cell cycle. By contrast, miR-486-5p downregulation showed
adverse effect on cell cycle progress (Fig. 3D and E). Therefore, miR-486-5p
regulated apoptosis and cell cycle in GC cells.
miR-486-5p suppresses migration and
epithelial-to-mesenchymal transition in GC cells
Metastasis is a major problem in GC (30). Transwell assay demonstrated that
miR-486-5p upregulation by mimics reduced cell migration and
invasion in AGS and HGC27 cells. By contrast, miR-486-5p
downregulation increased the migration and invasion ability of AGS
and HGC27 cells (Fig. 4A-D).
Epithelial-to-mesenchymal transition (EMT) is a characteristic of
metastasis (31). As immunoblotting
results showed, miR-486-5p overexpression upregulated E-cadherin
and downregulated N-cadherin and Vimentin in AGS cells, while
knockdown of miR-486-5p effectively inhibited E-cadherin,
upregulated N-cadherin and Vimentin expression in HGC27 cells
(Fig. 4E). Taken together,
miR-486-5p inhibited migration, invasion and EMT in GC.
miR-486-5p downregulates FGF9 by
binding to its 3′UTR sequence
To explore the downstream targets of miR-486-5p, the
negatively correlated genes of miR-486-5p from TCGA database were
analyzed and it was found that expression of FGF9 was inversely
associated with expression of miR-486-5p (Fig. 5A). To validate whether miR-486-5p
regulated FGF9, immunoblotting assays were performed to analyze
FGF9 in Ctrl mimics and miR-486-5p overexpressed cells. The results
showed that miR-486-5p overexpression suppressed the expression of
FGF9 in GC cells (Fig. 5B). The
target for miR-486-5p was analyzed by TargetScan and it was
observed that FGF9 was a potential target for miR-486-5p (Fig. 5C). Luciferase activity was then
assessed in 293T cells transfected with psi-CHECK containing NC,
