Introduction
Chronic stress is a non-specific complex
psychosomatic process that is a key risk factor for depression
(1), anxiety (2), drug abuse (3), cardiovascular disease (4), ulcers (5) and cancer (6). Therefore, investigating the positive
or negative responses to stressors is important to reveal the
pathogenesis of stress-associated disease. The body's response to
stressors depends on the nature, intensity and duration of the
stressor, forming an ‘inverted U’-shaped continuous quantitative
effect curve (7). In long-term
stress, early stress causes the body to produce compensatory
responses, such as increased alertness, sensitivity; as duration of
stress increases, the body systems act in concert to maintain
homeostasis, thus actively adapting to the stressor. The cumulative
effect of further stress leads to a disturbance of homeostasis in
the body, resulting in pathophysiological damage (1).
The hippocampus is responsible for learning, memory,
cognition and emotion; it is also one of the key brain areas that
mediate stress reactions (8).
Prolonged stress results in decreased hippocampal nerve cell
plasticity, disequilibrium between hippocampal apoptosis and
regeneration, leading to nerve cell atrophy and loss and causing
structural and functional damage in the hippocampus (9,10).
Studies have investigated the effects of different stress patterns
on gene expression in the hippocampus (11,12), but, to the best of our knowledge,
there are no reports on dynamic changes in gene expression in the
hippocampus during chronic stress.
To determine the effects of stress on hippocampal
structure, the present study investigated transcriptomic changes in
the rat hippocampus during chronic unpredictable mild stress (CUMS)
using mRNA sequencing (seq) technology. The CUMS rodent model uses
both physiologically and psychologically stressful stimulations to
mimic adverse stress from negative life events and induces
decreased locomotion and anhedonia, similar to symptoms of
depression in humans, which makes it a widely used animal model to
study the molecular basis of stress in the brain (13). The present study investigated gene
expression in the hippocampus of rats in the early, middle and late
stages of CUMS to clarify continuous gene expression changes in the
hippocampus in CUMS and reveal the pathophysiological mechanism of
stress injury.
Materials and methods
Animals
A total of 36 male Sprague Dawley (SD) rats (age, 2
months; weight, 180–220 g) were obtained from SBF (Beijing)
Biotechnology Co., Ltd. All animals were housed in groups of three
or four in plastic cages and maintained under standard laboratory
