Introduction
Cervical cancer (CC) has a high incidence in
developing countries and is one of the most common causes of
cancer-associated mortality in females (1–3),
seriously threatening their health, life and safety (4). CC is divided into two subtypes,
squamous cell carcinoma (SCC) and adenocarcinoma (5). High-risk human papillomavirus
(HR-HPV) infection is considered to be the most crucial risk factor
for CC and is closely related to the occurrence and development of
CC (6). Although the morbidity
and mortality of CC have decreased due to the implementation of
prevention programs in recent years, the prognosis of CC is still
not promising (7). Therefore,
exploring the factors associated with HPV infection is expected to
provide novel ideas for the diagnosis and treatment of CC.
Studies have pointed out that the differences in the
pathological process between HPV infection-positive (HPV+) and
-negative (HPV-) patients may be related to heterogeneous
epigenetic changes in non-coding RNAs, including long non-coding
RNAs (lncRNAs) and microRNAs (miRNAs/miRs) (8). To date, various miRNAs associated
with the development of HPV+ CC have been identified and reported
(9,10). HPV infection is associated with
the aberrant expression of certain lncRNAs (11), whilst reports on lncRNAs
associated with HPV infection are limited. LncRNAs have roles in
numerous diseases (12–16), including CC (17–19). The lncRNA deleted in lymphocytic
leukemia 1 (DLEU1) has been indicated to have a role in promoting
tumor progression in CC (20),
and in the present study, data from the Cancer Genome Atlas (TCGA)
database were analyzed to indicate that DLEU1 may be associated
with the prognosis of patients with CC. However, whether the
aberrant expression of DLEU1 in patients with CC is associated with
HPV infection and what role it has in HPV-infected CC had remained
elusive.
Therefore, the purpose of the present study was to
determine the expression levels of DLEU1 in patients with CC with
different HPV infection status, explore whether DLEU1 was related
to HPV infection and investigate the clinical significance of DLEU1
in HPV-infected patients with CC. The present study provided a
novel biomarker for the diagnosis and survival prognosis of
HPV-infected patients with CC and indicated a novel target for the
treatment of HPV-infected CC.
Materials and methods
Patients and tissue collection
In the present study, 128 patients with CC who were
admitted to Weifang People's Hospital (Weifang, China) for
treatment between March 2012 and May 2016 were included and the
patients' tumor tissues were collected. The inclusion criteria were
as follows: i) Patients were pathologically diagnosed with CC; and
ii) none of the patients with CC received chemotherapy,
radiotherapy or other adjuvant treatments. Patients were excluded
from the present study if they fulfilled the following criteria: i)
Presence of other malignant tumors; and ii) current pregnancy or
lactation. A total of 99 patients who underwent hysterectomy due to
hysteromyoma at the same hospital during the same period were
collected as controls and the participants in the control group
were confirmed to have no CC or precancerous lesions. Cervical
tissue samples were collected from controls at the time of
hysterectomy. After the surgery, all tissue samples were
immediately stored in liquid nitrogen until use. Samples of
patients with CC and controls were subjected to HPV-DNA testing to
determine the presence of HPV infection. There were 90 HPV+
patients and 38 HPV- patients among the 128 patients with CC; the
98 controls included 58 HPV+ samples and 41 HPV- samples. All
patients attended the 5-year survival follow-up and their survival
data were recorded. Each participant was signed an informed consent
form.
Cell culture
The HPV16+ CC cell line SiHa [cat. no. HTB-35;
American Type Culture Collection (ATCC)], the HPV18+ CC cell line
HeLa (cat. no. CRM-CCL-2; ATCC), the HPV- CC cell line C33A (cat.
no. HTB-31; ATCC) and the HPV- normal immortalized epithelial cell
line HaCaT (cat. no. 300493/p800_HaCaT; Cell Line Services GmnH)
were used in the present study. The cells were then cultured in
DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin. The conditions of the cell culture were
5% CO2 and 37°C.
Bioinformatics analysis
Gene Expression Profiling Interaction Analysis 2.0
(GEPIA 2.0; http://gepia2.cancer-pku.cn/#index) was used to
evaluate the expression of DLEU1 in CC tissues and analyze the
association of DLEU1 with overall survival of patients with CC
based on the TCGA database. The molecules that interacted with
DLEU1 were predicted using RNA Interactome Database (RNAInter;
http://www.rna-society.org/raid/home.html).
RNA extraction
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA. Each sample was
checked at least 3 times. The tissue was homogenized with a
homogenizer after the addition of the lysate, followed by the
addition of TRIzol reagent. To extract RNA from cells, the medium
was discarded, cells were washed extensively with PBS to remove
residual medium and then TRIzol was added directly into the cell
culture dish to lyse the cells. After complete lysis, two-phase
separation, RNA precipitation, RNA clean-up, RNA drying and
dissolving RNA precipitation were performed. The obtained RNA
solution was stored at −80°C for further use. The purity and
concentration of RNA were verified by a NanoDrop 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.) and RNA was used
for further analysis when the optical density ratio of 260/280 nm
was close to 2.0.
Reverse transcription-quantitative PCR
(RT-qPCR)
A total of 1 µg RNA was then reverse-transcribed
into cDNA using a Reverse Transcription Kit (Takara Biotechnology,
Co., Ltd.). SYBR-Green Real-time PCR Master Mix kit (Toyobo) in the
7900HT fast real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) were used for qPCR, which was performed to detect
the expression of DLEU1. The thermal cycling conditions were as
follows: 95°C for 5 min, and then 40 cycles of 95°C for 20 sec,
50°C for 30 sec and 72°C for 30 sec. The primers were all
synthesized by GenePharma and the primer sequences were as follows:
DLEU1 forward, 5′-CGTGCATTTAAAACCGCC-3′ and reverse,
5′-TGTCTGCATTGTGACTCAATTC-3′; GAPDH forward,
5′-ATGATGACATCAAGAAGGTGGTG-3′ and reverse,
5′-CCATGAGGTCCACCACCCTGTTG-3′. DLEU1 expression was normalized to
GAPDH and was calculated using the 2−ΔΔCq method
(21).
Statistical analysis
All analyses were performed by SPSS 22.0 (IBM
Corporation) and GraphPad Prism 7.0 software (GraphPad Software,
Inc.). Values are expressed as the mean ± standard deviation.
