Introduction
Breast cancer (BC) is a leading cause of
cancer-related mortality in women worldwide and remains the most
common malignancy among younger patients (1–3).
Although treatments have improved, including chemotherapy and
radiotherapy, BC prognoses remain poor (4). Therefore, it is important to
investigate the molecular mechanisms of BC growth and metastasis to
discover novel molecular targets for BC treatment (5).
Long non-coding (lnc)RNAs are defined as non-coding
transcripts that are >200 nucleotides in length. Dysregulated
lncRNA expression has be observed in numerous types of cancer,
which suggests that aberrant lncRNA expression may be a major
contributor to tumorigenesis (6–9).
lncRNAs also regulate various biological processes, including cell
proliferation, apoptosis, differentiation, migration and invasion
in the cancer microenvironment (9–11), and
therefore provide new opportunities for cancer diagnosis and
treatment (9,12). For example, lncRNA RUSC1-AS1
promotes the progression of BC through mediating cyclin-dependent
kinase inhibitor 1 and kruppel-like factor 2 (13). Overexpression of lncRNA HOTAIR has
been shown to promote the development of retinoblastoma via the
miR-613/c-met axis, which modulates the epithelial-mesenchymal
transition in retinoblastoma (14).
Furthermore, lncRNA in non-homologous end joining pathway 1 may
promote BC growth by regulating BC cell metastasis and increasing
the expression of epithelial-mesenchymal transition-related markers
(15).
lncRNA prostate cancer-associated transcript 1
(PCAT1) was first discovered in 2011 as a prostate-specific
regulator of cell proliferation in prostate cancer (16). Recent studies have indicated that
PCAT1 serves a role in numerous types of cancer (16–18).
It has also been reported that PCAT1 promotes epithelial ovarian
cancer by mediating the expression of cyclin D1/CDK4 (17). Furthermore, PCAT1 knockdown may
reduce the expression of cyclin B1 and CDC2 kinase activity,
inhibiting the growth of esophageal squamous cell carcinoma (ESCC)
by sponging miR-326 (19).
Therefore, the aim of the present study was to explore the function
and molecular mechanism of lncRNA PCAT1/microRNAs
(miRNAs/miRs)-134-3p/pituitary homeobox 2 (PITX2) in BC.
Materials and methods
Tissue specimens
This study involved 30 patients who underwent BC
resection at Liyang People's Hospital (Liyang, China) between March
2019 and March 2020. All patients were newly diagnosed with primary
BC. Patients <18 or >75 years of age, who presented with
first distant metastasis, combined with other malignancies, or who
received neoadjuvant therapy were excluded from the present study.
Tumor and matched adjacent normal tissues were collected and stored
in liquid nitrogen. The present study was approved by the Ethics
Committee of Liyang People's Hospital (approval no. 2018024), and
were conducted in accordance with the Declaration of Helsinki.
Written informed consent was obtained from all participants prior
to the study.
Cell culture and transfection
Human BC cell lines (MDA-MB-231, SKBr-3, MCF-7 and
ZR-75-30) and the normal human mammary epithelial MCF10A cell line
were purchased from the American Type Culture Collection.
MDA-MB-231, MCF10A, SKBr-3 and MCF-7 were cultured in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin and incubated at 37°C in an atmosphere
containing 5% CO2. ZR-75–30 cells were cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with
10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin and
incubated at 37°C in an atmosphere containing 5%
CO2.
For cell transfection, short hairpin RNAs (shRNAs)
targeting lncRNA PCAT1 (sh-PCAT1), PITX2 (sh-PITX2) and the
corresponding sh-negative control (NC) were subcloned into the
GV248 (hU6-MCS-UbiquitinEGFP-IRES-puromycin) vector (Shanghai
GeneChem Co., Ltd.). The following sequences were used: sh-PCAT1,
5′-ATACATAAGACCATGGAAAT-3′; sh-PITX2, 5′-GATGCAATGATGTTTCTGAAA-3′;
and sh-NC, 5′-TTCTCCGAACGTGTCACGT-3′. miR-134-3p mimic, miR-134-3p
inhibitor and their corresponding NCs (NC mimic and NC inhibitor)
were purchased from Thermo Fisher Scientific, Inc.
The sequences were as follows: miR-134-3p mimic,
5′-CCUGUGGGCCACCUAGUCACCAA-3′; miR-134-3p inhibitor,
5′-CCUGUGGGCCACCUAGUCACCAA-3′; NC mimic,
5′-UUCUCCGAACGUGUCACGUTT-3′; and NC inhibitor, 5′
UCACAACCUCCUAGAAAGAGUAGA 3′. For plasmid or miRNA mimic/inhibitor
transfection, SKBR-3 and MCF-7 cells were seeded in 6-well plates
at a density of 1×106 cells/well and cultured to 80%
confluence. Plasmids (1 µg) or miRNA mimic/inhibitor (20 nm) were
transfected into cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were then
cultured at 37°C for 24 h, and transfection efficiency was
assessed. Subsequent experiments were performed 48 h
post-transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from BC and adjacent normal
tissue samples or cells (MCF10A, MDA-MB-231, SKBr-3, MCF-7 and
ZR-75-30). To detect the expression levels of miR-134-3p, the
miRNeasy Kit (Shanghai Yeasen Biotechnology Co., Ltd.) was used to
extract total RNA. Total RNA was reverse transcribed using the
miScript II RT Kit (Qiagen, Inc.) into complementary DNA (cDNA)
according to the manufacturer's protocol. qPCR reaction systems
were prepared using the miScript SYBR Green PCR Kit (Qiagen, Inc.)
with U6 as the internal reference gene. To detect the mRNA
expression levels, total RNA was extracted using TRIzol®
reagent (Takara Bio, Inc.). Total RNA was reverse transcribed into
cDNA using the PrimeScript™ II 1st Strand cDNA Synthesis Kit
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
protocol. All PCR reactions were carried out using an ABI 7500
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH
and U6 were used as the internal reference genes. The following
primers were used for qPCR: PCAT1 forward (F),
5′-GCTGGCATTGGTCAACATAAC-3′ and reverse (R),
5′-GTGAATATGGCGGATGAGGAA-3′; miR-134-3p F,
5′-CTGTGGGCCACCTAGTCACCAA-3′ and R, 5′-GCTGTCAACGATACGCTACCTA-3′;
PITX2 F, 5′-GGCCGCCGCTTCTTACA-3′ and R,
5′-CACTGGCGATTTGGTTCTGATTT-3′; U6 F, 5′-GTGATCACTCCCTGCCTGAG-3′ and
R, 5′-GGACTTCACTGGACCAGACG-3′; and GAPDH F,
5′-CCGCATCTTCTTGTGCAGTG-3′ and R, 5′-CCCAATACGGCCAAATCCGT-3′. The
thermocycling conditions were as follows: 10 min at 95°C for 1
cycle, followed by denaturation at 95°C for 30 sec, annealing at
56°C for 1 min, and final extension at 72°C for 30 sec for 40
cycles. Relative gene expression was calculated using the
2−ΔΔCq method (20) and
were normalized to either GAPDH or U6 levels.
Cell proliferation assay
The proliferation rates of SKBr-3 and MCF-7 cells
were analyzed using the Cell Counting Kit-8 (CCK-8) assay (Dojindo
Laboratories, Inc.) according to the manufacturer's instructions.
Cells were seeded into 96-well plates at a density of ~4,000
cells/well. Cells were then incubated for 24, 48 and 72 h.
Subsequently, 10 µl CCK-8 reagent was added to each well. The
plates were incubated at 37°C for 2 h. The optical density was
measured at 450 nm using a ultraviolet spectrophotometer (Thermo
Fisher Scientific, Inc.).