wild type or mutant 3′UTR sequence of FGF9. The results showed that
miR-486-5p significantly decreased the luciferase activity in cells
transfected with wide type 3′UTR but had no effect in cells
transfected with NC or mutant vectors (Fig. 5D). Taken together the data suggested
that miR-486-5p bound to the 3′UTR sequence to suppress FGF9
expression.
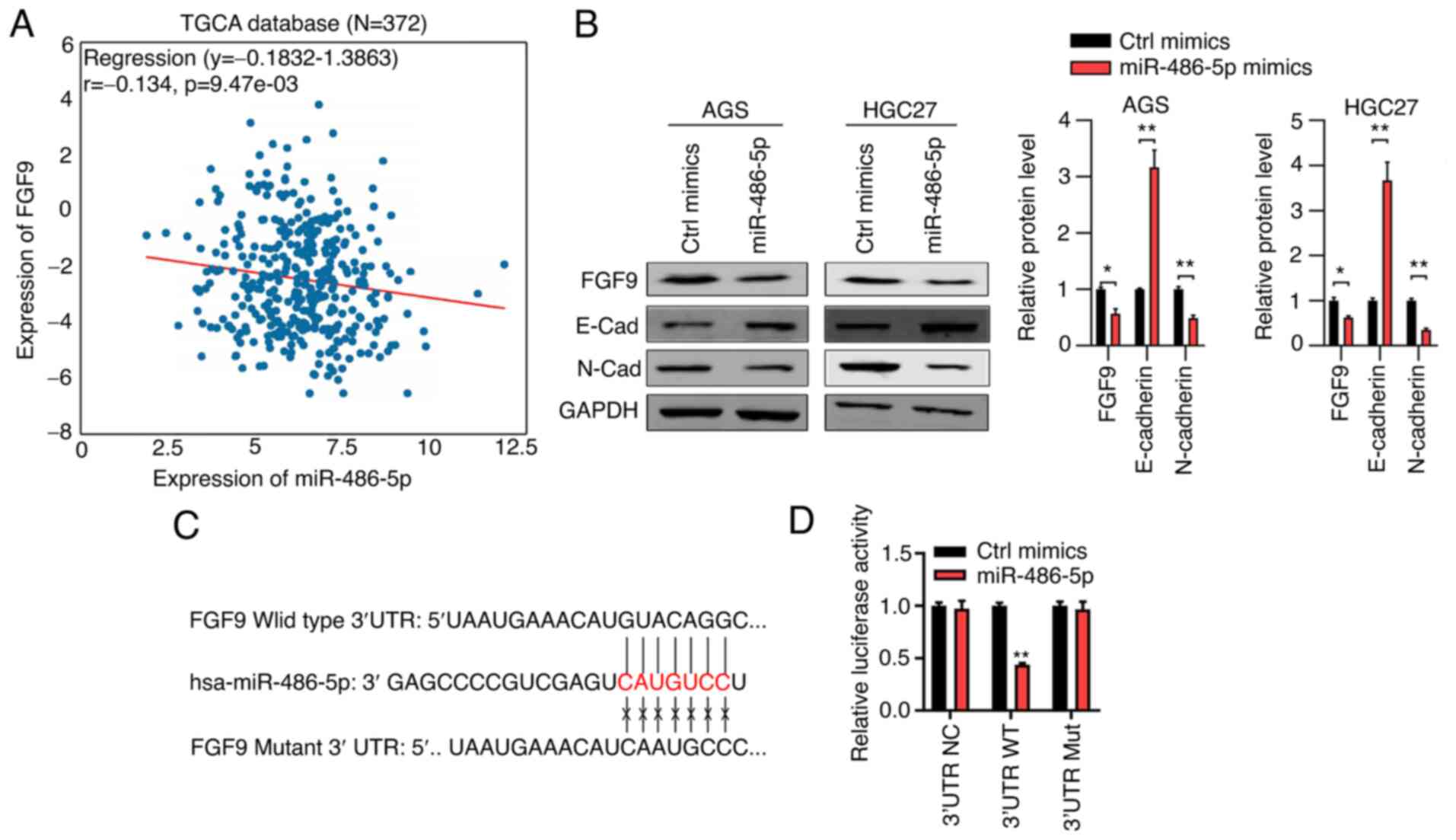 | Figure 5.miR-486-5p downregulates FGF9 in GC
tissues and cells. (A) Correlation between miR-486-5p expression
and FGF9 expression in stomach adenocarcinoma tissues, which was
analyzed from TCGA database. (B) Western blot analysis of FGF9
expression in Ctrl-mimics and miR-486-5p overexpressed AGS and
HGC27 cells. (C) A putative binding site of miR-486-5p in the 3′UTR
sequence of FGF9, as predicted by TargetScan. (D) Luciferase
activity was detected in cells which were co-transfected with
psiCHECK vectors, which were inserted with WT or MU 3′UTR sequence
of FGF9 and miR-486-5p mimics. *P<0.05, **P<0.01 Ctrl mimics
vs. miR-486-50p cotransfected with WT 3′UTR sequence of FGF9. miR,
microRNA; GC, gastric cancer; Ctrl, control; TCGA, The Cancer
Genome Atlas database; UTR, untranslated region; WT, wild-type; MU,
mutant; FGF9, fibroblast growth factor 9. |
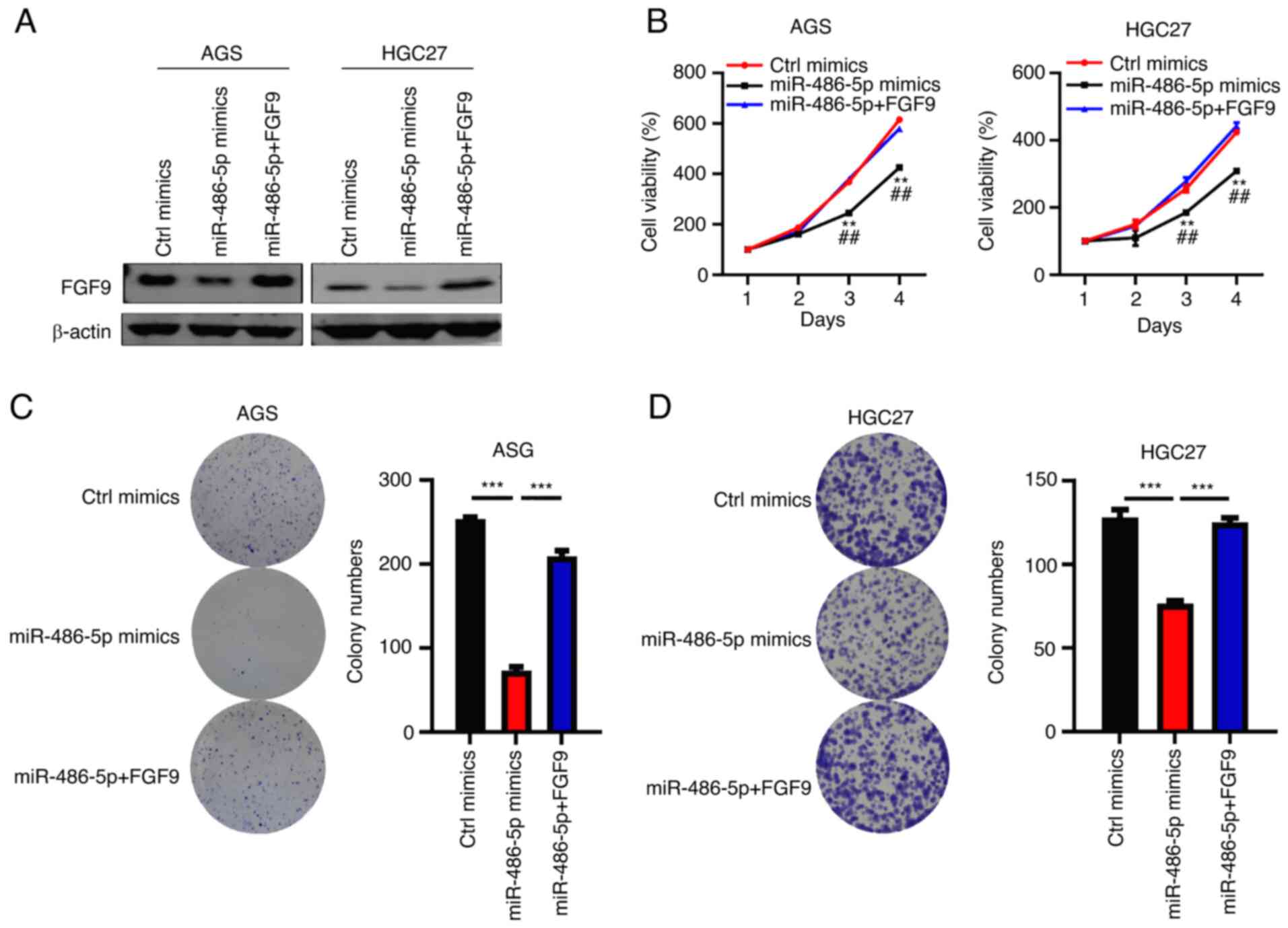
FGF9 restores cell function in
miR-486-5p Overexpressed GC cells
FGF9 has been shown to be an oncoprotein in cancers
(32) and the role of FGF9 in GC
should be determined. The present study overexpressed FGF9 by
lentivirus in AGS and HGC27 cells transfected with miR-486-5p
mimics. The results suggested that FGF9 was significantly
overexpressed in AGS and HGC27 cells transfected with
FGF9-overexpression vector (Fig.
S2). Western blotting results demonstrated that FGF9 was
