Introduction
Triple-negative breast cancer (TNBC) is the most
aggressive subtype of breast cancer. Higher proliferation rates and
a higher incidence of metastases with a lack of targeted therapies
have made TNBC an unmet clinical challenge (1). Currently, chemotherapy is regarded
as a systemic therapeutic alternative for TNBC, and taxanes such as
paclitaxel (PTX) are commonly used as antitumor agents against TNBC
(2,3). However, numerous types of TNBC
cells, especially the MDA-MB-231 cell line that carry mutant
K-Ras and TP53, are highly aggressive and invasive
breast cancer cell lines and often escape drug-induced apoptosis,
leading to poor therapeutic effects (4,5).
It has been reported that autophagy can protect tumor cells from
damage and undergoing apoptosis during PTX treatment (6). Therefore, treatment approaches
targeting autophagy may provide novel therapeutic value for
TNBC.
Autophagy is a classical self-digestive process that
maintains cellular homeostasis and ensures cell survival under
stress conditions by degrading and recycling damaged organelles and
protein aggregates (7). It plays
a pivotal role in tumor progression, during early stages of cancer
development it prevents tumor initiation, but in fully developed
tumors it protects tumor cells from various stressful stimuli
(4,8,9).
Various chemotherapy drugs induce autophagy by activating different
signaling pathways. For example, SKF-96365, a store-operated
calcium entry inhibitor, induces cytoprotective autophagy by
preventing the release of cytochrome C in colorectal cancer cells,
which is mechanistically involved in AKT-related signaling
(10). In addition,
apatinib-induced autophagy in anaplastic thyroid carcinoma cells is
related to the downregulation of phosphorylated (p)-AKT and
p-mechanistic target of rapamycin kinase (mTOR) signaling (11). PTX treatment often activates
different autophagy-related pathways depending on the tumor type.
In A549 lung cancer cells, PTX induces autophagy by regulating the
autophagy-related genes, including autophagy related 5
(ATG5) and beclin 1 (BECN1) (12). However, in ovarian cancer, PTX
induces autophagy by upregulating the thioredoxin domain-containing
protein 17 gene, which shortens the survival of patients (13). However, the key molecule or
mechanism associated with PTX-induced autophagy in TNBC cells
remains unclear.
Apoptosis is a terminal pathway of cell death and is
closely related to morphogenesis during the elimination of aged or
harmful cells to maintain adult tissue homeostasis (14). Some crosstalk between autophagy
and apoptosis has been reported in tumors; autophagy may be an
adaptive stress response prior to apoptotic cell death, either
enabling or antagonizing apoptosis (9). Therefore, the relationship between
apoptosis and autophagy is critical for tumor targeted therapy.
Given the high invasiveness of the MDA-MB-231 cell
line, which is commonly used to investigate the molecular basis of
TBNC and for the development of novel therapeutic approaches
(15), MDA-MB-231 cells were used
in the present study to investigate the molecular mechanism of PTX
against TNBC in vitro. The present study assessed whether
PTX induced cell apoptosis and autophagy at certain concentrations,
whether inhibition of autophagy contributed to PTX-induced
apoptosis and the underlying molecular mechanism of PTX. Finally,
it was investigated whether inhibition of the molecular mechanism
promotes PTX-induced apoptosis and may be a clinical chemotherapy
strategy of PTX for TNBC.
Materials and methods
Regents and antibodies
All experimental reagents and antibodies were
purchased from Sigma-Aldrich (Merck KGaA) unless otherwise stated.
Bafilomycin A1 (Baf A1) was purchased from Sangon Biotech Co., Ltd.
The antibodies used in this study included anti-FOXO1 antibody
(cat. no. 2880; Cell Signaling Technology, Inc.), anti-p-FOXO1
antibody (cat. no. WL03634; Wanleibio Co., Ltd.) and anti-Lamin AC
antibody (cat. no. 10298-1-AP; ProteinTech Group, Inc.).
Cell culture
MDA-MB-231 TNBC cell lines were purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences and HCC-1937 cell lines were provided by the Kunming
Institute of Zoology, Chinese Academy of Sciences. The cell lines
were maintained in Dulbecco's modified Eagle's medium (DMEM; cat.
no. C11995500BT; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; cat. no. 10099-141;
Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/ml) and
streptomycin (100 µg/ml) (Invitrogen; Thermo Fisher Scientific,
Inc.) in a humidified atmosphere of 5% CO2 at 37°C.
Cell morphological observation
Exponentially growing MDA-MB-231 and HCC-1937 cells
were transferred to 12-well plates at a density of 2×104
cells/well and cultured at 37°C in a 5% CO2 atmosphere.
The cells were treated with 0, 10, 20 and 30 nM PTX (cat. no.
48248; MedChemExpress) at 37°C. When the cells reached 60-70%
confluency, they were rinsed twice with PBS and supernatant was
discarded and cells were used for morphological observation. Images
were acquired using an Olympus IX 71 microscope (Olympus
Corporation). Next, MDA-MB-231 cells were treated with different
concentrations of PTX (0, 0.03, 0.1, 0.3, 1, 3, 10 and 30 nM) at
37°C, 5% CO2 for 24 h to perform cell proliferation
assay using cell counting kit (CCK-8) (cat. no. P10330; TransGen
Biotech) according to manufacturer's instruction.
Annexin V-FITC/PI staining for
apoptosis analysis
For detecting PTX-induced cell apoptosis, MDA-MB-231
cells were treated with 0, 10, 20 and 30 nM PTX at 37°C, 5%
CO2 for 24 h. For detecting PTX-induced cell apoptosis
under FOXO1 inhibition, MDA-MB-231 cells were transfected with
small interfering RNA at 37°C for 7 h, and then were treated with
or without 20 nM PTX at 37°C, 5% CO2 for 24 h. The
apoptosis assay was performed using an Annexin V-FITC/PI test kit
(cat. no. FXP018-100; Beijing 4A Biotech Co., Ltd.), according to
the manufacturer's instructions. Flow cytometry analysis was
performed using a Beckman CytoFLEX flow cytometer (Beckman Coulter,
Inc.). The cells undergoing apoptosis were defined as reported
previously (16) early (Annexin
V-FITC-positive/PI-negative, Q3) + late apoptotic cells (Annexin
V-FITC-positive/PI-positive; Q2).
Immunofluorescence
For detecting LC3, MDA-MB-231 cells were pretreated
with Baf A1 (0 and 10 nM) at 37°C, 5% CO2 for 24 h and
then treated with or without 20 nM PTX at 37°C, 5% CO2
for 24 h. Cells were treated with 0, 10 and 30 nM PTX combined with
or without 5 nM 3-methyladenine (3-MA; 5 nM; a PI3K inhibitor that
is a widely used inhibitor of autophagy via its inhibitory effect
on class III PI3K) at 37°C, 5% CO2 for 24 h. For
detecting FOXO1, MDA-MB-231 cells were treated with 0, 10 and 20 nM
PTX at 37°C, 5% CO2 for 24 h. Immunofluorescence
analysis of light chain 3 (LC3) and FOXO1 proteins was performed as
described in our previous study (17). In brief, the treated cells were
fixed in 95% methanol at room temperature for 10 min and blocked in
a buffer containing 1% BSA (cat. no. A8020; Beijing Solarbio
Science & Technology Co., Ltd.) and 0.1% Triton X-100 at RT for
1 h. Then, the fixed cells were incubated with primary antibodies
against LC3B (1:200) and FOXO1 (1:100) at 4°C overnight. Next, the
cells were incubated with secondary fluorescence-conjugated
antibodies (Green for LC3, 1:200, cat. no. 111-545-003; Red for
FOXO1, 1:200, cat. no. 711-605-152, Jackson ImmunoResearch Inc.) at
room temperature for 1 h for visualization via laser confocal
microscopy (Olympus FV 1000; Olympus Corporation).