Differences in measurement data between two groups and among
multiple groups were compared by Student's t-test and one-way
analysis of variance followed by Tukey's test, respectively.
Comparison between categorical variables was performed by the
χ2 test. Receiver operating characteristic (ROC)
analysis was used to evaluate the ability of DLEU1 to screen
patients with CC from controls, screen HPV+ controls from HPV-
controls and screen HPV+ patients with CC from HPV- patients with
CC. Kaplan-Meier survival analysis and the log-rank test were used
to investigate the relationship between DLEU1 and the overall
survival of patients with CC. Multivariate Cox regression analysis
was used to evaluate the prognostic value of DLEU1 in patients with
CC. All analyses were independently repeated at least 3 times.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Baseline characteristics of the
participants
Table I presents
the baseline characteristics of all participants included. There
were no significant differences in age, body mass index (BMI) and
HPV infection status between controls and patients with CC (all
P>0.05). In addition, the tumor size of patients with CC was
3.6±0.9 cm. Furthermore, patients with CC were divided into 106
cases with SCC and 22 cases with adenocarcinoma, 96 cases with
negative and 32 cases with positive lymph node metastasis or 90
cases with Federation of Gynecology and Obstetrics (FIGO) stage
I–II and 38 cases with FIGO stage III–IV.
 | Table I.Baseline characteristics of the
participants. |
Table I.
Baseline characteristics of the
participants.
| Characteristic | Normal controls
(n=99) | CC patients
(n=128) | P-value |
|---|
| Age, years | 46.4±3.0 | 47.1±2.8 | 0.104 |
| BMI,
kg/m2 | 23.6±4.4 | 23.7±3.9 | 0.934 |
| HPV infection |
|
| 0.066 |
|
Negative | 41 | 38 |
|
|
Positive | 58 | 90 |
|
| Tumor size
(cm) | - | 3.6±0.9 | - |
| Histological
type |
|
| - |
|
SCC | - | 106 |
|
|
Adenocarcinoma | - | 22 |
|
| Lymph node
metastasis |
|
| - |
|
Negative | - | 96 |
|
|
Positive | - | 32 |
|
| FIGO stage |
|
| - |
|
I–II | - | 90 |
|
|
III–IV | - | 38 |
|
Expression of DLEU1 in patients with
CC and cell lines
DLEU1 was expressed at a significantly higher level
in the tumor tissues of patients with CC compared with that in
normal controls (P<0.001; Fig.
1A). The results of the TCGA data analysis were consistent with
the results of the present study, indicating that DLEU1 was
significantly upregulated in CC tissue samples (P<0.05; Fig. 1B). As indicated in Fig. 1C, HPV+ controls had a
significantly higher level of DLEU1 than HPV- controls (P<0.05);
furthermore, patients with CC with HPV- status had higher levels of
DLEU1 than both HPV+ and HPV- controls, but the difference from
HPV+ controls was somewhat smaller (all P<0.05); in addition,
DLEU1 levels were the highest in patients with CC with HPV+ and the
difference of DLEU1 expression between HPV+ patients with CC and
HPV- patients with CC also reached statistical significance (all
P<0.01). Analysis of DLEU1 levels in cells similarly indicated
significantly increased DLEU1 in both HPV- CC cells and HPV+ CC