efficiently downregulated following treatment with miR-486-5p
mimics, while the expression of FGF9 was recovered in the cells
transfected with miR-486-5p mimics (Fig. 6A). When miR-486-5p mimics suppressed
the proliferation of AGS and HGC27 cells, FGF9 overexpression
restored the cell proliferation capacity (Fig. 6B). Consistent results were observed
in cell colony formation in AGS and HGC27 cells (Fig. 6C and D). Thus, it was concluded that
miR-486-5p downregulation of FGF9 inhibited GC cell growth and
proliferation.
Discussion
GC is one of the commonest malignancies and a large
number of cancer patients succumb to this deadly disease. The
present study found that miR-486-5p was downregulated in GC
tissues. Loss-of-function and gain-of-function assays demonstrated
that miR-486-5p acted as a tumor suppressive microRNA in GC, as
revealed by CCK8, colony formation, apoptosis, cell cycle and
migration assays. miR-486-5p also suppressed EMT in GC cells. Thus,
miR-486-5p downregulation in GC patients may contribute to GC
progression.
miR-486-5p was initially identified as a tumor
suppressor in lung cancer. Pim-1 kinase oncogene was the target for
miR-486-5p in lung cancer (24). In
addition, downregulation of miR-486-5p contributes to lung cancer
progression through targeting the proto-oncogene ARHGAP5 (33). miR-486-5p could cause deficient
metastasis and EMT in prostate cancer by targeting Snail (34). Downregulation of miR-486-5p is also
observed in colorectal cancer patients (35). DNA-methylation causes downregulation
of miR-486-5p-stimulated colorectal cancer cell growth and
migration, depending on the activation of the PLAGL2/IGF2/β-catenin
signaling pathway (36). However,
there is evidence that demonstrates miR-486-5p is an onco-miRNA.
