Introduction
Neurodegenerative diseases are generally progressive
and refractory because they involve irreversible degeneration of
brain neurons and many remain incurable. Treatments to slow
progression after disease onset do not compensate for neural
tissues that have already been lost and do not result in a
retrograde recovery of symptoms. Therefore, it is important to
implement therapeutic interventions and risk management as early as
possible in order to delay disease onset and progression and to
extend healthy life expectancy (1). However, this is difficult because
diagnosis is based on the detection of clinical symptoms and
neuroimaging abnormalities, which appear relatively late in the
disease course. Nonetheless, molecular responses to genetic and
environmental disorders begin much earlier, and non-coding RNA
networks have been found to be involved in these cellular
regulatory mechanisms. MicroRNAs (miRNAs) are small non-coding RNAs
of approximately 20–25 nucleotides, and more than 2,000 different
miRNAs have been suggested to exist in humans (2). MiRNAs repress the expression of
various genes in vivo and exhibit unique disease-specific
expression patterns. In addition, miRNAs are stable in body fluids
such as blood, urine, and saliva, which are easy to collect from
living organisms, and can be detected with high sensitivity
(3). Thus, profiling the miRNA
expression patterns may facilitate the detection of diseases at a
preliminary stage. Indirect evaluation of pathological processes is
particularly important in neurological diseases, where direct
access to tissues for molecular analysis is not possible. Living
neurons and other cells of the central nervous system (CNS) secrete
miRNAs wrapped in exosomes, microvesicles, and lipoprotein
complexes (4). Interactions
between external stimuli and pathological processes in the brain
have been reported to be reflected in peripheral tissues, and their
use as potential diagnostic markers is of interest. Cerebrospinal
fluid (CSF) has also been shown to contain circulating miRNAs. As
the CSF is in direct contact with the brain, it is said to reflect
changes in the brain and may be suitable as a biomarker (5). Recently, specific miRNAs in the CSF
have been reported in Alzheimer's disease (6) and Parkinson's disease (PD) (7) and are expected to be potential
diagnostic markers and therapeutic targets.
Progressive supranuclear palsy (PSP) is a
progressive neurodegenerative tauopathy characterized by postural
instability with falls, vertical supranuclear gaze palsy,
parkinsonism with poor levodopa response, pseudobulbar palsy, and
frontal release signs (8). The
mean age of onset of PSP is approximately 65 years and the
condition leads to death within approximately 6–9 years after
symptom onset (9,10). The factors involved in its
pathogenesis are still unknown, and no fundamental treatment has
been established. In addition, the diagnosis of PSP is currently
dependent on clinical and imaging findings, which makes early
diagnosis difficult.
It is difficult to differentiate PSP from PD and the
other Parkinsonian syndromes such as multiple system atrophy (MSA)
and corticobasal degeneration (CBD), especially in the early
stages. The differences in the Parkinsonian syndrome, which has
similar clinical symptoms to PSP, are presented in Table SI. To improve the accuracy of the
clinical diagnosis of Parkinsonian syndrome, CSF markers have been
explored (11). Therefore, we
focused on miRNAs as one candidate that could lead to early
diagnosis of PSP.
At present, there are very few reports of miRNA
expression profiling in PSP. It has been reported that miR-132
(12), miR-147a, and miR-518e
(13) are expressed in the brain
tissues of patients with PSP, but these miRNAs are rarely reported
in biofluids. We herein aimed to examine CSF miRNAs, which are
thought to be a sensitive indicator of changes in the brain, to
elucidate the pathophysiology of PSP and establish specific
biomarkers for diagnosis.
Materials and methods
Participants
Eleven patients with PSP (nine men and two women;
median age, 76 years; range, 68–82 years) and eight age- and
sex-matched controls (seven men and one woman; median age, 75.5
years; range, 63–89 years) were recruited for the present study
between January 2017 and December 2018 from the Department of
Neurology, Kagawa University Hospital. All participants in this
study were Japanese. We enrolled patients fulfilling the Movement
Disorder Society (MDS) clinical diagnostic criteria for PSP
(MDS-PSP criteria) (14) and for
probable PSP, Richardson's syndrome (PSP-RS) phenotype according to
their disease history and clinical information including
neuroimaging findings. All patients had sporadic PSP. Patient
background data (age, sex, disease duration, comorbidities, and
clinical parameters) at the time of sample collection were
retrospectively obtained from medical records. Clinical parameters
included scores on the PSP Rating Scale (PSPRS) (15), Mini-Mental State Examination
(MMSE) (16), Frontal Assessment
Battery (FAB) (17), and Montreal
Cognitive Assessment (MoCA) (18). We assessed brain magnetic
resonance imaging scans for findings suggestive of PSP.
Furthermore, we applied the magnetic resonance parkinsonism index
(MRPI) (19) and MRPI 2.0
(20). The threshold for the MRPI
and MRPI 2.0 was >13.88 and >2.50, respectively (20). Patients with PSP were divided into
two subgroups: The ‘early disease stage’ subgroup included those
who underwent neurological evaluation within 2 years of symptom
onset and the ‘advanced disease stage’ subgroup included patients
who underwent evaluation >2 years after symptom onset. Patients
with concurrent malignant tumors, psychiatric disorders, collagen
diseases, endocrine diseases, or infections were excluded because
these conditions may alter the expression profile of some miRNAs.
Owing to the above-mentioned reason, participants in the control
group with concurrent malignant tumors, psychiatric disorders,
collagen diseases, endocrine diseases, or infections were also
excluded. In addition, participants with neurological diseases such
as dementia and parkinsonism were excluded. The study was approved
by the Medical Ethics Committee of Kagawa University (Kagawa,
Japan) (Ethics approval Heisei 22–002). Participants and controls
received a full explanation of the study purpose and protocol and
provided written informed consent before inclusion in the
study.
