Introduction
Cervical cancer ranks as the fourth most common
cancer diagnosed among women, with ~570,000 cases in 2018 worldwide
(1). Squamous cell carcinoma and
adenocarcinoma constitute the main types of cervical cancer and are
associated with human papillomavirus (HPV) infection (2). Recently, increasing rates of cervical
cancer in young women have been reported (3). Surgery, radiotherapy, and chemotherapy
are the common therapeutic strategies for treating cervical cancer.
A nine-valent HPV vaccine has been developed to prevent HPV
infection (4). However, further
studies are urgently needed for designing effective diagnosis and
prognosis biomarkers and determining the underlying mechanisms.
Along with the mRNA coding proteins, other parts of
the transcript have important roles in regulating numerous
biological processes. Among these, long non-coding RNAs (lncRNAs)
have been identified as a key regulators of tumor progression
(5). For example, the lncRNA UCA1
is upregulated in numerous types of tumor and has been reported to
promote cancer cell migration, invasion, proliferation and immune
escape (6). In addition, the lncRNA
PSTAR suppresses liver cancer cell proliferation and
tumorigenesis via the p53 pathway, but does not affect apoptosis
(7). The roles of various lncRNAs
in cervical squamous cell carcinoma and endocervical adenocarcinoma
(CESC) have been extensively studied (8). In CESC, the lncRNA (HOX transcript
antisense RNA) HOTAIR is overexpressed and promotes the migration,
invasion and proliferation of tumor cells (9,10).
Furthermore, previous studies on biomarker-analysis have predicted
six candidate lncRNAs, including TMEM220-AS1, TRAM2-AS1,
C5orf66-AS1, RASSF8-AS1, AC126474 and AC004908, for cervical cancer
(11). However, these lncRNAs
should be validated by experimental and clinical investigation. A
few studies have investigated the role of the lncRNA MIR205HG. For
instance, it has been reported that MIR205HG was highly expressed
in p53-mutant head and neck squamous cell carcinoma compared with
p53-wild-type tumors, and promoted the proliferation of cancer
cells in head and neck squamous cell carcinoma (12). MIR205HG can also inhibit the
basal-luminal differentiation of human prostate basal cells by
binding to the interferon regulatory factor binding site (13) and MIR205HG was reannotated as Long
Epithelial Alu-interacting Differentiation-related RNA (LEDAR)
(14).
In order to determine the potential diagnostic and
therapeutic lncRNA targets in cervical cancer, the differentially
expressed lncRNAs in CESC were analyzed. The role of one lncRNA in
particular in regulating the proliferation and migratory and
invasive abilities of CESC cell lines was subsequently
investigated.
Materials and methods
Gene expression data of CESC
The transcriptome data of 306 CESC and 3 normal
samples were downloaded from The Cancer Genome Atlas (TCGA)
database (https://tcga-data.nci.nih.gov/tcga/). Among the CESC
patients, 24 cases had p53 mutation and 282 cases had no p53
mutation. Furthermore, the GSE27678 dataset, which includes 14
healthy and 30 squamous cell carcinomas of cervix (including two
premalignant lesions and squamous cell carcinomas cell lines), was
obtained from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27678).
All data were publicly available and were downloaded for research
purpose.
Differentially expressed lncRNA
analysis
The expression of MIR205HG was analyzed by GEPIA2
(http://gepia2.cancer-pku.cn/#analysis) in 306 CESC and
13 normal samples (3 normal samples from TCGA and 10 normal samples
from Genotype-Tissue Expression database). The differentially
expressed lncRNAs were analyzed using R software version 3.6.3
(https://www.r-project.org/), and a
|log2foldchange| >1 was used to determine significance. GSE27678
was analyzed by GEO2R. Pathway enrichment analysis was performed
using DAVID (https://david.ncifcrf.gov/). The top 20 enriched
pathways were selected (P<0.05). The bubble plots were designed
using ggplot2 package of R (http://had.co.nz/ggplot2/). Survival and correlation
analysis were performed using GEPIA2 (http://gepia2.cancer-pku.cn/#index). The network
analysis was performed using Cytoscape V3.6.1 (https://cytoscape.org/).
Cell culture
Ca Ski and C-33 A cell lines were purchased from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. Human cervical epithelial cell line (HCerEpiC) was
obtained from Shanghai Zhongqiaoxinzhou Biotechnology Co., Ltd.
(cat. no. 7060). Ca Ski was cultured in RPMI 1640 (cat. no.
10-040-CV; Corning, Inc.), C-33 A was cultured in MEM (cat. no.
E600020; Sangon Biotech Co., Ltd.) and HCerEpiC was cultured in
DMEM (cat. no. 10-013-CV; Corning, Inc.). All media were
supplemented with 10% FBS (cat. no. 10099-141-FBS; Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin-streptomycin (cat. no.
E607011; Sangon Biotech Co., Ltd.). All cells were placed at 37°C
in a humidified incubator containing 5% CO2.
Reverse transcription quantitative
(RT-q) PCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and reverse
transcription was performed using cDNA Synthesis kit (cat. no.
K1622; Thermo Fisher Scientific, Inc.) according to the
manufacturers' instructions. Quantitative PCR was carried out on an
ABI Q6 system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
RT-qPCR reactions were performed as follows: 95°C for 10 min, 45
cycles of 95°C for 15 sec, 60°C for 60 sec and a final dissociation
stage. The relative expression levels were normalized to endogenous
control GAPDH and were expressed as 2−ΔΔCq (15). The sequences of the primers used
were as follows: GAPDH, forward 5′-AGAAGGCTGGGGCTCATT-3′, reverse
5′-TGCTAAGCAGTTGGTGGTG-3; MIR205HG, forward
5′-GTTTCACCATGTTGCCCAGACT-3′, reverse 5′-CCTGTGCGGAACAGAAATGACT-3′;
fibroblast growth factor receptor 3 (FGFR3), forward
5′-GTGCTCAAGACGGCGGGC-3′, reverse 5′-GCCACGCAGAGTGATGAGAAAA-3′;
thymidine phosphorylase (TYMP), forward
5′-GAGTCTATTCCTGGATTCAATGTCA-3′, reverse
5′-AGAATGGAGGCTGTGATGAGTG-3′; and GTPase HRas (HRAS), forward
5′-CTGAGGAGCGATGACGGAAT-3′ and reverse
5′-GGAATCCTCTATAGTGGGGTCGT-3′.
