Introduction
Oral cancer is a major public health concern which
accounts for the fourth highest incidence of malignancy (1). Oral squamous cell carcinoma (OSCC)
is the most common malignant neoplasm of the oral cavity (2,3). A
large number of patients with OSCC are often diagnosed at advanced
stages and therefore face complex surgical procedures and a poor
prognosis (4). Although current
research focuses on novel treatment options for OSCC, the prognosis
remains poor (5). Thus,
investigations into the potential mechanisms underlying the
pathogenesis of OSCC may be beneficial for the discovery of novel
treatment options for OSCC.
GATA binding protein 6 (GATA6) is a member of the
GATA transcription factor family that binds to the (A/T)GATA(A/G)
consensus sequence for the activation or inhibition of expression
of genes involved in cell fate decision making and tissue
morphogenesis (6). GATA6 is an
essential factor in modulating cell differentiation and tumor
dissemination and is regarded as an independent risk factor for the
prognosis of ovarian cancer (7,8).
The results of previous studies demonstrate that GATA6 is expressed
at high levels in a number of types of cancer, including
cholangiocarcinoma, gastric and colorectal cancer (9–11).
Additionally, GATA6 is significantly increased in oral carcinoma
cell lines (12). The results of
a previous study also demonstrated that regulation of microRNA
(miRNA/miR)-506 suppresses the expression of GATA6, thus prolonging
the development of OSCC (13).
However, the specific effects of GATA6 on the proliferation,
invasion and migration of OSCC cells remain to be elucidated.
Fibronectin 1 (FN1), a glycoprotein present in the extracellular
matrix, is closely associated with cellular adhesion and migration
(14). The results of a previous
study demonstrated that FN1 is considered as a potential biomarker
in OSCC associated with the tongue, mouth floor and edentulous
ridge (15). Furthermore,
inhibition of FN1 suppresses the development of OSCC (16). Thus, the present study aimed to
determine whether GATA6 promoted the malignant development of OSCC
by inducing FN1 expression.
In the present study, the expression of GATA6 was
determined in a number of OSCC cell lines and the subsequent
effects on proliferation, invasion and migration of OSCC cells were
investigated. Furthermore, the role of FN1 in GATA6 expression and
the progression of OSCC was explored.
Materials and methods
Bioinformatics analyses
The expression of GATA6 and FN1 in OSCC tissues and
adjacent tissues was analyzed using the TNMplot database
(https://tnmplot.com; the parameters used were
Gene-Chip and unpaired samples) (17). The GSE dataset GSE146483
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146483)
consisting of 11 samples (8 OSCC and 3 human normal oral mucosal
epithelial keratinocytes) was downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/)
and analyzed using GEO2R. Relative gene expression analysis of
GATA6 in head and neck squamous cell carcinoma (HNSC) was conducted
through LinkedOmics (https://www.linkedomics.org/login.php/) using HiSeq
RNA Platform (18). Gene
enrichment analysis was conducted by WebGestalt (http://www.webgestalt.org/) using the Gene Set
Enrichment Analysis (GSEA) tool and Kyoto Encyclopedia of Genes and
Genomes (KEGG) functional database (FDR <0.25, adjusted P-value
<0.05). The potential transcription factor binding sites in the
promoter of FN1 and GATA6 were predicted by JASPAR (http://jaspar.genereg.net/) database.
Cell lines
Human normal oral mucosal epithelial keratinocytes
(HOK) and OSCC cell lines (HN4, HN6, SCC-9 and Cal-27), were
purchased from the American Type Culture Collection and cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Hyclone; Cytiva) at 37°C and 5% CO2.
Cell transfection
Cal-27 cells in the logarithmic phase were
inoculated into a 96-well plate and cultured until 70% confluence
was reached. Small interfering (si)RNA targeting GATA6 [si-GATA6-1
(TAGAGTTATTGTGTTAAGAAAGT) and si-GATA6-2
(TAGCGAAGTACTCATAATCTAAT)], the corresponding negative control
(si-NC), FN1 overexpression pcDNA3.1 plasmid (Oe-FN1) and the empty
vector plasmid (Oe-NC), at a concentration of ~20 µM, were obtained
from Shanghai GenePharma Co., Ltd. Cells were transfected using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Cells
were incubated with 5% CO2 at 37°C and were used in
subsequent experiments after 48 h of transfection. Successful
transfection was determined using reverse
transcription-quantitative (RT-q)PCR and western blot analysis.
Cell viability assay
A total of 4×104 cells/well were seeded
into a 96-well plate along with 10 µl Cell Counting Kit-8 solution
(Shanghai Yeasen Biotechnology Co., Ltd.) and incubated in cell
culture medium for 2 h. The viability was subsequently analyzed at
a wavelength of 450 nm in a microplate reader (Promega
Corporation).
Colony formation
Cal-27 cells were seeded into 96-well plates at a
density of 2×104 cells/well and cultured for 10 days for
the formation of colonies defined as >50 cells/colony. Cells
were washed three times with PBS and fixed using 4%
polyoxymethylene for 15 min at 37°C. Cells were subsequently
stained with crystal violet for 10 min at 37°C, images were
captured using a light microscope (magnification, ×10; Olympus
Corporation) and the colonies counted.
Transwell assay
For the evaluation of cell invasion, the upper
chamber was pretreated with 100 µl Matrigel (BD Biosciences) at
37°C for 30 min. A total of 1×105 Cal-27 cells suspended
in 200 µl serum-free DMEM were plated in the upper chamber of a
24-well 8-µm pore Transwell insert (Costar; Corning, Inc.).
Subsequently, DMEM containing 10% FBS was placed in the basolateral
chamber. Invaded cells were fixed with 4% paraformaldehyde for 20
min at room temperature, washed three times with PBS and stained
with crystal violet for 10 min at room temperature. The cells were
observed under a light microscope (Olympus Corporation)
(magnification, ×100) for statistical analysis.
Wound healing assay
Cal-27 cells were inoculated into 96-well plates and
incubated until 80% confluence was reached. A 10-µl pipette tip was
used to create a wound on the surface of the Cal-27 cells. Cells
were subsequently cultured in serum-free DMEM. Following an
indicated time after the scratch, the migratory ability of Cal-27
cells was observed using a light microscope (Olympus
Corporation).
Dual-luciferase reporter assay
Wild-type (WT) or mutant (MUT) promoter site of FN1
was introduced into the pGL3-Basic vector (Promega Corporation) to
make WT-FN1 promoter and MUT-FN1 promoter plasmids. The luciferase
reporter plasmids and GATA6-expressing plasmid or empty vector were
co-transfected into Cal-27 cells using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Luciferase activities were measured 48 h
after transfection using the Dual-Luciferase Reporter Assay kit
(Promega Corporation). The relative luciferase activity was
normalized to Renilla luciferase activity.