CSF miRNA expression profiling
CSF samples were obtained during lumbar puncture
between the L3 and L5 vertebrae with the participant in a lateral
supine position. Lumbar punctures were performed between 9 and 12
am, using a sterile technique and a 21–23G spinal needle in
accordance with local guidelines. At least 4 ml of CSF was
collected from each participant. CSF was collected in polypropylene
tubes and cytologically controlled to be free of erythrocytes. The
sample was centrifuged (2,000 × g, 4°C, 10 min), frozen
immediately, and stored at −80°C until use. The CSF was at no time
thawed/refrozen. Total RNA was extracted from 900 µl of frozen CSF
using a 3D-Gene® RNA extraction reagent (Toray)
according to the manufacturer's specifications. The BioAnalyzer
2100 system (Agilent Technologies, Inc.) was used to determine the
RNA integrity number (RIN) for RNA quality assessment. No
difference in total RNA concentration or RIN was observed between
the PSP group and controls. After labeling with a 3D-Gene miRNA
labeling kit (Toray), the extracted total RNA was hybridized onto
the 3D-Gene® Human miRNA oligo chip Version 22.0 (Toray)
with 2,632 fluorescent probes. Hybridization signals were obtained
using a 3D-Gene 3000 miRNA microarray scanner (Toray). The raw data
of each detected signal were normalized by the global normalization
method, subtracting the mean intensity of the background signal in
each microarray, and the median of the detected signal intensity
was adjusted to 25. Microarray data have been deposited in NCBI's
Gene Expression Omnibus (21) and
are accessible through GEO Series accession number GSE186921
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186921).
Heatmap
Hierarchical clustering was performed on the PSP and
control groups, using a subset of miRNAs determined to be
significantly differentially expressed between the two groups. The
Ward agglomerative method was used to cluster both the samples and
miRNAs. The heatmap was color-coded according to the expression
levels. The center level of the color code was set as the median
value over all of the values used in the heatmap. Gray color
represented mean values, red indicated an increase, and blue
represented a decrease in expression. The heatmap was created using
JMP® Pro version 15.1.0 (SAS Institute Inc.).
miRNA target gene bioinformatics
analysis
Using in silico analysis, the target genes of
miRNAs were predicted using the miRDB (http://mirdb.org/miRDB/), Targetscan 7.2 (http://www.targetscan.org/), and mirTarBase version
8.0 (http://mirtarbase.mbc.nctu.edu.tw/) databases. To
enhance the reliability of the bioinformatics analysis, overlapping
target genes were identified in this analysis.
Statistical analysis
For comparisons of clinical characteristics between
the groups, the Mann-Whitney U test was used to compare all
variables except for sex and age. The Fisher's exact test and
Kruskal-Wallis test were used to compare sex and age, respectively.
The median (interquartile range) values for each parameter are
presented. Statistical analysis of miRNA expression levels in each
group was conducted using a non-parametric Mann-Whitney U test and
the Kruskal-Wallis test as appropriate. Benjamini-Hochberg
procedures for adjusting the false discovery rate (FDR) in multiple
comparisons were also applied. Multiple comparisons of continuous
variables among multiple groups were performed using the
Kruskal-Wallis test, followed by the Steel-Dwass post-hoc test.
P-values of <0.05 were regarded as statistically significant.
Statistical analyses were performed using JMP® Pro
version 15.1.0 (SAS Institute Inc.).
Results
Clinical characteristics of
participants
The clinical characteristics are presented in
Table I. There were no
significant differences in the sex ratio and age between the PSP
and control groups. The PSPRS scores and disease severity were
significantly higher in the advanced stage group than in the early
stage group. The scores on the cognitive assessment scale, MMSE,
MoCA, and FAB were significantly lower in the advanced stage group
than in the early stage group. There was also a significant
difference in the imaging scale scores between the early and
advanced stage groups.
 | Table I.General and clinical characteristics
of the participants. |
Table I.
General and clinical characteristics
of the participants.
| Variable | Control (n=8) | PSP early stage
(n=5) | PSP advanced stage
(n=6) | P-value |
|---|
| Sex, n |
|
|
|
|
|
Male | 7 | 3 | 6 | 0.316a |
|
Female | 1 | 2 | 0 |
|
| Median (IQR) age at
test, years | 75.5
(64.8-79.0) | 75.0
(69.0-76.0) | 76.5
(70.8-79.8) | 0.516b |
| Median (IQR)
disease duration, years | ND | 2 (2) | 5 (4–6) | 0.005c |
| Median (IQR) PSPRS
score,/100 | ND | 26.0
(24.5-31.0) | 58.0
(51.8-62.3) | 0.008c |
| Median (IQR) MMSE
score,/30 | ND | 26.0
(25.0-28.0) | 19.0
(11.5-20.3) | 0.008c |
| Median (IQR) FAB
score,/18 | ND | 12.0
(10.5-13.5) | 6.0 (5.0-9.3) | 0.042c |
| Median (IQR) MoCA
score,/30 | ND | 21.0
(19.0-23.0) | 8.5 (5.0-12.8) | 0.008c |
| Median (IQR)
MRPI | ND | 30.6
(23.0-31.2) | 42.6
(37.9-48.1) | 0.014c |
| Median (IQR) MRPI
2.0 | ND | 6.8 (5.0-7.9) | 10.9
(9.7-14.2) | 0.008c |
CSF miRNA expression profile
Of the 2,632 miRNAs detectable on the
3D-Gene® Human miRNA OligoChip (Version 22.0), 1,104
were detected in all CSF samples from both the PSP and control
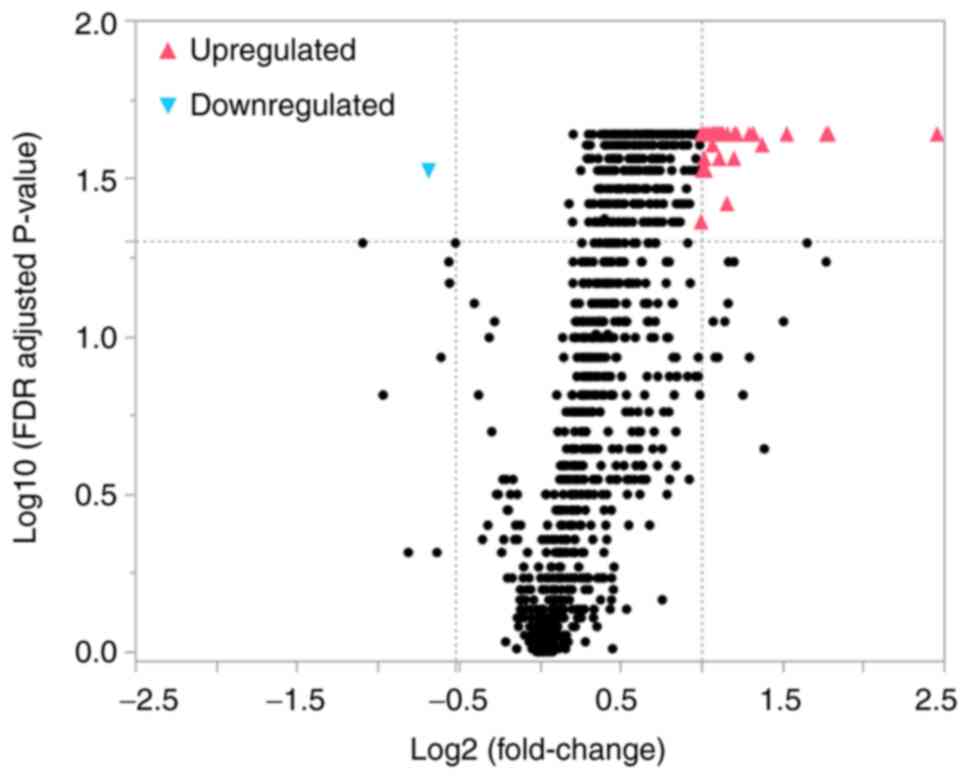
groups. The volcano plot (Fig. 1)
shows the relationship between the fold change and significance of
1,104 detectable miRNAs between the two groups. Thirty-eight miRNAs
had significantly higher (upregulated) and one miRNA had
significantly lower (downregulated) normalized values in the PSP
group than in the control group, as assessed by a fold change ratio
of >2 or <0.7 and an FDR-adjusted P-value of <0.05. These
miRNAs are indicated in Table
II. The chromosomal locations of the miRNAs and known disease
associated with the miRNAs (22–25) are also presented in Table II.