Wound healing assay
The migratory abilities of SKBr-3 and MCF-7 cells
were analyzed using wound healing assays. Transfected cells
(5×105 cells/well) were inoculated into 6-well plates
and incubated in serum-free medium. Subsequently, the confluent
cell monolayer (95–100%) was scratched using a 200 µl pipette tip.
Cell migration was observed and quantified at 0 and 48 h. Migrated
cells were counted in three randomly selected fields using an IX70
inverted optical microscope (magnification, ×100; Olympus
Corporation). Cell migration was calculated according to the
following formula: Cell migration (%) = (width at 0 h - width at 48
h) / width at 0 h ×100.
Transwell assay
The migration and invasion of SKBr-3 and MCF-7 cells
were analyzed using Transwell assays. The upper chamber was filled
with serum-free DMEM containing 5×104 cells, and the
lower chamber was filled with DMEM containing 10% FBS. Following
incubation for 24 h at 37°C, cells in the lower chamber were
collected and stained with 0.5% crystal violet for 20 min at room
temperature. Stained cells were counted using an IX70 inverted
optical microscope (magnification, ×100; Olympus Corporation). The
invasion assay was carried out using the aforementioned Transwell
assay protocol, but the upper chamber was precoated with Matrigel
(37°C for 30 min).
Colony formation assay
The colony forming ability of SKBr-3 and MCF-7 cells
was assessed using a colony formation assay. The SKBr-3 or MCF-7
cells were seeded into 6-well plates at a density of
1×103 cells/well. Colonies were formed for 10 days at
room temperature and then the culture medium was removed. Following
which, the colonies were washed with PBS three times and fixed with
methyl alcohol for 10 min at room temperature and stained with 0.5%
crystal violet solution for a further 10 min at room temperature.
After washing with water, the colonies were imaged and counted
under a light microscope (magnification, ×10; Olympus
Corporation).
Cell cycle analysis
Flow cytometry was used for SKBr-3 and MCF-7 cells
(1×105) cycle analysis. Cells were collected and
centrifuged at 100 × g at room temperature for 5–10 min. Cells were
digested using trypsin and fixed in 70% ethanol and incubated at
4°C for 1 h. Subsequently, the cells were incubated with 25 µg/ml
PI, 25 µl/ml RNase A and Triton X-100 for 30 min at 4°C. The cell
cycle results were detected using a flow cytometer (FACSCalibur; BD
Biosciences) and analyzed with FlowJo software (version 10.9;
FlowJo LLC).
Flow cytometry
Flow cytometry was employed for early and late
apoptosis analysis, which was completed using an Annexin V-FITC kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. SKBr-3 and MCF-7 cells (1×105) were collected
and centrifuged at 100 × g at room temperature for 5–10 min. Cells
were first stained with 5 µl Annexin V-FITC and 5 µl PI
simultaneously at 4°C for 15 min in the dark. Cell apoptosis was
detected using a flow cytometer (FACSCalibur; BD Biosciences). The
apoptosis results were analyzed with FlowJo software (version 10.9;
FlowJo LLC).
Western blotting
Total protein from SKBr-3 and MCF-7 cells after
transfection was extracted using RIPA buffer (Invitrogen; Thermo
Fisher Scientific, Inc.), followed by quantification using the BCA
Assay Kit (Santa Cruz Biotechnology, Inc.). Then, 25 µg
protein/lane was separated by SDS-PAGE on an 8–12% gel. The
separated proteins were transferred onto a 0.22-µm PVDF membrane
and blocked in Tris-buffered saline with 1% Tween-20 and 5% non-fat
milk for 1 h at room temperature. Subsequently, the membranes were
incubated overnight at 4°C with primary antibodies (all from Abcam)
targeted against: Anti-Bax (1:1,000; cat. no. ab32503), anti-Bcl-2
(1:1,000; cat. no. ab32124), anti-cleaved caspase-3 (1:500; cat.
no. ab32042), anti-cleaved caspase-9 (1:500; cat. no. ab2324),
anti-cyclooxygenase 2 (Cox-2; 1:1,000; cat. no. ab179800),
anti-MMP-2 (1:1,000; cat. no. ab92536), anti-MMP-9 (1:1,000; cat.
no. ab76003), anti-PITX2 (1:1,000; cat. no. ab221142) and
anti-GAPDH (1:2,000; cat. no. ab181602). Following which, membranes
were incubated with a HRP-conjugated anti-rabbit secondary antibody
(1:10,000; cat. no. ab6721; Abcam) for 2 h at room temperature.
Protein bands were visualized via enhanced chemiluminescence
(Pierce; Thermo Fisher Scientific, Inc.) and were semi-quantified
using ImageJ software (version 1.42; National Institutes of
Health).
Dual-luciferase reporter assay
The starBase database (version 2.0; http://starbase.sysu.edu.cn/starbase2/)
was used to predict the potential miRNA and miR-134-3p target
genes. Wild-type (WT) and mutant (Mut) PCAT1 3′ untranslated
regions (3′UTRs) were integrated into the pGL3 vector (Promega
Corporation) to synthesize pGL3-PCAT1-WT and pGL3-PCAT1-Mut. The WT
and Mut 3′UTRs of PITX2 were also integrated into the luciferase
reporter plasmid psiCHECK-2 (Promega Corporation) to synthesize
psiCHECK-2-PITX2-WT and psiCHECK-2-PITX2-Mut. SKBr-3 and MCF-7
cells at 5×104 cells/well in 24-well plates were
co-transfected with WT or Mut PCAT1 3′UTR or Mut PITX2 3′UTR
reporter plasmids and miR-134-3p mimic or mimic NC using
Lipofectamine 2000 at 37°C for 48 h. Luciferase activity was
detected using the Dual-Luciferase Reporter Assay System (Promega
Corp.). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Statistical analysis
All presented data were obtained from at least three
independent experiments. All of the data were carried out using
GraphPad Prism 10.0. (GraphPad Software, Inc.). Data are presented
as the mean ± standard deviation (SD). Statistical comparisons were
determined using unpaired/paired Student's t-tests, Mann-Whitney U
test or one-way ANOVA followed by Bonferroni's post hoc test.
Pearson's correlation coefficient was used to analyze the
correlation between the expression levels of PCAT1 and miR-134-3p
in clinical BC samples. P<0.05 was considered to indicate a
statistically significant difference.
Results
Knockdown of PCAT1 inhibits BC cell
proliferation
The expression patterns of PCAT1 were determined to
investigate its biological function in human BC cell lines. PCAT1
expression levels were evaluated in 30 paired human BC specimens
and normal adjacent tissues. The results demonstrated that PCAT1
expression levels were significantly higher in BC specimens
compared with those in normal adjacent tissues (Fig. 1A). Furthermore, PCAT1 expression
levels in multiple human BC cell lines were upregulated compared
with those in MCF10A cells, especially in SKBr-3 and MCF-7 cells
(Fig. 1B). Therefore, to further
investigate the role of PCAT1 in human BC, sh-PCAT1 and its
scrambled control (sh-NC) were transfected into SKBr-3 and MCF-7
cells. The knockdown efficiency of sh-PCAT1 in SKBr-3 and MCF-7
cells was validated using RT-qPCR analysis. The results
demonstrated that sh-PCAT1 significantly decreased PCAT1 expression
levels compared with sh-NC (Fig.
1C). CCK-8 and colony formation assays demonstrated that PCAT1
knockdown significantly inhibited the proliferation of SKBr-3 and
MCF-7 cells compared with the sh-NC group (Fig. 1D and E). Cell proliferation is
closely connected with the cell cycle. Therefore, the effects of
PCAT1 knockdown on the cell cycle in SKBr-3 and MCF-7 cells were
investigated. The results demonstrated that PCAT1 knockdown
significantly induced cell cycle arrest in the G1 phase
compared with in the sh-NC group (Fig.