RNAi interference
MIR205HG-homo-474 was knocked down using small
interfering (si) RNA, which was transfected into Ca Ski and C-33 A
cell lines using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Ca Ski and C-33 A cells were seeded into a
6-well plate with 30×104 cells/well 1 day before
transfection. Cells were transfected with MIR205HG siRNA or control
siRNA with a final concentration of 50 nM, and culture for 24 h.
The transfection efficiency was confirmed by RT-qPCR. The siRNA was
purchased from Suzhou GenePharma Co., Ltd. The MIR205HG siRNA
sequence was 5′-GCUGAACUGGGUGCUUUAUTT-3′;
5′-GCUGAACUGGGUGCUUUAUTTAUAAAGCACCCAGUUCAGCTT-3′, and that of the
siRNA control was 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-ACGUGACACGUUCGGAGAAT-3′.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using CCK-8 assay
(Beyotime Institute of Biotechnology). Cells were cultured at the
density of 1,000 cells/well and culture for 24 h before
transfection in a 6-well plate. Each sample was assessed in six
duplicates. Subsequently, 10 µl CCK-8 was added to each well for 1
h, and absorbance was detected at 450 nm on a microplate reader
(Infinite M1000; Tecan Group, Ltd.).
Transwell assay
The migratory and invasive ability of cells was
analyzed using Transwell assay. The 8.0-µm pore size membranes
(cat. no. 353097; Falcon®; BD Biosciences) were used for
migration assay whereas the BioCoat™ Matrigel® 0.8-µm
pore size membranes (cat. no. 354480; Corning, Inc.) were used for
invasion assay. The membranes were placed in a 24-well plate, and a
total of 75,000 cells were seeded in the upper chamber containing
serum-free medium. A volume of 700 µl medium containing 10% FBS was
loaded into the lower chamber at the bottom of 24-well plate. The
filters were stained with crystal violet (Sangon Biotech Co., Ltd.)
after 24 h, at 20°C for 30 min. Cells were observed and counted
under a light microscope at ×200 magnification (Nikon Corporation;
SMZ1000). Three random fields were counted for each microscopic
field.