Colony formation assay
This assay was performed according to our previous
study (18). Briefly, adherent
MDA-MB-231 cells (250 cells/plate) were treated with or without
3-MA and/or PTX (0, 10, 30 nM) at 37°C for 72 h and then cultured
for 15 days. Thereafter, the cells were fixed with 4%
paraformaldehyde (cat. no. P1110; Solarbio) at RT for 30 min and
stained with 10% Giemsa (cat. no. G1015; Beijing Solarbio Science
& Technology Co., Ltd.) at RT for 15 min. The colonies were
washed, air dried, imaged and counted (>100 cells as one
colony). Finally, the colony formation ratio was calculated
according to the following formula: Colony formation ratio=number
of colonies/number of seeded cells ×100%.
Small interfering RNA (siRNA) and
transient transfection
MDA-MB-231 cells (2×106 cells/well) were
seeded into 96- or 6-well plates. Then, control random siRNA (cat.
no. sc-37007; proprietary sequence) or FOXO1-targeted siRNA (cat.
no. sc-35382) that included a pool of three sequences are in
Table SI (Santa Cruz
Biotechnology, Inc.; 100 pmol/well) were transfected into
MDA-MB-231 cells using an siRNA transfection reagent (Santa Cruz
Biotechnology, Inc.) according to the manufacturer's protocol. The
cells were transfected at 37°C for 7 h, washed with PBS and then
were immediately treated with 0 and 20 nM PTX at 37°C for 24 h.
Then, the cells were immediately collected, and cell lysates were
prepared for reverse transcription-quantitative PCR (RT-qPCR) and
western blotting. Cells were also used for apoptosis analyses.
RT-qPCR
The MDA-MB-231 cells treated with 0, 10 and 20 nM
PTX at 37°C for 24 h were used for the analysis of mRNA expression
of the autophagy-regulated genes. The MDA-MB-231 cells transfected
with siRNA at 37°C for 7 h were treated with 0 and 20 nM PTX at
37°C, 5% CO2 for 24 h and used for the analysis of mRNA
expression of the autophagy-related genes (ATG). The mRNA
expression levels of genes were assessed via qPCR, including the
autophagy-regulated genes, sestrin 1 (SESN1), phosphatase
and tensin homolog (PTEN), mTOR, ectopic P-granules
autophagy protein 5 homolog (EPG5), TSC complex subunit 1
(TSC1), serine/threonine kinase 1 and 11 (AKT and
LKB1), FOXO1, lamin A/C (LMNA), autophagy and
beclin 1 regulator 1 (AMBRA1), DNA damage regulated
autophagy modulator 1 and 2 (DRAM1 and DRAM2), as
well as, the ATGs, ATG5, class III phosphoinositide 3-kinase
vacuolar protein sorting 34 (VPS34), autophagy related 4B
cysteine peptidase (ATG4B), BECN1 and microtubule
associated protein 1 light chain 3β (MAP1LC3B). Total RNA
was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was synthesized from total RNA using a
Prime-Script RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. The obtained cDNA was used as a template
for TB Green-based qPCR kit (cat. no. RR820B; Takara Bio, Inc.) on
a CFX-96 system (Bio-Rad Laboratories, Inc.). GAPDH was used for
normalization. The primer sequences are shown in Table SII. Thermocycling conditions were
as follows: Initial denaturation, 95°C for 30 sec followed by 40
cycles of 95°C for 5 sec, 60°C for 30 sec, and 72°C for 30 sec.
Finally, the data was analyzed using 2−∆∆Cq method
(19).
Western blotting
MDA-MB-231 and HCC-1937 cells were treated with 0,
10, 20 and 30 nM PTX at 37°C, 5% CO2 unless otherwise
stated for 24 h. At the end of the designed treatment time, the
cells were washed twice with PBS and collected. The cells were
lysed in RIPA lysis buffer (cat. no. BB-3201-2; Bestbio) with
protease inhibitors (cat. no. D1201-2; TransGen Biotech) at 4°C.
Then, the total protein concentrations of the cell lysates were
determined using a BCA Protein Assay kit (cat. no. P0009-1 & 2;
Beyotime Institute of Biotechnology). The cytosolic and nuclear
proteins were extracted using Nuclear and Cytoplasmic Protein
Extraction kit (cat. no. P0028-1; Beyotime Institute of
Biotechnology) according to manufacturer's instruction. Protein
samples (50 µg/lane) were separated via 12% SDS-PAGE and
subsequently transferred onto PVDF membranes. The membranes were
incubated at RT for 30 min in 5% BSA buffer (Beijing Solarbio
Science & Technology Co., Ltd.) with gentle shaking to block
non-specific binding before incubation with the diluted primary
antibody (Bcl-2, 1:1,000, cat. no. AF6139, Affinity; Bax, 1:1,000,