 | Table II.Statistical results, chromosomal
locations and known associated diseases of miRNAs which were
significantly altered in patients with progressive supranuclear
palsy. |
Table II.
Statistical results, chromosomal
locations and known associated diseases of miRNAs which were
significantly altered in patients with progressive supranuclear
palsy.
| MiRNA | Fold change | P-value | FDR adjusted
P-value | Chromosomal
localization | miRNA-disease
association |
|---|
| Upregulated |
|
|
|
|
|
|
hsa-miR-204-3p | 5.50 | 0.0003 | 0.0230 | 9q21.12 | Retinal dystrophy
and iris coloboma with or without cataract (22) |
|
hsa-miR-4476 | 3.46 | 0.0034 | 0.0230 | 9p13.2 |
|
|
hsa-miR-6132 | 3.42 | 0.0015 | 0.0230 | 7q31.2 |
|
|
hsa-miR-4638-5p | 2.89 | 0.0003 | 0.0230 | 5q35.3 |
|
|
hsa-miR-7110-5p | 2.60 | 0.0044 | 0.0249 | 3q21.1 |
|
|
hsa-miR-3679-5p | 2.50 | 0.0015 | 0.0230 | 2q21.2 |
|
|
hsa-miR-1236-5p | 2.46 | 0.0011 | 0.0230 | 6p21.33 |
|
|
hsa-miR-6867-3p | 2.33 | 0.0026 | 0.0230 | 17q21.1 |
|
|
hsa-miR-6761-3p | 2.31 | 0.0011 | 0.0230 | 12q24.12 |
|
|
hsa-miR-423-5p | 2.30 | 0.0057 | 0.0275 | 17q11.2 |
|
|
hsa-miR-7111-3p | 2.25 | 0.0020 | 0.0230 | 6p21.31 |
|
|
hsa-miR-3156-3p | 2.24 | 0.0015 | 0.0230 | 21q11.2 |
|
|
hsa-miR-12114 | 2.24 | 0.0118 | 0.0382 | 22q13.33 |
|
|
hsa-miR-6889-5p | 2.20 | 0.0026 | 0.0230 | 22q13.2 |
|
|
hsa-miR-6740-3p | 2.20 | 0.0011 | 0.0230 | 1q32.1 |
|
|
hsa-miR-885-5p | 2.18 | 0.0020 | 0.0230 | 3p25.3 |
|
|
hsa-miR-6894-3p | 2.18 | 0.0020 | 0.0230 | Xp11.22 |
|
|
hsa-miR-487b-5p | 2.16 | 0.0015 | 0.0230 | 14q32.31 | Neuroblastomas
(23) |
|
hsa-miR-6820-5p | 2.16 | 0.0057 | 0.0275 | 22q13.1 |