1F). At the molecular level, PCAT1 knockdown resulted in
significantly downregulated cyclin D1 and significantly upregulated
p21 protein expression levels compared with in the sh-NC group
(Fig. 1G). Flow cytometry revealed
that PCAT1 knockdown significantly promoted the apoptosis of SKBr-3
and MCF-7 cells compared with in the sh-NC group (Fig. 1H). Furthermore, the protein
expression levels of apoptosis-related proteins were examined by
western blotting. The results demonstrated that PCAT1 knockdown
significantly reduced the protein expression levels of
antiapoptotic protein Bcl-2, and significantly upregulated the
protein expression levels of proapoptotic proteins, Bax, cleaved
caspase-3 and cleaved caspase-9 in SKBr-3 and MCF-7 cells (Fig. 1I). Overall, these results indicated
that knockdown of PCAT1 may inhibit proliferation and promote the
apoptosis of human BC cells.
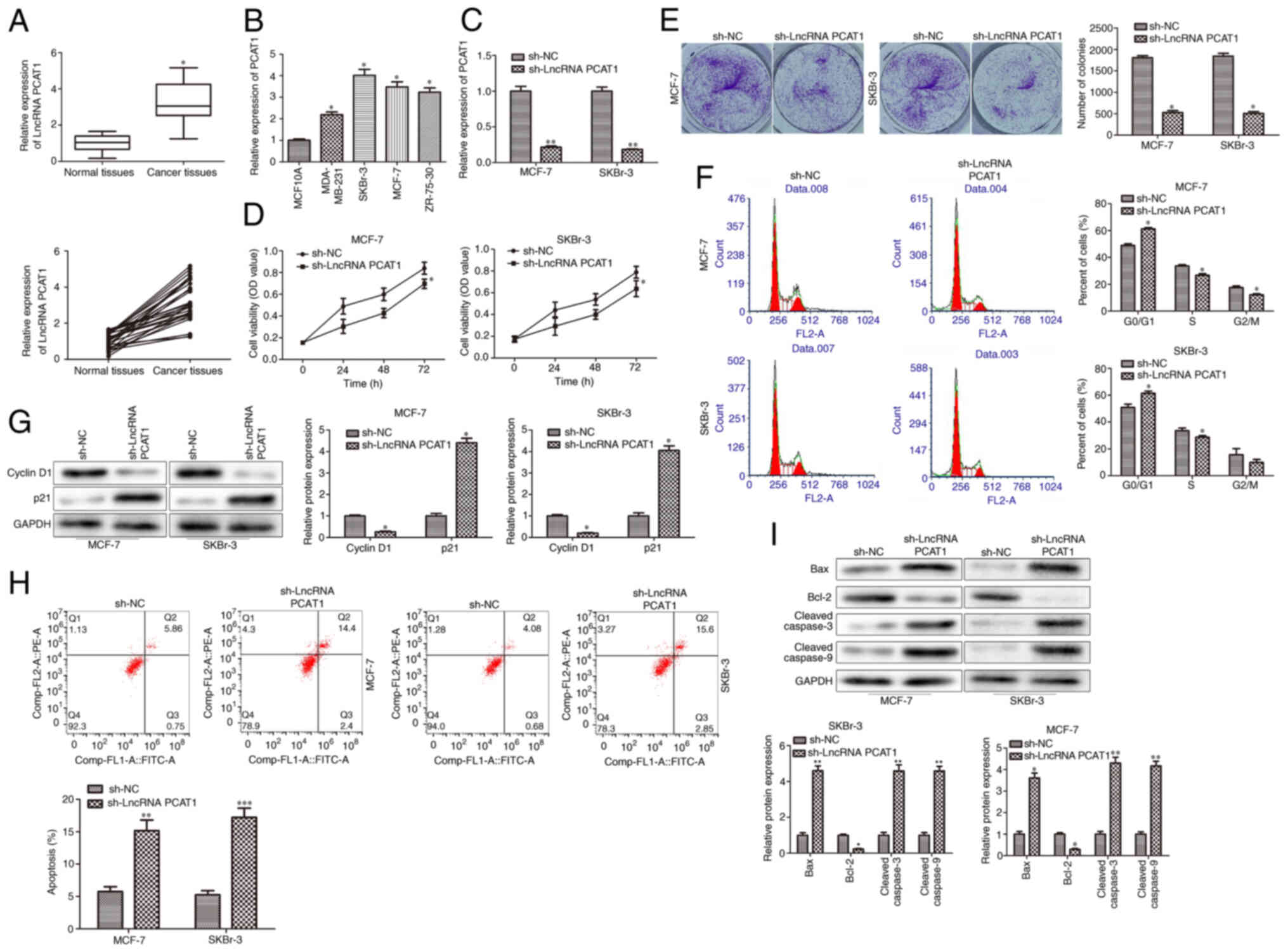 | Figure 1.PCAT1 is highly expressed in BC cells
and its knockdown inhibits BC cell proliferation. (A) RT-qPCR was
performed to determine PCAT1 expression levels in BC specimens and
normal adjacent tissues. Data are presented as the mean ± SD.
*P<0.05 vs. normal adjacent tissues. n=30. (B) RT-qPCR was
performed to determine PCAT1 expression levels in BC cell lines
(MDA-MB-231, SKBr-3 and MCF-7 and ZR-75-30) and the human mammary
epithelial MCF10A cell line. Data are presented as the mean ± SD.
*P<0.05 vs. MCF10A. n=3. BC SKBr-3 and MCF-7 cell lines were
transfected with sh-PCAT1. (C) RT-qPCR was performed to detect the
knockdown efficiency of sh-PCAT1. (D) Cell Counting Kit-8 assays
were performed to determine cell proliferation at 24, 28 and 72 h
in SKBr-3 and MCF-7 cells. (E) Colony formation assays were
performed to demonstrate cell proliferation in SKBr-3 and MCF-7
cells. (F) Flow cytometry was used to analyze the cell cycle. (G)
Western blotting was performed to determine cyclin D1 and p21
protein expression levels. (H) Flow cytometry was used to analyze
the apoptotic rate. (I) Western blotting was performed to determine
the protein expression levels of apoptosis-associated proteins,
Bax, Bcl-2, cleaved caspase-3 and cleaved caspase-9. Data are
presented as the mean ± SD. *P<0.05, **P<0.01 and