cat. no. WL01637, Wanlei, China; LC3B, 1:2,000, cat. no. L7543;
Sigma-Aldrich; Merck KGaA; P62; 1:2,000, cat. no. P0067;
Sigma-Aldrich; Merck KGaA; FOXO1, 1:1,000; p-FOXO1, 1:1,000; AKT,
1:1,000, cat. no. 4691S; Cell Signaling Technology, Inc.; p-AKT,
1:1,000, cat. no. 9271S; Cell Signaling Technology, Inc.; Lamin AC,
1:2,000; β-actin, 1:5,000, cat. no. A5441; Sigma-Aldrich; Merck
KGaA) overnight at 4°C. Subsequently, membranes were incubated with
horseradish peroxidase-conjugated secondary antibody (rabbit IgG,
1:5,000, cat. no. HAF008; mouse IgG, 1:10,000, cat. no. HAF007;
R&D Systems) for 90 min at RT. Membranes were washed three
times in PBS for 10 min each time. Then, the membranes were treated
for 3 min in the dark with reagent from an Easysee Western Blot kit
(TransGen Biotech) and visualized using an imaging system (Bio-Rad
Laboratories, Inc.). The densitometry of protein bands was
performed using Image Lab Software (version 6.1; Bio-Rad
Laboratories, Inc.).
Statistical analysis
All numerical data are representative of at least
three independent experiments and are presented as the mean ±
standard deviation (SD) on the basis of three independent
experiments. The data from the different concentrations of PTX were
analyzed by one-way ANOVA, while the data from the combined
treatment with PTX + Baf A1 or PTX + si-FOXO1 were analyzed by
two-way ANOVA followed by a Tukey's post hoc test. The data from
PTX and PTX + 3-MA treatment were analyzed using unpaired Student's
t-test. Analysis of all numerical data was performed using SPSS
version 22 software (IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
PTX induces cytotoxicity and apoptosis
of MDA-MB-231 cells
PTX is a chemotherapeutic agent that promotes the
polymerization of tubulin, which disrupts normal microtubule
dynamics, leading to cell death (20). To determine the effect of PTX on
MDA-MB-231 cell viability, morphological observations, and CCK-8
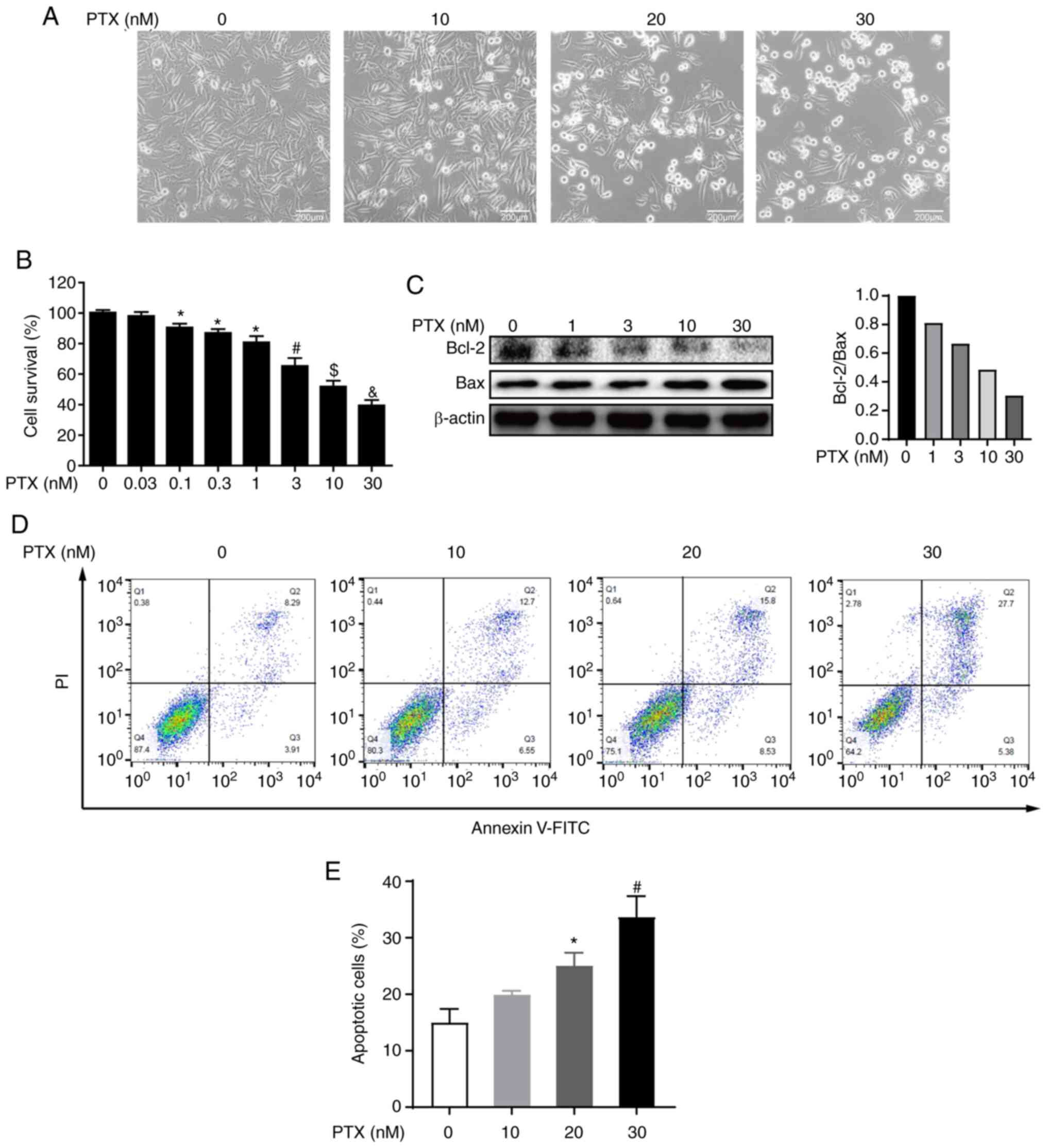
assays were performed after PTX treatment. Morphological changes
revealed that MDA-MB-231 cell death increased with increasing doses
of PTX (Fig. 1A). Furthermore,
the CCK-8 assay showed that the cell survival rate also decreased
with increasing PTX doses, and the half maximal inhibitory
concentration (IC50) of PTX fell between 10 and 30 nM
(Fig. 1B). These results
indicated that PTX induced MDA-MB-231 cell death in a
dose-dependent manner.
To further determine whether the observed cell death
was caused by PTX inducing the apoptotic pathway, the expression of
apoptotic markers was evaluated and the results indicated that the
expression levels of the apoptosis markers Bcl-2 and Bax,
especially expression levels of Bcl-2, were decreased in a
dose-dependent manner (Fig. 1C).
Further, PI and Annexin V staining coupled with flow cytometry were
also performed and it was observed that PTX exposure resulted in a
dose-dependent increase in the apoptosis rate (Fig. 1D and E). Collectively, these
results indicated that PTX induced the apoptosis of MDA-MB-231
cells.
PTX induces autophagy in MDA-MB-231
cells
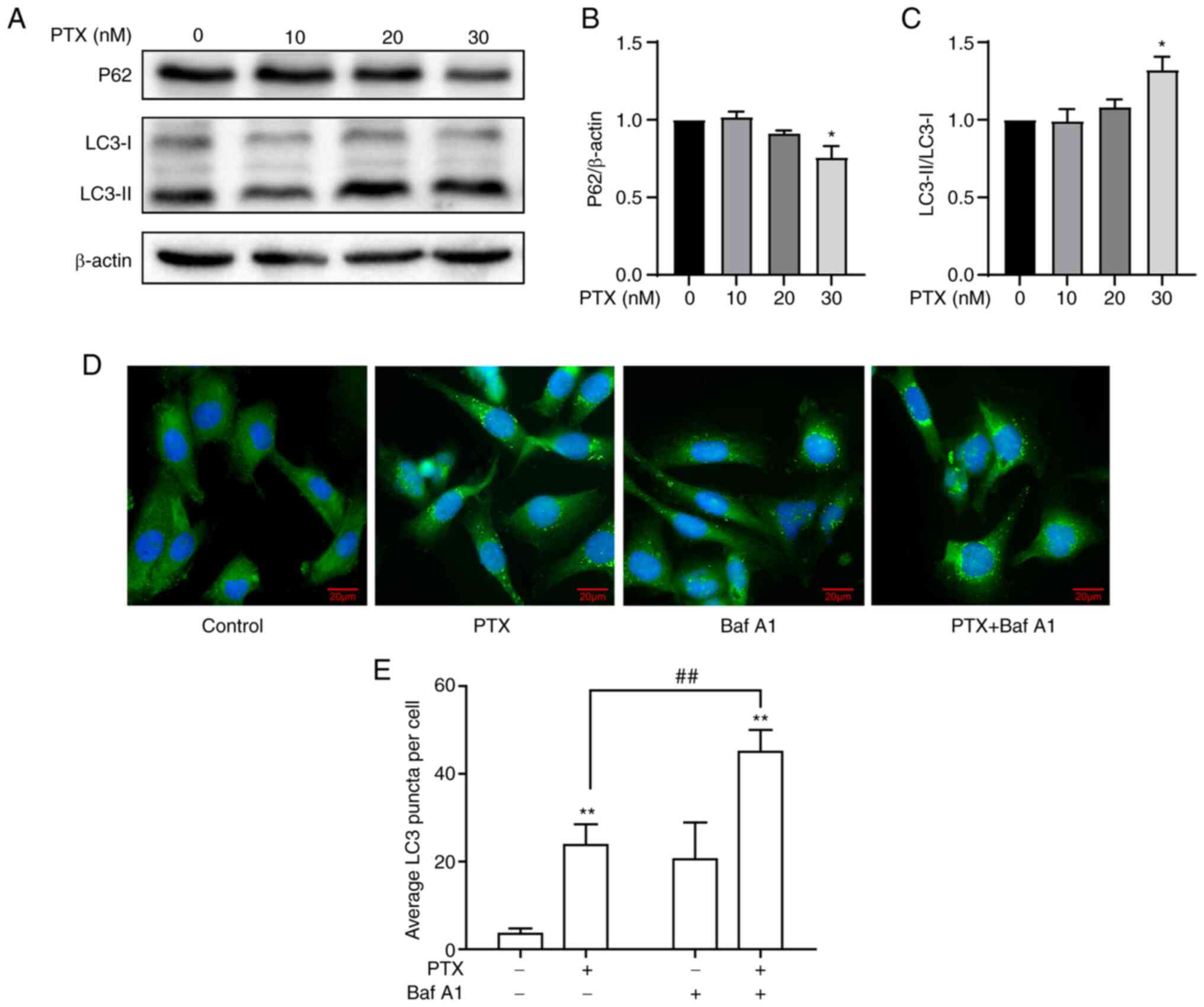
In response to PTX-induced MDA-MB-231 cell
apoptosis, the effect of PTX on autophagy was investigated. A
significant increase in LC3-II/LC3-I and a decrease in P62 in
response to 30 nM PTX was observed (Fig. 2A-C), indicating an increased level
of autophagy. Considering that the IC50 value of PTX was
close to 20 nM, it was speculated whether 20 nM PTX could induce
autophagy. Therefore, autophagic flux levels in MDA-MB-231 cells
treated with 20 nM PTX were assessed after blocking
autophagosome-lysosome fusion and degradation with Baf A1.
Immunoblotting results revealed a significant increase in LC3
puncta in the presence of PTX + Baf A1 (Fig. 2D and E), suggesting that 20 nM PTX
induced increased autophagic flux. Taken together, 20 and 30 nM PTX
induced autophagy, while simultaneously inducing apoptosis in
MDA-MB-231 cells.
In addition, another TNBC cell line (HCC-1937) were
also treated with 0, 10, 20 and 30 nM PTX and it was observed that
HCC-1937 cells were more sensitive to PTX treatment than MDA-MB-231
cells because more HCC-1937 cells died compared with MDA-MB-231
cells at the same dose of PTX (Fig.