cells compared with that in normal cells; furthermore, DLEU1
expression was higher in HPV+ CC cells than that in HPV- CC cells
(all P<0.01; Fig. 1D). The raw
data of DLEU1 expression are provided in Table SI.
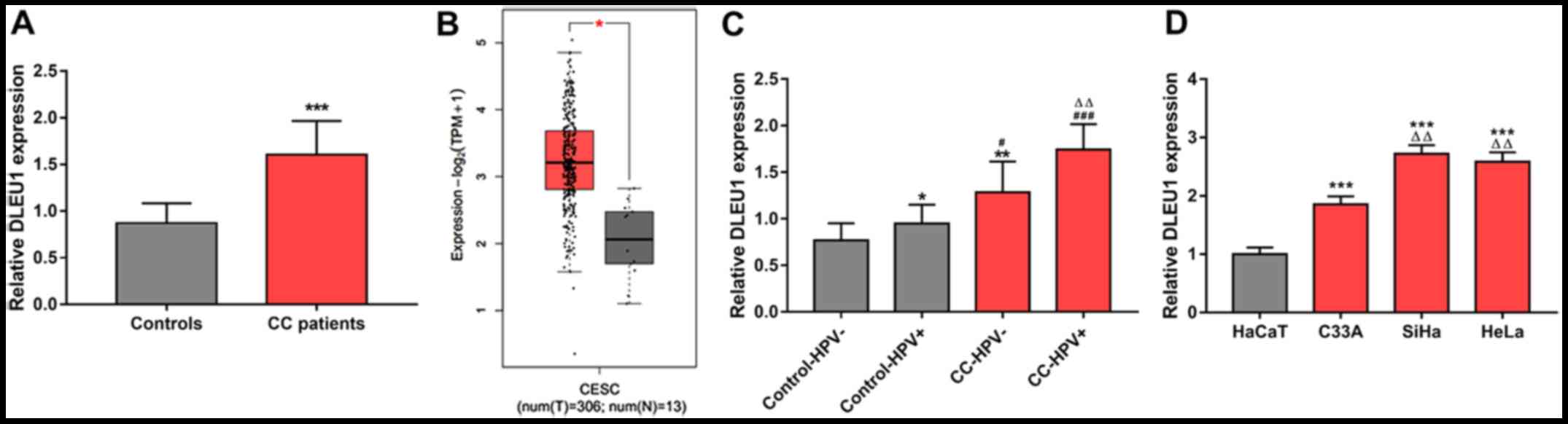 | Figure 1.Expression of DLEU1 in patients with
CC and cell lines. (A) Expression of DLEU1 in patients with CC and
controls. (B) Expression of DLEU1 was increased in CC tissues
according to the analysis results of TCGA data. (C) Expression of
DLEU1 in HPV+ and HPV- controls and in patients with CC with HPV+
and HPV- status. (D) Expression of DLEU1 in a normal cell line
(HaCaT), two HPV+ CC cell lines (SiHa and HeLa) and an HPV- CC cell
line (C33A). *P<0.05, **P<0.01, ***P<0.001 vs. Controls or
CC tissues from TCGA data or Control-HPV- or HaCaT;
#P<0.05, ###P<0.001 vs. Control-HPV+;
△△P<0.01 vs. CC-HPV- or C33A. CC, cervical cancer;
CESC, cervical squamous cell carcinoma; TCGA, the Cancer Genome
Atlas; HPV, human papillomavirus; DLEU1, long noncoding RNA deleted
in lymphocytic leukemia 1; T, tumor; N, normal; num, number. |
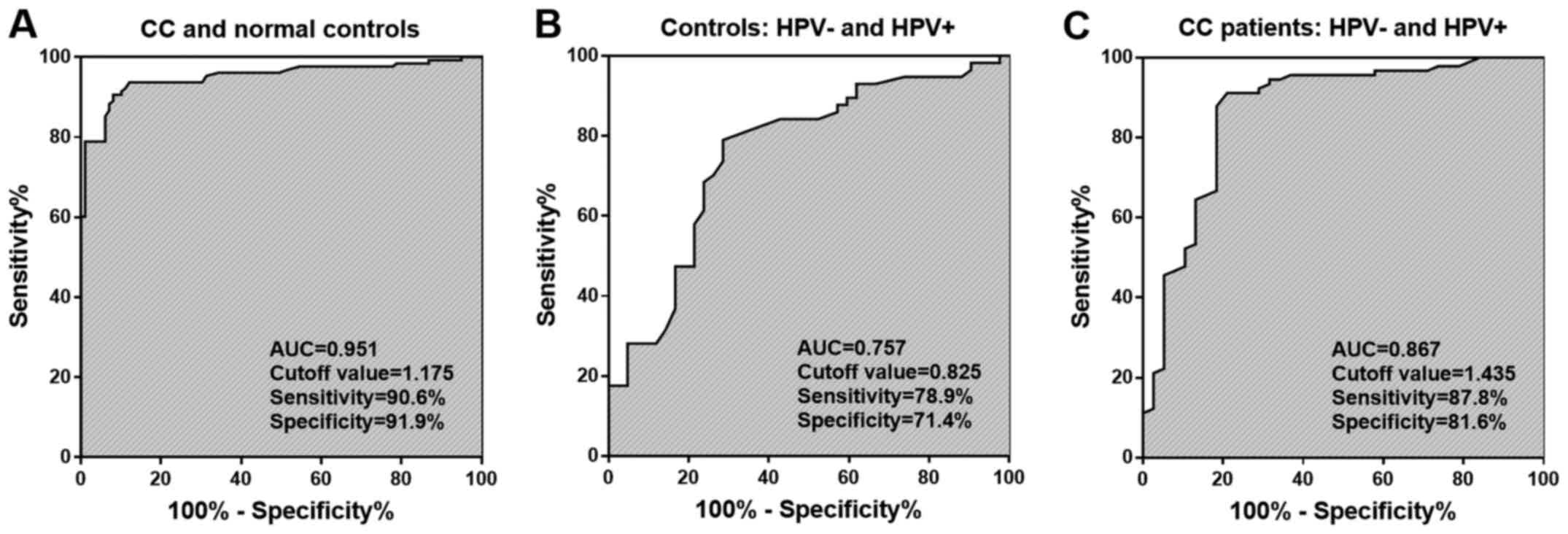
Differentially expressed DLEU1 used to distinguish
patients with CC with different HPV infection conditions. Through
ROC analysis, the diagnostic value of DLEU1 was explored. The
results of Fig. 2A suggested that
DLEU1 was able to screen patients with CC from controls with an
area under the ROC curve (AUC) of 0.951. In the control group,
DLEU1 had a certain utility in differentiating between HPV+ and
HPV- populations (Fig. 2B). The
results of Fig. 2C suggested that
DLEU1 had an ability to discriminate between HPV+ and HPV- patients
with CC with an AUC of 0.867.
Association between DLEU1 expression
and clinicopathological characteristics of patients with CC
As presented in Table
II, DLEU1 was indicated to be associated with tumor size, HPV
infection, lymph node metastasis and FIGO stage in patients with CC
(all P<0.05). However, no association was obtained between DLEU1
expression and age, BMI or histological type (all P>0.05).
 | Table II.Association between DLEU1 expression
and clinicopathological characteristics of patients with cervical
cancer. |
Table II.
Association between DLEU1 expression
and clinicopathological characteristics of patients with cervical
cancer.
| Characteristic | Total (n=128) | Low DLEU1
(n=62) | High DLEU1
(n=66) | P-value |
|---|
| Age, years |
|
|
| 0.973 |
|
<47 | 58 | 28 | 30 |
|
|
≥47 | 70 | 34 | 36 |
|
| BMI,
kg/m2 |
|
|
| 0.715 |
|
<24 | 66 | 33 | 33 |
|
|
≥24 | 62 | 29 | 33 |
|
| HPV infection |
|
|
| 0.003 |
|
Negative | 38 | 26 | 12 |
|
|
Positive | 90 | 36 | 54 |
|
| Tumor size, cm |
|
|
| 0.034 |
|
<4 | 81 | 44 | 37 |
|
| ≥4 | 47 | 18 | 29 |
|
| Histological
type |
|
|
| 0.437 |
|
SCC | 106 | 53 | 53 |
|
|
Adenocarcinoma | 22 | 9 | 13 |
|
| Lymph node
metastasis |
|
|
| 0.008 |
|
Negative | 96 | 53 | 43 |
|
|
Positive | 32 | 9 | 23 |
|
| FIGO stage |
|
|
| 0.001 |
|
I–II | 90 | 52 | 38 |
|
|
III–IV | 38 | 10 | 28 |
|
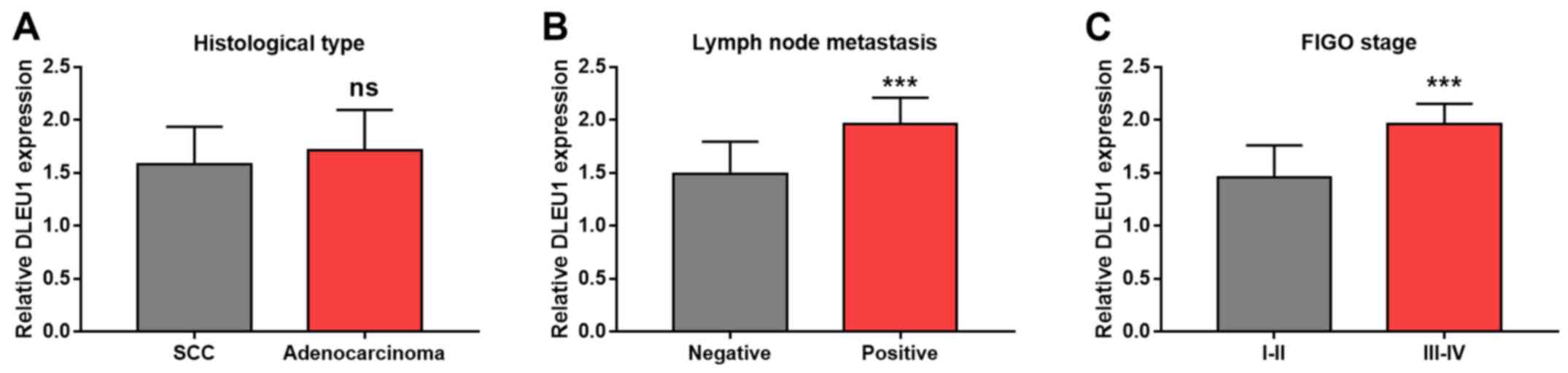
As indicated in Fig.