***P<0.001 vs. sh-NC. n=3. PCAT1, prostate cancer-associated
transcript 1; BC, breast cancer; RT-qPCR, reverse
transcription-quantitative PCR; lncRNA, long non-coding RNA; sh,
short hairpin; NC, negative control; PE, phycoerythrin. |
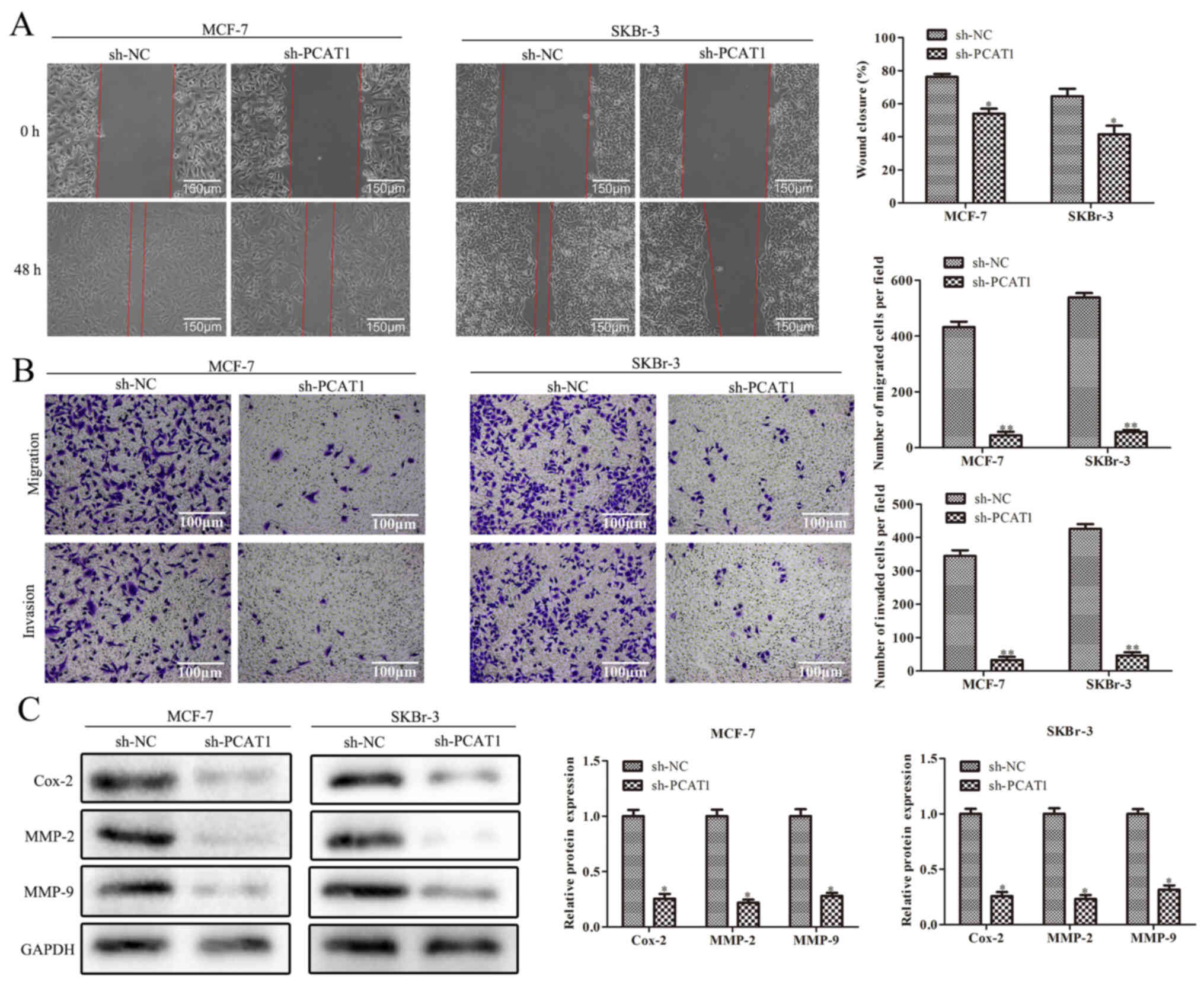
Knockdown of PCAT1 inhibits the
migration and invasion of BC cells
Wound healing and Transwell assays demonstrated that
the migration and invasion of SKBr-3 and MCF-7 cells were
significantly reduced in cells transfected with sh-PCAT1 compared
with sh-NC (Fig. 2A and B). Western
blotting also revealed that the protein expression levels of
migration- and invasion-related proteins, Cox-2, MMP-2 and MMP-9,
were significantly downregulated by sh-PCAT1 in SKBr-3 and MCF-7
cells compared with in the sh-NC group (Fig. 2C). Overall, these results indicated
that sh-PCAT1 may inhibit the migration and invasion of human BC
cells.
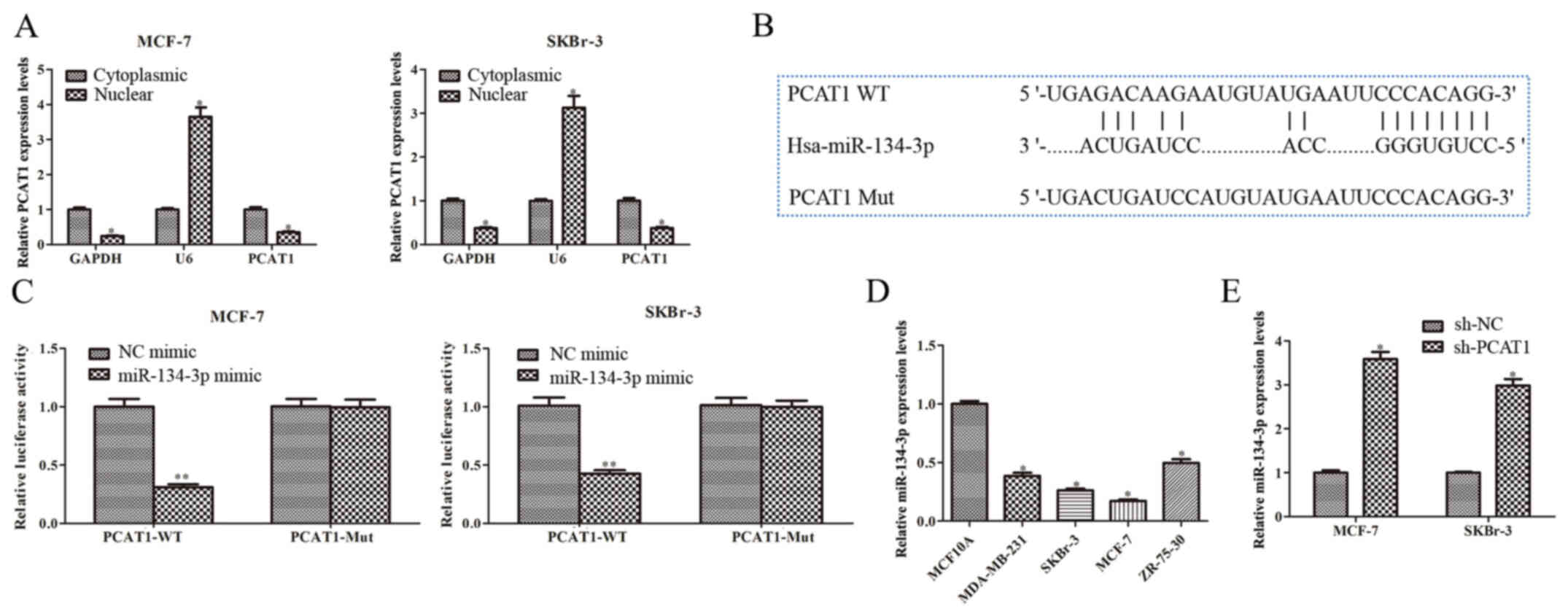
PCAT1 targets miR-134-3p in human BC
cells
Subsequently, the molecular mechanism of PCAT1 in BC
was investigated. Cytoplasmic and nuclear RNA were extracted from
SKBr-3 and MCF-7 cells. The RT-qPCR results demonstrated that PCAT1
was significantly more highly expressed in the cytoplasmic fraction
compared with in the nuclear fraction (Fig. 3A). Therefore, the starBase database
was used to predict the potential miRNA PCAT1 interacted with,
identifying miR-134-3p as a potential target (Fig. 3B). To evaluate the interaction
between miR-134-3p and PCAT1, the dual-luciferase reporter assay
was performed. The results demonstrated a significant decrease in
luciferase activity in cells co-transfected with PCAT1-WT and
miR-134-3p mimic compared with the NC-mimic (Fig. 3C). However, no significant
difference was observed in luciferase activity between the
miR-134-3p mimic group and NC mimic group when the putative binding
sites were mutated (Fig. 3C).
Furthermore, miR-134-3p expression levels in multiple human BC cell
lines were significantly downregulated compared with those in
MCF10A cells (Fig. 3D). miR-134-3p
expression levels were upregulated when SKBr-3 and MCF-7 cells were
transfected with sh-PCAT1 compared with sh-NC (Fig. 3E). These results indicated that
PCAT1 may function as a sponge for miR-134-3p in SKBr-3 and MCF-7
cells.