S1A). A significant decrease in the Bcl-2/Bax ratio (Fig. S1B and C) and a significant
increase in the LC3-II/LC3-I ratio (Fig. S1F and G) was found, although no
change in P62 protein expression (Fig. S1D and E) was found at 30 nM PTX,
suggesting that PTX-induced apoptosis and autophagy in TNBC cells
is a common phenomenon.
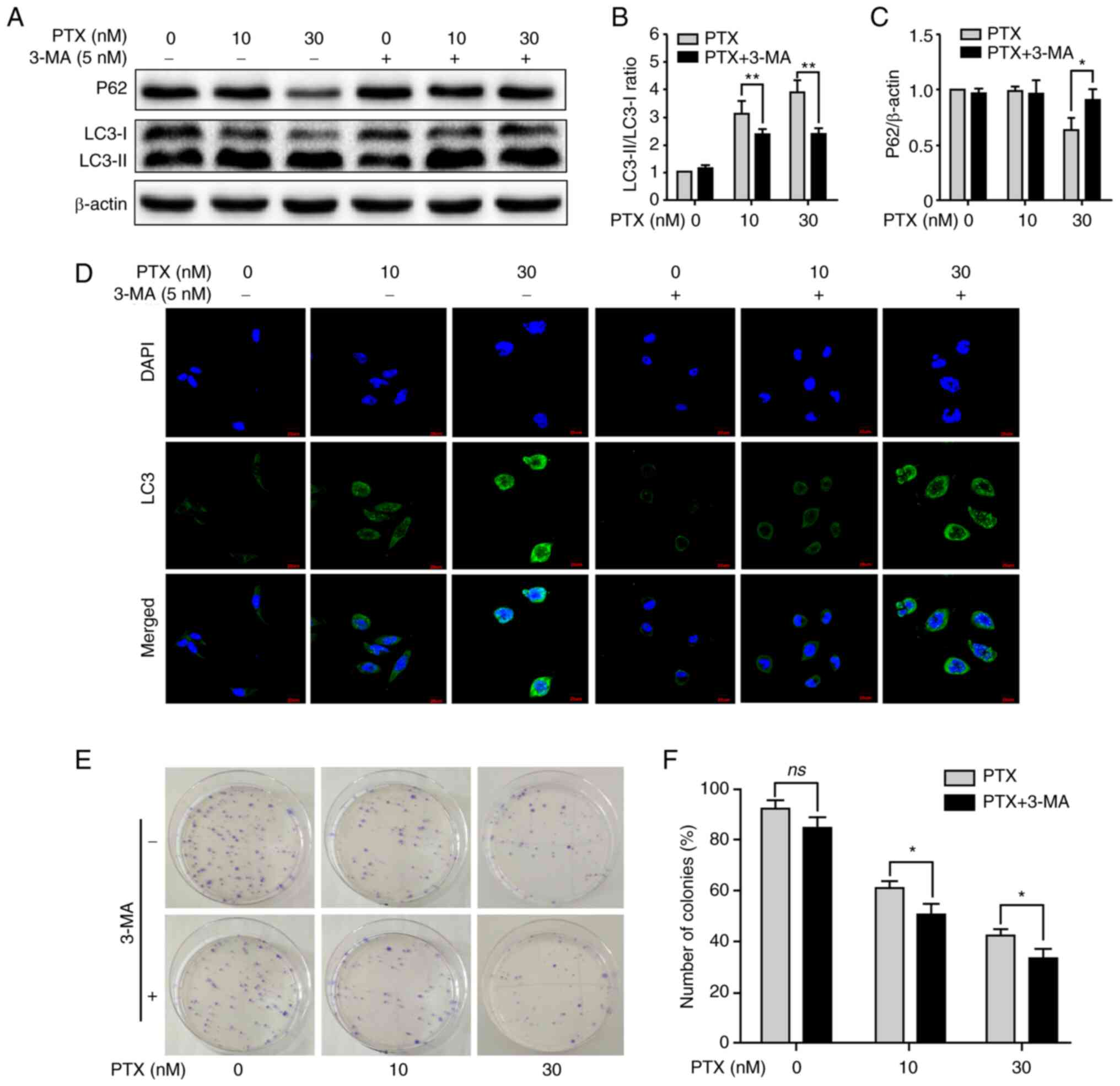
Inhibition of autophagy increases
PTX-induced apoptotic cell death in MDA-MB-231 cells
Given that PTX enabled the simultaneous induction of
apoptosis and autophagy in MDA-MB-231 cells, it was next
investigated whether inhibition of autophagy contributed to
improving PTX-induced apoptotic cell death. After treatment with
PTX + 3-MA, inhibition of autophagy was clearly observed, including
a significant decrease in the LC3-II/LC3-I ratio at 10 and 30 nM
PTX (Fig. 3A and B) and an
increase in P62 expression at 30 nM (Fig. 3A and C). In addition, LC3 puncta
also markedly decreased at 10 and 30 nM PTX after treatment with
3-MA (Fig. 3D). Furthermore, the
formation of cell colonies was effectively inhibited at 10 and 30
nM PTX + 3-MA (Fig. 3E and F).
These results indicated that inhibition of autophagy with 3-MA
promoted PTX-induced apoptosis in MDA-MB-231 cells.
PTX increases FOXO1 expression in
MDA-MB-231 cells
Autophagy is regulated by autophagy-regulated genes,
such as the FOXO1, PTEN, LKB1, mTOR, SESN1, EPG5, TSC1, AKT,
LMNA, AMBRA1 and DRAM1 genes, which are essential for
autophagy signaling pathways (21–23). The mRNA levels of these genes were
measured via RT-qPCR and it was found that 20 nM PTX induced a
2.7-fold increase in FOXO1 mRNA in MDA-MB-231 cells (Fig. 4A). Therefore, the role of FOXO1 in
20 nM PTX-treated MDA-MB-231 cells was the focus of subsequent
assays. First, FOXO1 and p-FOXO1 protein expression in response to
PTX treatment was assessed. The results showed that PTX induced a
significant decline in the p-FOXO1/FOXO1 ratio in a dose-dependent
manner, suggesting that FOXO1 rather than p-FOXO1 played an
important role in PTX-induced autophagy in MDA-MB-231 cells
(Fig. 4B and C). In addition, the
upstream inhibitor of FOXO1, p-ATK, exhibited a significantly
downregulated pattern (Fig. 4D and
E). Thus, it was speculated that the decreased p-FOXO1/FOXO1
ratio was related to decreased p-AKT levels. Since FOXO1 is a
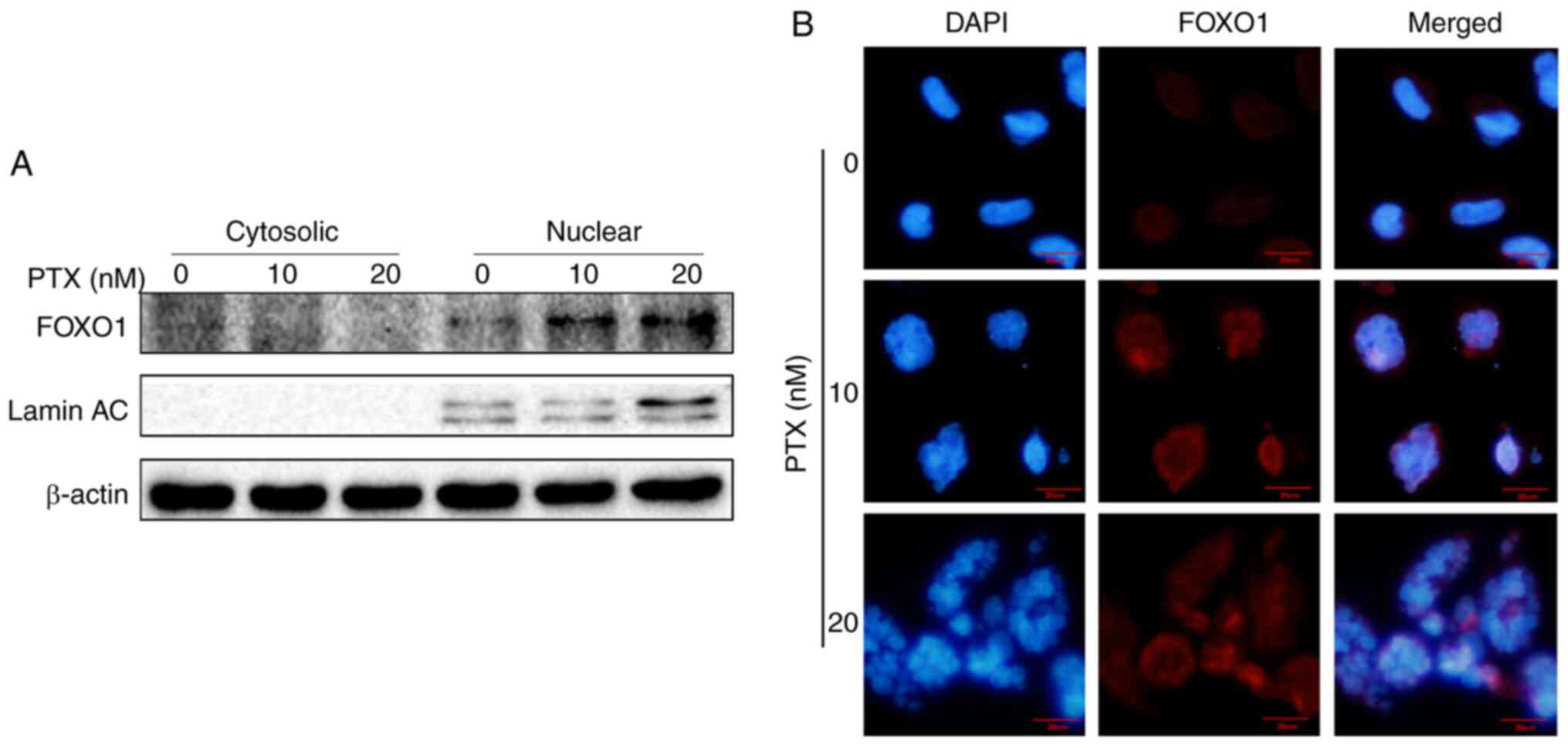
transcription factor, it was assessed whether FOXO1 exerted a