|
|
hsa-miR-873-3p | 2.15 | 0.0008 | 0.0230 | 9p21.1 | Multiple sclerosis
(24) |
|
hsa-miR-7109-3p | 2.14 | 0.0026 | 0.0230 | 22q12.2 |
|
|
hsa-miR-5193 | 2.12 | 0.0015 | 0.0230 | 3p21.31 |
|
|
hsa-miR-4648 | 2.10 | 0.0034 | 0.0230 | 7p22.3 |
|
|
hsa-miR-10398-5p | 2.10 | 0.0044 | 0.0249 | 6p21.1 |
|
|
hsa-miR-1825 | 2.10 | 0.0015 | 0.0230 | 20q11.21 |
|
|
hsa-miR-6870-5p | 2.10 | 0.0026 | 0.0230 | 20p12.2 |
|
|
hsa-miR-6825-5p | 2.07 | 0.0034 | 0.0230 | 3q21.3 |
|
|
hsa-miR-4700-3p | 2.07 | 0.0020 | 0.0230 | 12q24.31 |
|
|
hsa-miR-3622a-3p | 2.05 | 0.0011 | 0.0230 | 8p21.1 |
|
|
hsa-miR-5001-5p | 2.04 | 0.0073 | 0.0300 | 2q37.1 |
|
|
hsa-miR-6510-5p | 2.04 | 0.0073 | 0.0300 | 17q21.2 |
|
|
hsa-miR-4505 | 2.04 | 0.0073 | 0.0300 | 14q24.3 |
|
|
hsa-miR-4665-5p | 2.03 | 0.0057 | 0.0275 | 9p24.1 |
|
|
hsa-miR-8485 | 2.02 | 0.0008 | 0.0230 | 2p16.3 |
|
|
hsa-miR-7110-3p | 2.01 | 0.0073 | 0.0300 | 3q21.1 |
|
|
hsa-miR-6862-3p | 2.00 | 0.0015 | 0.0230 | 16p12.1 |
|
|
hsa-miR-6886-3p | 2.00 | 0.0034 | 0.0230 | 19p13.2 |
|
|
hsa-miR-328-5p | 2.00 | 0.0149 | 0.0436 | 16q22.1 | Myelogenous
leukemia (25) |
| Downregulated |
|
|
|
|
|
|
hsa-miR-6840-5p | 0.62 | 0.0073 | 0.0300 | 7q22.1 |
|
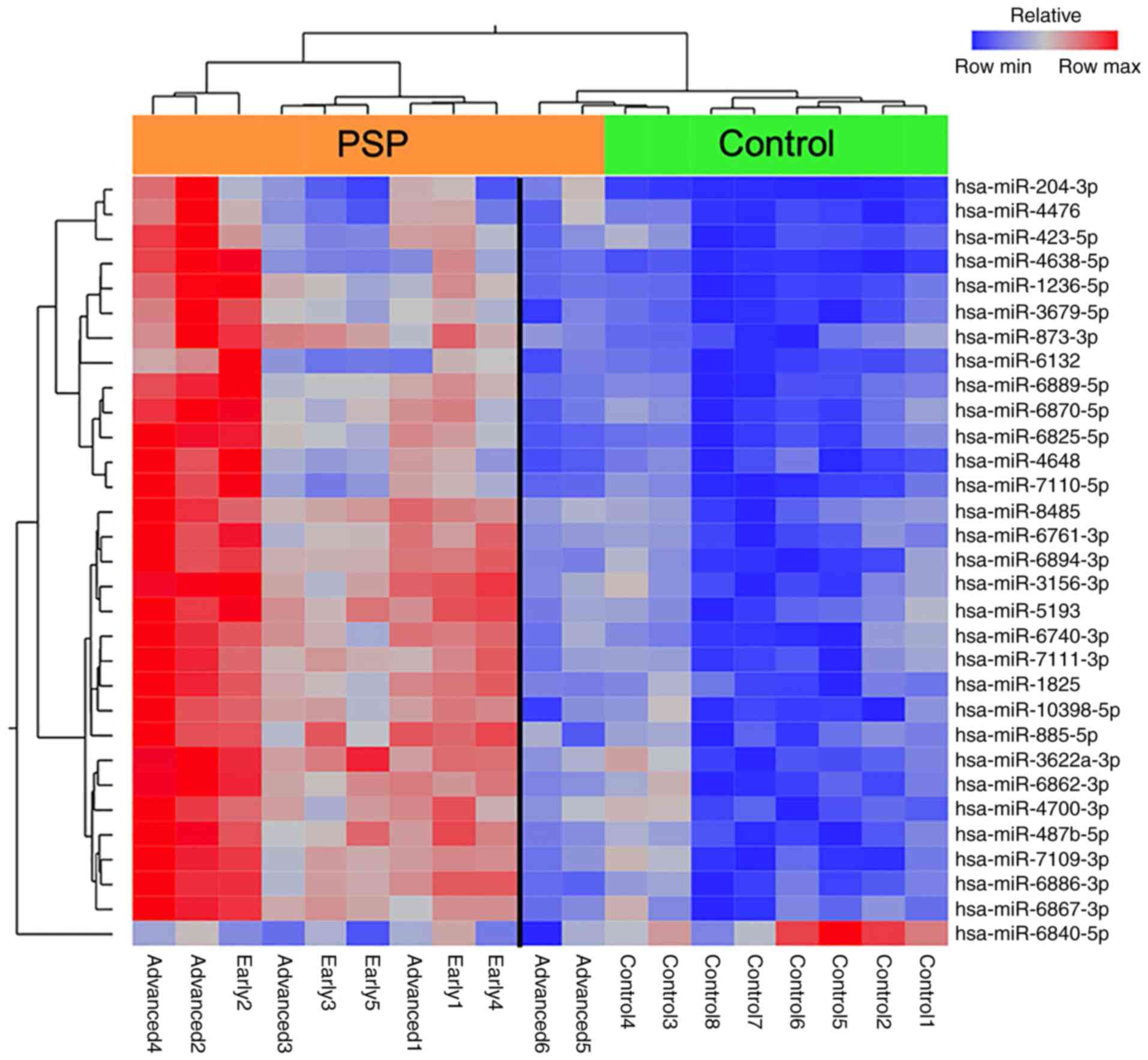
The extent to which the upper 30 miRNAs with high
expression levels and one with low expression levels in patients
with PSP separated from controls was further explored by
hierarchical clustering, which revealed a clear partition between
the PSP and control groups when the two patients with the longest
disease duration were excluded from the analysis (Fig. 2).