Immunofluorescence staining
Immunofluorescence staining was performed using the
conditions suggested by the primary antibody suppliers. Briefly,
coverslips were placed into the 24-well plate, and the digested
cells were inoculated to the 24-well plate with a cell density of
~50,000 cells/well and 500 µl medium, which were then cultured at
37°C for 24 h. After the cell fusion rate was 70%, cells were
transfected with MIR205HG or control siRNA (50 nM) and cultured for
24 h. Cells were washed with PBS, fixed with 4% paraformaldehyde
for 15 min at 20–25°C, and permeabilized at 20°C using 0.1% Triton
X-100 and 5% BSA (cat. no. A8020; Beijing Solarbio Science &
Technology Co., Ltd.) in PBS for 5–15 min. After permeation, the
cells were washed with PBS three times/5 min. Cells were incubated
with 200 µl primary antibodies against Ki-67 (Cell Signaling
Technology, Inc.; cat. no. 9449; 1:100; mouse mAb) and p16
(Beyotime Institute of Biotechnology; cat. no. AF1672; 1:300;
rabbit mAb) at 4°C overnight. The cells were then incubated with
200 µl Alexa Fluor® 488 labeled goat anti-mouse IgG
secondary antibody (1:500; Abcam; cat. no. ab150113) and
Cy3-labeled goat anti-rabbit IgG secondary antibodies (1:500;
Institute of Biotechnology; cat. no. A0516) for 30 min at 20°C. The
nuclei were counterstained with DAPI for 5 min. Images were
obtained using fluorescence microscopy at ×400 magnification (Leitz
Orthoplan; Leica Microsystems GmbH).
Statistical analysis
Comparison between two groups was performed using
two-tailed Student's t-test and comparison between three groups was
performed by one-way ANOVA followed by Tukey's post hoc test.
Statistical analyses were made using SPSS package 17.0 (SPSS,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Differentially expressed lncRNAs in
CESC tissues
Expression data of 306 CESC samples and 13 normal
samples were retrieved from the TCGA. The cut-off
|log2fold-change| >1 was used. A total of 28
upregulated and 175 downregulated lncRNAs in clinical cancer types