transcriptional function in PTX-mediated autophagy of MDA-MB-231
cells. Therefore, the localization of FOXO1 was further detected
via western blotting and immunofluorescence. The results showed
that FOXO1 was primarily distributed in the nucleus, further
confirming that FOXO1 exerted its autophagy-regulated function in a
transcriptional activation pattern (Fig. 5A and B).
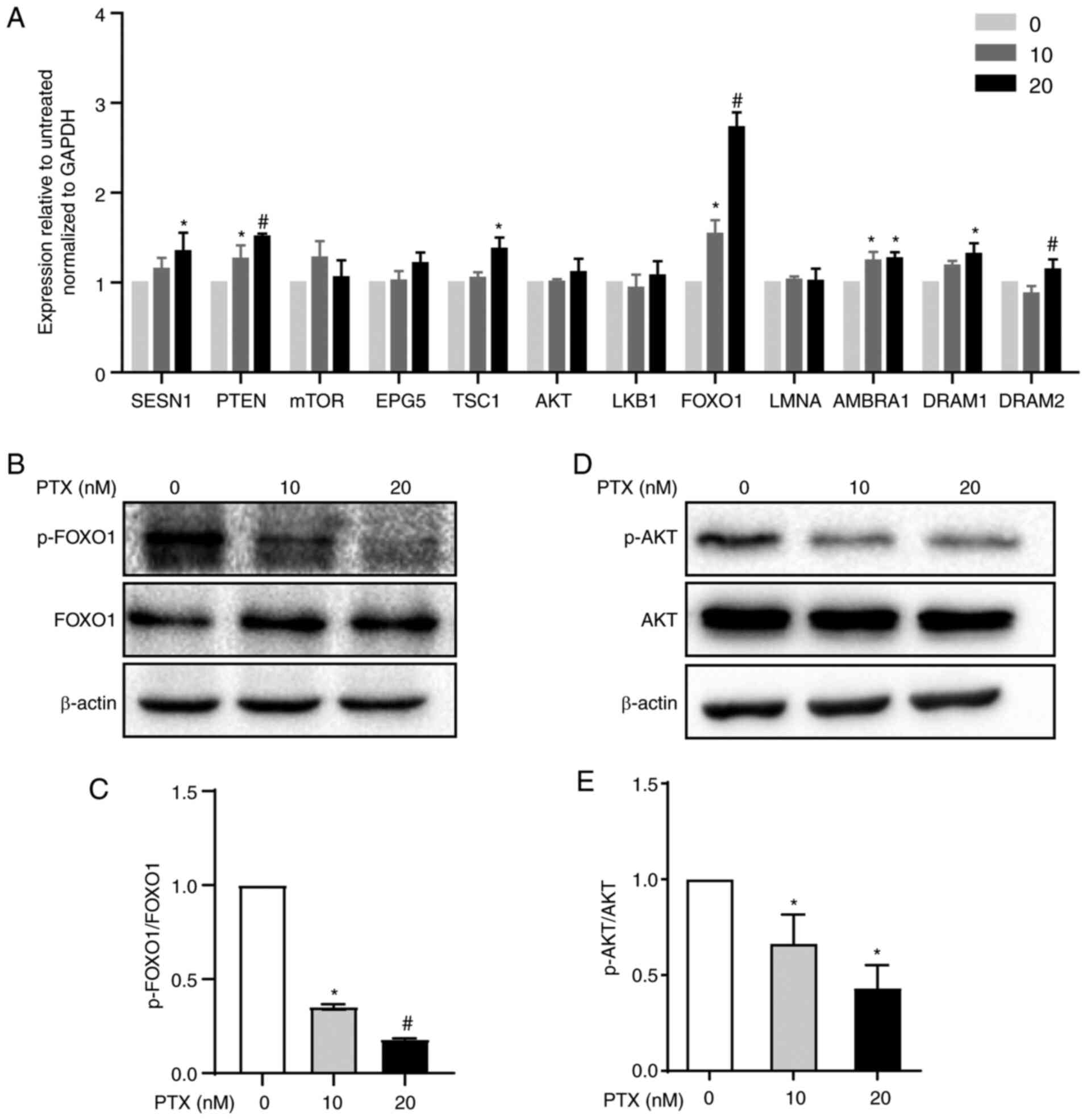 | Figure 4.Autophagy regulatory pathway
induction by PTX in MDA-MB-231 cells. (A) mRNA expression levels of
autophagy signaling pathway-related genes in response to treatment
with 10 and 20 nM PTX. GAPDH was used as an internal control.
(*P<0.05 vs. 0 nM PTX; #P<0.05 vs. 10 nM PTX). (B)
Protein expression of FOXO1 and p-FOXO1 and (C) semi-quantification
of the p-FOXO1/FOXO1 ratio (*P<0.05 vs. 0 nM PTX;
#P<0.05 vs. 10 nM PTX). (D) Protein expression of AKT
and p-AKT and (E) semi-quantification of the p-AKT/AKT ratio
(*P<0.05 vs. 0 nM PTX). β-actin was used as an internal control.
Data are presented as the means ± SD of three independent
experiments. PTX, paclitaxel; FOXO1, forkhead box transcription
factor O1; p-, phosphorylated; SESN1, sestrin 1;
PTEN, phosphatase and tensin homolog; mTOR,
mechanistic target of rapamycin kinase; EPG5, ectopic
P-granules autophagy protein 5 homolog; TSC1, TSC complex
subunit 1; AKT, serine/threonine kinase 1; LKB1,
serine/threonine kinase 11; LMNA, lamin A/C; AMBRA1,
autophagy and beclin 1 regulator 1; DRAM1, DNA damage
regulated autophagy modulator 1; DRAM2, DNA damage regulated
autophagy modulator 2. |
FOXO1 is required for PTX-induced
autophagy in MDA-MB-231 cells
To investigate the function of FOXO1 in PTX-induced
autophagy in MDA-MB-231 cells, FOXO1 expression was knocked down
using siRNA. At the protein level, the expression of FOXO1 was
successfully inhibited after treatment with siRNA (Fig. 6A and B). Knocking down the FOXO1
gene led to significantly decreased LC3II and significantly
increased P62 protein expression levels in PTX-treated MDA-MB-231
cells, suggesting reduced autophagy levels (Fig. 6A, C and D). A number of
autophagy-related genes, such as ATG5, VPS34
(PI3KC3), ATG4B, BECN1 and MAP1LC3B, are
transcriptionally regulated by FOXO1 (24). In this study, it was also found
that these genes were suppressed in response to FOXO1 knockdown in
PTX-treated MDA-MB-231 cells (Fig.