3A, there was no significant difference in DLEU1 expression
between patients with CC with SCC and adenocarcinoma (P>0.05).
Furthermore, the expression of DLEU1 was significantly increased in
patients with CC with positive vs. negative lymph node metastasis
(P<0.001; Fig. 3B) and
patients with CC with FIGO stage III–IV vs. I–II (P<0.001;
Fig. 3C).
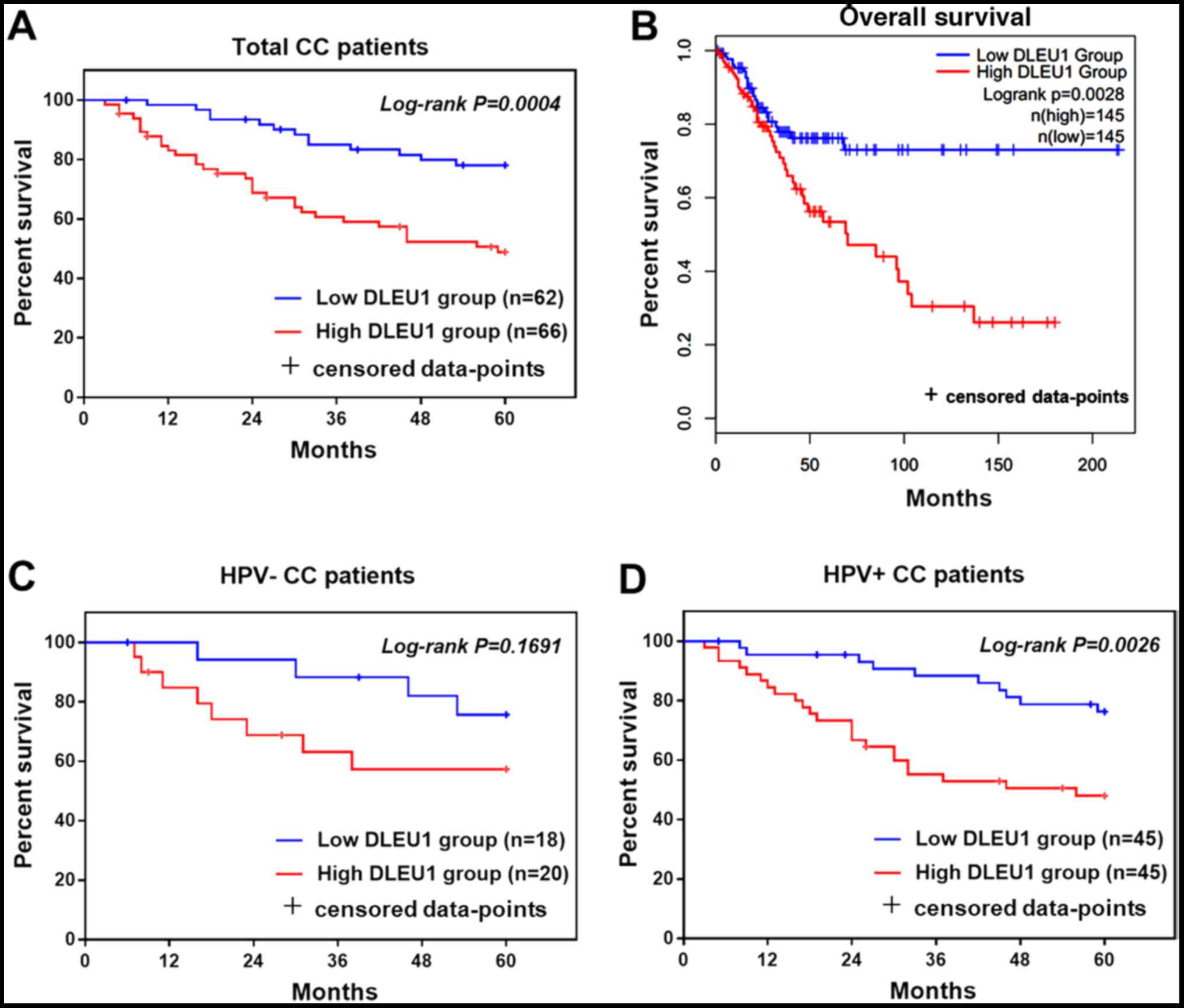
High DLEU1 predicts poor prognosis of
patients with CC
Analysis of the 5-year follow-up data of the present
study indicated that patients with CC with high levels of DLEU1 had
poor survival prognosis (log-rank P=0.0004; Fig. 4A). Similarly, analysis of the TCGA
database indicated that patients with CC with high levels of DLEU1
had poor prognosis (log-rank P=0.0028; Fig. 4B). After dividing the patients
into HPV+ and HPV- groups, the relationship between DLEU1 and
survival was analyzed separately. The results indicated that after
grouping, the relationship between DLEU1 and survival of HPV-
patients was not obvious (log-rank P=0.1691; Fig. 4C), but DLEU1 was still associated
with survival of HPV+ patients (log-rank P=0.0026; Fig. 4D). As presented in Table III, lymph node metastasis
[hazard ratio (HR)=1.692, 95% confidence interval (CI)=1.115-2.388,
P=0.042], FIGO stage (HR=2.124, 95% CI=1.560-2.974, P=0.006) and
DLEU1 (HR=2.437, 95% CI=1.612-3.329, P=0.004) were independently
associated with the survival prognosis of patients with CC.
 | Table III.Multivariate Cox regression analysis
for the survival of patients with cervical cancer. |
Table III.