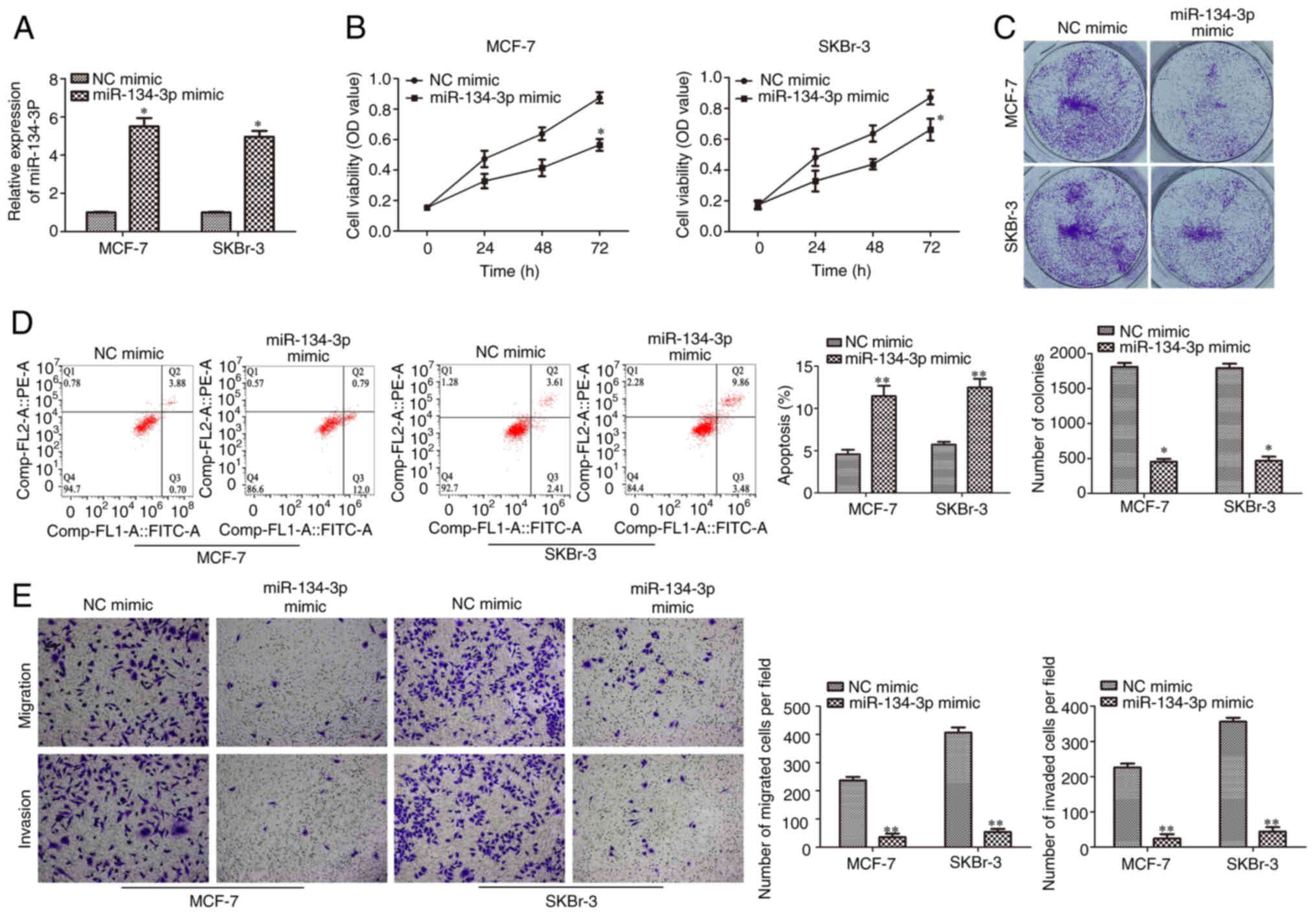
miR-134-3p overexpression inhibits
cell proliferation, migration and invasion, and promotes apoptosis
in human BC cells
In order to investigate the biological function of
miR-134-3p in BC progression, miR-134-3p mimic and NC mimic were
transfected into SKBr-3 and MCF-7 cells. RT-qPCR was performed to
confirm the overexpression efficiency of miR-134-3p mimic in SKBr-3
and MCF-7 cells. Compared with the NC mimic, miR-134-3p expression
levels were significantly upregulated by miR-134-3p mimic
transfection (Fig. 4A). CCK-8 and
colony formation assays demonstrated that miR-134-3p mimic
significantly inhibited the proliferation of SKBr-3 and MCF-7 cells
compared with the NC mimic (Fig. 4B and
C). Furthermore, flow cytometry elucidated that miR-134-3p
overexpression significantly promoted cell apoptosis in SKBr-3 and
MCF-7 cells compared with the NC mimic (Fig. 4D). The Transwell assay further
demonstrated that migration and invasion were significantly reduced
in SKBr-3 and MCF-7 cells transfected with miR-134-3p mimic
compared with the NC mimic (Fig.
4E). These data indicated that miR-134-3p overexpression may
inhibit cell proliferation, migration and invasion but promote
apoptosis in human BC cells.
PITX2 is a potential target of
miR-134-3p
To explore the regulatory mechanism of miR-134-3p in
BC progression, the starBase database was used to predict
miR-134-3p target genes and identified PITX2 as a potential target
(Fig. 5A). The dual-luciferase
reporter assay was used to determine the interaction between
miR-134-3p and PITX2. The results demonstrated that luciferase
activity was significantly reduced when the cells were
co-transfected with PITX2-WT and miR-134-3p mimics. However, there
was no significant difference in luciferase activity of PITX2-Mut
between cells transfected with miR-134-3p mimic or NC mimic
(Fig. 5B). To further analyze the
association between PITX2 and miR-134-3p, RT-qPCR and western
blotting were used. The results demonstrated that PITX2 mRNA and
protein expression levels were significantly decreased by
miR-134-3p mimic in SKBr-3 and MCF-7 cells compared with the NC
mimic (Fig. 5C and D). Furthermore,
PITX2 mRNA expression levels were significantly increased in BC
cells (MDA-MB-231, ZR-75-30, SKBr-3 and MCF-7) compared with those
in MCF10A cells (Fig. 5E).
Pearson's correlation coefficient analysis demonstrated a negative
correlation between PCAT1 and miR-134-3p expression levels, and
between miR-134-3p and PITX2 expression levels in clinical BC
samples (Fig. 5F and G). These
results indicated that PITX2 may be a downstream target gene of
miR-134-3p and that PCAT1 may regulate PITX2 expression via
miR-134-3p.