6E). Importantly, knockdown of FOXO1 enhanced PTX-induced
apoptotic cell death (Fig. 6F and
G). These results illustrated that FOXO1 played a role in
PTX-induced autophagy in MDA-MB-231 cells and that targeting FOXO1
may improve the efficacy of PTX in TNBC therapy.
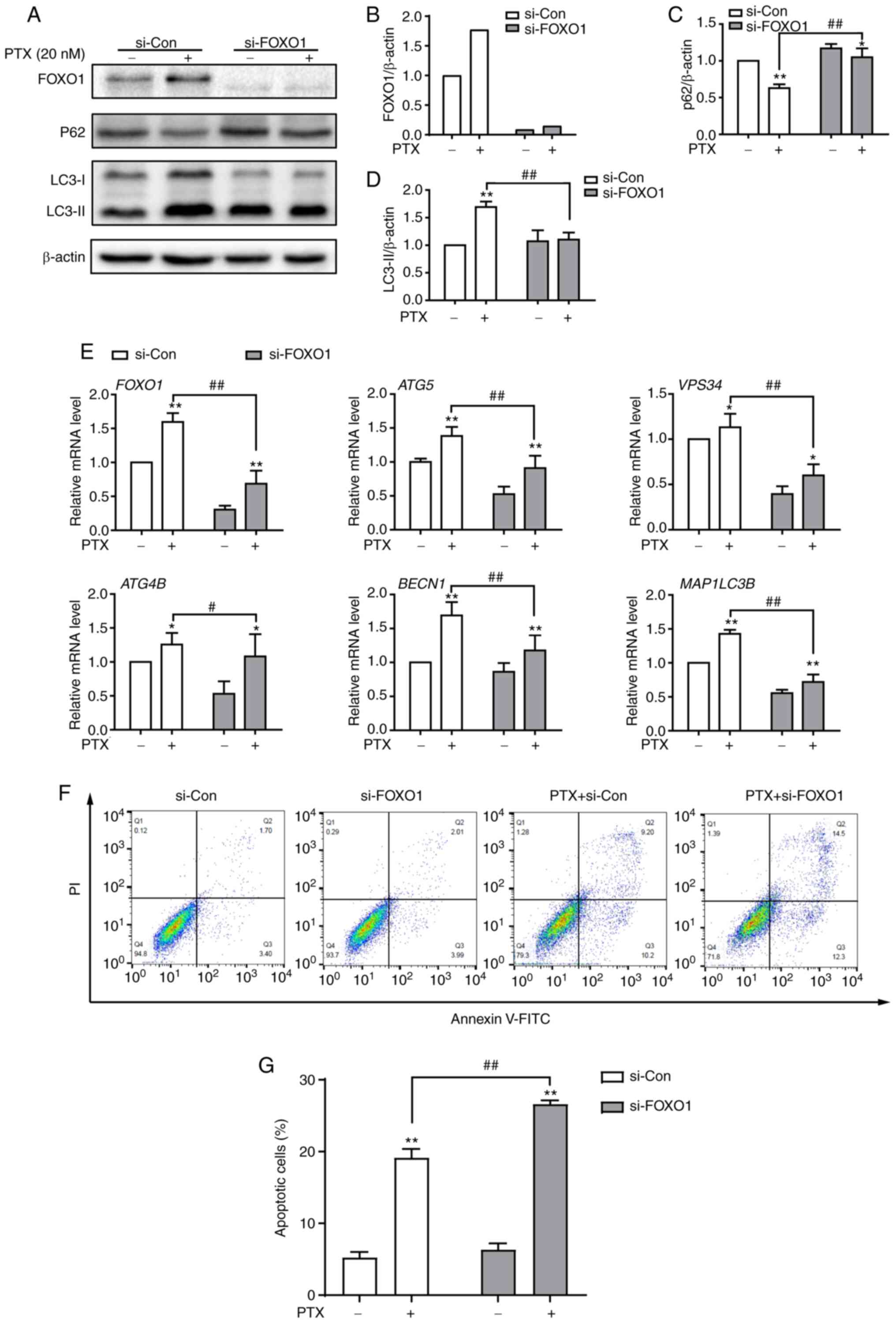 | Figure 6.Knockdown of FOXO1 attenuates
PTX-induced autophagy and promotes apoptotic cell death in
MDA-MB-231 cells. (A) Protein expression of the autophagy markers
P62, LC3-I and LC3-II after treatment with si-FOXO1, with β-actin
as an internal control. Semi-quantification of (B) FOXO1, (C) P62
and (D) LC3-II protein expression. (E) The mRNA expression levels
of FOXO1 and its downstream target genes after treatment with siRNA
and/or PTX. (F) The effect of FOXO1 knockdown on PTX-induced
apoptosis was analyzed by flow cytometry. (G) Quantification of the
ratio of apoptotic cells after the knockdown of FOXO1. PTX was used
at a concentration of 20 nM. Data are presented as the means ± SD
of three independent experiments. *P<0.05 vs. 0 nM PTX;
**P<0.01 vs. 0 nM PTX; #P<0.05 vs. 20 nM PTX +
si-Con; ##P<0.01 vs. 20 nM PTX + si-Con. PTX,
paclitaxel; FOXO1, forkhead box transcription factor O1; LC3, light
chain 3; si, small interfering; ATG5, autophagy related 5;
VPS34, class III phosphoinositide 3-kinase vacuolar protein
sorting 34; ATG4B, autophagy related 4B cysteine peptidase;
BECN1, beclin 1; MAP1LC3B, microtubule associated
protein 1 light chain 3β. |
Discussion
Although PTX has been widely used for the treatment
of various solid tumors, including ovarian, lung and breast cancer
(25), its chemotherapeutic
efficacy varies among different types of cancer, particularly in
cases where resistance has been developed against it. Previously,
it has also been found that TNBC frequently acquires resistance
against PTX through different regulatory pathways (26,27). For example, PTX can trigger breast
cancer type 1 (BRCA1)-IRIS expression, a product of the oncogene
BRCA1, which enhances AKT-related signaling and results in a
PTX resistance phenotype in TNBC (28). Another study demonstrated that
aurora kinase A can also promote PTX resistance in TNBC by
stabilizing FOXM1 (3). Therefore,
targeting these pathways or inhibiting tumor-associated factors may
be helpful for improving PTX efficacy in TNBC (29–33). Additionally, it has been
demonstrated that inhibition of autophagy enhances PTX-induced cell
death in TNBC MDA-MB-231 cells with PTX resistance (34). Consistently, inhibition of
autophagy also contributed to promoting PTX-induced apoptosis in
MDA-MB-231 cells in the present study. Elevated autophagic flux was
implicated in the significant increase in nuclear FOXO1.
Furthermore, knockdown of FOXO1 enhanced PTX-induced apoptosis.
These findings may provide a potential anticancer target and be
important for improving PTX efficacy in TNBC treatment.
Autophagy and apoptosis are two antagonistic and
interconnected molecular mechanisms of various cellular stresses.
Autophagy, a prosurvival regulatory process, often attenuates
apoptotic cell death (35,36).