miR-486-5p was highly expressed in and promoted myeloid leukemias
of Down syndrome by cooperating with GATA1s (37). Although the expression of miR-486-5p
is reduced in gastric adenocarcinoma patients (38), the function and molecular mechanism
of miR-486-5p in GC are poorly investigated. Consistent with this
finding, the present study found that miR-486-5p was downregulated
in GC tissues and cells. miR-486-5p mimics and inhibitors
transfection revealed that miR-486-5p suppressed cell growth,
proliferation and migration in AGS and HGC27 cells. Cell cycle was
arrested at G0/G1 phase and apoptosis was
promoted by miR-486-5p. miR-486-5p also suppressed EMT in GC. These
results indicated that miR-486-5p functioned as a tumor suppressive
miRNA in GC, which was similar with the function of miR-486-5p
observed in other malignancies, such as lung cancer, colorectal
cancer and prostate cancer (39,40).
The FGF family comprises 24 different polypeptides.
FGF9 gene is mapped on chromosome 13q11-q12 and the protein is
abundant in multiple mammal organs (41). Overexpression of FGF9 contributes to
the development of a wide variety of types of cancer. The first
evidence revealing the oncogenic function of FGF9 was reported in
1999 by Giri et al (42).
Subsequently, a number of studies on cancer research provided
evidence that supported the oncogenic function of FGF9. For
example, FGF9 is regulated by Wnt signaling and exhibits oncogenic
function in ovarian endometrioid adenocarcinomas (43). FGF9 is also induced by hypoxia and
contributes to colon cancer development (44). In GC, FGF9 can be targeted by
miR-26a, which is identified as a tumor suppressive miRNA (45). Nevertheless, the correlation between
miR-486-5p and FGF9 remained to be elucidated. To explore the
downstream targets of miR-486-5p in GC, immunoblotting and
luciferase activity assays were performed and it was found that
miR-486-5p could bind to the 3′UTR sequence of fibroblast growth
factor 9 (FGF9). miR-486-5p downregulated FGF9 in GC cells. There
was a negative correlation between miR-486-5p expression and FGF9
expression in STAD samples based on TCGA database. Notably, FGF9
ectopic expression significantly restored the proliferation and
colony formation ability of AGS and HGC27 cells which had been
transfected with miR-486-5p mimics. These results indicated that
miR-486-5p suppresses GC growth through downregulating of FGF9.
In summary, the present study provided evidence that
the miR-486-5p/FGF9 axis promoted GC cell growth and proliferation.
As miR-486-5p was downregulated in GC patients, the results
suggested that miR-486-5p had tumor suppressive function in GC.
Targeting FGF9 might be an option for the patients with
dysregulated miR-486-5p.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The research was financially supported by the
Heilongjiang Health and Family Planning Commission Scientific
Research Project (grant no. 2018-351) and Heilongjiang Provincial
Health Commission scientific research project (grant no.
2019-314).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HT and QW designed the study. WW and CL performed
most of the experiments and wrote the manuscript. RY and QT
performed the western blot assay. HT and QW confirm the
authenticity of all the raw data. All authors reviewed and approved
the final manuscript.
Ethics approval and consent to
participate
The experiments were carried out according to World
Medical Association Declaration of Helsinki and were supported by
the Ethics Committee of First Affiliated Hospital of Jiamusi
University (Ethic approval number: 202067).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smyth E, Nilsson M, Grabsch H, van Grieken
N and Lordick F: Gastric cancer. Lancet. 396:635–648. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
O'Connor A, O'Morain C and Ford A:
Population screening and treatment of Helicobacter pylori
infection. Nat Rev Gastroenterol Hepatol. 14:230–240. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schulz C, Schutte K, Mayerle J and
Malfertheiner P: The role of the gastric bacterial microbiome in
gastric cancer: Helicobacter pylori and beyond. Therap Adv
Gastroenterol. 12:17562848198940622019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zang Z, Cutcutache I, Poon SL, Zhang SL,
McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sethi NS, Kikuchi O, Duronio GN, Stachler
MD, McFarland JM, Ferrer-Luna R, Zhang Y, Bao C, Bronson R, Patil
D, et al: Early TP53 alterations engage environmental exposures to
promote gastric premalignancy in an integrative mouse model. Nat
Genet. 52:219–230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ebert M, Yu J, Hoffmann J, Rocco A, Röcken
C, Kahmann S, Müller O, Korc M, Sung JJ and Malfertheiner P: Loss
of beta-catenin expression in metastatic gastric cancer. J Clin
Oncol. 21:1708–1714. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zarkavelis G, Boussios S, Papadaki A,
Katsanos KH, Christodoulou DK and Pentheroudakis G: Current and
future biomarkers in colorectal cancer. Ann Gastroenterol.