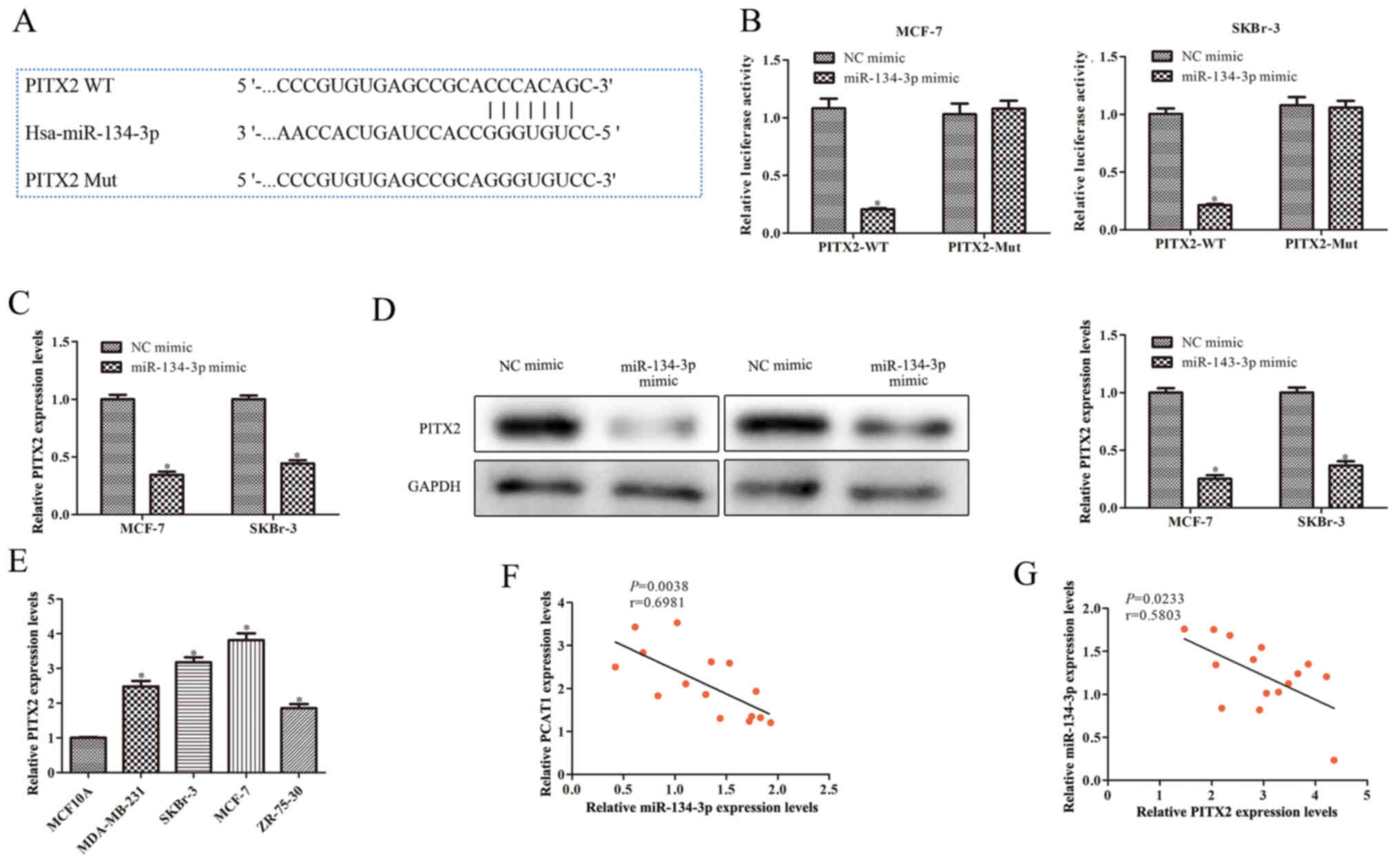 | Figure 5.PCAT1 regulates the expression of
PITX2 via miR-134-3p. (A) Bioinformatics analysis using starBase
predicted the binding site between PITX2 and miR-134-3p. (B)
Dual-luciferase reporter assays were performed following
co-transfection of SKBr-3 and MCF-7 cells with the luciferase
reporter plasmid, containing WT or Mut forms of the PITX2
3′untranslated region, and NC mimic or miR-134-3p mimic. (C)
RT-qPCR was performed to determine PITX2 mRNA expression levels in
SKBr-3 and MCF-7 cells transfected with miR-134-3p mimic. (D)
Western blotting was performed to analyze PITX2 protein expression
levels. *P<0.05 vs. NC mimic. (E) RT-qPCR was performed to
determine PITX2 mRNA expression levels in human BC cell lines and
the human mammary epithelial MCF10A cell line. *P<0.05 vs.
MCF10A cells. Correlation coefficient analysis demonstrated a
negative correlation between (F) PCAT1 and miR-134-3p expression
levels and between (G) miR-134-3p and PITX2 expression levels in
clinical BC samples. Data are presented as the mean ± SD. n=3.
PCAT1, prostate cancer-associated transcript 1; PITX2, pituitary
homeobox 2; miR, microRNA; WT, wild-type; Mut, mutant; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR; BC,
breast cancer. |
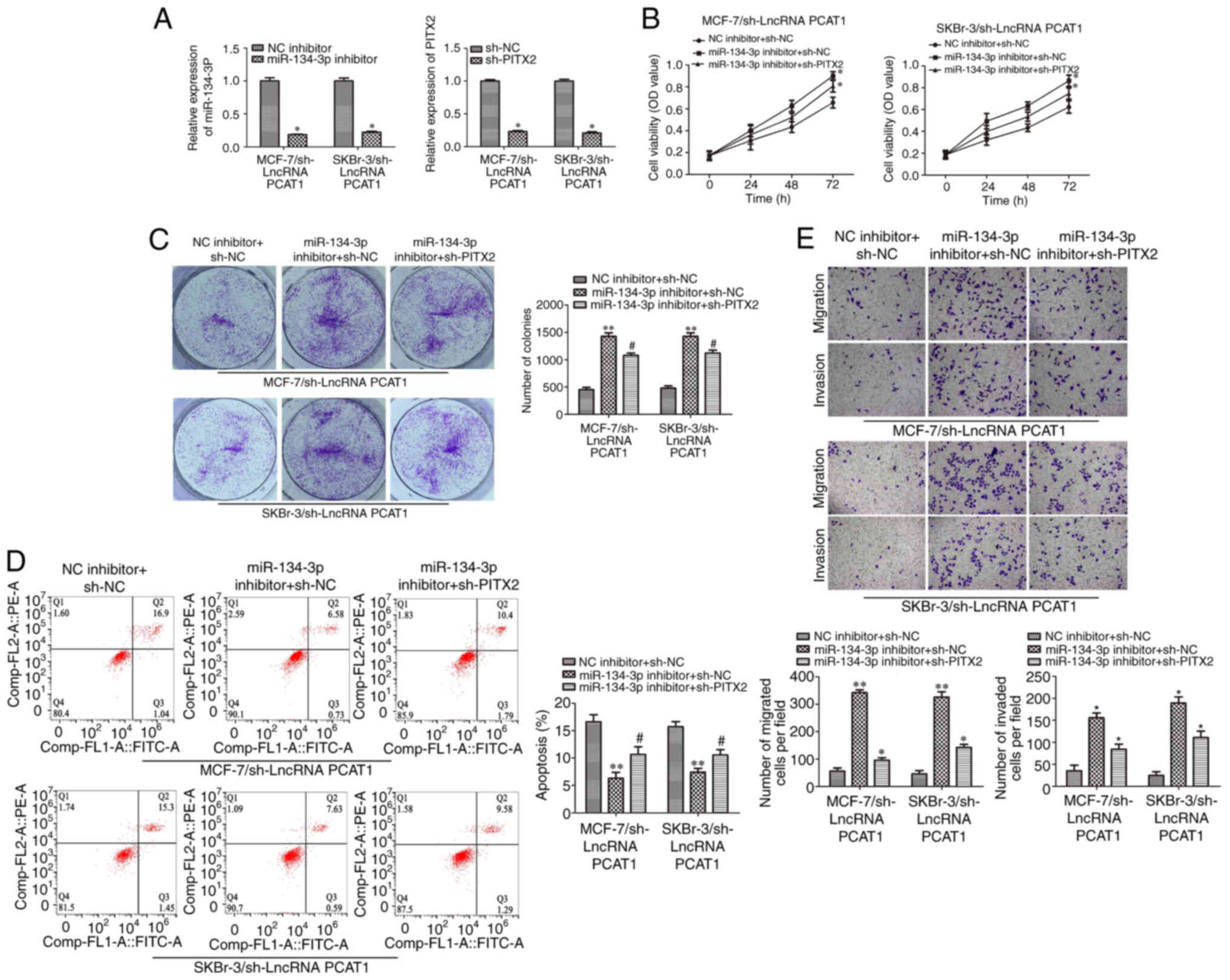
sh-PITX2 rescues the function of the
miR-134-3p inhibitor in human BC cells
Rescue experiments were performed to investigate the
functions of PCAT1, miR-134-3p and PITX2 in human BC cells.