Differences between the early and
advanced stage groups
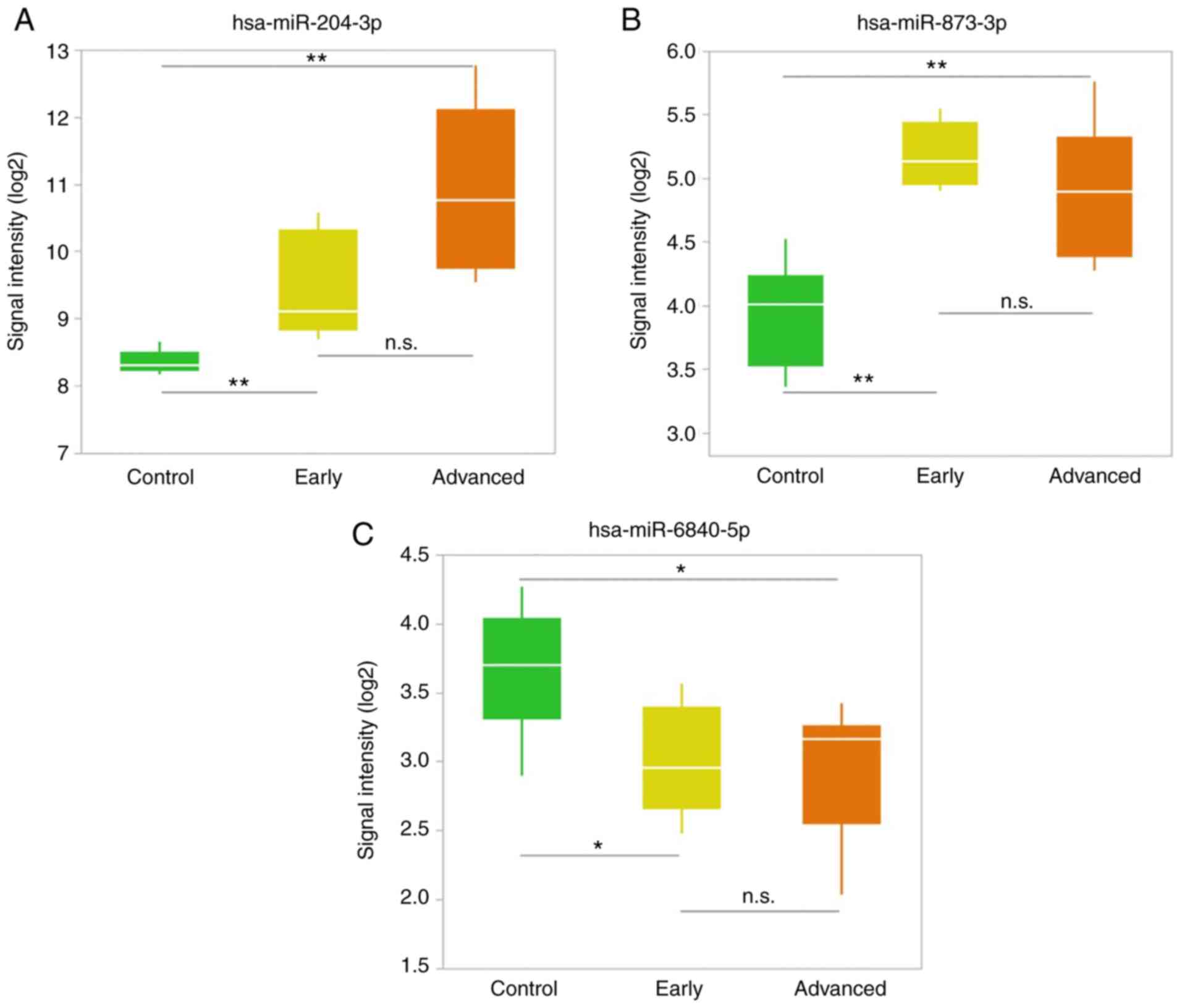
We selected up- and downregulated miRNAs with the
most significant difference in the early stage group. The top two
upregulated miRNAs and the top downregulated miRNA that were
selected are presented in Table
III and in Fig. 3. The
expression level of hsa-miR-204-3p in the CSF was higher than that
of other miRNAs. Hsa-miR-204-3p expression levels were
significantly higher in the early stage group than in the control
group (P=0.0043). Although the difference was not significant,
hsa-miR-204-3p was also increased in the advanced stage group
compared to in the early stage group (P=0.0552) (Fig. 3A). Hsa-miR-873-3p was also
significantly elevated in the early stage group (P=0.0043) and
remained high in the advanced stage group (Fig. 3B). In contrast, hsa-miR-6840-5p
was downregulated in the early stage group compared to controls
(P=0.0338) but did not significantly differ between the early and
advanced stage groups (Fig.
3C).
 | Table III.Upregulated and downregulated miRNAs
in the subgroups of patients with progressive supranuclear
palsy. |
Table III.
Upregulated and downregulated miRNAs
in the subgroups of patients with progressive supranuclear
palsy.
| MiRNA | P-value
(Kruskal-Wallis) | Post hoc
(Steel-Dwass) |
|---|
| Upregulated |
|
|
|
hsa-miR-204-3p |
|
|
|
Control vs. PSP
early | 0.0043 | 0.0120 |
|
Control vs. PSP
advanced | 0.0024 | 0.0068 |
|
PSP early vs. PSP
advanced | 0.0552 | 0.1338 |
|
hsa-miR-873-3p |
|
|
|
Control vs. PSP
early | 0.0043 | 0.0120 |
|
Control vs. PSP
advanced | 0.0081 | 0.0222 |
|
PSP early vs. PSP
advanced | 0.4113 | 0.6896 |
| Downregulated |
|
|
|
hsa-miR-6840-5p |
|
|
|
Control vs. PSP
early | 0.0338 | 0.0853 |
|
Control vs. PSP
advanced | 0.0239 | 0.0617 |
|
PSP early vs. PSP
advanced | 0.9273 | 0.9954 |
Discussion
In the present study, we investigated changes in the
expression of a wide range of miRNAs via microarray analysis in
samples from patients with PSP and healthy controls. In previous
studies, the analysis of miRNA expression profiles among patients
with PSP showed that miR-132 is specifically downregulated in the
brain tissue (12). miR-147a and
miR-518e were also significantly upregulated in the forebrain
tissue of patients with PSP (13). In this study, miR-132 was detected
at low levels in the CSF of all participants. However, miR-147a was
not detected in the control group but was detected in some patients
with PSP (n=8/11, 73%). miR-518e-5p expression was significantly
downregulated in patients with PSP relative to controls (fold
change ratio=0.76, P=0.03, data not shown), and there was no
significant difference in the expression between the early and
advanced stage groups. The profile of circulating miRNAs in the CSF
in our study differed slightly from that of miRNAs in the brain
tissue reported in previous studies, indicating that not all
altered miRNAs in the tissue are released as circulating
miRNAs.
In addition, miRNAs with altered expression levels
were divided into early and advanced disease stages to understand
the pathophysiology of the disease process by following the changes
in miRNAs over the course of disease. We found that in the early
stages of the disease, hsa-miR-204-3p and hsa-miR-873-3p expression
levels were significantly elevated in the PSP group compared to
those in the control group, and hsa-miR-6840-5p expression was
significantly lower in the PSP group than in the control group.
Genes that change from an early stage of PSP and show similar
changes in the advanced stage were thought to be more involved in
the pathogenesis of PSP. Genes that show a gradual increase from
early to advanced stages may reflect the process of
neurodegeneration.
In silico analysis was performed for each
miRNA, and the significance of miRNA expression changes was
examined from the predicted target genes. The target genes were
predicted using multiple databases, particularly for the miRNAs
that were significantly altered in the early stage group
(miR-204-3p, miR-873-3p, and miR-6840-5p). To further enhance the
reliability of the bioinformatics analysis, overlapping target
genes were identified (Table
IV).
 | Table IV.Prediction of the genes targeted by
the three miRNAs. |
Table IV.