PTX, as a broad-spectrum anticancer agent, often induces cytotoxic
apoptosis or inhibits autophagy (37,38). However, it has also been reported
that PTX induces autophagy in tumor cells (39), which might have an adverse effect
on PTX efficacy. Another previous study indicated that PTX induces
both autophagy and apoptosis in several cancer cells. The
upregulated levels of autophagy are related to the
autophagosome-regulatory genes ATG5 and BECN1. After
3-MA treatment or BECN1 knockdown, the tumor cell response
switches from autophagic to apoptotic (12). Similarly, the current study also
demonstrated that autophagy was induced in MDA-MB-231 cells
accompanied by PTX-induced apoptosis, and autophagic inhibition
with 3-MA enhanced apoptotic cell death. In addition, to assess
that whether PTX generally induced apoptosis and autophagy in TNBC
cells, another TNBC cell line, HCC-1937, was treated with PTX
(Fig. S1) and the downregulated
value of Bcl-2/Bax and the upregulated value of LC3-II/LC3-I showed
that PTX induced apoptosis and autophagy (Fig. S1B-G). Therefore, we speculated
that PTX treatment in different TNBC cells would have similar
effects. However, HCC-1937 cells seemed to be more sensitive to PTX
treatment than MDA-MB-231 cells (Fig. S1A). Therefore, HCC-1937 cells
were not selected for subsequent assays.
FOXO1 is a representative member of the forkhead
transcription factor (FOX) family, which plays a crucial role in
the inhibition of tumor proliferation and the induction of cellular
responses (7). In the present
study, it was found that FOXO1 played an important role in
PTX-induced autophagy in MDA-MB-231 cells. The increase in FOXO1
was accompanied by attenuated AKT1 phosphorylation, suggesting that
PTX induces elevated FOXO1 expression by inhibiting AKT1-related
signaling. Previously, it was also determined that p-ATK1 catalyzed
the phosphorylation of FOXO1, resulting in the loss of
transcriptional activity and translocation from the nucleus to the
cytoplasm (40). However, in the
current study, it was observed that FOXO1 accumulated in the
nucleus rather than translocating to the cytoplasm. Therefore, the
expression of core autophagy-related genes that are
transcriptionally regulated by FOXO1 were analyzed. The results
revealed that ATG5, BECN1 and MAP1LC3B were
significantly upregulated in response to PTX treatment, consistent
with previous reports that nuclear FOXO1 transcriptionally
activates ATG5 and BECN1 to promote autophagy
(41). After inhibition of FOXO1
in combination with PTX treatment, ATG5, VPS34, ATG4B, BECN1
and MAP1LC3B were markedly downregulated. These findings
suggested that FOXO1-mediated autophagy in PTX-treated MDA-MB-231
cells regulates its downstream target genes through
transactivation, which is different than cytosolic FOXO1 inducing
autophagy by binding to the ATG7 gene (42). In addition, FOXO1
transcriptionally inhibits mTOR activity by increasing
SENSE3 expression to promote autophagy (43). Importantly, in the present study
autophagy was inhibited after FOXO1 knockdown; at the same time,
the rate of apoptotic MDA-MB-231 cell death induced by PTX was
increased. This finding was consistent with the concept that
regulation of apoptosis by autophagy enhances cancer therapy
(9) and provides a potential
therapeutic target for TNBC. However, the current study only
explored the in vitro role of FOXO1 in MDA-MB-231 cells,
thus whether PTX treatment would lead to elevated FOXO1 expression
in vivo is still unclear. Therefore, we are creating a
tumor-bearing mouse model by injecting MDA-MB-231 cells into nude
mice. In future studies, we will treat these mice with PTX and
investigate whether FOXO1 plays the same role in vivo.
Clinically, it has been reported that FOXO1 expression is not
observed in tissues from patients with TNBC (44). Therefore, it is important to
determine whether the tissues of patients with TNBC treated with
PTX exhibit increased FOXO1 expression, which may provide novel
insight into the clinicopathological significance.
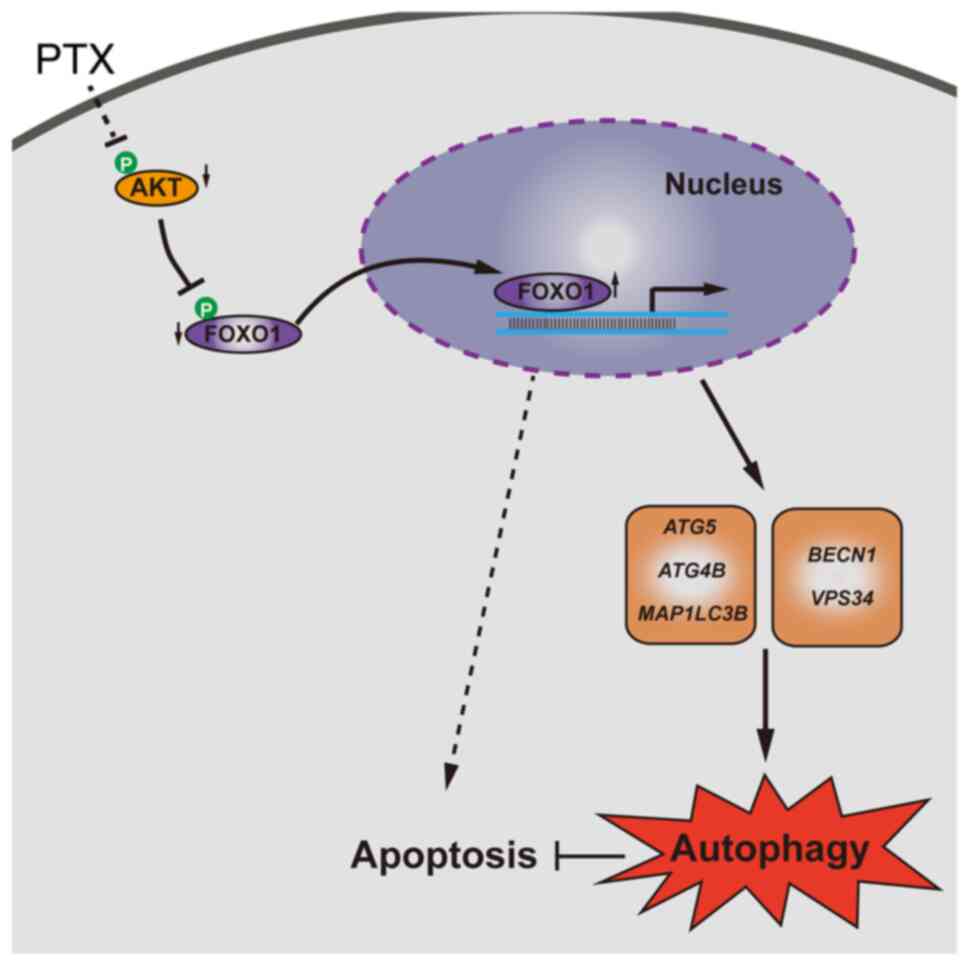
In summary, PTX induced both autophagy and
apoptosis, and inhibition of autophagy promoted apoptosis in
MDA-MB-231 cells. Furthermore, elevated FOXO1 was associated with
increased autophagic flux. Knocking down FOXO1 enhanced PTX-induced
cell death, which might be important for improving PTX efficacy in
TNBC (Fig. 7).
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key R&D Program of
China (grant no. 2019YFA0110700) and National Natural Science
Foundation of China (grant no. 81872452).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HYZ and HJW conceived and designed the study. KX,
WZ, AX, ZX and DJ performed the cell culture and morphological
observation. KX, WZ and AX performed the apoptotic assay and
immunofluorescence. ZX, DZ, HZ and YQ performed the PCR and western
blotting assays. KX, MAJ and HYZ analyzed the data. KX, WZ and AX
organized the data. KX, WZ, MAJ and HYZ wrote the manuscript. HYZ
and HJW proofread the manuscript and confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mustacchi G and De Laurentiis M: The role
of taxanes in triple-negative breast cancer: Literature review.
Drug Des Devel Ther. 9:4303–4318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang N, Wang C, Wang J, Wang Z, Huang D,
Yan M, Kamran M, Liu Q and Xu B: Aurora kinase A stabilizes FOXM1
to enhance paclitaxel resistance in triple-negative breast cancer.
J Cell Mol Med. 23:6442–6453. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lock R, Kenific CM, Leidal AM, Salas E and
Debnath J: Autophagy-dependent production of secreted factors
facilitates oncogenic RAS-driven invasion. Cancer Discov.
4:466–479. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu CC, Chan ML, Chen WY, Tsai CY, Chang FR
and Wu YC: Pristimerin induces caspase-dependent apoptosis in
MDA-MB-231 cells via direct effects on mitochondria. Mol Cancer
Ther. 4:1277–1285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu Q, Wu W, Fu B, Shi L, Wang X and Kuca
K: JNK signaling in cancer cell survival. Med Res Rev.