Multivariate Cox regression analysis
for the survival of patients with cervical cancer.
| Variable | HR | 95% CI | P-value |
|---|
| Age (≥47 vs.
<47) | 1.231 | 0.812-1.698 | 0.329 |
| BMI (≥24 vs.
<24) | 1.189 | 0.703-1.777 | 0.632 |
| HPV infection
(positive vs. negative) | 1.121 | 0.728-1.803 | 0.417 |
| Tumor size (≥4 vs.
<4) | 1.437 | 0.841-2.046 | 0.223 |
| Histological type
(adenocarcinoma vs. SCC) | 1.237 | 0.938-1.603 | 0.129 |
| Lymph node
metastasis (positive vs. negative) | 1.692 | 1.115-2.388 | 0.042 |
| FIGO stage (III–IV
vs. I–II) | 2.124 | 1.560-2.974 | 0.006 |
| DLEU1 (high vs.
low) | 2.437 | 1.612-3.329 | 0.004 |
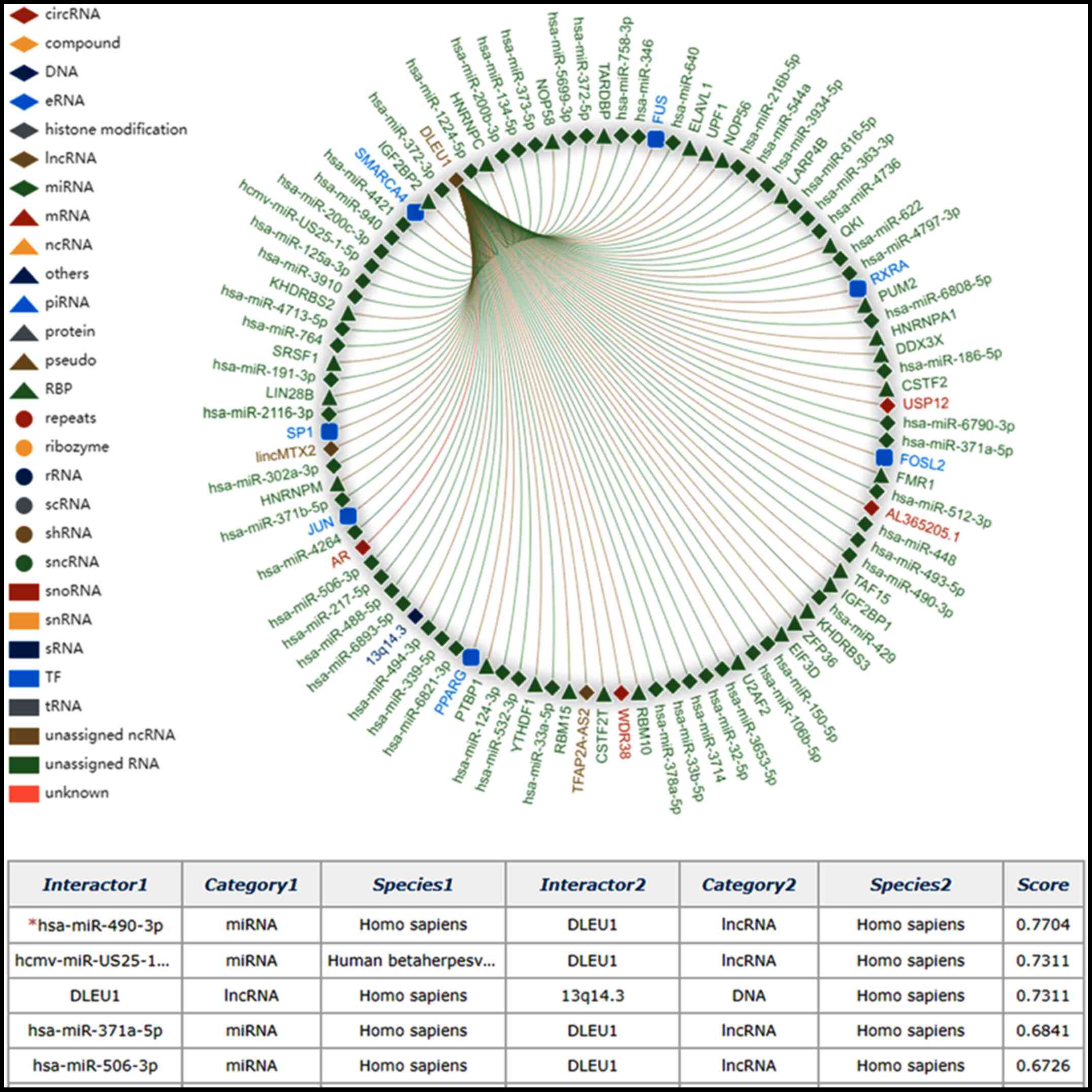
Predicted molecules interacting with
DLEU1
Molecules interacting with DLEU1 were predicted
using the RNAInter database, most of which were miRNAs. The top 5
predicted interacting molecules were miR-490-3p, human
cytomegalovirus (hcmv)-miR-US25-1-5p, 13q14.3, miR-371a-5p and
miR-506-3p (Fig. 5).
Discussion
DLEU1 has been indicated to have a promoting role in
CC progression (20).