conditions (12/12-h light/dark cycle, 22±2°C, relative humidity
40–70%, ad libitum access to food and water).
CUMS procedure
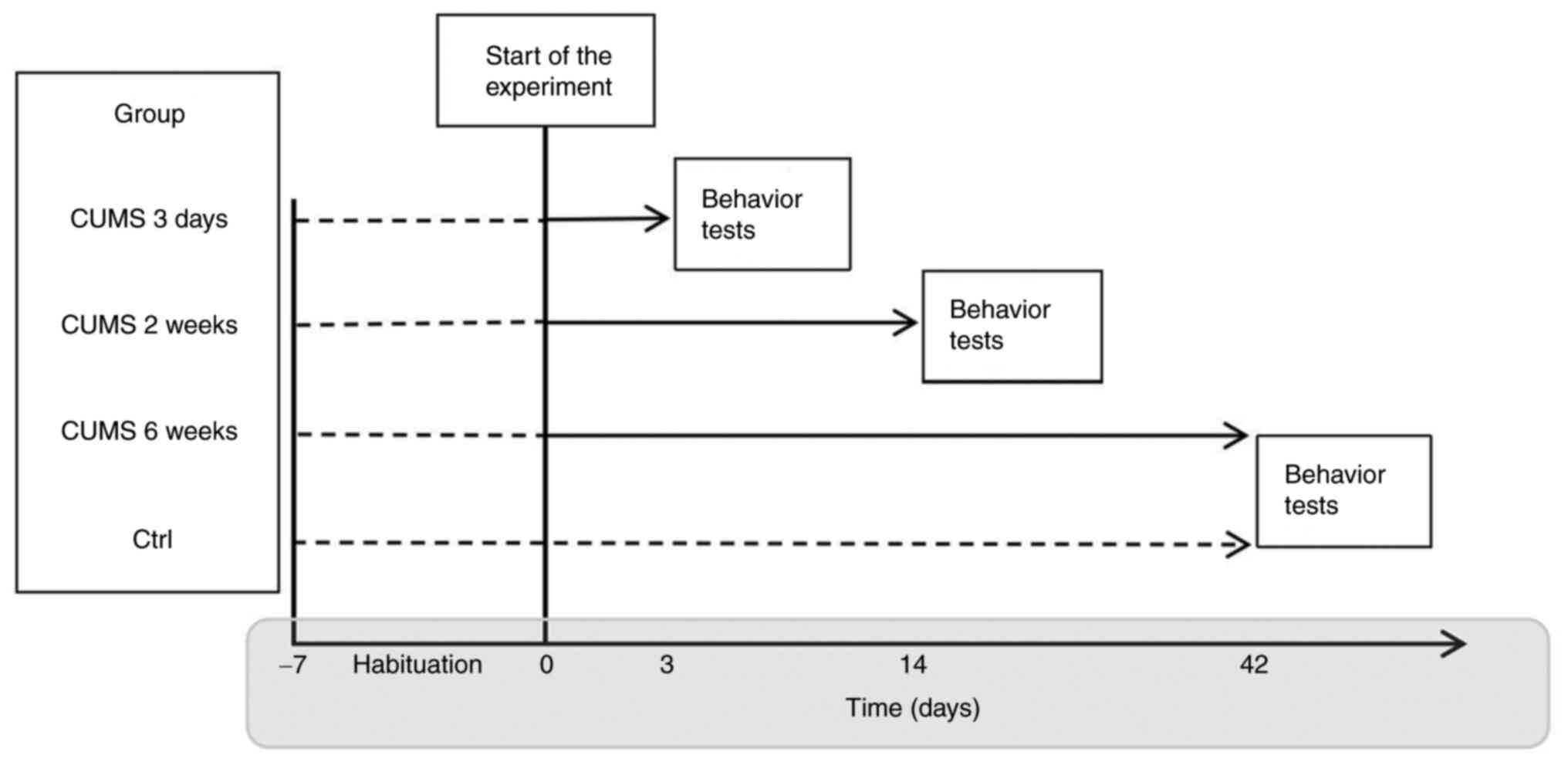
In brief, SD male rats were randomly divided into
four groups (n=9 per group) as follows: i) Control and CUMS for ii)
3 days and iii) 2and iv) 6 weeks (Fig. 1). The CUMS model was established
as previously reported (14): i)
Overnight light exposure, ii) darkness for 12 h, iii) wet bedding
for 12 h, iv) removal of bedding for 12 h, v) deprivation of food
for 12 h, vi) deprivation of water for 12 h, vii) soaking for 3 min
in cold water (4°C), viii) physical restraint for 4 h, ix) cage
vibration (70 rpm) for 2 h and x) electrical shock to the feet for
5 min (0.2 mA). Stressors were randomly scheduled to produce an
unexpected mild stress effect. A combination of two different
stressors was applied separately each day and the same stressor was
not repeated within 3 days. Control animals were left uninterrupted
except for regular cage cleaning. The effectiveness of the applied
stress procedure for induction of the stress response was confirmed
by cognitive behavior tests.
Open field test
The test was performed in a black wooden box
measuring 100×100×45 cm. The floor was divided into 25 equal
squares (20×20 cm). At the beginning of the experiment, rats were
placed individually in the center of the open field and ambulation
(number of squares crossed) and head-rise frequency were observed
for 5 min using an ANY-maze behavior analysis system (Stoelting
Co.). To ensure that the test results were free from any previous
residual effect, the inside of the box was cleaned before each
test. An open field test score was calculated as the sum of
ambulation and immobility frequency.
Novel object recognition test
The experimental device was a black wooden box
(60×60×45 cm). The test comprised three steps: Adaptation, training
and testing. On the day before the experiment, rats were allowed to
move freely in the experimental setup (without objects) for 10 min.
During the training trial, two identical objects were placed on the
left and right ends of a sidewall, and the rats were placed in the
field with their back facing the two objects. The animal was placed
in the center of the open field and allowed to explore freely for
10 min. During the test, one of the two objects was replaced with
another object of different color and shape and the rat was
returned to the same box for 10 min. The ANY-maze behavior analysis
system recorded the time rats spent exploring the new and old
object, denoted as Tnew and Told,
respectively. The cognitive index (CI) was expressed as follows:
CI=(Tnew-Told)/(Tnew +
Told).