Transfection efficiency is shown in Fig. S1A and B. miR-134-3p expression
levels were significantly downregulated when cells were
co-transfected with miR-134-3p inhibitor and sh-PCAT1, in SKBr-3
and MCF-7 cells, compared with NC inhibitor and sh-PCAT1. PITX2
mRNA expression levels were also significantly downregulated when
cells were co-transfected with sh-PITX2 and sh-PCAT1 in SKBr-3 and
MCF-7 cells, compared with sh-NC and sh-PCAT1 (Fig. 6A). Colony formation and CCK-8 assays
demonstrated PITX2 knockdown significantly reversed the effect of
miR-134-3p inhibition on the proliferation of SKBr-3 and MCF-7
cells transfected with sh-PCAT1 compared with the miR-134-3p
inhibitor + sh-NC group (Fig. 6B and
C). Subsequently, an apoptotic assay was performed via flow
cytometry. Compared with miR-134-3p inhibitor + sh-NC, the results
demonstrated that sh-PITX2 significantly reversed the suppressive
effects of miR-134-3p inhibitor on cell apoptosis in SKBr-3 and
MCF-7 cells following PCAT1 knockdown (Fig. 6D). Moreover, Transwell assays
demonstrated that sh-PITX2 significantly reversed miR-134-3p
inhibitor-enhanced cell migration and invasion of PCAT1-knockdown
SKBr-3 and MCF-7 cells compared with the miR-134-3p inhibitor +
sh-NC group (Fig. 6E). Overall,
these results demonstrated that sh-PITX2 could reverse the function
of the miR-134-3p inhibitor on SKBr-3 and MCF-7 cells, suggesting
that PITX2 serves a role in BC tumorigenesis via the miRNA ‘sponge’
mechanism.
Discussion
BC is one of the most common types of cancer in
women (21,22), with ~500,000 BC-related deaths each
year. Traditional therapeutic approaches, such as surgery and
chemotherapy, rarely completely cure BC and the treatment process
is painful; in addition, most patients will develop recurrence
following treatment (23).
Therefore, the identification of novel treatment strategies is
important and urgently needed. Previous studies have reported that
lncRNAs are crucial regulators involved in cancer-related gene
expression and cancer development (8,17,24).
Consequently, lncRNAs may become new potential therapeutic targets
in clinical applications (25).
lncRNA PCAT1 was first discovered to be upregulated
in patients with prostate cancer, whereby it was demonstrated to
promote prostate cell proliferation (17). Previous studies have reported that
PCAT1 knockdown may inhibit malignant phenotypes in numerous types
of human cancer, including gastric cancer, lung cancer and
hepatocellular carcinoma (26–28).
lncRNA PCAT1 has been shown to be upregulated in human non-small
cell lung cancer (NSCLC), and knockdown of PCAT1 can markedly
suppress cell growth by inducing cell cycle arrest and apoptosis in
NSCLC cells (28). In the present
study, PCAT1 expression levels were found to be significantly
upregulated in BC cell lines in comparison with the non-tumorigenic
epithelial MCF10A cell line. Downregulation of PCAT1 expression
levels was significantly associated with the inhibition of BC cell
proliferation and an increase in apoptosis-related protein
expression levels. These data demonstrated that PCAT1 possibly
promotes the progression of BC.
lncRNAs are considered to play an important
regulatory role in carcinogenesis and cancer development. They
regulate the interaction between proteins and genes as a scaffold
or guide, and act as a bait for binding proteins or miRNA (29). The interaction between lncRNA and
miRNA functional networks has attracted much attention (30). lncRNAs can function by sponging
miRNAs to regulate the expression of specific genes (19) and can therefore affect cancer
progression. For example, lncRNA nuclear paraspeckle assembly
transcript 1 has been reported to be upregulated in BC and to
inhibit the activation of the miR-133b/translocase of inner
mitochondrial membrane 17A axis (29). lncRNA PCAT1 was also shown to
promote ESCC cell proliferation by sponging miR-326 and may
therefore serve as a non-invasive biomarker for ESCC (19). Increasing evidence has indicated
that miR-134 is essential for human carcinoma, including lung, BC
and colorectal cancer, and that it participates in tumor cell
proliferation, apoptosis, invasion and metastasis (31–35).
miR-134-3p, as a member of the miR-134 family, has been reported to
have a significant regulatory role in non-small cell lung cancer
proliferation and invasion (31).
Although the downstream mechanism of miR-134-3p in cancer
progression has been extensively studied, to the best of our
knowledge, the upstream regulatory mechanism of miR-134-3p has not
been elucidated until now. In the present study, miR-134-3p was
identified as a target of PCAT1 and miR-134-3p expression levels
were significantly downregulated in BC cell lines, which was
negatively associated with PCAT1 expression levels. Moreover,
miR-134-3p mimic significantly inhibited cell proliferation,
migration and invasion, but significantly increased the apoptotic
rate of BC cell lines. Together, these results demonstrated that
PCAT1 may regulate BC progression by sponging miR-134-3p.
PITX2 was predicted to be a possible target of
miR-134-3p by bioinformatics analysis, but the mechanisms of
miR-134-3p and PITX2 in BC remain unclear. Previous studies have
highlighted the association of PITX2 with the progression of
numerous types of human cancer, including gonadotroph tumors and
colorectal cancer (36–38). In the present study, PITX2 was
experimentally confirmed as a target of miR-134-3p. PITX2 mRNA
expression levels were negatively associated with miR-134-3p
expression levels. Moreover, sh-PITX2 partially significantly
reduced the lncRNA PCAT1/miR-134-3p knockdown-mediated effects on
cell proliferation, migration, invasion and apoptosis in BC cell
lines.
Overall, these findings demonstrated that PCAT1 may
induce the downregulation of PITX2 expression levels in BC
progression possibly via the upregulation of miR-134-3p expression
levels. Each of the three molecules in the PCAT1/miR-134-3p/PITX2
axis may become a novel therapeutic target for the treatment of BC,
which is of crucial significance for the clinical prevention and
diagnosis of BC. However, the detailed mechanisms of how PITX2
functions in BC requires further investigation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YX, WT, GL and YJ contributed to the conception and
design of the present study, and were responsible for the
acquisition, analysis and interpretation of the data. YX and WT
confirm the authenticity of all the raw data. YX and WT drafted and
critically revised the work for important intellectual content. YX
gave final approval of the version to be published. YX and WT agree
to be accountable for all aspects of the work in ensuring that
questions related to the accuracy or integrity of any part of the
work are appropriately investigated and resolved. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol of the present study was
in accordance with the Declaration of Helsinki and was approved by
the Ethics Committee of the Liyang People's Hospital (Liyang,
China; approval no. 2018024). Written informed consent was obtained
from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q,
Qin Q, Zhao L, Huang Q, Luo Z, et al: Long noncoding RNA LINC00511
contributes to breast cancer tumourigenesis and stemness by
inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res.
37:2892018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia W, Liu Y, Cheng T, Xu T, Dong M and Hu
X: Down-regulated lncRNA SBF2-AS1 inhibits tumorigenesis and
progression of breast cancer by sponging microRNA-143 and
repressing RRS1. J Exp Clin Cancer Res. 39:182020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corey B, Smania MA, Spotts H and Andersen
M: Young Women With Breast Cancer: Treatment, Care, and Nursing
Implications. Clin J Oncol Nurs. 24:139–147. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo Q, Lv S, Wang B, Li Y, Cha N, Zhao R,
Bao W and Jia B: Long non-coding RNA PRNCR1 has an oncogenic role
in breast cancer. Exp Ther Med. 18:4547–4554. 2019.PubMed/NCBI
|
|
5
|
Zhou Y, Meng X, Chen S, Li W, Li D, Singer
R and Gu W: IMP1 regulates UCA1-mediated cell invasion through
facilitating UCA1 decay and decreasing the sponge effect of UCA1
for miR-122-5p. Breast Cancer Res. 20:322018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang Z, Zhou JK, Peng Y, He W and Huang
C: The role of long noncoding RNAs in hepatocellular carcinoma. Mol
Cancer. 19:772020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gugnoni M and Ciarrocchi A: Long Noncoding
RNA and Epithelial Mesenchymal Transition in Cancer. Int J Mol Sci.