Consistently, the present study demonstrated that the expression of
DLEU1 was increased in CC tissues from the present cohort and TCGA
database. In addition, in patients with CC, DLEU1 expression was
related to tumor size, HPV infection, lymph node metastasis and
FIGO stage. Furthermore, DLEU1 has been reported to be involved in
the progression of other cancer types, such as glioma (22), endometrial cancer (23) and oral squamous cell carcinoma
(OSCC) (24). Thus, DLEU1 is
involved in the progression of patients with CC.
It is known that infection by HPV is closely related
to the progression of patients with CC. Of note, abnormal lncRNAs
have been reported to be associated with HPV infection in cancers,
including CC. For instance, lncRNA psoriasis-susceptibility-related
RNA gene induced by stress (PRINS) has been indicated to be
markedly higher in HPV+ patients with head and neck squamous cell
carcinoma (HNSCC) than that in HPV- patients with HNSCC, and the
expression of PRINS was significantly related to HPV-infected HNSCC
(25). Zhou et al
(26) reported that lncRNA
oncogene-induced senescence 1 (lncRNA-OIS1) is decreased in HPV+
patients with cervical squamous cell carcinoma (CSCC) and HPV+ CSCC
cells and may enhance the proliferation of HPV+ CSCC cells. A study
by Zhang et al (27)
determined upregulated expression of lncRNA human ovarian
cancer-specific transcript 2 (lncRNA HOST2) in HPV+ CC tissues and
cells and the promoting effect of HOST2 on HPV+ CC cell function.
The levels of long intergenic non-protein coding RNA (LINC)01101
and LINC00277 have been indicated to be decreased in HR-HPV+
samples (28). In the present
study, DLEU1 expression was higher in HPV+ controls than that in
HPV- controls and it was higher in HPV+ patients with CC than that
in HPV- patients with CC. Furthermore, DLEU1 expression in HPV+ CC
cells was significantly higher when compared with that in HPV- CC
cells. In addition, by using the χ2 test, DLEU1 was
indicated to be associated with HPV infection in patients with CC.
Thus, DLEU1 is associated with HPV infection and may be involved in
the progression of HPV-infected CC.
Accumulating evidence has indicated that lncRNAs may
serve as diagnostic and prognostic biomarkers in different types of
cancer (29–31). In addition, certain lncRNAs, such
as lncRNA SOX21 antisense divergent transcript 1 (32) and lncRNA LIPE antisense RNA 1
(LIPE-AS1) (33), have been
indicated to be of use as diagnostic and prognostic biomarkers for
patients with CC. The present study also suggested that DLEU1 had
the ability to screen patients with CC from normal controls. In
addition, DLEU1 was closely related to survival of patients with CC
according to analyses of the present study cohort and TCGA
database, and was able to independently predict the prognosis of
patients with CC. Thus, DLEU1 may be used as a biomarker for the
diagnosis of CC and prognostic survival prediction for affected
patients.
It has been indicated that lncRNA loc285194
(34), lncRNA heart and neural
crest derivatives expressed 2-antisense RNA 1 (35) and lncRNA-OIS1 (26) were able to screen HPV+ patients
with CSCC from normal controls. In addition, lncRNA steroid
receptor RNA activator 1 was indicated to have high diagnostic
value in screening HPV+ patients with CSCC from HPV- CSCC patients
(36). Furthermore, PRINS
expression was reported to correlate with overall survival in HPV+
HNSCC patients (25). In the
present study, DLEU1 had a high diagnostic value in screening HPV+
patients with CC and had a certain ability to screen HPV+ controls.
In addition, there was a significant correlation between DLEU1 and
survival of HPV+ patients with CC. However, there was no
significance regarding the Kaplan-Meier curve results in HPV-
patients with CC. This nonsignificant result may be due to the
small number of HPV- patients with CC in the present study. Of
note, DLEU1 may be used to diagnose and predict the prognosis of
other cancer types, such as OSCC (24), glioma (37) and breast cancer (38). The above results indicated that
DLEU1 may be a biomarker for the diagnosis and prognosis of HPV+
patients with CC.
Finally, the molecules interacting with DLEU1, which
may be involved in DLEU1 exerting its biological function, were
predicted. Most of the molecules predicted were miRNAs,
illustrating that DLEU1 as an lncRNA may bind miRNAs to regulate
key genes to exert its function. Certain studies have reported the
role of the lncRNA/miRNA axis in HPV+ CC. For instance, Zhang et
al (39) revealed that
LINC00511 may aggravate HPV+ and HPV- CC by targeting the
miR-324-5p/DNA damage regulated autophagy modulator 1 axis. The
lncRNA metastasis-associated long adenocarcinoma transcript
1/miR-124/root rake brush grapple 2 axis was reported to be
involved in the function of HR-HPV+ CC cells (40). The present study found that
hcmv-miR-US25-1-5p is a molecule interacting with DLEU1. In
addition, it is worth noting that hcmv-miR-US25-1-5p encoded miRNA
and is associated with cytomegalovirus (41). More importantly, a study suggested
that human herpesviruses-6 together with cytomegalovirus may
promote the development of CC (42). Thus, it was hypothesized that
DLEU1 may have a role in HPV-infected CC by regulating
hcmv-miR-US25-1-5p, which will be the subject for future research.
The prediction of molecules interacting with DLEU1 may serve as a
basis for subsequent studies and offers suggestions for future
investigations.
The present study was the first, to the best of our
knowledge, to explore the association between DLEU1 and HPV
infection status and to explore its clinical significance in
HPV-infected CC. However, the study sample was small, which is a
limitation of the present study, and a large cohort is required in
further studies. In addition, as a non-coding RNA, DLUE1 has no
ability to code proteins, and thus, it was not possible to measure
any protein distribution and levels of DLEU1 in tissue samples.
However, the exploration of the downstream miRNAs and target genes
may be able to compensate for this deficiency, thus illustrating
the expression patterns and function of DLEU1 in CC.
In conclusion, the present study indicated that
DLEU1 expression was increased in HPV-infected patients with CC and
cells, and was associated with HPV infection in patients with CC.