Morris water maze
The experimental device was a black circular plastic
tank (diameter, 160 cm; depth, 75 cm) containing water at 22±2°C.
The curtains around the tank were labeled with stickers of
different shapes and colors serving as spatial reference cues. Rats
were first required to complete 5 half-days of training at a
frequency of once a day for 3 min each. During training, rats were
placed into the maze in one of four quadrants facing the curtain
and were trained to find a hidden platform submerged 2 cm beneath
the surface of the water. If rats could not find the platform
within 3 min, they were guided to the platform and kept there for
10 sec. During the test phase, the platform was removed and rats
were placed into the water on the opposite quadrant. Each rat was
allowed to swim in the water for 3 min, during which, the first
time the rats found the original platform location and the number
of platform passes within 3 min were recorded.
Sample preparation
Following behavioral observation, rats were
decapitated following anesthesia with sodium pentobarbital (1%, 50
mg/kg, intraperitoneal). The whole brain was removed and washed
with cold 0.9% saline. The hippocampus was dissected on an ice
plate on a super-clean bench. The dissected hippocampi were placed
into freezing EP tubes, frozen in liquid nitrogen and transferred
into a −80°C low-temperature refrigerator for storage and use. RNA
was extracted from hippocampal tissue using TRI reagent
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. Three hippocampal tissue samples were pooled as a single
sample for the total transcript array analysis to increase the
quantity of extracted RNA. RNA purity and quantification were
evaluated using the NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.). RNA integrity was assessed using the Agilent
2100 Bioanalyzer (Agilent Technologies, Inc.). All samples selected
for further analysis were of high quality (RNA integrity number
>9, 260/280-2.1). Then the libraries were constructed using
VAHTS Universal V6 RNA-seq Library Prep Kit for Illumina (catalog
no. NR604-02; Nanjing Vazyme Biotech Co.) according to the
manufacturer's instructions. The concentration of the library used
for sequencing was ≥3.75 fmol/µl and was measured using a Qubit
ssDNA Assay Kit (catalog no. Q10212) and Qubit Fluorometer (catalog
no. Q33238) (both; Invitrogen; Thermo Fisher Scientific, Inc.).
Transcriptome sequencing and analysis were performed by Shanghai Oe
Biotech Co., Ltd.
RNA-seq and differentially expressed
gene (DEG) analysis
The libraries were sequenced on MGI DNBSEQ-T7
platform using DNBSEQ-T7RS high-throughput sequencing kit
(1000016102, MGI), and 150 bp paired-end reads were generated. A
total of ~49.5 million raw reads were generated for each sample.
Raw data (raw reads) of FASTQ format were processed using
Trimmomatic (version 0.36) (15)
and low-quality reads (adaptor and data of Q30 <85%) were
removed to obtain the clean reads. Then ~49 million clean reads
were retained for each sample for subsequent analysis.
The clean reads were mapped to the rat genome
(mRatBN7.2) using Hierarchical Indexing for Spliced Alignment of
Transcripts 2 (version 2.2.1.0) (16). Fragments per kilobase of
transcript per million mapped reads (FPKM) of each gene was
calculated using Cufflinks (version 2.2.1) (17) and the read count of each gene was
obtained by HTSeqcount (version 0.9.1) (18). Differential expression analysis
was performed using DESeq (2012) R package (19). P<0.05 and fold-change >2 or
<0.5 was set as the threshold for significantly differential
expression.
To identify the functional classes associated with
DEGs, the online analysis tool Database for Annotation,
Visualization, and Integrated Discovery 6.8
(david.ncifcrf.gov/summary.jsp) and Gene Ontology (GO) database
were used to classify the biological process, molecular function,
and cellular components of DEGs. GO annotations were obtained from
GO database version 2.2 (20).
Hypergeometric distributions were used to detect over- or
under-represented biological process terms in the studied set
compared with the population set. Terms with P<0.05 were
considered to be significant.