30:613–621. 2017.PubMed/NCBI
|
|
9
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R, Calin G, Lopez-Berestein G
and Sood A: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Wang S, Lai Q, Fang Y, Wu C, Liu
Y, Li Q, Wang X, Gu C, Chen J, et al: Cancer-associated
fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer
aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive
feedback loop. Cancer Lett. 491:22–35. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Q, Luo G, Yang Z, Zhu F, An Y, Shi Y
and Fan D: miR-17-5p promotes proliferation by targeting SOCS6 in
gastric cancer cells. FEBS Lett. 588:2055–2062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song J, Liu Y, Wang T, Li B and Zhang S:
miR-17-5p promotes cellular proliferation and invasiveness by
targeting RUNX3 in gastric cancer. Biomed Pharmacother.
128:1102462020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Liu G, Li S and Liu X: miR-17-5p
knockdown inhibits proliferation, autophagy and promotes apoptosis
in thyroid cancer via targeting PTEN. Neoplasma. 67:249–258. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Jia X, Liu Y, Dong JH, Ren XM, Xu
O, Liu SH and Shan CG: Silencing of miR-17-5p suppresses cell
proliferation and promotes cell apoptosis by directly targeting
PIK3R1 in laryngeal squamous cell carcinoma. Cancer Cell Int.
20:142020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Xu W, Wang Y, Xu X, Lv S and Dong
X: miR-17-5p promotes migration and invasion in breast cancer cells
by repressing netrin 4. Int J Clin Exp Pathol. 12:1649–1657.
2019.PubMed/NCBI
|
|
17
|
Wang G, Zhou Y, Chen W, Yang Y, Ye J, Ou H
and Wu H: miR-21-5p promotes lung adenocarcinoma cell
proliferation, migration and invasion via targeting WWC2. Cancer
Biomark. 28:549–559. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiao T and Jie Z: MiR-21 promotes the
invasion and metastasis of gastric cancer cells by activating
epithelial-mesenchymal transition. Eur Surg Res. 60:208–218. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun L, Tian D, Yang Z and Li J: Exosomal
miR-21 promotes proliferation, invasion and therapy resistance of
colon adenocarcinoma cells through its target PDCD4. Sci Rep.
10:82712020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim K, Kim HH, Lee CH, Kim S, Cheon GJ,
Kang KW, Chung JK and Youn H: Therapeutic efficacy of modified
anti-miR21 in metastatic prostate cancer. Biochem Biophys Res
Commun. 529:707–713. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boussios S, Ozturk MA, Moschetta M,
Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, Christodoulou
DK and Pavlidis N: The developing story of predictive biomarkers in
colorectal cancer. J Pers Med. 9:122019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valenti MT, Deiana M, Cheri S, Dotta M,
Zamboni F, Gabbiani D, Schena F, Dalle Carbonare L and Mottes M:
Physical exercise modulates miR-21-5p, miR-129-5p, miR-378-5p, and
miR-188-5p expression in progenitor cells promoting osteogenesis.
Cells. 8:7422019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boussios S, Abson C, Moschetta M, Rassy E,
Karathanasi A, Bhat T, Ghumman F, Sheriff M and Pavlidis N: Poly
(ADP-Ribose) polymerase inhibitors: Talazoparib in ovarian cancer
and beyond. Drugs R D. 20:55–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pang W, Tian X, Bai F, Han R, Wang J, Shen
H, Zhang X, Liu Y, Yan X, Jiang F and Xing L: Pim-1 kinase is a
target of miR-486-5p and eukaryotic translation initiation factor
4E, and plays a critical role in lung cancer. Mol Cancer.
13:2402014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rask L, Balslev E, Søkilde R, Høgdall E,
Flyger H, Eriksen J and Litman T: Differential expression of
miR-139, miR-486 and miR-21 in breast cancer patients
sub-classified according to lymph node status. Cell Oncol (Dordr).