In addition, DLEU1 may be used as a biomarker to distinguish HPV+
patients with CC from HPV- patients with CC and predict the
prognosis of HPV-infected patients with CC. The present study may
provide novel diagnostic and prognostic biomarkers for the
treatment of HPV-infected patients with CC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a Scientific Research Project
of Weifang Health Commission (grant no. WFWSJK-2021-066).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
XM and BX were responsible for the design and
conception of the study. ZW and AD analyzed and interpreted the
data. AD and BX performed the cell experiments and the other
analyses. ZW and XM contributed essential reagents or tools. All
authors wrote and revised the manuscript. All authors read and
approved the final manuscript. AD and XM confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The experimental procedures were all in accordance
with the guidelines of the Ethics Committee of Weifang People's
Hospital (Weifang, China) and were approved by the Ethics Committee
of Weifang People's Hospital (Weifang, China; approval no.
0012064). The present study complies with the Declaration of
Helsinki. Written informed consent was obtained from each
participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pourhanifeh MH, Darvish M, Tabatabaeian J,
Fard MR, Mottaghi R, Azadchehr MJ, Jahanshahi M, Sahebkar A and
Mirzaei H: Therapeutic role of curcumin and its novel formulations
in gynecological cancers. J Ovarian Res. 13:1302020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nahand JS, Vandchali NR, Darabi H,
Doroudian M, Banafshe HR, Moghoofei M, Babaei F, Salmaninejad A and
Mirzaei H: Exosomal microRNAs: Novel players in cervical cancer.
Epigenomics. 12:1651–1660. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sadri Nahand J, Moghoofei M, Salmaninejad
A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh
MH, Bokharaei-Salim F, Mirzaei H, et al: Pathogenic role of
exosomes and microRNAs in HPV-mediated inflammation and cervical
cancer: A review. Int J Cancer. 146:305–320. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu F, Liu J, Dong W, Xie J and Zhao X: The
diagnostic value of miR-145 and miR-205 in patients with cervical
cancer. Am J Transl Res. 13:1825–1832. 2021.PubMed/NCBI
|
|
5
|
Yang PM, Chou CJ, Tseng SH and Hung CF:
Bioinformatics and in vitro experimental analyses identify the
selective therapeutic potential of interferon gamma and apigenin
against cervical squamous cell carcinoma and adenocarcinoma.
Oncotarget. 8:46145–46162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park S, Eom K, Kim J, Bang H, Wang HY, Ahn
S, Kim G, Jang H, Kim S, Lee D, et al: MiR-9, miR-21, and miR-155
as potential biomarkers for HPV positive and negative cervical
cancer. BMC Cancer. 17:6582017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Causin RL, da Silva LS, Evangelista AF,
Leal LF, Souza KC, Pessôa-Pereira D, Matsushita GM, Reis RM,
Fregnani JH and Marques MM: MicroRNA biomarkers of high-grade
cervical intraepithelial neoplasia in liquid biopsy. BioMed Res
Int. 2021:66509662021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boscolo-Rizzo P, Furlan C, Lupato V,
Polesel J and Fratta E: Novel insights into epigenetic drivers of
oropharyngeal squamous cell carcinoma: Role of HPV and lifestyle
factors. Clin Epigenetics. 9:1242017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zamani S, Sohrabi A, Hosseini SM,
Rahnamaye-Farzami M and Akbari A: Deregulation of miR-21 and
miR-29a in Cervical Cancer Related to HPV Infection. MicroRNA.
8:110–115. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Chu ZP, Han H, Zhang Y, Tian F,
Zhang JQ and Huang XH: Suppression of miR-93-5p inhibits high-risk
HPV-positive cervical cancer progression via targeting of BTG3. Hum
Cell. 32:160–171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nohata N, Abba MC and Gutkind JS:
Unraveling the oral cancer lncRNAome: Identification of novel
lncRNAs associated with malignant progression and HPV infection.
Oral Oncol. 59:58–66. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Razavi ZS, Asgarpour K, Mahjoubin-Tehran
M, Rasouli S, Khan H, Shahrzad MK, Hamblin MR and Mirzaei H:
Angiogenesis-related non-coding RNAs and gastrointestinal cancer.
Mol Ther Oncolytics. 21:220–241. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mahjoubin-Tehran M, Rezaei S, Jesmani A,
Birang N, Morshedi K, Khanbabaei H, Khan H, Piranviseh A, Nejati M,
Aschner M, et al: New epigenetic players in stroke pathogenesis:
From non-coding RNAs to exosomal non-coding RNAs. Biomed
Pharmacother. 140:1117532021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shafabakhsh R, Arianfar F, Vosough M,
Mirzaei HR, Mahjoubin-Tehran M, Khanbabaei H, Kowsari H, Shojaie L,
Azar ME, Hamblin MR, et al: Autophagy and gastrointestinal cancers:
The behind the scenes role of long non-coding RNAs in initiation,
progression, and treatment resistance. Cancer Gene Ther. Jan
11–2021.(Epub ahead of print). doi: 10.1038/s41417-020-00272-7.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mirzaei H and Hamblin MR: Regulation of
glycolysis by non-coding RNAs in cancer: Switching on the Warburg
effect. Mol Ther Oncolytics. 19:218–239. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hashemian SM, Pourhanifeh MH, Fadaei S,
Velayati AA, Mirzaei H and Hamblin MR: Non-coding RNAs and
exosomes: their role in the pathogenesis of sepsis. Mol Ther
Nucleic Acids. 21:51–74. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hashemipour M, Boroumand H, Mollazadeh S,
Tajiknia V, Nourollahzadeh Z, Rohani Borj M, Pourghadamyari H,
Rahimian N, Hamblin MR and Mirzaei H: Exosomal microRNAs and
exosomal long non-coding RNAs in gynecologic cancers. Gynecol
Oncol. 161:314–327. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rahimian N, Razavi ZS, Aslanbeigi F,
Mirkhabbaz AM, Piroozmand H, Shahrzad MK, Hamblin MR and Mirzaei H:
Non-coding RNAs related to angiogenesis in gynecological cancer.
Gynecol Oncol. 161:896–912. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Razavi ZS, Tajiknia V, Majidi S, Ghandali
M, Mirzaei HR, Rahimian N, Hamblin MR and Mirzaei H: Gynecologic
cancers and non-coding RNAs: Epigenetic regulators with emerging
roles. Crit Rev Oncol Hematol. 157:1031922021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Tian X, Zhang J and Jiang L: Long
Non-coding RNA DLEU1 promotes proliferation and invasion by
interacting with miR-381 and enhancing HOXA13 expression in
cervical cancer. Front Genet. 9:6292018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv QL, Wang LC, Li DC, Lin QX, Shen XL,
Liu HY, Li M, Ji YL, Qin CZ and Chen SH: Knockdown lncRNA DLEU1
inhibits gliomas progression and promotes temozolomide
chemosensitivity by regulating autophagy. Front Pharmacol.