Chromatin immunoprecipitation (CHIP)
assay
The binding of GATA6 to the FN1 promoter was
examined using a CHIP assay kit (Beyotime Institute of
Biotechnology), according to the manufacturer's protocol. Cells
were cross-linked using 1% formaldehyde for 15 min at room
temperature and cell lysates in SDS lysis buffer (GBCBIO
Technologies, Inc.) were sonicated (10 sec, five times) on ice to
achieve 200-1,000 bp chromatin fragments and centrifuged at 4°C,
16,100 × g for 10 min. CHIP was conducted following incubation with
the anti-GATA6 antibody (cat. no. 5851T; 1:50; Cell Signaling
Technology, Inc.). The recuperated DNA fragments were evaluated via
qPCR. The primer sequences were: FN1, forward
5′-CCGAAGAGAGGTGACGCAAT-3′ and reverse
5′-GAGTGGCTGGACTTGTGTGA-3′.
RT-qPCR
Total RNA was extracted from 1×104 cells
using TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Total RNA was reverse
transcribed into cDNA using a reverse transcriptase cDNA synthesis
kit (PrimeScript RT Reagent kit; Takara Bio, Inc.) according to the
manufacturer's protocol. The temperature protocol was as follows:
70°C for 5 min, 37°C for 5 min and 42°C for 1 h. qPCR was
subsequently performed using a SYBR-Green PCR kit (Takara Bio,
Inc.) on an ABI 7500 Real-Time PCR detection instrument (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The following thermocycling conditions were used: pre-denaturation
at 95°C for 10 min, denaturation at 95°C for 15 sec, annealing at
60°C for 1 min (40 cycles) and extension at 72°C for 1 min. The
primer sequences for PCR were: GATA6 5′-TGCAATGCTTGTGGACTCTA-3′
(forward) and 5′-GTGGGGGAAGTATTTTTGCT-3′ (reverse); FN1
5′-CGGTGGCTGTCAGTCAAAG-3′ (forward) and 5′-AAACCTCGGCTTCCTCCATAA-3′
(reverse); GAPDH 5′-GGCTCATGACCACAGTCCATG-3′ (forward) and
5′-TCAGCTCTGGGATGACCTTG-3′ (reverse). mRNA levels were quantified
using the 2−ΔΔCq method and normalized to the internal
reference gene, GAPDH (19). All
experiments were performed in triplicate.
Western blot analysis
Total proteins were obtained from Cal-27 cells
(1×104) using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Total protein was quantified using a BCA kit
(Beyotime Institute of Biotechnology) and 40 µg protein sample was
separated by SDS-PAGE on a 10% gel. The separated proteins were
subsequently transferred onto PVDF membranes and blocked for 1 h at
room temperature with 5% skimmed milk. The membranes were incubated
overnight at 4°C with the following primary antibodies: Anti-GATA6
(cat. no. 5851T; 1:1,000; Cell Signaling Technology, Inc.),
anti-FN1 (cat. no. 26836S; 1:1,000; Cell Signaling Technology,