Confirmation of gene expression levels
using reverse transcription-quantitative (RT-q)PCR
All selected hippocampal RNA specimens were chosen
from the animals used in the mRNA-seq study and were analyzed
individually. Meeting three strict conditions [P<0.05,
log2(fold-change)>1 and 2<average FPKM<50],
seven genes were selected to verify the accuracy and reliability of
mRNA-seq data. These genes were tested using SYBR-Green-based
RT-qPCR in 96-well plates on a LightCycler 96 thermocycler (Roche
Diagnostics GmbH). Primers (Table
I) were designed using Primer-BLAST tool (National Center for
Biotechnology Information) with the Rattus norvegicus RefSeq
database (reference genome mRatBN7.2). The primers produced
amplicons spanning exon-exon junctions and including all known
alternatively spliced mRNA variants. A total of 1,000 ng total RNA
from each sample was reverse-transcribed to cDNA according to the
manufacturer's instructions (ABScript II RT Mix for qPCR, ABclonal
Biotech Co., Ltd.). RT-qPCR was run using TB Green Premix Ex Taq II
(Takara Bio, Inc.) according to the manufacturer's instructions.
The volume of the PCR reaction system, cDNA sample, forward and
reverse prime, and TB Green II were 10.0, 1.0, 0.2, 0.2 and 5.0 µl,
respectively. The thermocycling conditions were as follows: Initial
denaturation for 2 min at 95°C followed by 50 cycles of 95°C for 10
sec, 60°C for 10 sec and 72°C for 15 sec. All experiments were
performed in triplicate. Raw cycle threshold values were calculated
on LightCycler. Samples were analyzed by the ΔΔCq method. ΔCq
values represent normalized target genes levels with respect to the
internal control. Normalization was based on a single reference
gene (β-actin). ΔΔCq values were calculated as the ΔCq of each test
sample (CUMS) minus the mean ΔCq of the calibrator samples (ctrl)
for each target gene. The fold change was calculated using the
equation 2−ΔΔCq (21).
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) | Annealing
temperature, °C | Product size,
bp |
|---|
| Slc6a20 | CTA CAT CCT CAC GGG
AAC GC | GTG GCT GAC TTC GGT
CTT TG | 60 | 231 |
| Bmp7 | CAT GGA CCC CAG AAC
AAG CA | CTT TGG AGT CTT GGA
GCG GT | 60 | 123 |
| Ptgds | GTG CAG CCC AAC TTT
CAA CA | CCG GAA CCA GCT TGA
ATT GG | 60 | 81 |
| Hbb-b1 | TTG GCA GCC TCA GTG
AAC TCC | GAC AGA AGC TCT CTT
GGG AAC A | 60 | 248 |
| Pdk4 | AAT GTG GTC CCT ACG
ATG GC | ATC GCA GTT GGG GTC
GAT AC | 60 | 213 |
| Sccpdh | CGA AAC CAG ATG AAC
GGC AC | TGC CCT TAT CGC CAA
AAC CA | 60 | 127 |
| Npas4 | GTG GCA GCA CTA CCT
GGA TT | GTT TGT TGC CTG CAC
TCT GG | 60 | 265 |
| Actb | CTT CCT GGG TAT GGA
ATC CT | TCT TTA CGG ATG TCA
ACG TC | 60 | 80 |
Statistical analysis
The results of behavioral tests were compared using
Shapiro-Wilk test. The expression of each mRNA in the hippocampus
of CUMS and control groups was compared using Mann-Whitney U test
as data did not show a normal distribution. Nine independent
experiments were carried out in each group in behavioral tests,
while three independent experiments were conducted in each group in
RT-qPCR experiments. All data are presented as the mean ± standard
deviation. GraphPad Prism 8 software (ver. 8.4.2; GraphPad
Software, Inc.) was used for statistical analysis and Spearman's
correlation was used for the correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Chronic stress induced time-dependent
cognitive decline in rats
Three different behavioral tests were used to
evaluate cognitive function in rats. The behavioral results showed
that the open field score, object recognition CI and number of
platform passes decreased with longer stress time, while the escape
latency in the Morris water maze test increased gradually. The
difference (P<0.05) first occurred in the open-field test at 2
weeks of CUMS. All behavioral performance associated with cognitive
function decreased significantly (P<0.01) by week 6 of CUMS,
suggesting that chronic stress induced cognitive dysfunction in
rats (Fig. S1).