11:5605432020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shan L, Zhao T and Wang Y: Upregulation of
Serum lncRNA DLEU1 predicts progression of premalignant endometrial
lesion and unfavorable clinical outcome of endometrial cancer.
Technol Cancer Res Treat. Dec 17–2020.(Epub ahead of print). doi:
10.1177/1533033820965589. View Article : Google Scholar
|
|
24
|
Lv T, Liu H, Wu Y and Huang W: Knockdown
of lncRNA DLEU1 inhibits the tumorigenesis of oral squamous cell
carcinoma via regulation of miR-149 5p/CDK6 axis. Mol Med Rep.
23:4472021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kopczyńska M, Kolenda T, Guglas K,
Sobocińska J, Teresiak A, Bliźniak R, Mackiewicz A, Mackiewicz J
and Lamperska K: PRINS lncRNA is a new biomarker candidate for HPV
infection and prognosis of head and neck squamous cell carcinomas.
Diagnostics (Basel). 10:E7622020. View Article : Google Scholar
|
|
26
|
Zhou D, Wu F, Cui Y, Wei F, Meng Q and Lv
Q: Long non-coding RNA-OIS1 inhibits HPV-positive, but not
HPV-negative cervical squamous cell carcinoma by upregulating
MTK-1. Oncol Lett. 17:2923–2930. 2019.PubMed/NCBI
|
|
27
|
Zhang Y, Jia LG, Wang P, Li J, Tian F, Chu
ZP and Kang S: The expression and significance of lncRNA HOST2 and
microRNA let-7b in HPV-positive cervical cancer tissues and cell
lines. Eur Rev Med Pharmacol Sci. 23:2380–2390. 2019.PubMed/NCBI
|
|
28
|
Iancu IV, Anton G, Botezatu A, Huica I,
Nastase A, Socolov DG, Stanescu AD, Dima SO, Bacalbasa N and Plesa
A: LINC01101 and LINC00277 expression levels as novel factors in
HPV-induced cervical neoplasia. J Cell Mol Med. 21:3787–3794. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fattahi S, Nikbakhsh N, Taheri H, Ghadami
E, Ranaee M and Akhavan-Niaki H: LINC02688 and PP7080 as novel
biomarkers in early diagnosis of gastric cancer. Noncoding RNA Res.
6:86–91. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Sun Y, Ding L, Shi W, Ding K and
Zhu Y: Long non-coding RNA LINC00467 correlates to poor prognosis
and aggressiveness of breast cancer. Front Oncol. 11:6433942021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chao Y and Zhou D: lncRNA-D16366 is a
potential biomarker for diagnosis and prognosis of hepatocellular
carcinoma. Med Sci Monit. 25:6581–6586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du P, Zhi Y, Wang R, Li Y, Li H, Zhang X,
Cheng G and Li X: Aberrant methylation of the SOX21-AS1 promoter
region promotes gene expression and its clinical value in cervical
cancer. Reprod Sci. 28:532–540. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Jiang P, Wang S, Cheng W and Fu
S: LncRNA LIPE-AS1 predicts poor survival of cervical cancer and
promotes its proliferation and migration via modulating
miR-195-5p/MAPK pathway. Front Oncol. 11:6399802021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Zhang Y, Lin R, Mao B, Wang W, Bai
Y, He W and Liu Q: Long noncoding RNA loc285194 expression in human
papillomavirus-positive and -negative cervical squamous cell
carcinoma, C33A, and SiHa cells and transforming growth factor-β1.
Med Sci Monit. 25:9012–9018. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin L, Ji J, Shi L, Jin S and Pei L:
lncRNA HAND2-AS1 inhibits cancer cell proliferation, migration and
invasion by downregulating ROCK1 in HPV-positive and negative
cervical squamous cell carcinoma. Exp Ther Med. 18:2512–2518.
2019.PubMed/NCBI
|
|
36
|
Liu Y, Li M, Yu H and Piao H: LncRNA SRA1
is down-regulated in HPV-negative cervical squamous cell carcinoma
and regulates cancer cell behaviors. Biosci Rep. Aug 15–2019.(Epub
ahead of print). doi: 10.1042/BSR20191226.
|
|
37
|
Cao Y, Yang R, Lee I, Zhang W, Sun J, Meng
X and Wang W: Prediction of LncRNA-encoded small peptides in glioma
and oligomer channel functional analysis using in silico
approaches. PLoS One. 16:e02486342021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang C, Xie XX, Li WJ and Jiang DQ: LncRNA
DLEU1/microRNA-300/RAB22A axis regulates migration and invasion of
breast cancer cells. Eur Rev Med Pharmacol Sci. 23:10410–10421.
2019.PubMed/NCBI
|
|
39
|
Zhang X, Wang Y, Zhao A, Kong F, Jiang L
and Wang J: Long Non-coding RNA LINC00511 accelerates proliferation
and invasion in cervical cancer through targeting miR-324-5p/DRAM1
axis. OncoTargets Ther. 13:10245–10256. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu S, Song L, Zeng S and Zhang L:
MALAT1-miR-124-RBG2 axis is involved in growth and invasion of
HR-HPV-positive cervical cancer cells. Tumour Biol. 37:633–640.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang S, Qi Y, He R, Huang Y, Liu Z, Ma Y,
Guo X, Shao Y, Sun Z and Ruan Q: Human cytomegalovirus microRNA
miR-US25-1-5p inhibits viral replication by targeting multiple
cellular genes during infection. Gene. 570:108–114. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Broccolo F, Cassina G, Chiari S,
Garcia-Parra R, Villa A, Leone BE, Brenna A, Locatelli G, Mangioni
C and Cocuzza CE: Frequency and clinical significance of human
beta-herpesviruses in cervical samples from Italian women. J Med
Virol. 80:147–153. 2008. View Article : Google Scholar : PubMed/NCBI
|