were analyzed. Using the same standard, 1,542 upregulated and 2,726
downregulated mRNAs were identified. The expression of the top 20
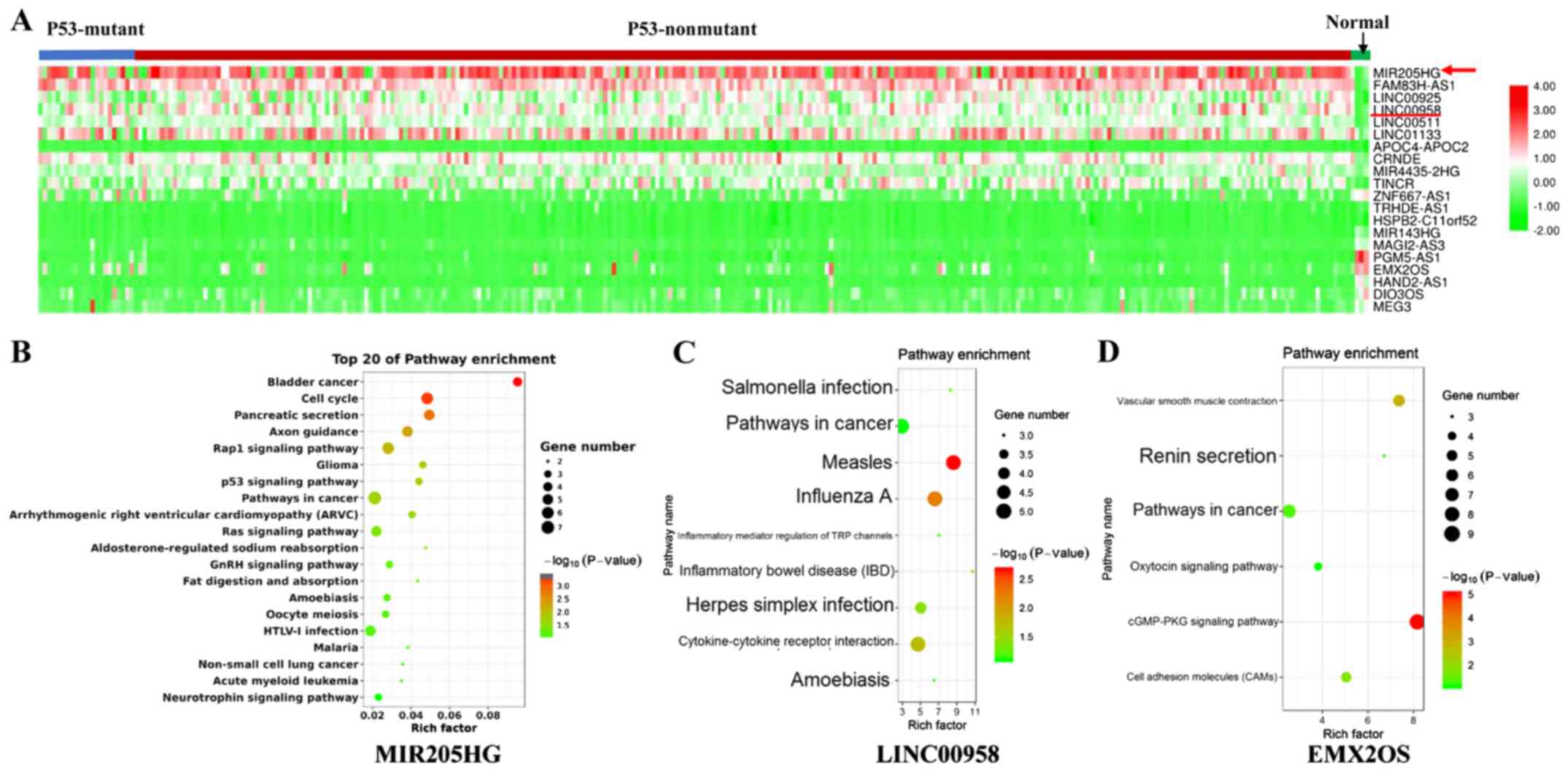
differentially expressed lncRNAs in each sample are presented in
the heat map of Fig. 1A. The red
arrow corresponds to MIR205HG, which was the most commonly
upregulated lncRNA. Each column represents a sample in the CESC
data retrieved from the TCGA.
Pathway enrichment analysis of the
co-expression mRNA of lncRNAs
To predict the function of these lncRNAs, the top
400 coexpressed genes were selected by Spearman's correlation
analysis (>0.2). Subsequently, overlaps of coexpressed and
differentially expressed genes were selected. A pathway enrichment
analysis was performed using DAVID. The results including MIR205HG,
LINC00925 and EMX2OS are presented in Fig. 1B-D, respectively. MIR205HG-related
genes were enriched in cancer-related pathways, such as ‘cell
cycle’, ‘p53 signaling pathway’, ‘Ras signaling pathway’ and
‘bladder cancer’ (Fig. 1B). The
LINC00958 coexpression genes were significantly enriched in
pathways related to virus infection (Fig. 1C).
Overall survival rate analysis for the
candidate lncRNAs
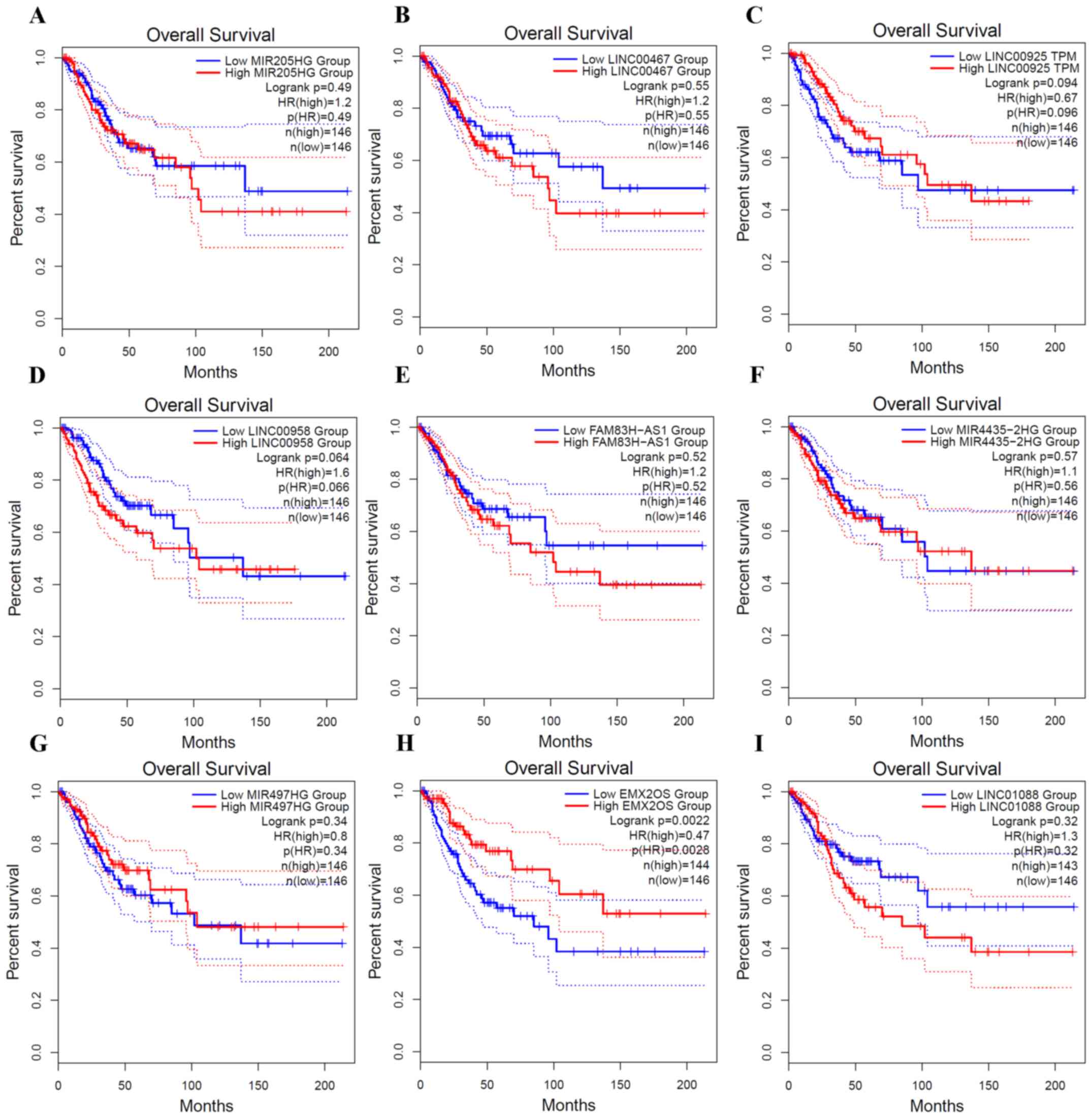
To investigate the clinical outcome of these
lncRNAs, survival analysis was performed using GEPIA (Fig. 2). Nine lncRNAs, including six
upregulated (Fig. 2A-F) and three
downregulated (Fig. 2G-I) lncRNAs,
were analyzed. Most of these lncRNAs have no significant
association with overall survival (log rank P<0.05). EMX2OS,
which is one of the most downregulated lncRNAs, had a significant
association with the overall survival. In addition, the EMX2OS high
expression group had an improved overall survival compared with the
EMX2OS low expression group (Fig.
2H). The coexpressed genes of EMX2OS were enriched in the
‘cGMP-PKG signaling pathway’ (Fig.
1D).
MIR205HG promotes cell migratory and
invasive abilities and proliferation of CESC cells
To validate the previous results, the lncRNA