Prediction of the genes targeted by
the three miRNAs.
| miRNA | Target genes |
|---|
| miR-204-3p | UNG, NUP50,
TBL1XR1, PIK3C2B, CNNM4, NFASC, KRAS, APOLD1, METTL1, ACAP2, MOB3A,
TGFBR1, SEPT6, XKR7, AJAP1 |
| miR-873-3p | MIDN, ZFX, USP36,
CC2D2A, CD40LG, MXRA7 |
| miR-6840-5p | ENPP1, PFKFB3,
ICAM5, IL1RAPL1, FBXL18, PITX1, MGRN1, PPP1R14B |
miR-204-3p was abundantly expressed in the PSP and
control groups, and the expression level showed a small dispersion
and equivalency in the control group. Further, the expression level
in the PSP group increased from the early to advanced stages of
disease. miR-204-3p upregulation in the CSF has not been reported
previously, while miR-204-5p expression level was demonstrated to
be low in the CSF of patients with frontotemporal dementia
(26) or MSA (27). In this study, miR-204-3p was
significantly upregulated in the CSF of patients with PSP.
Activating transcription factor 2 (ATF2) (28), regulator of G-protein signaling 5
(RGS5) (29), protein
phosphatase, Mg2+/Mn2+ dependent 1K (30), and K homology domain-containing,
RNA-binding, signal transduction-associated protein 1
(KHDRBS1) (31) are
confirmed targets of miR-204. miR-204 has been reported to be
involved in the process of autophagy (32). ATF2 is involved in the
regulation of cell proliferation, apoptosis, or autophagy of
several cancers, such as glioblastoma (28), non-small cell lung cancer
(33), and cervical cancer
(34). KHDRBS1 is also
known as Sam68 or p62 and is widely known as the central regulator
between the ubiquitin-proteasome system and autophagy (35). The predicted target genes of
miR-204-3p are listed in Table
IV. Among them, miR-204-3p was predicted to target adherens
junctions associated protein 1 (AJAP1) (36). Interestingly, AJAP1 was
found to be hypermethylated in patients with PSP in an
epigenome-wide association study (EWAS) (37). These data suggest that miR-204-3p
may be involved in the pathogenesis of PSP.
miR-873-3p is reportedly downregulated in the CSF of
patients with PD, but upregulated miR-873-3p expression in the CSF
of patients with other neurological disorders has not been reported
previously (7). In the present
analysis, miR-873-3p was significantly elevated in the early stage
group, suggesting that miR-873-3p may be useful in differentiating
PSP and PD, which can be difficult to distinguish in the early
stages. miR-873-3p was predicted to target midbrain nucleolar
protein (midnolin, MIDN) (38–40). MIDN has been confirmed as a
genetic risk factor for PD (41)
and is suggested to be a novel regulator of parkin expression
via the promotion of parkin E3 ubiquitin ligase expression
(42). In this regard,
hsa-miR-873-3p may be involved in the differences in the
pathogenesis of PSP and PD.
In this study, miR-6840-5p expression was
downregulated in patients with PSP. As shown in Table IV, miR-6840-5p was predicted to
target genes such as the gene encoding F-Box and leucine-rich
repeat protein 18 (FBXL18) (43) and Mahogunin ring finger-1
(MGRN1) (43,44), as previously reported.
Interestingly, FBXL18 was found to be hypomethylated in
patients with PSP in an EWAS (37). FBXL18 has been reported to
bind directly to leucine-rich repeat kinase 2 (LRRK2),
regulate LRRK2 stability, and control LRRK2-mediated toxicity
(45). In contrast, MGRN1,
an E3 ubiquitin ligase of the really interesting new gene finger
family, is a crucial component in autophagy and the
ubiquitin-proteasome system and is involved in abnormal protein
degradation and quality control. Previous findings have indicated
that dysfunctional MGRN1 can cause neurodegeneration and
mitochondrial dysfunction in mouse brains (46). In summary, we found that many of
the genes targeted by miRNAs with significantly altered expression
levels in this study were involved in the ubiquitin-proteasome
system and autophagy pathway.
Recently, Ramaswamy's group proposed a role for
miRNAs in the expression of key pathological features of
Parkinsonian movement disorders. Disruption of protein homeostasis
mechanisms such as autophagy due to miRNA dysregulation has been
suggested to be involved in neurodegeneration. According to them,
miRNAs, such as miR-204, play a role in the regulation of the
autophagy pathway (47).
Interestingly, the expression of miRNAs, which may be involved in
the autophagy pathway, was also found to be variable in this
study.
There were a number of limitations to this study.
The miRNA expression levels could not be validated, and individual
miRNA expression levels should be determined using reverse
transcription quantitative polymerase chain reaction in the future.
In addition, it is necessary to investigate the expression of these
miRNAs in serum, which is very accessible and will make it easier
to use as a biomarker. Furthermore, the sample size was small as
PSP is a rare disease; moreover, this was a single-center study and
patients with concurrent malignant tumors, psychiatric diseases,
collagen diseases, endocrine diseases, or infectious diseases were
excluded. In addition, the collection of CSF samples is more
invasive than that of other body fluid samples such as blood;
therefore, the number of participants was limited, and the study
was conducted on a small sample size. There is only one previous
report of profiling data of CSF miRNAs in PSP patients. An miRNA
panel including 372 miRNAs was used (21). In this study, we used a microarray
chip containing 2632 miRNAs to perform a comprehensive analysis
that included many miRNAs that had not been analyzed in previous
studies. Therefore, most of the miRNAs that showed changes in this
study were relatively newly discovered, and their target genes and
functions were not fully understood. Thus, there was a limited
number of miRNAs for which in silico analysis was possible.
Although this study is based on data from a small sample size, we
believe that the results of this analysis are significant in terms
of their novelty and originality. In future studies, we would like
to examine miRNA expression in a greater number of patients with
PSP.
In conclusion, we identified miRNAs that were
significantly altered in the CSF of patients with PSP using
microarrays. Among them, some miRNAs (hsa-miR-204-3p, 873-3p, and
6840-5p) were significantly altered from an early stage of the
disease. We performed bioinformatics analysis of the genes
associated with these miRNAs and found that they are involved in
processes such as autophagy and the ubiquitin-proteasome system,
which are relevant to cellular quality control. Interestingly, we
found altered expression of miRNAs targeting genes that have been
reported to show epigenetic changes following EWAS of DNA from
brain tissues of PSP patients. This suggests that these miRNAs and
genes may have some involvement in the pathogenesis of PSP. In
future studies, we will analyze how miRNAs interact with such
molecules and how they contribute to the pathogenesis of
disease.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The microarray datasets generated and/or analyzed
during the current study are available in the NCBI Gene Expression
Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186921).