37:215–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma X, Wei J, Zhang L, Deng D, Liu L, Mei
X, He X and Tian J: miR-486-5p inhibits cell growth of papillary
thyroid carcinoma by targeting fibrillin-1. Biomed Pharmacother.
80:220–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang RA, Li ZS, Yan QG, Bian XW, Ding YQ,
Du X, Sun BC, Sun YT and Zhang XH: Resistance to apoptosis should
not be taken as a hallmark of cancer. Chin J Cancer. 33:47–50.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu H, Xu H, Lu F, Zhang J, Xu L, Xu S,
Jiang H, Zeng Q, Chen E and He Z: Exosome-derived miR-486-5p
regulates cell cycle, proliferation and metastasis in lung
adenocarcinoma via targeting NEK2. Front Bioeng Biotechnol.
8:2592020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saragoni L: Upgrading the definition of
early gastric cancer: Better staging means more appropriate
treatment. Cancer Biol Med. 12:355–361. 2015.PubMed/NCBI
|
|
31
|
Yeung KT and Yang J:
Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol.
11:28–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang MM, Lai MS, Hong SY, Pan BS, Huang
H, Yang SH, Wu CC, Sun HS, Chuang JI, Wang CY and Huang BM:
FGF9/FGFR2 increase cell proliferation by activating ERK1/2,
Rb/E2F1, and cell cycle pathways in mouse Leydig tumor cells.
Cancer Sci. 109:3503–3518. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Zhang T, Yang K, Zhang M and Wang
K: miR-486-5p suppresses prostate cancer metastasis by targeting
Snail and regulating epithelial-mesenchymal transition. Onco
Targets Ther. 9:6909–6914. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shindo Y, Hazama S, Nakamura Y, Inoue Y,
Kanekiyo S, Suzuki N, Takenouchi H, Tsunedomi R, Nakajima M, Ueno
T, et al: miR-196b, miR-378a and miR-486 are predictive biomarkers
for the efficacy of vaccine treatment in colorectal cancer. Oncol
Lett. 14:1355–1362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Chen X, Zeng K, Xu M, He B, Pan Y,
Sun H, Pan B, Xu X, Xu T, et al: DNA-methylation-mediated silencing
of miR-486-5p promotes colorectal cancer proliferation and
migration through activation of PLAGL2/IGF2/β-catenin signal
pathways. Cell Death Dis. 9:10372018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shaham L, Vendramini E, Ge Y, Goren Y,
Birger Y, Tijssen MR, McNulty M, Geron I, Schwartzman O, Goldberg
L, et al: MicroRNA-486-5p is an erythroid oncomiR of the myeloid
leukemias of Down syndrome. Blood. 125:1292–1301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen H, Ren C, Han C, Wang D, Chen Y and
Fu D: Expression and prognostic value of miR-486-5p in patients
with gastric adenocarcinoma. PLoS One. 10:e01193842015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX and Ma ZL: Direct repression
of the oncogene CDK4 by the tumor suppressor miR-486-5p in
non-small cell lung cancer. Oncotarget. 7:34011–34021. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pisano A, Grinan-Lison C, Farace C,
Fiorito G, Fenu G, Jiménez G, Scognamillo F, Peña-Martin J,
Naccarati A, Pröll J, et al: The inhibitory role of miR-486-5p on
CSC phenotype has diagnostic and prognostic potential in colorectal
cancer. Cancers (Basel). 12:34322020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mattei MG, Penault-Llorca F, Coulier F and
Birnbaum D: The HumanFGF9Gene Maps to chromosomal region 13q11-q12.
Genomics. 29:811–812. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Giri D, Ropiquet F and Ittmann M: FGF9 is
an autocrine and paracrine prostatic growth factor expressed by
prostatic stromal cells. J Cell Physiol. 180:53–60. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hendrix N, Wu R, Kuick R, Schwartz D,
Fearon E and Cho K: Fibroblast growth factor 9 has oncogenic
activity and is a downstream target of Wnt signaling in ovarian
endometrioid adenocarcinomas. Cancer Res. 66:1354–1362. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen TM, Shih YH, Tseng JT, Lai MC, Wu CH,
Li YH, Tsai SJ and Sun HS: Overexpression of FGF9 in colon cancer
cells is mediated by hypoxia-induced translational activation.
Nucleic Acids Res. 42:2932–2944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu
YX, Liu JF and He ZM: miR-26a suppresses tumor growth and
metastasis by targeting FGF9 in gastric cancer. PLoS One.
8:e726622013. View Article : Google Scholar : PubMed/NCBI
|