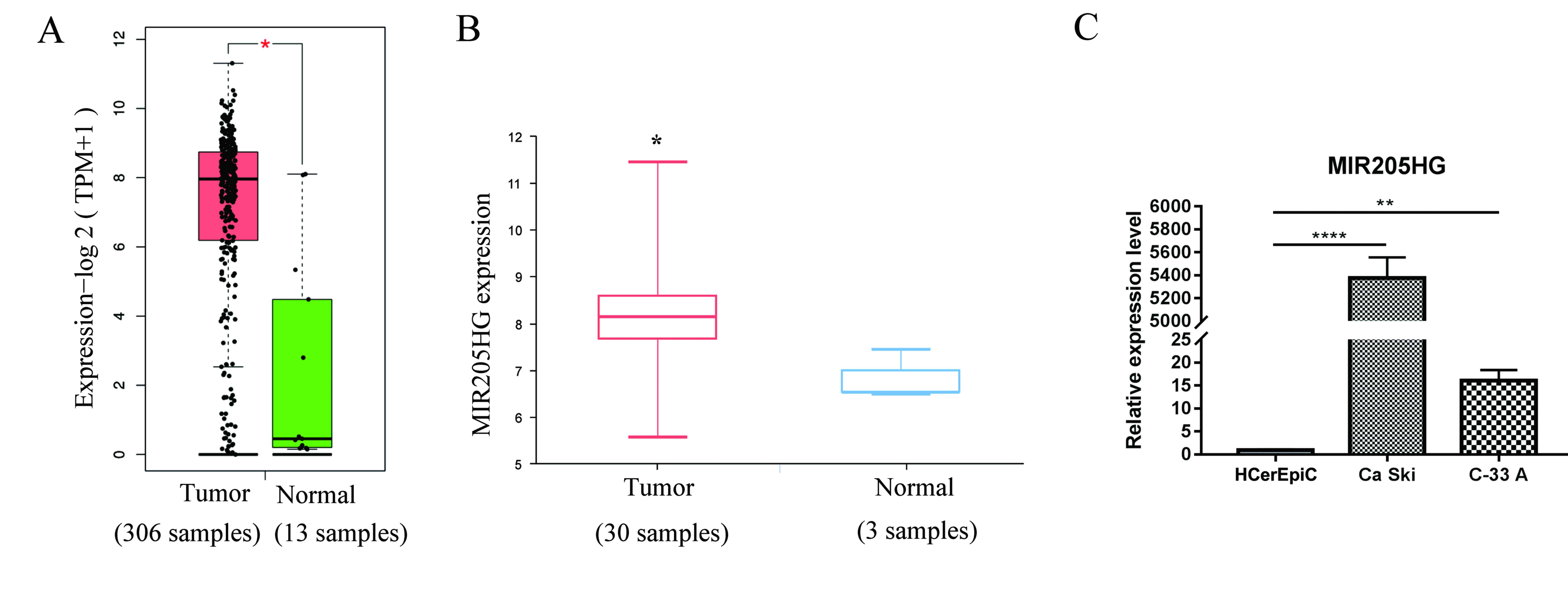
MIR205HG was further studied. We analyzed the expression of
MIR205HG in GSE27678 dataset, which contained 30 tumor samples and
3 normal samples. The results demonstrated that MIR205HG had higher
expression in tumors samples compared with normal samples (Fig. 3B), which was in accordance with TCGA
data (Fig. 3A and B). Subsequently,
the expression of MIR205HG was detected in the two CESC cell lines
Ca Ski and C-33 A. The results from RT-qPCR demonstrated that
MIR205HG was overexpressed in Ca Ski and C-33 A cell lines compared
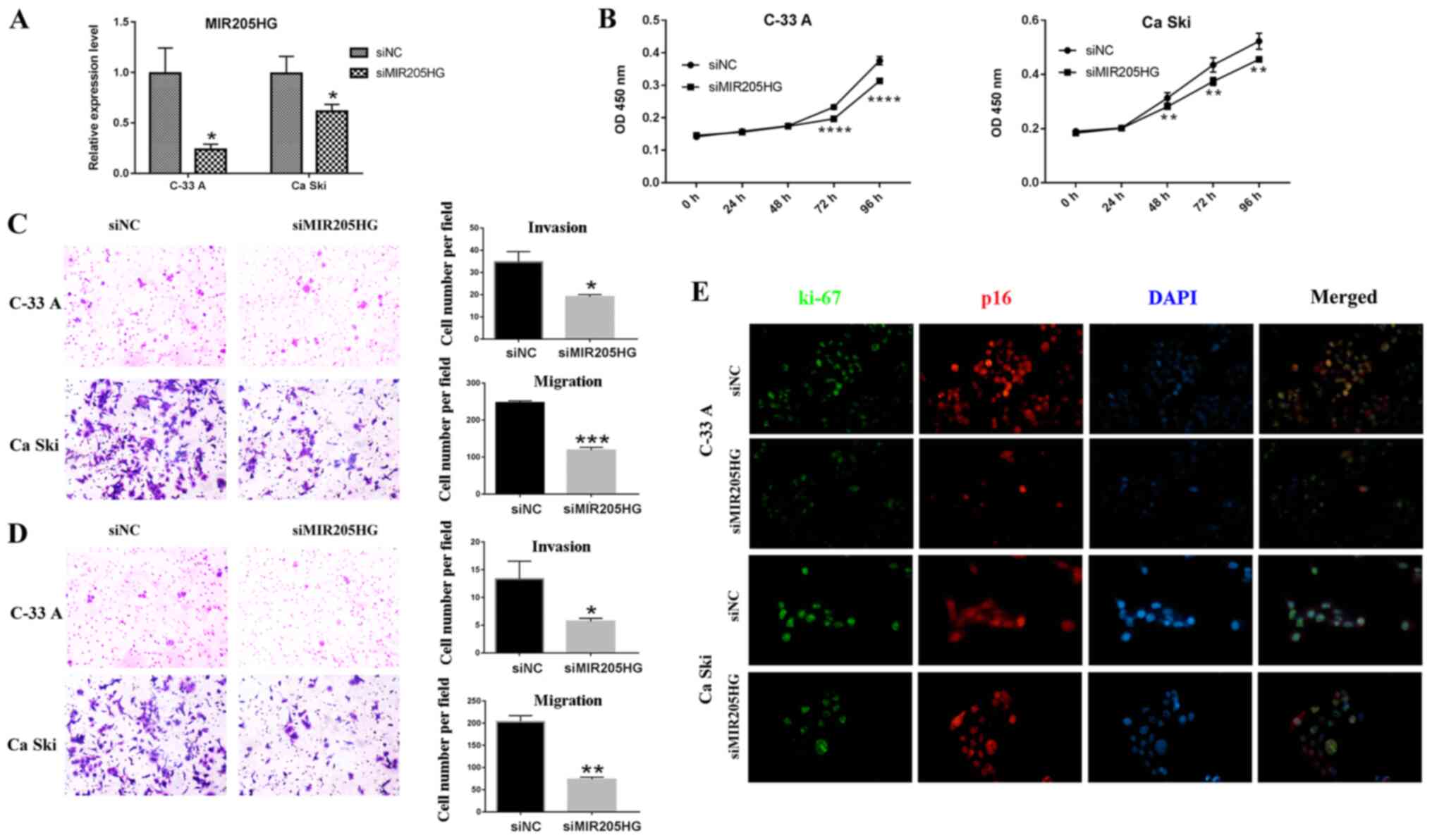
with the normal cervix cell line HCerEpiC (Fig. 3C). Then, MIR205HG was knockdown in
Ca Ski and C-33 A cells (Fig. 4A).
C-33 A and Ca Ski cell proliferation was significantly decreased
following MIR205HG knockdown (Fig.
4B). Furthermore, the migratory and invasive abilities were
significantly inhibited following MIR205HG knockdown (Fig. 4C and D). The migratory and invasive
abilities of Ca Ski cells appeared to be higher than that of C-33 A
cells. Immunofluorescence staining of p16 and Ki-67 was then
performed (Fig. 4E). Ki-67
staining, which is the marker of proliferation, was decreased in
C-33 A following MIR205HG knockdown and was partially decreased in
Ca Ski. In addition, p16 fluorescence intensity was lower in C-33 A
cells after MIR205HG knockdown, whereas no change was observed in
Ca Ski cells.
Network analysis of MIR205HG
A network between MIR205HG and its coexpressed genes
were analyzed by cytoscape3.6.1 (Fig.
5A). Genes in enriched pathways were selected. A total of 49
related genes are presented. Subsequently, the expression of three
selected genes, FGFR3, TYMP and HRAS, was detected in
MIR205HG knocked down CESC cells. In C-33 A cells, all these genes
were significantly downregulated after MIR205HG knockdown (Fig. 5B-D), whereas HRAS showed no
significant change (Fig. 5D).
Furthermore, the expression of these three genes was positively
correlated with MIR205HG expression in CESC data of TCGA (Fig. 5E-G).