Inc.), anti-Ki67 (cat. no. ab16667; 1:1,000; Abcam),
anti-proliferating cell nuclear antigen (PCNA; 13110T; 1:1,000;
Cell Signaling Technology, Inc.), anti-matrix metalloproteinase-2
(MMP2; cat. no. 40994S; 1:1,000; Cell Signaling Technology, Inc.),
anti-MMP9 (cat. no. 13667T; 1:1,000; Cell Signaling Technology,
Inc.) and anti-GAPDH (cat. no. 5174T; 1:1,000; Cell Signaling
Technology, Inc.). Following the primary antibody incubation, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibody (cat. no. 7074S; 1:3,000; Cell Signaling
Technology, Inc.) for 1 h at room temperature and washed three
times with PBS. Proteins bands were visualized using enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.). The relative
intensity of each band was semi-quantified using ImageJ software
(version 1.52r; National Institutes of Health). GAPDH was used as
the loading control.
Statistical analysis
Data were presented as the mean ± standard deviation
of three independent experiments simultaneously. Results were
analyzed using GraphPad Prism software (version 8.0; GraphPad
Software, Inc.). Statistical differences were evaluated using an
unpaired Student's t-test or one-way ANOVA followed by a Tukey's
post-hoc test. Pearson's correlation analysis was utilized to
confirm the correlation between GATA6 and FN1. P<0.05 was
considered to indicate a statistically significant difference.
Results
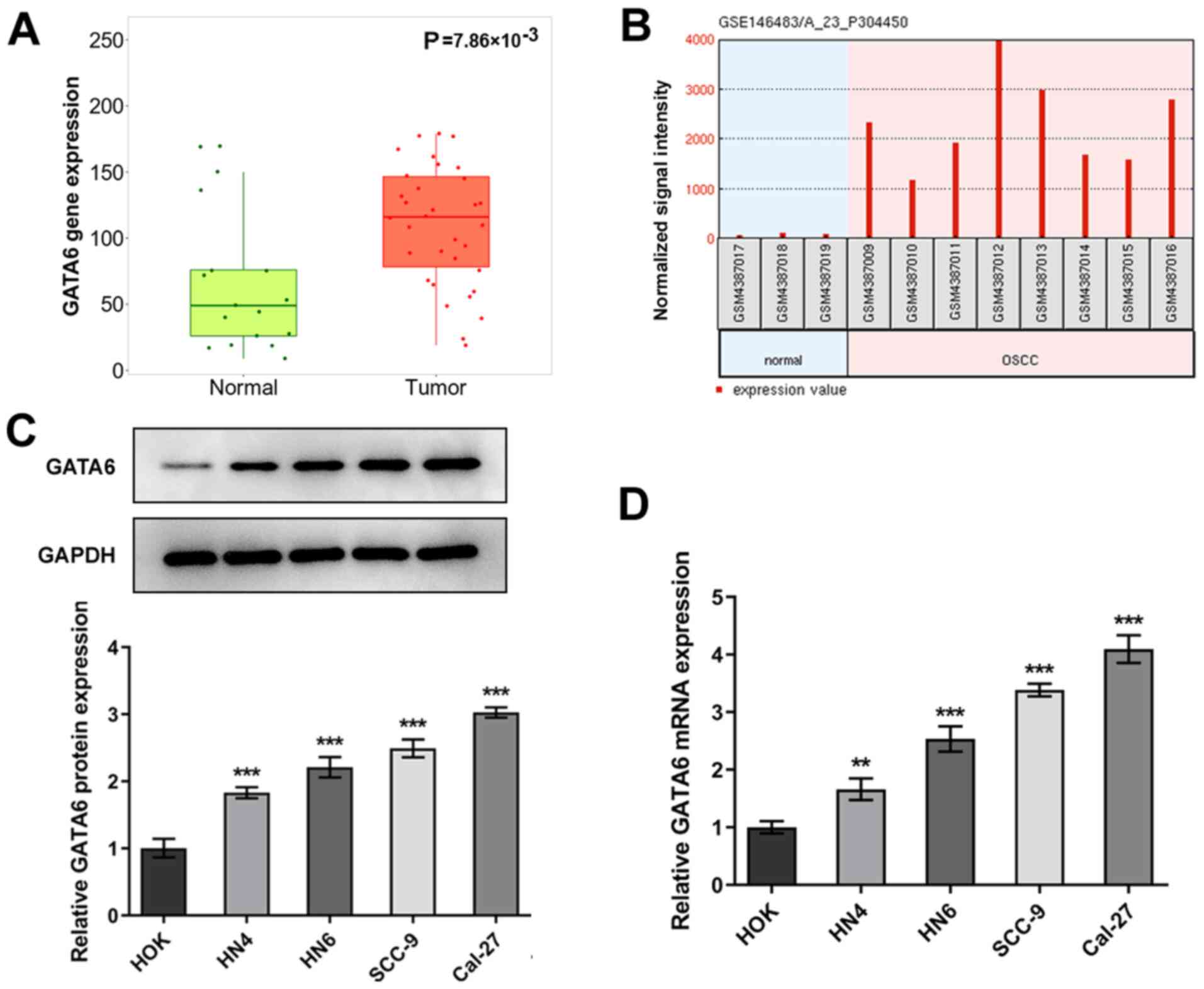
GATA6 is markedly upregulated in OSCC
tissues and cells
To investigate the role of GATA6 in OSCC
development, TNMplot data was employed to analyze the expression of
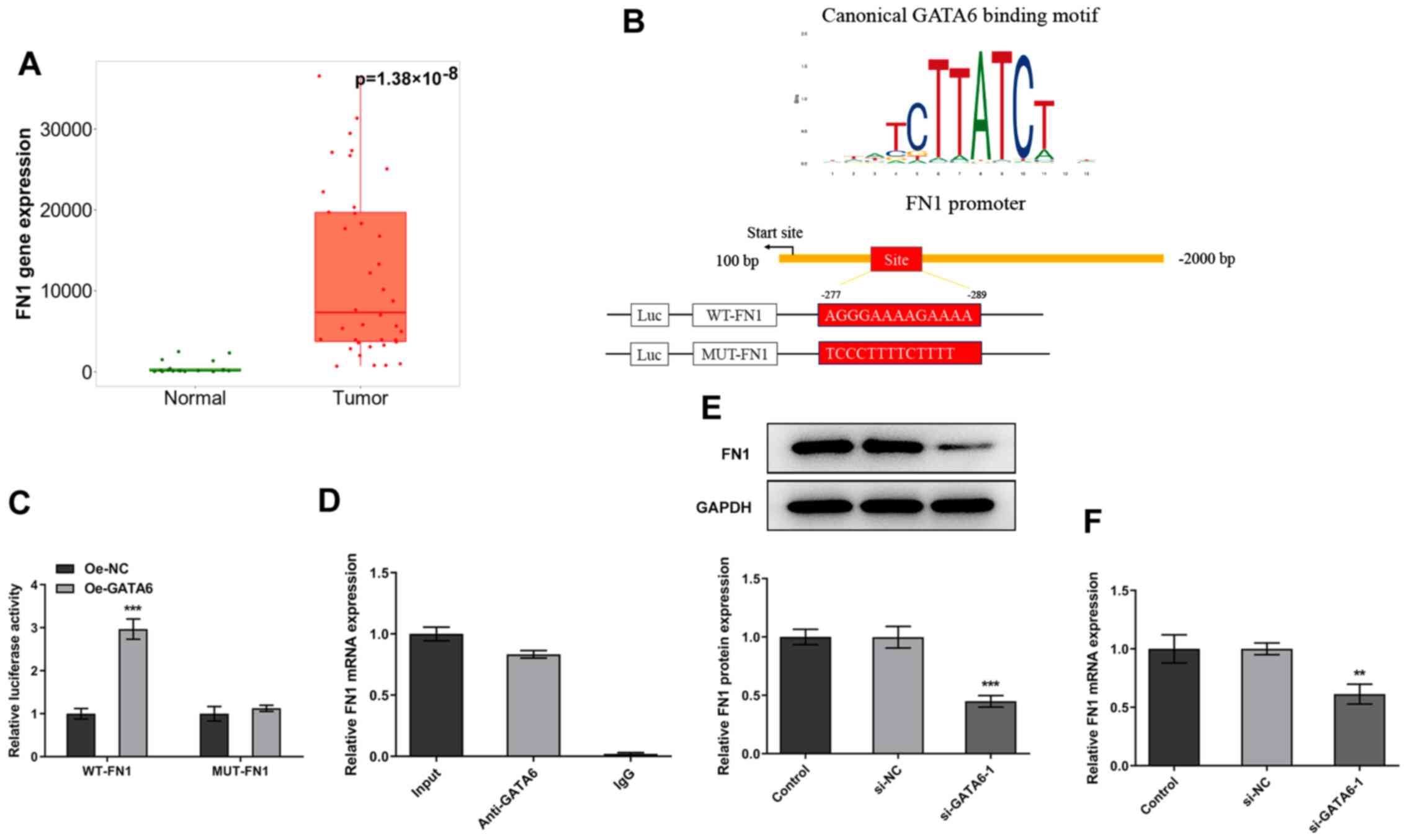
GATA6 in OSCC tissues and adjacent tissues. As shown in Fig. 1A, GATA6 was highly expressed in
tumor group as comparison to the normal group. The expression
levels of GATA6 in eight OSCC cells and three human normal oral
mucosal epithelial keratinocytes and were determined using the
GEO2R analysis in an independent cohort (GSE146483). As
demonstrated in Fig. 1B, GATA6
expression was markedly upregulated in OSCC group compared with the
human normal oral mucosal epithelial keratinocytes (normal) group.
Subsequently, RT-qPCR and western blot analyses were used to assess
the expression levels of GATA6 in OSCC and HOK cell lines. As
demonstrated in Fig. 1C and D,
the expression levels of GATA6 protein and mRNA were high in OSCC
cells compared with the HOK group. The highest expression levels of
GATA6 were observed in Cal-27 cells, which were therefore used for
subsequent analyses. These results indicated that GATA6 was
upregulated in OSCC tissues and cells.