39:2082–2104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xing YQ, Li A, Yang Y, Li XX, Zhang LN and
Guo HC: The regulation of FOXO1 and its role in disease
progression. Life Sci. 193:124–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheon SY, Kim H, Rubinsztein DC and Lee
JE: Autophagy, cellular aging and age-related human diseases. Exp
Neurobiol. 28:643–647. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tompkins KD and Thorburn A: Regulation of
apoptosis by autophagy to enhance cancer therapy. Yale J Biol Med.
92:707–718. 2019.PubMed/NCBI
|
|
10
|
Jing Z, Sui X, Yao J, Xie J, Jiang L, Zhou
Y, Pan H and Han W: SKF-96365 activates cytoprotective autophagy to
delay apoptosis in colorectal cancer cells through inhibition of
the calcium/CaMKIIγ/AKT-mediated pathway. Cancer Lett. 372:226–238.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng H, Cheng X, Kuang J, Chen L, Yuen S,
Shi M, Liang J, Shen B, Jin Z, Yan J and Qiu W: Apatinib-induced
protective autophagy and apoptosis through the AKT-mTOR pathway in
anaplastic thyroid cancer. Cell Death Dis. 9:10302018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xi G, Hu X, Wu B, Jiang H, Young CYF, Pang
Y and Yuan H: Autophagy inhibition promotes paclitaxel-induced
apoptosis in cancer cells. Cancer Lett. 307:141–148. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang SF, Wang XY, Fu ZQ, Peng QH, Zhang
JY, Ye F, Fu YF, Zhou CY, Lu WG, Cheng XD and Xie X: TXNDC17
promotes paclitaxel resistance via inducing autophagy in ovarian
cancer. Autophagy. 11:225–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagata S: Apoptosis and clearance of
apoptotic cells. Annu Rev Immunol. 36:489–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mohammed F, Rashid-Doubell F, Taha S,
Cassidy S and Fredericks S: Effects of curcumin complexes on
MDA-MB-231 breast cancer cell proliferation. Int J Oncol.
57:445–455. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiao D, Cheng W, Zhang X, Zhang Y, Guo J,
Li Z, Shi D, Xiong Z, Qing Y, Jamal MA, et al: Improving porcine
SCNT efficiency by selecting donor cells size. Cell Cycle.
20:2264–2277. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu W, Qu H, Xu K, Jia B, Li H, Du Y, Liu
G, Wei HJ and Zhao HY: Differences in the starvation-induced
autophagy response in MDA-MB-231 and MCF-7 breast cancer cells.
Anim Cells Syst (Seoul). 21:190–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv C, Qu H, Zhu W, Xu K, Xu A, Jia B, Qing
Y, Li H, Wei HJ and Zhao HY: Low-dose paclitaxel inhibits tumor
cell growth by regulating glutaminolysis in colorectal carcinoma
cells. Front Pharmacol. 8:2442017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mekhail TM and Markman M: Paclitaxel in
cancer therapy. Expert Opin Pharmacother. 3:755–766. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alers S, Loffler AS, Wesselborg S and
Stork B: Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy:
Cross talk, shortcuts, and feedbacks. Mol Cell Biol. 32:2–11. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Onorati AV, Dyczynski M, Ojha R and
Amaravadi RK: Targeting autophagy in cancer. Cancer. 124:3307–3318.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Füllgrabe J, Ghislat G, Cho DH and
Rubinsztein DC: Transcriptional regulation of mammalian autophagy
at a glance. J Cell Sci. 129:3059–3066. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weaver BA: How taxol/paclitaxel kills
cancer cells. Mol Biol Cell. 25:2677–2681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Orr GA, Verdier-Pinard P, McDaid H and
Horwitz SB: Mechanisms of taxol resistance related to microtubules.
Oncogene. 22:7280–7295. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yusuf R, Duan Z, Lamendola D, Penson R and
Seiden M: Paclitaxel resistance: Molecular mechanisms and
pharmacologic manipulation. Curr Cancer Drug Targets. 3:1–19. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Blanchard Z, Paul BT, Craft B and ElShamy
WM: BRCA1-IRIS inactivation overcomes paclitaxel resistance in
triple negative breast cancers. Breast Cancer Res. 17:52015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhola NE, Balko JM, Dugger TC, Kuba MG,
Sánchez V, Sanders M, Stanford J, Cook RS and Arteaga CL: TGF-β
inhibition enhances chemotherapy action against triple-negative
breast cancer. J Clin Invest. 123:1348–1358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wee ZN, Yatim SMJ, Kohlbauer VK, Feng M,
Goh JY, Bao Y, Lee PL, Zhang S, Wang PP, Lim E, et al: IRAK1 is a
therapeutic target that drives breast cancer metastasis and
resistance to paclitaxel. Nat Commun. 6:87462015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sha L, Zhang Y, Wang W, Sui X, Liu SK,
Wang T and Zhang H: MiR-18a upregulation decreases dicer expression
and confers paclitaxel resistance in triple negative breast cancer.
Eur Rev Med Pharmacol Sci. 20:2201–2208. 2016.PubMed/NCBI
|
|
32
|
Yuan Z, Jiang H, Zhu X, Liu X and Li J:
Ginsenoside Rg3 promotes cytotoxicity of Paclitaxel through
inhibiting NF-κB signaling and regulating Bax/Bcl-2 expression on
triple-negative breast cancer. Biomed Pharmacother. 89:227–232.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou YF, Sun Q, Zhang YJ, Wang GM, He B,
Qi T, Zhou Y, Li XW, Li S and He L: Targeted inhibition of notch1
gene enhances the killing effects of paclitaxel on triple negative
breast cancer cells. Asian Pac J Trop Med. 10:179–183. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wen J, Yeo S, Wang C, Chen S, Sun S, Haas
MA, Tu W, Jin F and Guan JL: Autophagy inhibition re-sensitizes
pulse stimulation-selected paclitaxel-resistant triple negative
breast cancer cells to chemotherapy-induced apoptosis. Breast
Cancer Res Treat. 149:619–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang K: Autophagy and apoptosis in liver
injury. Cell Cycle. 14:1631–1642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Veldhoen R, Banman S, Hemmerling D, Odsen
R, Simmen T, Simmonds AJ, Underhill DA and Goping IS: The
chemotherapeutic agent paclitaxel inhibits autophagy through two
distinct mechanisms that regulate apoptosis. Oncogene. 32:736–746.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie S, Ogden A, Aneja R and Zhou J:
Microtubule-binding proteins as promising biomarkers of paclitaxel
sensitivity in cancer chemotherapy. Med Res Rev. 36:300–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eum KH and Lee M: Crosstalk between
autophagy and apoptosis in the regulation of paclitaxel-induced
cell death in v-Ha-ras-transformed fibroblasts. Mol Cell Biochem.
348:61–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou J, Liao W, Yang J, Ma K, Li X, Wang
Y, Wang D, Wang L, Zhang Y, Yin Y, et al: FOXO3 induces
FOXO1-dependent autophagy by activating the AKT1 signaling pathway.
Autophagy. 8:1712–1723. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu P, Das M, Reilly J and Davi RJ: JNK
regulates FoxO-dependent autophagy in neurons. Genes Dev.
25:310–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao Y, Yang J, Liao W, Liu X, Zhang H,
Wang S, Wang D, Feng J, Yu L and Zhu WG: Cytosolic FoxO1 is
essential for the induction of autophagy and tumour suppressor
activity. Nat Cell Biol. 12:665–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang J, Ng S, Wang J, Zhou J, Tan SH,
Yang N, Lin Q, Xia D and Shen HM: Histone deacetylase inhibitors
induce autophagy through FOXO1-dependent pathways. Autophagy.
11:629–642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rehman A, Kim Y, Kim H, Sim J, Ahn H,
Chung MS, Shin SJ and Jang K: FOXO3a expression is associated with
lymph node metastasis and poor disease-free survival in
triple-negative breast cancer. J Clin Pathol. 71:806–813. 2018.
View Article : Google Scholar : PubMed/NCBI
|