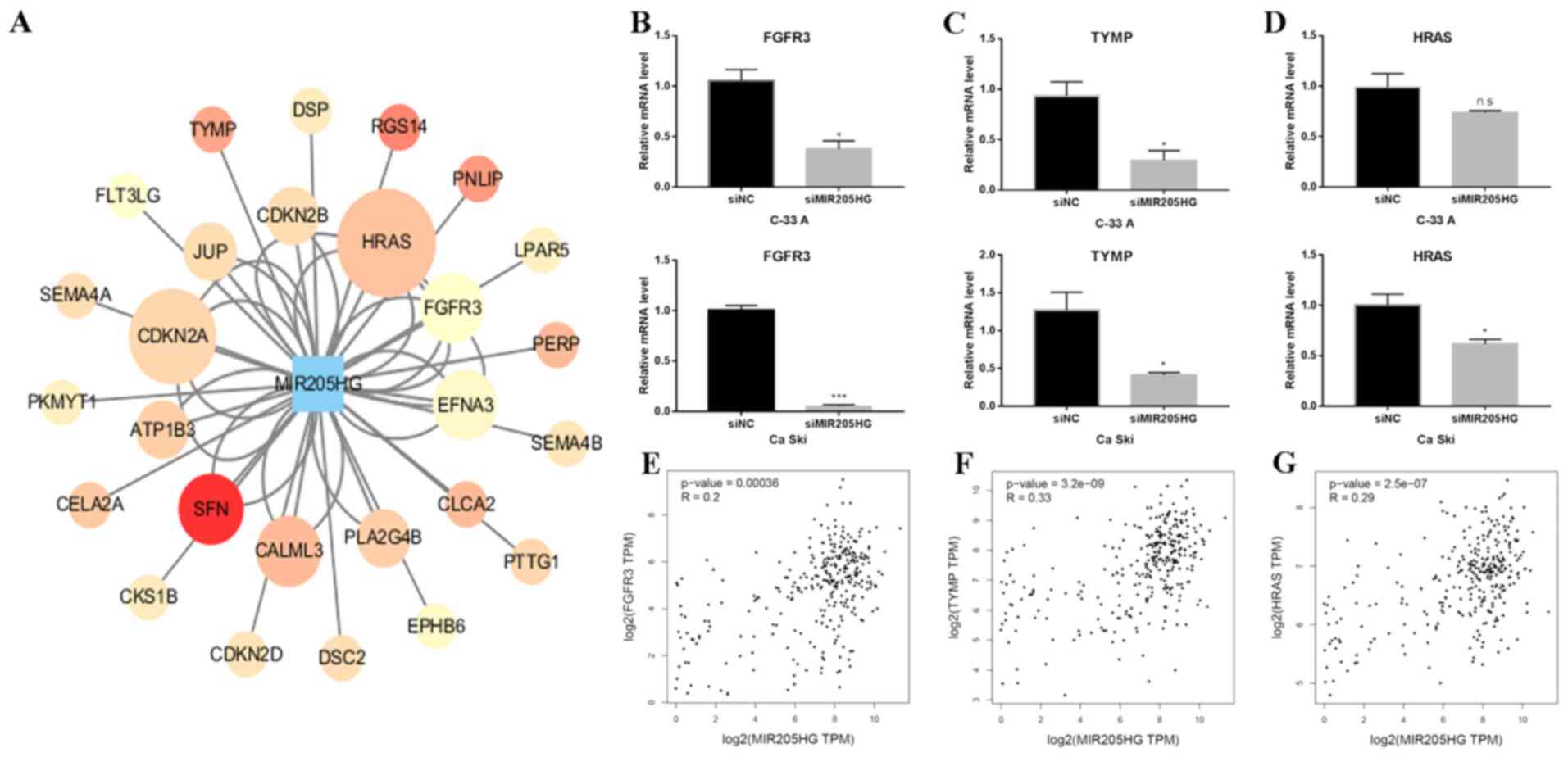 | Figure 5.Network analysis of MIR205HG
co-expression genes. (A) Network analysis showed the relationship
between MIR205HG and co-expressed genes. Red nodes indicate a high
correlation. (B) FGFR3 mRNA level was significantly
decreased following MIR205HG knockdown in C-33 A and Ca Ski cells.
(C) RT-qPCR showed that the expression of TYMP in C-33 A and
Ca Ski transfected with siMIR205HG was significantly downregulated.
(D) RT-qPCR showed that expression of HRAS in Ca Ski cells
transfected with siMIR205HG was significantly downregulated,
whereas it was not significant in C-33 A. Two-tailed student's
t-test in B, C and D. *P<0.01 and ***P<0.001. (E-G)
Correlation between MIR205HG and FGFR3, TYMP, and
HRAS using TCGA CESC data, respectively. NC, negative
control; si, small interfering; ns, non-significant; FGFR3,
fibroblast growth factor receptor 3; TYMP, thymidine phosphorylase;
HRAS, GTPase HRas; RT-qPCR, reverse transcription quantitative
PCR. |
Discussion
Previous studies have revealed various functions of
lncRNAs in regulating numerous complex biological processes
(8–10). For example, HOTAIR enhances cervical
cancer aggressiveness by increasing the expression levels of
vascular endothelial growth factor, matrix metallopeptidase 9 and
epithelial-mesenchymal transition-associated genes (9). The data from TCGA in the present study
provided an insight into the differentially expressed lncRNAs in
cancer and normal tissues, which were previously reported as
promising therapeutic targets, diagnosis biomarkers or prognosis
biomarkers. For instance, Gong et al (16) selected several lncRNAs
differentially expressed in TCGA and RNA-seq data and analyzed them
using survival rate. They identified LINC01537 as having a role in
the regulation of energy metabolism via phosphodiesterase 2A in
lung cancer.
In the present study, 203 differentially expressed
lncRNAs were identified in the CESC data retrieved from the TCGA.
The top 15 upregulated and downregulated lncRNAs are listed in
Table I. LINC00958, the fourth
upregulated lncRNA, was reported to promote tumor progression in
various types of cancer, including CESC (17,18).
EMX2OS, the most downregulated lncRNA, has been speculated to be a
prognostic biomarker for thyroid cancer (19). The roles of other lncRNAs in
cervical cancer remain unclear. Luo et al (11) analyzed the expression pattern of
differentially expressed lncRNAs and their role in cervical cancer
progression, which provides a novel insight into the diagnosis and
treatment of cervical cancer. The lncRNA candidates form the
present study were different from what they studied.
 | Table I.Top 15 upregulated and 15
downregulated long non-coding RNAs in cervical squamous cell
carcinoma and endocervical adenocarcinoma. |
Table I.