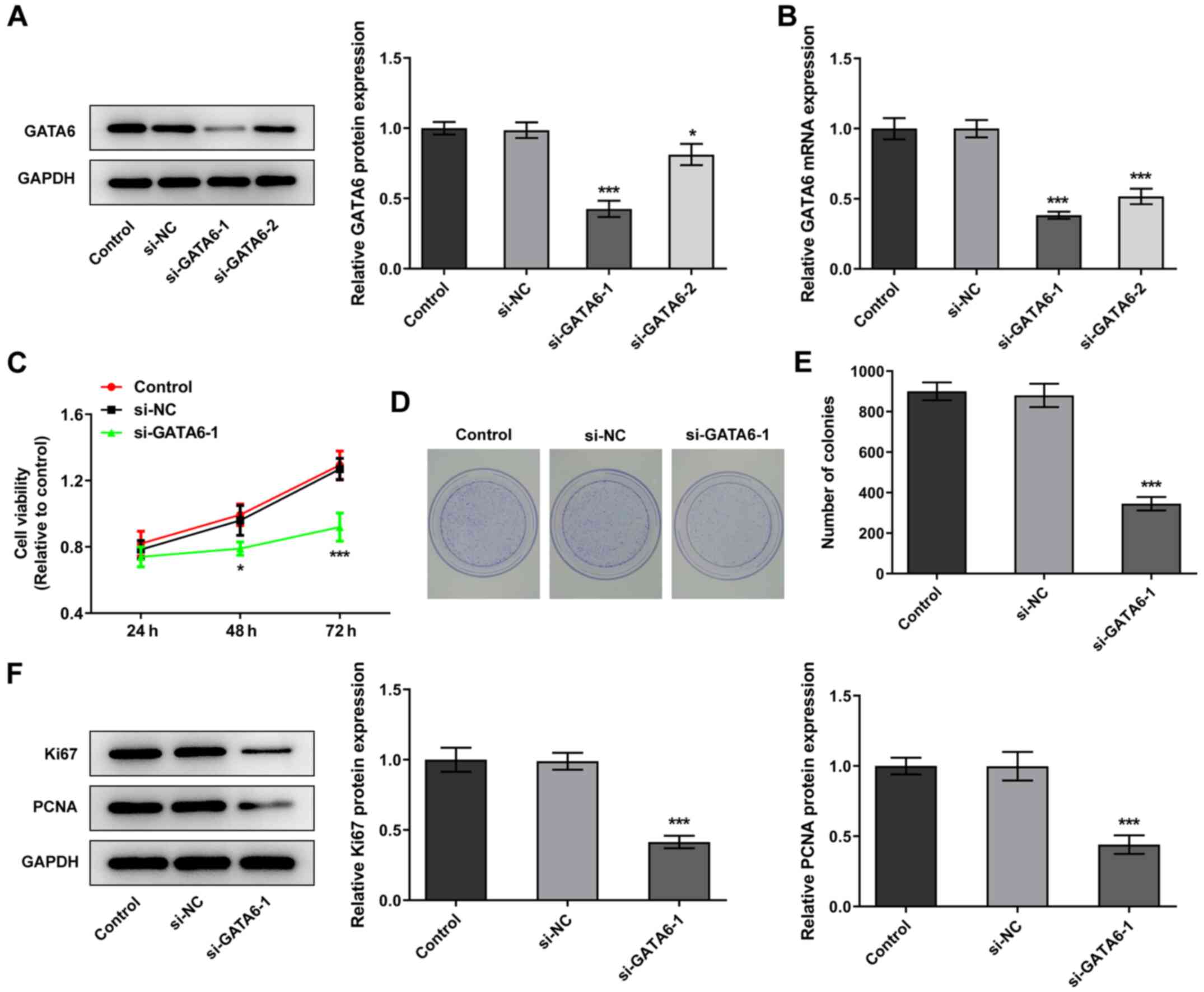
GATA6 silencing inhibits the
proliferation, invasion and migration of OSCC cells
Changes in cellular function were examined in Cal-27
cells following transfection with siRNA for the knockdown of GATA6.
si-GATA6-1 was selected for subsequent transfections due to a high
knockdown efficiency (Fig. 2A and
B). Notably, Cal-27 cells transfected with si-GATA6-1
demonstrated a suppressed ability to proliferate and form colonies
and downregulation of proliferation-related proteins, Ki67 and
PCNA, compared with the si-NC group (Fig. 2C-F). Furthermore, the levels of
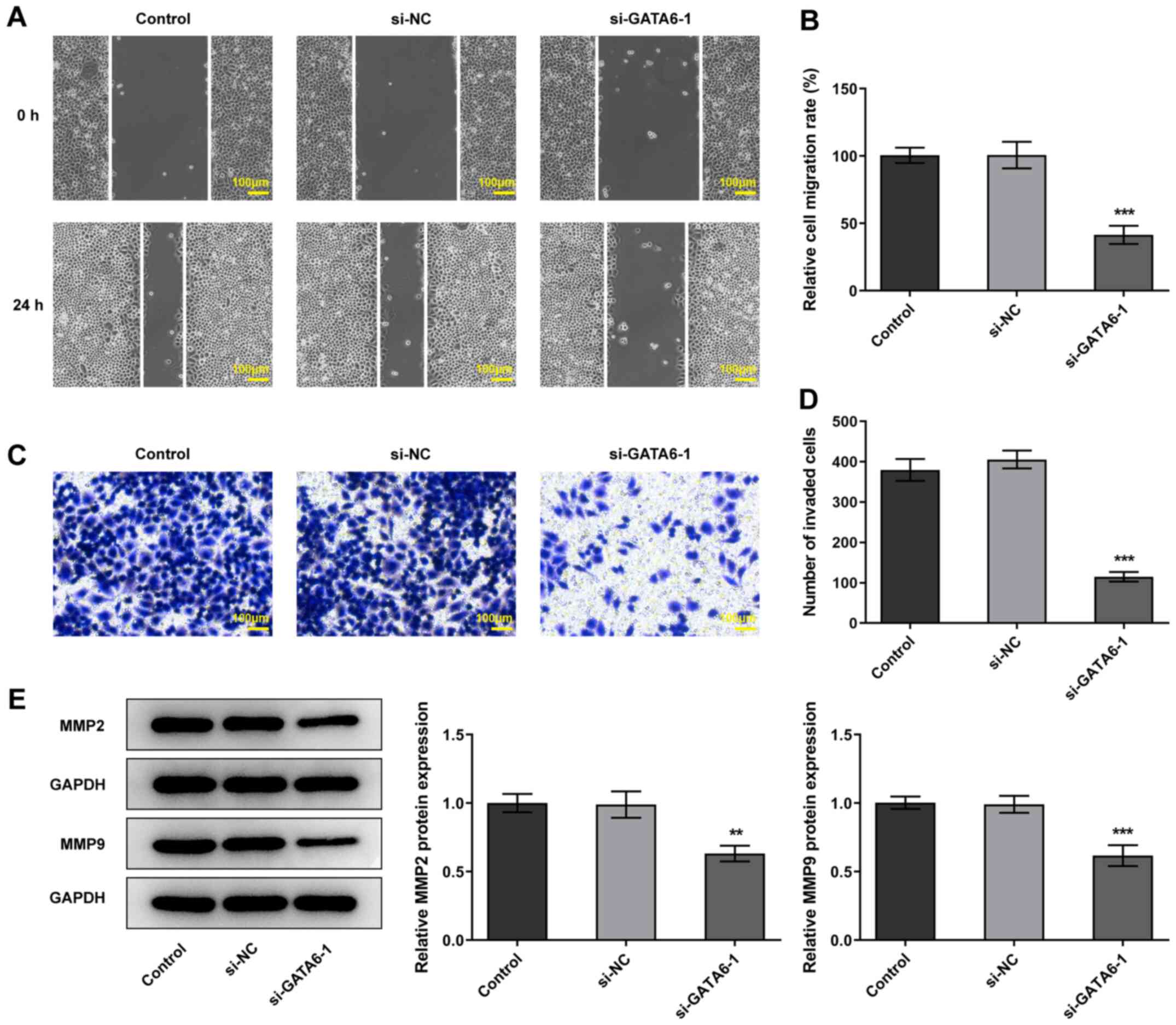
invasion and migration were reduced in Cal-27 cells transfected
with si-GATA6-1 and the expression levels of MMP2 and MMP9 were
also reduced, compared with the si-NC group (Fig. 3). Collectively, the results of the
present study suggested that GATA6 silencing inhibited the
proliferation, invasion and migration of OSCC cells.