Authors' contributions
WN, TTa, TI and TM conceived the study. WN, TTa, HI,
TI and TM curated data. WN, HI and TI performed formal analysis. KD
and TTo conceived the study and curated data. WN, SK, MK and HK
performed the experiments. WN, HI, OM and TN developed the
methodology. KD, TTo, TI and TM were involved in project
administration. SK, HK, MK, KD, TTo and TM provided resources. HK,
KD, TTo, OM, TN and TM supervised the study. WN, TTa, KD, TTo, OM,
TN and TI were involved in validation. WN and HI were involved in
visualization. WN and TI wrote the original draft. TTa, HI, KD,
TTo, OM, TN, TI and TM reviewed and edited the manuscript. All
authors have read and approved the final manuscript. TTa and TI
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The procedures were approved by the ethics committee
of Kagawa University Faculty of Medicine (Miki-cho, Kita-gun,
Kagawa, Japan). All patients and/or their relatives provided
written informed consent. All clinical investigations have been
conducted according to the principles expressed in the Declaration
of Helsinki.
Patient consent for publication
All patients and/or their relatives provided written
informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Agrawal M and Biswas A: Molecular
diagnostics of neurodegenerative disorders. Front Mol Biosci.
2:542015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee RC, Feinbaum RL and Ambrost V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Quinn JF, Patel T, Wong D, Das S, Freedman
JE, Laurant LC, Carter BS, Hochberg F, van Keuren-Jensen K,
Huentelmann M, et al: Extracellular RNAs: Development as biomarkers
of human disease. J Extracell Vesicles. 4:274952015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rao P, Benito E and Fischer A: MicroRNAs
as biomarkers for CNS disease. Front Mol Neurosci. 6:392013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roser AE, Caldi Gomes L, Schünemann J,
Maass F and Lingor P: Circulating miRNAs as diagnostic biomarkers
for Parkinson's disease. Front Neurosci. 12:6252018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Denk J, Boelmans K, Siegismund C, Lassner
D, Arlt S and Jahn H: MicroRNA profiling of CSF reveals potential
biomarkers to detect Alzheimer's disease. PLoS One.
10:e01264232015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Burgos K, Malenica I, Metpally R,
Courtright A, Rakela B, Beach T, Shill H, Adler C, Sabbagh M, Villa
S, et al: Profiles of extracellular miRNA in cerebrospinal fluid
and serum from patients with Alzheimer's and Parkinson's diseases
correlate with disease status and features of pathology. PLoS One.
9:e948392014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Steele JC, Richardson JC and Olszewski J:
Progressive Supranuclear palsy: A heterogeneous degeneration
involving the brainstem, basal ganglia and cerebellum with vertical
gaze and pseudobulbar palsy, nuchal dystonia and dementia. Arch
Neurol. 10:333–359. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coyle-Gilchrist IT, Dick KM, Patterson K,
Vázquez Rodríquez P, Wehmann E, Wilcox A, Lansdall CJ, Dawson KE,
Wiggins J, Mead S, et al: Prevalence, characteristics, and survival
of frontotemporal lobar degeneration syndromes. Neurology.
86:1736–1743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Respondek G, Stamelou M, Kurz C, Ferguson
LW, Rajput A, Chiu WC, van Swieten JC, Troakes C, Al Sarraj S,
Glepi E, et al: The phenotypic spectrum of progressive supranuclear
palsy: A retrospective multicenter study of 100 definite cases. Mov
Disord. 29:1758–1766. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Magdalinou NK, Paterson RW, Schott JM, Fox
NC, Mummery C, Blennow K, Bhatia K, Morris HR, Giunti P, Warner TT,
et al: A panel of nine cerebrospinal fluid biomarkers may identify
patients with atypical parkinsonian syndromes. J Neurol Neurosurg
Psychiatry. 86:1240–1247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith PY, Delay C, Girard J, Papon MA,
Planel E, Sergeant N, Buée L and Hébert SS: MicroRNA-132 loss is
associated with tau exon 10 inclusion in progressive supranuclear
palsy. Hum Mol Genet. 20:4016–4024. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tatura R, Buchholz M, Dickson DW, van
Swieten J, McLean C, Höglinger G and Müller U: MicroRNA profiling:
Increased expression of miR-147a and miR-518e in progressive
supranuclear palsy (PSP). Neurogenetics. 17:165–171. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Höglinger GU, Respondek G, Stamelou M,
Kurz C, Josephs KA, Lang AE, Mollenhauer B, Müller U, Nilsson C,
Whitwell JL, et al: Clinical diagnosis of progressive supranuclear
palsy: The movement disorder society criteria. Mov Disord.
32:853–864. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Golbe LI and Ohman-Strickland PA: A
clinical rating scale for progressive supranuclear palsy. Brain.
130:1552–1565. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Folstein MF, Folstein SE and McHugh PR:
‘Mini-mental state’: A practical method for grading the cognitive
state of patients for the clinician. J Psychiatr Res. 12:189–198.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dubois B, Slachevsky A, Litvan I and
Pillon B: The FAB: A Frontal assessment battery at bedside.
Neurology. 55:1621–1626. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nasreddine ZS, Phillips NA, Bédirian V,
Charbonneau S, Whitehead V, Collin I, Cummings JL and Chertkow H:
The montreal cognitive assessment, MoCA: A brief screening tool for
mild cognitive impairment. J Am Geriatr Soc. 53:695–699. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Quattrone A, Morelli M, Williams DR,
Vescio B, Arabia G, Nigro S, Nicoletti G, Salsone M, Novellino F,
Nisticò R, et al: MR parkinsonism index predicts vertical
supranuclear gaze palsy in patients with PSP-parkinsonism.
Neurology. 87:1266–1273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quattrone A, Morelli M, Nigro S, Quattrone
A, Vescio B, Arabia G, Nicoletti G, Nisticò R, Salsone M, Novellino
F, et al: A new MR imaging index for differentiation of progressive
supranuclear palsy-parkinsonism from Parkinson's disease.
Parkinsonism Relat Disord. 54:3–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Conte I, Hadfield KD, Barbato S, Pizzo M,
Bhat RS, Carissimo A, Karali M, Porter LF, Urquhart J, Hateley S,
et al: miR-204 is responsible for inherited retinal dystrophy
associated with ocular coloboma. Proc Natl Acad Sci USA.