Top 15 upregulated and 15
downregulated long non-coding RNAs in cervical squamous cell
carcinoma and endocervical adenocarcinoma.
| lncRNA | log2
(fold-change) | P-value | Description |
|---|
| MIR205HG | 7.508 |
3.08×10−07 | MIR205 host gene
(non-protein coding) |
| FAM83H-AS1 | 4.328 |
2.67×10−12 | FAM83H antisense RNA
1 (head to head) |
| LINC00925 | 3.691 |
7.19×10−10 | Long intergenic
non-protein coding RNA 925 |
| LINC00958 | 3.544 |
3.77×10−08 | Long intergenic
non-protein coding RNA 958 |
| LINC00511 | 3.512 |
6.06×10−22 | Long intergenic
non-protein coding RNA 511 |
| LINC01133 | 2.673 |
1.05×10−3 | Long intergenic
non-protein coding RNA 1133 |
| MALAT1 | 2.525 |
2.39×10−3 | Metastasis
associated lung adenocarcinoma transcript 1 (non-protein
coding) |
| APOC4-APOC2 | 2.365 |
2.29×10−04 | APOC4-APOC2
readthrough (NMD candidate) |
| CRNDE | 2.112 |
6.53×10−04 | Colorectal
neoplasia differentially expressed (non-protein coding) |
| MIR4435-2HG | 2.025 |
2.89×10−11 | MIR4435-2 host
gene |
| TINCR | 1.976 |
7.06×10−3 | Tissue
differentiation-inducing non-protein coding RNA |
| DGUOK-AS1 | 1.919 |
9.88×10−07 | DGUOK antisense RNA
1 |
| LINC00467 | 1.833 |
7.61×10−09 | Long intergenic
non-protein coding RNA 467 |
| UNC5B-AS1 | 1.781 |
6.64×10−04 | UNC5B antisense RNA
1 |
| CDKN2B-AS1 | 1.715 |
1.35×10−11 | CDKN2B antisense
RNA 1 |
| WT1-AS | −3.407 |
2.11×10−32 | WT1 antisense
RNA |
| LINC01088 | −3.456 |
4.83×10−23 | Long intergenic
non-protein coding RNA 1088 |
| FRMD6-AS2 | −3.516 |
1.03×10−61 | FRMD6 antisense RNA
2 |
| SOCS2-AS1 | −3.537 |
2.51×10−33 | SOCS2 antisense RNA
1 |
| MIR497HG | −3.567 |
1.63×10−38 | mir-497-195 cluster
host gene (non-protein coding) |
| ZNF667-AS1 | −3.601 |
2.57×10−06 | ZNF667 antisense
RNA 1 (head to head) |
| TRHDE-AS1 | −3.644 |
1.65×10−32 | TRHDE antisense RNA
1 |
| HSPB2-C11orf52 | −3.813 |
2.21×10−52 | HSPB2-C11orf52
readthrough (NMD candidate) |
| MIR143HG | −4.401 |
2.07×10−73 | MIR143 host gene
(non-protein coding) |
| MAGI2-AS3 | −4.46 |
7.03×10−42 | MAGI2 antisense RNA
3 |
| PGM5-AS1 | −4.569 |
1.63×10−55 | PGM5 antisense RNA
1 |
| EMX2OS | −4.652 |
6.45×10−21 | EMX2 opposite
strand/antisense RNA |
| HAND2-AS1 | −4.956 |
6.91×10−67 | HAND2 antisense RNA
1 (head to head) |
| DIO3OS | −5.252 |
1.1×10−24 | DIO3 opposite
strand/antisense RNA (head to head) |
| MEG3 | −6.572 |
6.13×10−30 | Maternally
expressed 3 (non-protein coding) |
MIR205HG was the host gene of microRNA (miR)-205 and
has not been thoroughly studied to the best of our knowledge.
miR-205 has been reported to be commonly downregulated in tumors,
in particular in bladder cancer (20). Di Agostino et al (12) reported that MIR205HG can promote
tumor progression in head and neck squamous cell carcinoma. The
present study demonstrated that MIR205HG was overexpressed in CESC
tissues compared with normal tissues and promoted the proliferation
and migratory and invasive abilities of CESC cells. These findings
suggested that MIR205HG may act as a pro-tumor lncRNA in CESC. As a
prognostic marker of cervical cancer, p16 is abnormally
overexpressed in HPV-positive or negative cervical cancer types
(21). MIR205HG knockdown
downregulated the expression of p16 in C-33 A cells, whereas no
significant change was observed in Ca Ski cells. C-33 A is an
HPV-negative cell line, whereas Ca Ski is an HPV-positive cell
line. It was reported that p16 is overexpressed in benign tumor and
high-grade malignant tumor (21).
These findings suggested that the inhibitory effect of MIR205HG on
cervical cancer cells might be dependent of the malignancy;
however, further investigation is required.
The present study investigated the potential
underlying mechanisms of MIR205HG on the regulation of
proliferation and invasive ability of CESC cells. The network
analysis of MIR205HG related differentially expressed mRNAs was
therefore completed and it was found that MIR205HG was co-expressed
with 49 genes, including FGFR3, TYMP and HRAS. A
recent study revealed the mechanism of MIR205HG which acts as a
ceRNA to promote tumor progression by sponging miR-122-5p in
cervical cancer (22). The results
from network analysis provided additional candidates regulated by
MIR205HG in the present study. FGFR3, TYMP and HRAS
were demonstrated to be positively correlated with MIR205HG. In
addition, the expression of these three genes was downregulated
following MIR205HG knockdown. FGFR3 serves an essential role
in the regulation of progenitor cell proliferation, differentiation
and apoptosis during the development of the embryo (23). It was reported that FGFR3 is
overexpressed or mutated in numerous types of cancer and can act as
an oncogene, in particular in bladder cancer (24–26).
HRAS is a small GTPase belonging to the Ras family of
proteins, which has been broadly studied in cancer (27,28).
The results from the present study revealed a significant
association between MIR205HG and FGFR3, TYMP and HRAS
following bioinformatic analysis and experimental results. However,
the underlying mechanism of MIR205HG regulating these genes remains
to be elucidated.
In conclusion, the present study determined
differentially expressed lncRNAs in CESC and reported MIR205HG as
being upregulated in CESC tissues compared with normal tissues.