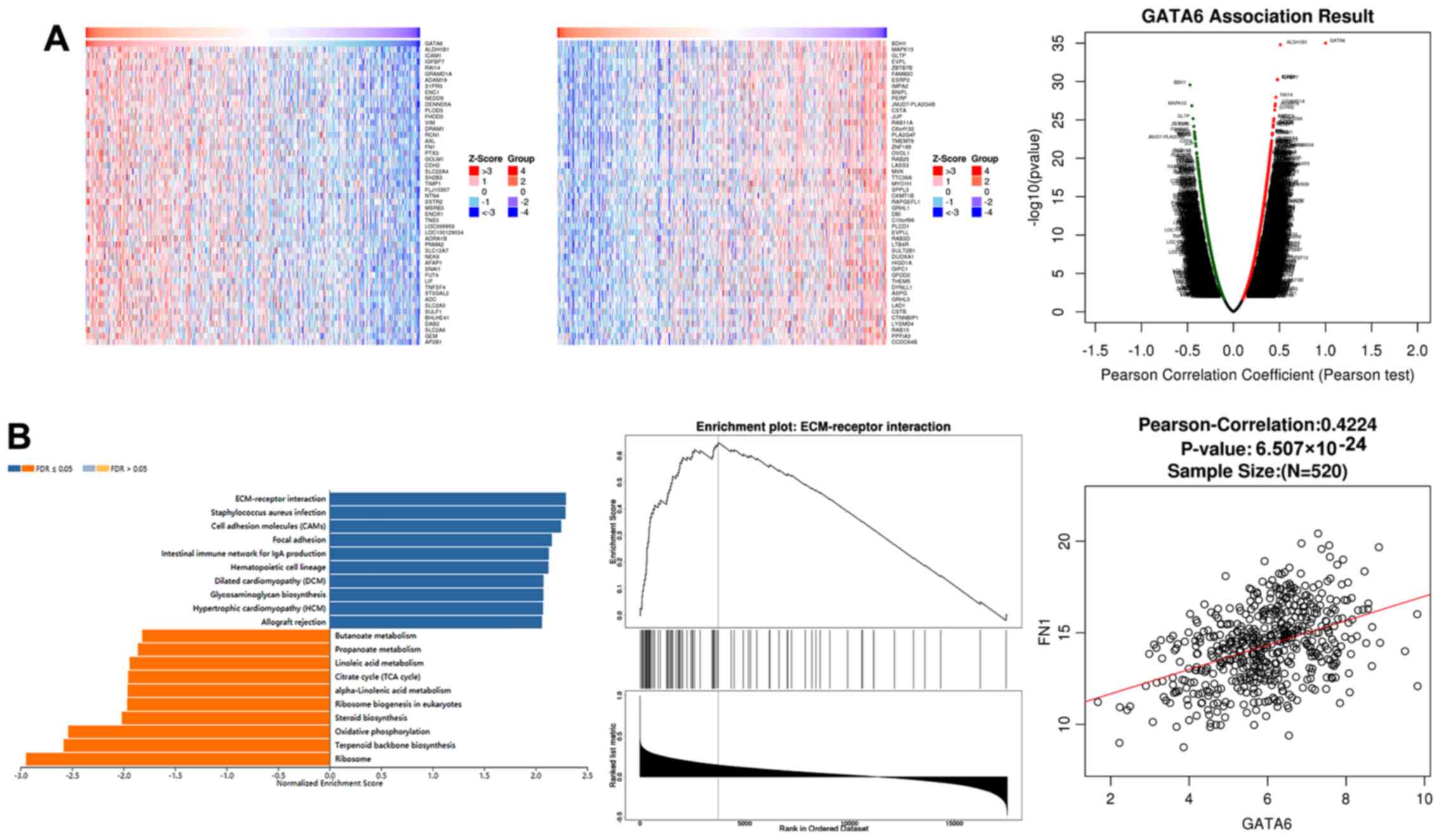
GATA6 transcriptionally regulates FN1
levels by binding to the FN1 promoter
To examine the transcriptional role of GATA6 in
OSCC, the LinkedOmics database was used to predict genes associated
with GATA6. A squamous cell carcinoma of the head and neck dataset
was used as a substitute as no OSCC dataset currently exists. Genes
positively associated with GATA6 were consistent with those
negatively associated with GATA6 (Fig. 4A). Furthermore, gene set
enrichment analysis (GSEA) revealed that GATA6 was positively
associated with extracellular matrix (ECM)-receptors and the
closest association was observed between FN1 and GATA6 (Fig. 4B). Subsequently, the expression of
FN1 in OSCC tissues and adjacent tissues was evaluated with TNMplot
data. It was found that FN1 level was markedly elevated in the
tumor group when compared to the normal group (Fig. 5A). The binding site of GATA6 on
the promoter region of FN1 was presented in Fig. 5B. Furthermore, dual-luciferase
reporter plasmids were activated following GATA6 overexpression
(Fig. 5C). CHIP assays performed
using Cal-27 cell extracts demonstrated a notable enrichment of the
FN1 promoter sequence through immunoprecipitation with an
anti-GATA6 antibody, compared with the control IgG antibody
(Fig. 5D). Additionally, western
blotting and RT-qPCR revealed that the expression levels of FN1
were markedly decreased following GATA6 silencing (Fig. 5E and F). Collectively, these
results demonstrated that GATA6 transcriptionally activates FN1
expression by binding to the FN1 promoter.