20:202019. View Article : Google Scholar
|
|
8
|
Guo Q, Zhang Q, Lu L and Xu Y: Long
noncoding RNA RUSC1-AS1 promotes tumorigenesis in cervical cancer
by acting as a competing endogenous RNA of microRNA-744 and
consequently increasing Bcl-2 expression. Cell Cycle. 19:1222–1235.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmitt AM and Chang HY: Long Noncoding
RNAs in Cancer Pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan S, Yang Z, Ke Z, Huang K, Liu N, Fang
X and Wang K: Downregulation of the long non-coding RNA TUG1 is
associated with cell proliferation, migration, and invasion in
breast cancer. Biomed Pharmacother. 95:1636–1643. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng H, Wang J, Chen T, Zhang K, Chen J,
Wang L, Li H, Tuluhong D, Li J and Wang S: Downregulation of long
non-coding RNA Opa interacting protein 5-antisense RNA 1 inhibits
breast cancer progression by targeting sex-determining region Y-box
2 by microRNA-129-5p upregulation. Cancer Sci. 110:289–302.
2019.PubMed/NCBI
|
|
12
|
Davalos V and Esteller M: Disruption of
Long Noncoding RNAs Targets Cancer Hallmark Pathways in Lung
Tumorigenesis. Cancer Res. 79:3028–3030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu CC, Liang YW, Hu JL, Liu LF, Liang JW
and Wang R: lncRNA RUSC1-AS1 promotes the proliferation of breast
cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur Rev Med
Pharmacol Sci. 23:6602–6611. 2019.PubMed/NCBI
|
|
14
|
Yang G, Fu Y, Lu X, Wang M, Dong H and Li
Q: lncRNA HOTAIR/miR-613/c-met axis modulated
epithelial-mesenchymal transition of retinoblastoma cells. J Cell
Mol Med. 22:5083–5096. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang Y, Li Y, Song X, Zhang N, Sang Y,
Zhang H, Liu Y, Chen B, Zhao W, Wang L, et al: Long noncoding RNA
LINP1 acts as an oncogene and promotes chemoresistance in breast
cancer. Cancer Biol Ther. 19:120–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang
B, Dong Q, Jiang N, Flores-Morales A, Chang C, et al: lncRNA PCAT1
activates AKT and NF-κB signaling in castration-resistant prostate
cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids
Res. 47:4211–4225. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ding C, Wei R, Rodríguez RA and Del Mar
Requena Mullor M: lncRNA PCAT-1 plays an oncogenic role in
epithelial ovarian cancer by modulating cyclinD1/CDK4 expression.
Int J Clin Exp Pathol. 12:2148–2156. 2019.PubMed/NCBI
|
|
18
|
Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX,
Zhao XJ, Lv BL and Liu JB: lncRNA PCAT-1 promotes tumour growth and
chemoresistance of oesophageal cancer to cisplatin. Cell Biochem
Funct. 36:27–33. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang L, Wang Y, Chen J, Wang Y, Zhao Y,
Wang Y, Ma Y, Chen X, Liu W, Li Z, et al: Long noncoding RNA PCAT1,
a novel serum-based biomarker, enhances cell growth by sponging
miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis.
10:5132019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng RM, Zong YN, Cao SM and Xu RH:
Current cancer situation in China: Good or bad news from the 2018
Global Cancer Statistics? Cancer Commun (Lond). 39:222019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang R, Wu JC, Zheng LM, Li ZR, Zhou KL,
Zhang ZS, Xu DF and Chen C: Long noncoding RNA RUSC1-AS-N indicates
poor prognosis and increases cell viability in hepatocellular
carcinoma. Eur Rev Med Pharmacol Sci. 22:388–396. 2018.PubMed/NCBI
|
|
24
|
Shen WJ, Zhang F, Zhao X and Xu J: lncRNAs
and Esophageal Squamous Cell Carcinoma - Implications for
Pathogenesis and Drug Development. J Cancer. 7:1258–1264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hou J, Wang L, Wu Q, Zheng G, Long H, Wu
H, Zhou C, Guo T, Zhong T, Wang L, et al: Long noncoding RNA H19
upregulates vascular endothelial growth factor A to enhance
mesenchymal stem cells survival and angiogenic capacity by
inhibiting miR-199a-5p. Stem Cell Res Ther. 9:1092018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bi M, Yu H, Huang B and Tang C: Long
non-coding RNA PCAT-1 over-expression promotes proliferation and
metastasis in gastric cancer cells through regulating CDKN1A. Gene.
626:337–343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wen J, Xu J, Sun Q, Xing C and Yin W:
Upregulation of long non coding RNA PCAT-1 contributes to cell
proliferation, migration and apoptosis in hepatocellular carcinoma.
Mol Med Rep. 13:4481–4486. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Li Y, Wang B, Ma Y and Chen P:
lncRNA-PCAT-1 promotes non-small cell lung cancer progression by
regulating miR-149-5p/LRIG2 axis. J Cell Biochem. Dec 19–2018.(Epub
ahead of print). doi: 10.1002/jcb.28046.
|
|
29
|
Li X, Deng S, Pang X, Song Y, Luo S, Jin L
and Pan Y: lncRNA NEAT1 Silenced miR-133b Promotes Migration and
Invasion of Breast Cancer Cells. Int J Mol Sci. 20:202019.
|
|
30
|
Chen L, Zhou Y and Li H: lncRNA, miRNA and
lncRNA-miRNA interaction in viral infection. Virus Res. 257:25–32.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qin Q, Wei F, Zhang J and Li B: miR-134
suppresses the migration and invasion of non small cell lung cancer
by targeting ITGB1. Oncol Rep. 37:823–830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qin Q, Wei F, Zhang J, Wang X and Li B:
miR-134 inhibits non-small cell lung cancer growth by targeting the
epidermal growth factor receptor. J Cell Mol Med. 20:1974–1983.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie Y, Song J, Zong Q, Wang A, Yang Y, Liu
F and Meng X: Decreased Expression of MIR-134 and its Clinical
Significance in Human Colorectal Cancer. Hepatogastroenterology.
62:615–619. 2015.PubMed/NCBI
|
|
34
|
Zhang J, Ma Y, Wang S, Chen F and Gu Y:
C/EBPα inhibits proliferation of breast cancer cells via a novel
pathway of miR-134/CREB. Int J Clin Exp Pathol. 8:14472–14478.
2015.PubMed/NCBI
|
|
35
|
Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong
FY, Bo T, He J, Hua RX, Hu WD, et al: miR-134: A Human Cancer
Suppressor? Mol Ther Nucleic Acids. 6:140–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Acunzo J, Roche C, Defilles C, Thirion S,
Quentien MH, Figarella-Branger D, Graillon T, Dufour H, Brue T,
Pellegrini I, et al: Inactivation of PITX2 transcription factor
induced apoptosis of gonadotroph tumoral cells. Endocrinology.
152:3884–3892. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hirose H, Ishii H, Mimori K, Tanaka F,
Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y,
et al: The significance of PITX2 overexpression in human colorectal
cancer. Ann Surg Oncol. 18:3005–3012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang JX, Chen ZH, Xu Y, Chen JW, Weng HW,
Yun M, Zheng ZS, Chen C, Wu BL, Li EM, et al: Downregulation of
MicroRNA-644a Promotes Esophageal Squamous Cell Carcinoma
Aggressiveness and Stem Cell-like Phenotype via Dysregulation of
PITX2. Clin Cancer Res. 23:298–310. 2017. View Article : Google Scholar : PubMed/NCBI
|