MIR205G knockdown decreased the proliferation and migratory and
invasive abilities of CESC cells. Furthermore, the expression of
MIR205HG was positively correlated with expression of the oncogenes
HRAS, FGFR3 and TYMP. The findings from this study suggested
that MIR205HG may have pro-tumor function in CESC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on request.
Authors' contributions
YZ and LZ conceived and designed the study. LY, YZ
and LZ performed the experiments and analyzed data. LY wrote the
paper. YZ and LZ reviewed and edited the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wentzensen N, Clarke MA, Bremer R, Poitras
N, Tokugawa D, Goldhoff PE, Castle PE, Schiffman M, Kingery JD,
Grewal KK, et al: Clinical evaluation of human papillomavirus
screening with p16/Ki-67 dual stain triage in a large organized
cervical cancer screening program. JAMA Intern Med. 179:881–888.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arun G, Diermeier SD and Spector DL:
Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol
Med. 24:257–277. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qin G, Tu X, Li H, Cao P, Chen X, Song J,
Han H, Li Y, Guo B, Yang L, et al: lncRNA PSTAR promotes p53
signaling by inhibiting hnRNP K deSUMOylation and suppresses
hepatocellular carcinoma. Hepatology. 2019.
|
|
8
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim
SW and Kim YT: Long non-coding RNA HOTAIR is associated with human
cervical cancer progression. Int J Oncol. 46:521–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W, Wang M, Liu J, Cui X and Wang H:
Identification of a six lncRNAs signature as novel diagnostic
biomarkers for cervical cancer. J Cell Physiol. 235:993–1000. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Agostino S, Valenti F, Sacconi A,
Fontemaggi G, Pallocca M, Pulito C, Ganci F, Muti P, Strano S and
Blandino G: Long non-coding MIR205HG depletes Hsa-miR-590-3p
leading to unrestrained proliferation in head and neck squamous
cell carcinoma. Theranostics. 8:1850–1868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Profumo V, Forte B, Percio S, Rotundo F,
Doldi V, Ferrari E, Fenderico N, Dugo M, Romagnoli D, Benelli M, et
al: LEADeR role of miR-205 host gene as long noncoding RNA in
prostate basal cell differentiation. Nat Commun. 10:3072019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Percio S, Rotundo F and Gandellini P: Gene
expression dataset of prostate cells upon MIR205HG/LEADR
modulation. Data Brief. 29:1051392020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gong W, Yang L, Wang Y, Xian J, Qiu F, Liu
L, Lin M, Feng Y, Zhou Y and Lu J: Analysis of survival-related
lncRNA landscape identifies a role for LINC01537 in energy
metabolism and lung cancer progression. Int J Mol Sci. 20:37132019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Zhu X, Dong P and Cai J: Long
noncoding RNA LINC00958 promotes the oral squamous cell carcinoma
by sponging miR-185-5p/YWHAZ. Life Sci. 242:1167822019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao H, Zheng GH, Li GC, Xin L, Wang YS,
Chen Y and Zheng XM: Long noncoding RNA LINC00958 regulates cell
sensitivity to radiotherapy through RRM2 by binding to
microRNA-5095 in cervical cancer. J Cell Physiol. 234:23349–23359.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu Y, Feng C, Liu T, Zhang B and Yang L:
The downregulation of lncRNA EMX2OS might independently predict
shorter recurrence-free survival of classical papillary thyroid
cancer. PLoS One. 13:e02093382018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gulìa C, Baldassarra S, Signore F, Rigon
G, Pizzuti V, Gaffi M, Briganti V, Porrello A and Piergentili R:
Role of non-coding RNAs in the etiology of bladder cancer. Genes
(Basel). 8:3392017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Romagosa C, Simonetti S, López-Vicente L,
Mazo A, Lleonart ME, Castellvi J and Cajal SR: p16(Ink4a)
overexpression in cancer: A tumor suppressor gene associated with
senescence and high-grade tumors. Oncogene. 30:2087–2097. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Wang H and Huang H: Long non-coding
RNA MIR205HG function as a ceRNA to accelerate tumor growth and
progression via sponging miR-122-5p in cervical cancer. Biochem
Biophys Res Commun. 514:78–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Inglis-Broadgate SL, Thomson RE, Pellicano
F, Tartaglia MA, Pontikis CC, Cooper JD and Iwata T: FGFR3
regulates brain size by controlling progenitor cell proliferation
and apoptosis during embryonic development. Dev Biol. 279:73–85.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kandimalla R, Masius R, Beukers W, Bangma
CH, Orntoft TF, Dyrskjot L, van Leeuwen N, Lingsma H, van Tilborg
AAG and Zwarthoff EC: A 3-plex methylation assay combined with the
FGFR3 mutation assay sensitively detects recurrent bladder cancer
in voided urine. Clin Cancer Res. 19:4760–4769. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Javidi-Sharifi N, Traer E, Martinez J,
Gupta A, Taguchi T, Dunlap J, Heinrich MC, Corless CL, Rubin BP,
Druker BJ and Tyner JW: Crosstalk between KIT and FGFR3 promotes
gastrointestinal stromal tumor cell growth and drug resistance.
Cancer Res. 75:880–891. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pouessel D, Neuzillet Y, Mertens LS, van
der Heijden MS, de Jong J, Sanders J, Peters D, Leroy K, Manceau A,
Maille P, et al: Tumor heterogeneity of fibroblast growth factor
receptor 3 (FGFR3) mutations in invasive bladder cancer:
Implications for perioperative anti-FGFR3 treatment. Ann Oncol.
27:1311–1316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He F, Melamed J, Tang MS, Huang C and Wu
XR: Oncogenic HRAS activates epithelial-to-mesenchymal transition
and confers stemness to p53-deficient urothelial cells to drive
muscle invasion of basal subtype carcinomas. Cancer Res.
75:2017–2028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murugan AK, Grieco M and Tsuchida N: RAS
mutations in human cancers: Roles in precision medicine. Semin
Cancer Biol. 59:23–35. 2019. View Article : Google Scholar : PubMed/NCBI
|