GATA6 silencing inhibits the
proliferation, invasion and migration of OSCC cells by regulating
FN1 expression
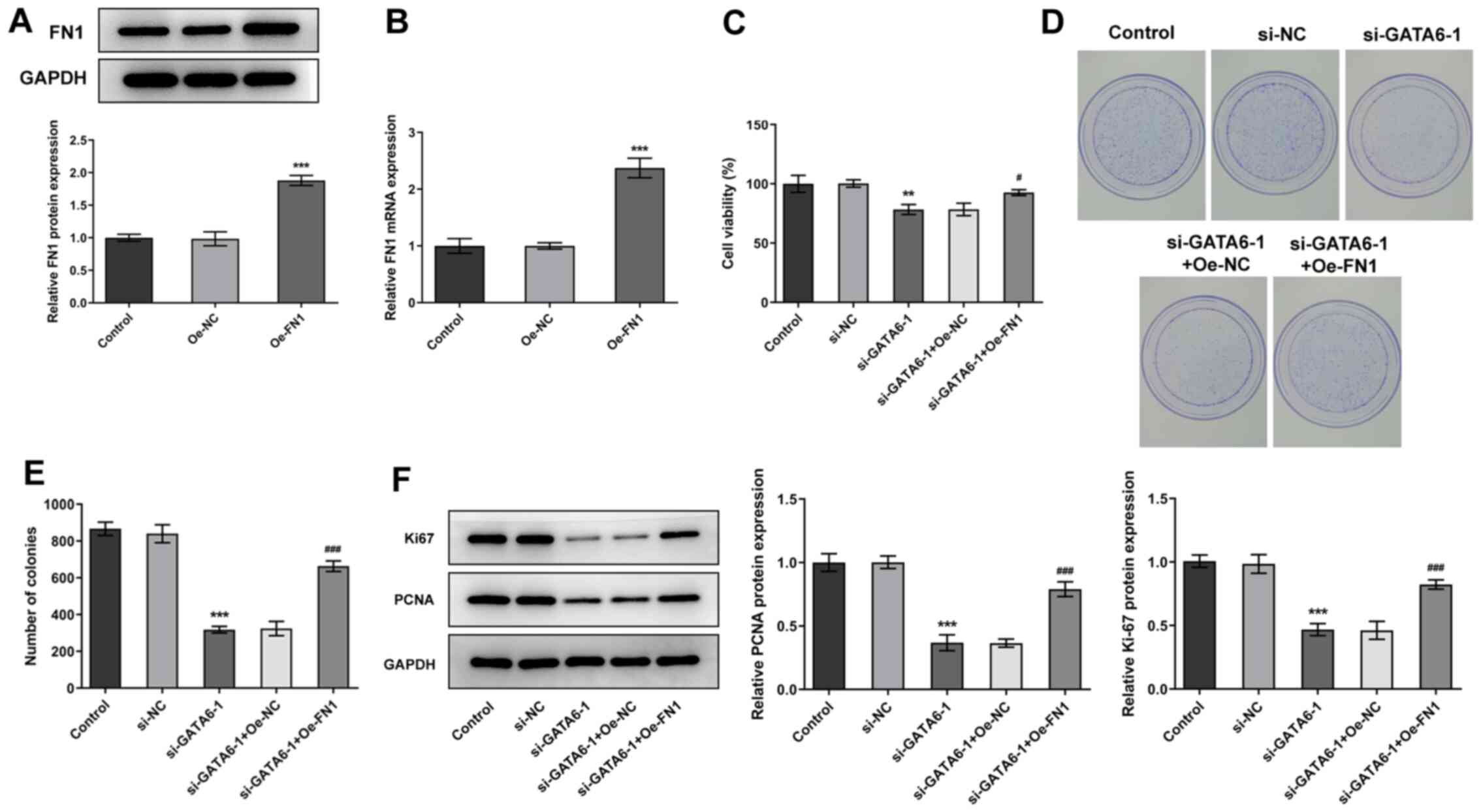
To further understand the mechanism underlying GATA6
in the progression of OSCC, FN1 was overexpressed in Cal-27 cells.
Transfection efficiency was indicated using western blotting and
RT-qPCR (Fig. 6A and B). Cal-27
cells transfected with both si-GATA6-1 and Oe-FN1 exhibited high
levels of cell viability, proliferation and colony formation and
high expression levels of Ki67 and PCNA, compared with Cal-27 cells
transfected with si-GATA6-1 + Oe-NC. These results highlighted that
Oe-FN1 reversed the effects of si-GATA6-1 on the proliferation of
Cal-27 cells (Fig. 6C-F).
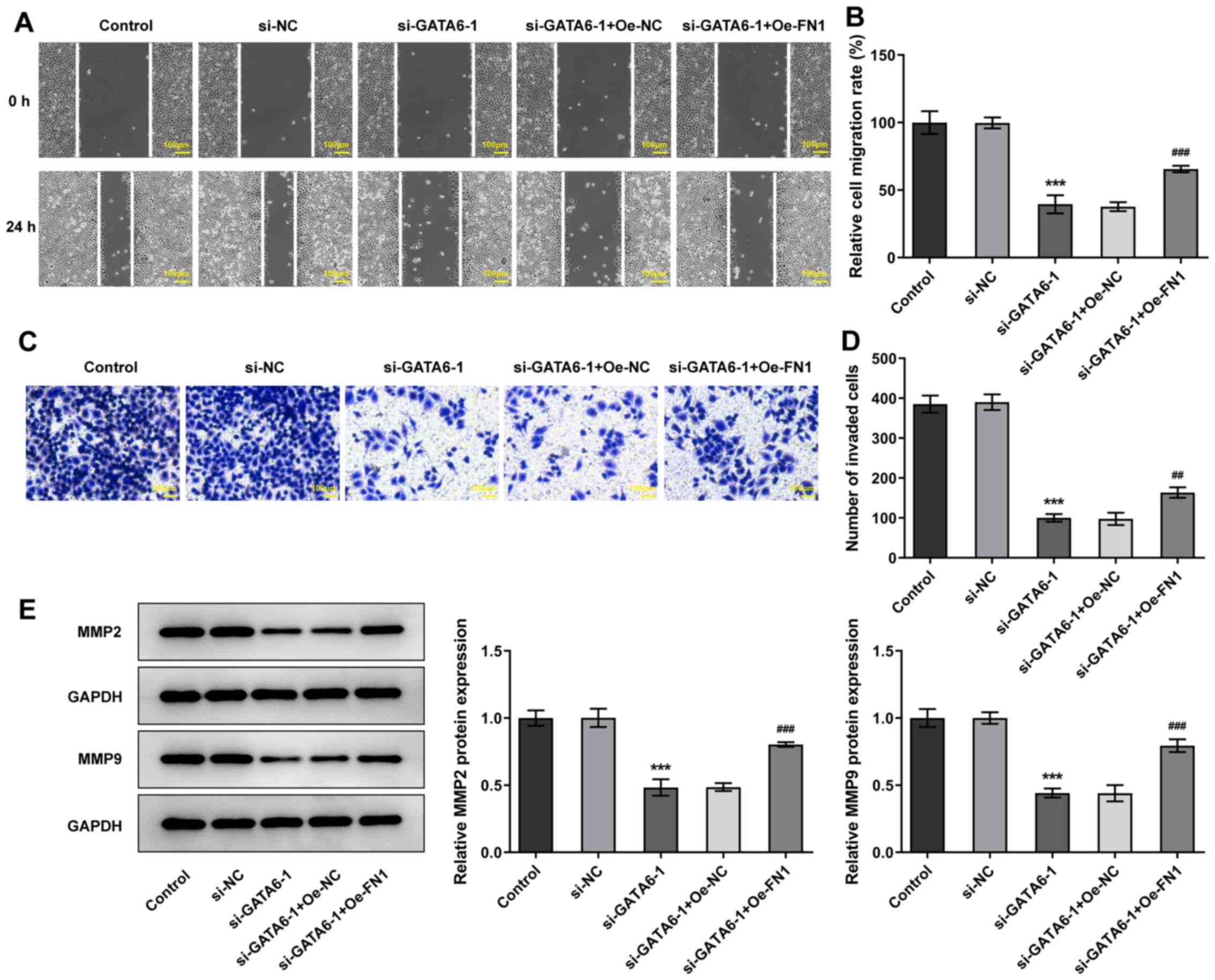
Additionally, transfection with Oe-FN1 abolished the suppressive
effects of si-GATA6-1 on the migration and invasion of Cal-27 cells
relative to the si-GATA6-1 + Oe-NC group (Fig. 7). The results of the present study
demonstrated that GATA6 silencing suppressed the proliferation,
invasion and migration of OSCC cells by regulating FN1.