112:E3236–E3245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gattolliat CH, Thomas L, Ciafrè SA,
Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P,
Valteau-Couanet D, et al: Expression of miR-487b and miR-410
encoded by 14q32.31 locus is a prognostic marker in neuroblastoma.
Br J Cancer. 105:1352–1361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, He F, Pang R, Zhao D, Qiu W, Shan
K, Zhang J, Lu Y, Li Y and Wang Y: Interleukin-17 (IL-17)-induced
MicroRNA 873 (miR-873) contributes to the pathogenesis of
experimental autoimmune encephalomyelitis by targeting A20
Ubiquitin-editing enzyme. J Biol Chem. 289:28971–28986. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eiring AM, Harb JG, Neviani P, Garton C,
Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al:
miR-328 Functions as an RNA decoy to modulate hnRNP E2 Regulation
of mRNA translation in leukemic blasts NIH Public Access. Cell.
140:652–665. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schneider R, McKeever P, Kim TH, Graff C,
van Swieten JC, Karydas A, Boxer A, Rosen H, Miller BL, Laforce R
Jr, et al: Downregulation of exosomal miR-204-5p and miR-632 as a
biomarker for FTD: A GENFI study. J Neurol Neurosurg Psychiatry.
89:851–858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Starhof C, Hejl AM, Heegaard NHH, Carlsen
AL, Burton M, Lilje B and Winge K: The biomarker potential of
cell-free microRNA from cerebrospinal fluid in Parkinsonian
syndromes. Mov Disord. 34:246–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song S, Fajol A, Tu X, Ren B and Shi S:
miR-204 suppresses the development and progression of human
glioblastoma by targeting ATF2. Oncotarget. 7:70058–70065. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Banaei-Esfahani A, Moazzeni H, Nosar PN,
Amin S, Arefian E, Soleimani M, Yazdani S and Elahi E: MicroRNAs
that target RGS5. Iran J Basic Med Sci. 18:108–114. 2015.PubMed/NCBI
|
|
30
|
Pan BF, Gao C, Ren SX, Wang YB, Sun HP and
Zhou MY: Regulation of PP2Cm expression by miRNA-204/211 and
miRNA-22 in mouse and human cells. Acta Pharmacol Sin.
36:1480–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Tian H, Yuan J, Wu H, Wu J and Zhu
X: CONSORT: Sam68 is directly regulated by miR-204 and promotes the
self-renewal potential of breast cancer cells by activating the
wnt/beta-catenin signaling pathway. Medicine (Baltimore).
94:e22282015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Y and Liang C: MicroRNAs: An emerging
player in autophagy. ScienceOpen Res. Dec 22–2015.(Epub ahead of
print). doi: 10.14293/S2199-1006.1.SOR-LIFE.A181CU.v1. PubMed/NCBI
|
|
33
|
Zhang S, Gao L, Thakur A, Shi P, Liu F,
Feng J, Wang T, Liang Y, Liu JJ, Chen M and Ren H: MiRNA-204
suppresses human non-small cell lung cancer by targeting ATF2.
Tumor Biol. 37:11177–11186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li N, Guo X, Liu L, Wang L and Cheng R:
Molecular mechanism of miR-204 regulates proliferation, apoptosis
and autophagy of cervical cancer cells by targeting ATF2. Artif
Cells, Nanomed Biotechnol. 47:2529–2535. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Katsuragi Y, Ichimura Y and Komatsu M:
P62/SQSTM1 functions as a signaling hub and an autophagy adaptor.
FEBS J. 282:4672–4678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weber A, Schwarz SC, Tost J, Trümbach D,
Winter P, Busato F, Tacik P, Windhorst AC, Fagny M, Arzberger T, et
al: Epigenome-wide DNA methylation profiling in progressive
supranuclear palsy reveals major changes at DLX1. Nat Commun.
9:29292018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hafner M, Landthaler M, Burger L, Khorshid
M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC,
Munschauer M, et al: Transcriptome-wide identification of
RNA-binding protein and microRNA target sites by PAR-CLIP. Cell.
141:129–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang
H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al: Direct conversion of
fibroblasts to neurons by reprogramming PTB-regulated microRNA
circuits. Cell. 152:82–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Krell J, Stebbing J, Carissimi C,
Dabrowska AF, de Giorgio A, Frampton AE, Harding V, Fulci V, Macino
G, Colombo T and Castellano L: TP53 regulates miRNA association
with AGO2 to remodel the miRNA-mRNA interaction network. Genome
Res. 26:331–341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Obara Y, Sato H, Nakayama T, Kato T and
Ishii K: Midnolin is a confirmed genetic risk factor for
Parkinson's disease. Ann Clin Transl Neurol. 6:2205–2211. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Obara Y, Imai T, Sato H, Takeda Y, Kato T
and Ishii K: Midnolin is a novel regulator of parkin expression and
is associated with Parkinson's disease. Sci Rep. 7:58852017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Karginov FV and Hannon GJ: Remodeling of
Ago2-mRNA interactions upon cellular stress reflects miRNA
complementarity and correlates with altered translation rates.
Genes Dev. 27:1624–1632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Spengler RM, Zhang X, Cheng C, McLendon
JM, Skeie JM, Johnson FL, Davidson BL and Boudreau RL: Elucidation
of transcriptome-wide microRNA binding sites in human cardiac
tissues by Ago2 HITS-CLIP. Nucleic Acids Res. 44:7120–7131.
2016.PubMed/NCBI
|
|
45
|
Ding X, Barodia SK, Ma L and Goldberg MS:
Fbxl18 targets LRRK2 for proteasomal degradation and attenuates
cell toxicity. Neurobiol Dis. 98:122–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Upadhyay A, Amanullah A, Chhangani D,
Mishra R, Prasad A and Mishra A: Mahogunin ring finger-1 (MGRN1), a
multifaceted ubiquitin ligase: Recent unraveling of neurobiological
mechanisms. Mol Neurobiol. 53:4484–4496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ramaswamy P, Christopher R, Pal PK and
Yadav R: MicroRNAs to differentiate Parkinsonian disorders:
Advances in biomarkers and therapeutics. J Neurol Sci. 394:26–37.
2018. View Article : Google Scholar : PubMed/NCBI
|