Discussion
GATA6 serves an essential role in enhancing and
suppressing the development of tumors (20). The differentiation and
self-renewal of lung and colon cancer stem cells is promoted by
GATA6, thereby inducing tumorigenesis (11). The results of previous studies
also demonstrated the involvement of GATA6 in lung cancer (21) and the effectiveness of GATA6 in
the reprogramming of hepatocellular carcinoma cells (22). Deletion of GATA6 aided lymphatic
metastasis in bladder cancer, thus indicating that GATA6 serves a
key role in the lymphatic dissemination of bladder cancer (23). The expression levels of GATA6 vary
in a number of types of cancer, for example, expression levels are
markedly reduced in liver, ovarian and gastric cancers, compared
with notably high expression levels detected in breast cancer and
head and neck cancer (24).
Additionally, GATA6 expression is markedly elevated in oral
carcinoma cell lines (12). The
dysregulation of GATA6 in a number of types of cancer is a result
of varying GATA6 gene targets (23). Furthermore, regulation of miR-506
expression represses the expression levels of GATA6, thus
suppressing the development of OSCC, and overexpression of GATA6
can attenuate the inhibitory effects of miR-506 on cell viability,
colony formation, migration and invasion of OSCC cells (13). Additionally, downregulation of
GATA6 markedly decreases the expression levels of MMP2 and MMP9 in
a murine model of asthma (25).
In the present study, GATA6 expression levels were markedly
upregulated in OSCC cells. In addition, Cal-27 cells transfected
with si-GATA6-1 demonstrated decreased cell proliferation, colony
formation, migration and invasion, accompanied by downregulated
expression of Ki67, PCNA, MMP2 and MMP9.
In the present study, GSEA revealed that GATA6 was
positively associated with FN1. According to the JASPAR database
analyses, GATA6 has the ability to bind to the promoter of FN1.
FN1, a glycoprotein present in the extracellular matrix, serves a
key role in cellular adhesion, migration and tissue remodeling
(26). Elevated expression of FN1
was demonstrated in renal and colorectal cancer cells, indicating
that FN1 may serve a role in the progression of cancer to an
advanced stage (27,28). The results of a previous study
further demonstrated the role of FN1 in cell metastasis,
differentiation and adhesion in a number of types of cancer
(29). In addition, previous
studies have labeled FN1 as an oncogene in tumorigenesis and tumor
progression and a regulator in physiological processes (30–32). Notably, inhibition of FN1
suppresses the aggressiveness of hepatocellular carcinoma and
cervical cancer cells (33). The
results of a previous study demonstrate an association between
abnormally high expression levels of FN1 and the tumor size and
poor prognosis associated with gastric cancer (34). High expression levels of FN1 are
also demonstrated in lung tumor growth and survival and resistance
to therapy (35). Quantitative
proteomics analyses revealed FN1 as a candidate biomarker of
sporadic medullary thyroid cancer (36) and FN1 is also considered as a
potential biomarker in OSCC regarding the tongue, mouth floor and
edentulous ridge (15). Ji et
al (37) revealed that the
protein expression levels of Ki67 and PCNA is decreased by the
silencing of FN1 in human trophoblasts. FN1 knockdown represses
cell proliferation, invasion and migration of nasopharyngeal
carcinoma cells, coupled with downregulated expression of MMP2 and
MMP9 (26). Furthermore,
downregulation of FN1 inhibits the development of OSCC (16). Although the role of FN1 in cancer
progression has previously been identified, the association between
GATA6 and FN1 expression in Cal-27 cells remains to be elucidated,
to the best of the authors' knowledge. Thus, the results of the
present study demonstrated that silencing of GATA6 suppressed the
proliferation, migration and invasion of Cal-27 cells, which was
partly abolished following FN1 overexpression. Therefore, it was
hypothesized that silencing GATA6 suppressed the proliferation,
migration and invasion of Cal-27 cells by regulating FN1
expression.
In conclusion, the present study is the first, to
the best of the authors' knowledge, to demonstrate that GATA6
silencing inhibits the proliferation, migration and invasion of
OSCC cells. GATA6 was determined to have a direct regulatory effect
on the transcription of FN1 by binding to the FN1 promotor. The
results of the present study may contribute to further
understanding the pathogenesis of OSCC and provide potential
therapeutic targets for the clinical treatment of OSCC.
Nevertheless, there are also some limitations to the current study.
For instance, whether signaling pathways in the downstream of FN1
are involved in the development of OSCC remains to be elucidated,
and whether other potential regulatory mechanisms exist in the
regulation of the proliferation, migration and invasion of OSCC
cells also needs to be further explored.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ and GL designed the experimental study. JZ
analyzed the experimental data and wrote the manuscript. GL helped
to correct the manuscript. JZ carried out the experiments. JZ and
GL confirmed the authenticity of all the raw data. Both the authors
have read and approved the final manuscript for submission.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chou CH, Chou YE, Chuang CY, Yang SF and
Lin CW: Combined effect of genetic polymorphisms of AURKA and
environmental factors on oral cancer development in Taiwan. PLoS
One. 12:e01715832017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reyes-Gibby CC, Anderson KO, Merriman KW,
Todd KH, Shete SS and Hanna EY: Survival patterns in squamous cell
carcinoma of the head and neck: Pain as an independent prognostic
factor for survival. J Pain. 15:1015–1022. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee WH, Chen HM, Yang SF, Liang C, Peng
CY, Lin FM, Tsai LL, Wu BC, Hsin CH, Chuang CY, et al: Bacterial
alterations in salivary microbiota and their association in oral
cancer. Sci Rep. 7:165402017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dave K, Ali A and Magalhaes M: Increased
expression of PD-1 and PD-L1 in oral lesions progressing to oral
squamous cell carcinoma: A pilot study. Sci Rep. 10:97052020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haddad RI and Shin DM: Recent advances in
head and neck cancer. N Engl J Med. 359:1143–1154. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tremblay M, Sanchez-Ferras O and Bouchard
M: GATA transcription factors in development and disease.
Development. 145:dev1643842018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang X, Li D, Shen W, Shen X and Liu Y:
LncRNA NEAT1 promotes hypoxia-induced renal tubular epithelial
apoptosis through downregulating miR-27a-3p. J Cell Biochem.
120:16273–16282. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martinelli P, Carrillo-de Santa Pau E, Cox
T, Sainz B Jr, Dusetti N, Greenhalf W, Rinaldi L, Costello E,
Ghaneh P, Malats N, et al: GATA6 regulates EMT and tumour
dissemination, and is a marker of response to adjuvant chemotherapy
in pancreatic cancer. Gut. 66:1665–1676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng X, Jiang P, Chen J, Li J, Li D, He Y,
Jiang Y, Zhang Y, Xu S, Li X, et al: GATA6 promotes
epithelial-mesenchymal transition and metastasis through
MUC1/β-catenin pathway in cholangiocarcinoma. Cell Death Dis.
11:8602020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sulahian R, Casey F, Shen J, Qian ZR, Shin
H, Ogino S, Weir BA, Vazquez F, Liu XS, Hahn WC, et al: An
integrative analysis reveals functional targets of GATA6
transcriptional regulation in gastric cancer. Oncogene.
33:5637–5648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsuji S, Kawasaki Y, Furukawa S, Taniue K,
Hayashi T, Okuno M, Hiyoshi M, Kitayama J and Akiyama T: The
miR-363-GATA6-Lgr5 pathway is critical for colorectal
tumourigenesis. Nat Commun. 5:31502014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu CL, Guan WQ and Wang XY: The expression
of the GATA6 gene in oral carcinoma cell lines. World J Surg Oncol.
19:1532021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng L and Liu H: MicroRNA-506 suppresses
growth and metastasis of oral squamous cell carcinoma via targeting
GATA6. Int J Clin Exp Med. 8:1862–1870. 2015.PubMed/NCBI
|
|
14
|
Gao W, Liu Y, Qin R, Liu D and Feng Q:
Silence of fibronectin 1 increases cisplatin sensitivity of
non-small cell lung cancer cell line. Biochem Biophys Res Commun.
476:35–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yen CY, Huang CY, Hou MF, Yang YH, Chang
CH, Huang HW, Chen CH and Chang HW: Evaluating the performance of
fibronectin 1 (FN1), integrin α4β1 (ITGA4), syndecan-2 (SDC2), and
glycoprotein CD44 as the potential biomarkers of oral squamous cell
carcinoma (OSCC). Biomarkers. 18:63–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Z, Tao Q, Qiao B and Zhang L:
Silencing of LINC01116 suppresses the development of oral squamous
cell carcinoma by up-regulating microRNA-136 to inhibit FN1. Cancer
Manag Res. 11:6043–6059. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartha Á and Győrffy B: TNMplot.com: A web
tool for the comparison of gene expression in normal, tumor and
metastatic tissues. Int J Mol Sci. 22:26222021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Allison TF, Smith AJH, Anastassiadis K,
Sloane-Stanley J, Biga V, Stavish D, Hackland J, Sabri S, Langerman
J, Jones M, et al: Identification and single-cell functional
characterization of an endodermally biased pluripotent substate in
human embryonic stem cells. Stem Cell Reports. 10:1895–1907. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang G, Meng W, Huang X, Zhu W, Yin C,
Wang C, Fassan M, Yu Y, Kudo M, Xiao S, et al: miR-196b-5p-mediated
downregulation of TSPAN12 and GATA6 promotes tumor progression in
non-small cell lung cancer. Proc Natl Acad Sci USA. 117:4347–4357.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan HW, Leung CO, Chan KK, Ho DW, Leung
MS, Wong CM, Ng IO and Lo RC: Deregulated GATA6 modulates stem
cell-like properties and metabolic phenotype in hepatocellular
carcinoma. Int J Cancer. 145:1860–1873. 2019.PubMed/NCBI
|
|
23
|
Wang C, Liu Q, Huang M, Zhou Q, Zhang X,
Zhang J, Xie R, Yu Y, Chen S, Fan J, et al: Loss of GATA6
expression promotes lymphatic metastasis in bladder cancer. FASEB
J. 34:5754–5766. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheung WK, Zhao M, Liu Z, Stevens LE, Cao
PD, Fang JE, Westbrook TF and Nguyen DX: Control of alveolar
differentiation by the lineage transcription factors GATA6 and HOPX
inhibits lung adenocarcinoma metastasis. Cancer Cell. 23:725–738.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang P, Shi HY, Wu XM, Zhang YH, Zhong YJ,
Deng WJ, Zhang YP and Xie M: Targeted inhibition of GATA-6
attenuates airway inflammation and remodeling by regulating
caveolin-1 through TLR2/MyD88/NF-κB in murine model of asthma. Mol
Immunol. 75:144–150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding Y, Pan Y, Liu S, Jiang F and Jiao J:
Elevation of MiR-9-3p suppresses the epithelial-mesenchymal
transition of nasopharyngeal carcinoma cells via down-regulating
FN1, ITGB1 and ITGAV. Cancer Biol Ther. 18:414–424. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Steffens S, Schrader AJ, Vetter G, Eggers
H, Blasig H, Becker J, Kuczyk MA and Serth J: Fibronectin 1 protein
expression in clear cell renal cell carcinoma. Oncol Lett.
3:787–790. 2012.PubMed/NCBI
|
|
28
|
Wu J, Wang Y, Xu X, Cao H, Sahengbieke S,
Sheng H, Huang Q and Lai M: Transcriptional activation of FN1 and
IL11 by HMGA2 promotes the malignant behavior of colorectal cancer.
Carcinogenesis. 37:511–521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi H, Dong Z and Gao H: LncRNA TUG1
protects against cardiomyocyte ischaemia reperfusion injury by
inhibiting HMGB1. Artif Cells Nanomed Biotechnol. 47:3511–3516.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai X, Liu C, Zhang TN, Zhu YW, Dong X and
Xue P: Down-regulation of FN1 inhibits colorectal carcinogenesis by
suppressing proliferation, migration, and invasion. J Cell Biochem.
119:4717–4728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie Y, Liu C, Qin Y, Chen J and Fang J:
Knockdown of IRE1a suppresses metastatic potential of colon cancer
cells through inhibiting FN1-Src/FAK-GTPases signaling. Int J
Biochem Cell Biol. 114:1055722019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song G, Liu K, Yang X, Mu B, Yang J, He L,
Hu X, Li Q, Zhao Y, Cai X, et al: SATB1 plays an oncogenic role in
esophageal cancer by up-regulation of FN1 and PDGFRB. Oncotarget.
8:17771–17784. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu X, Liu Z, Zhou L, Xie H, Cheng J, Ling
Q, Wang J, Guo H, Wei X and Zheng S: Characterization of
genome-wide TFCP2 targets in hepatocellular carcinoma: Implication
of targets FN1 and TJP1 in metastasis. J Exp Clin Cancer Res.
34:62015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Zhao C, Ye Y, Wang Z, He Y, Li Y
and Mao H: High expression of fibronectin 1 indicates poor
prognosis in gastric cancer. Oncol Lett. 19:93–102. 2020.PubMed/NCBI
|
|
35
|
Han S, Khuri FR and Roman J: Fibronectin
stimulates non-small cell lung carcinoma cell growth through
activation of Akt/mammalian target of rapamycin/S6 kinase and
inactivation of LKB1/AMP-activated protein kinase signal pathways.
Cancer Res. 66:315–323. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhan S, Li J, Wang T and Ge W:
Quantitative proteomics analysis of sporadic medullary thyroid
cancer reveals FN1 as a potential novel candidate prognostic
biomarker. Oncologist. 23:1415–1425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ji J, Chen L, Zhuang Y, Han Y, Tang W and
Xia F: Fibronectin 1 inhibits the apoptosis of human trophoblasts
by activating the PI3K/Akt signaling pathway. Int J Mol Med.
46:1908–1922. 2020.PubMed/NCBI
|