Number of DEGs decreases with the
increase in stress time
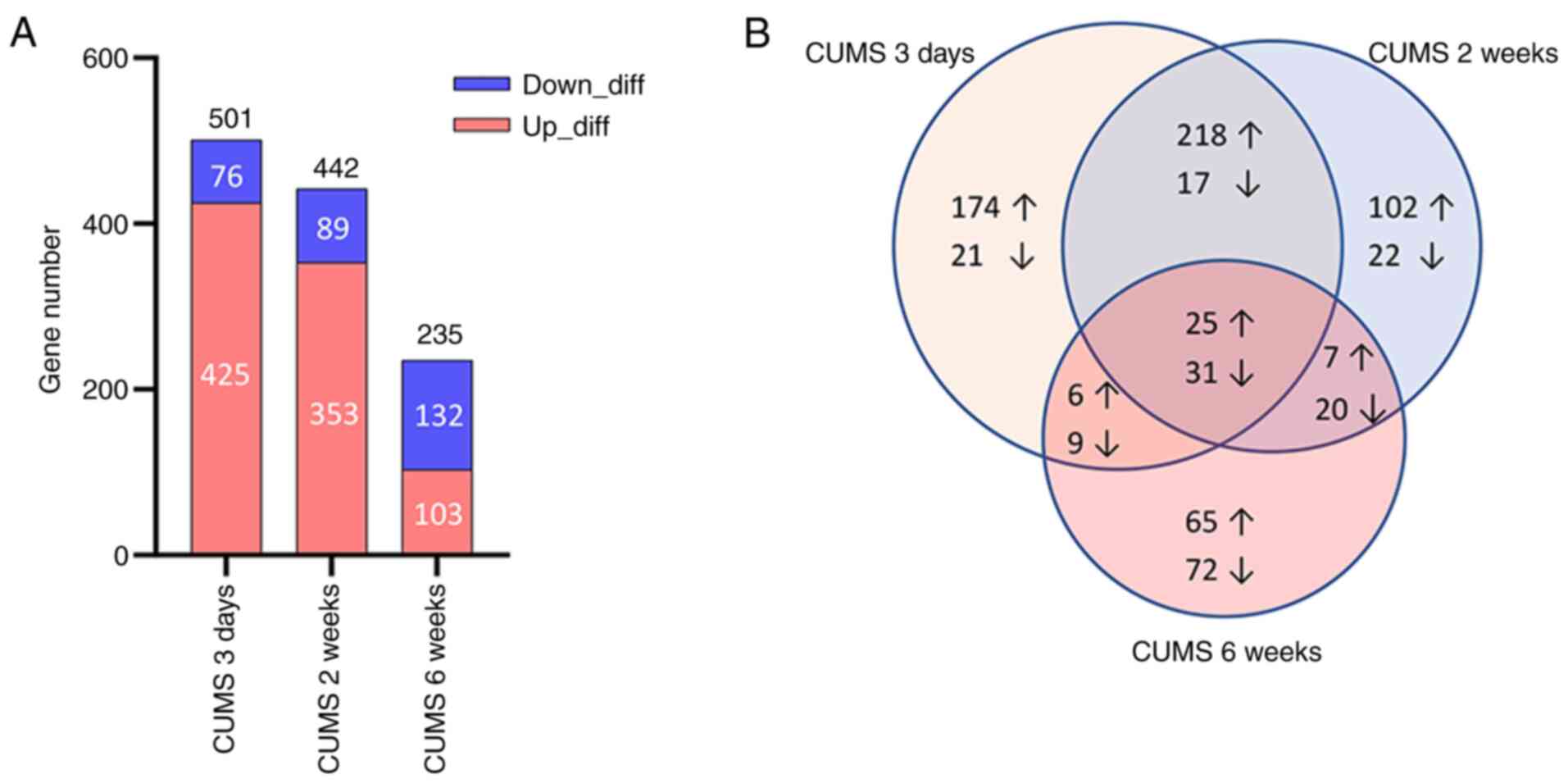
Compared with the control, 501 DEGs were identified
in CUMS (3 days) rats, of which 425 were up- and 76 were
downregulated. A total of 442 DEGs in the CUMS (2 weeks) group were
identified, of which 353 were up- and 89 were downregulated. A
total of 235 DEGs in the CUMS (6 weeks) group were identified, of
which 103 were up- and 132 were downregulated (Fig. 2; Table SI). As the duration of stress
increased, the total number of DEGs decreased; this was accompanied
by a decrease in up- and an increase in downregulated genes.
GO function analysis of DEGs suggests
a functional change from adaptation to damage in the early and late
stages of chronic stress
DEGs were divided into up- and downregulated groups
for GO enrichment analysis. GO terms were divided into three
categories: Biological processes, cellular components and molecular
functions. The top 10 terms with the most significant P-values
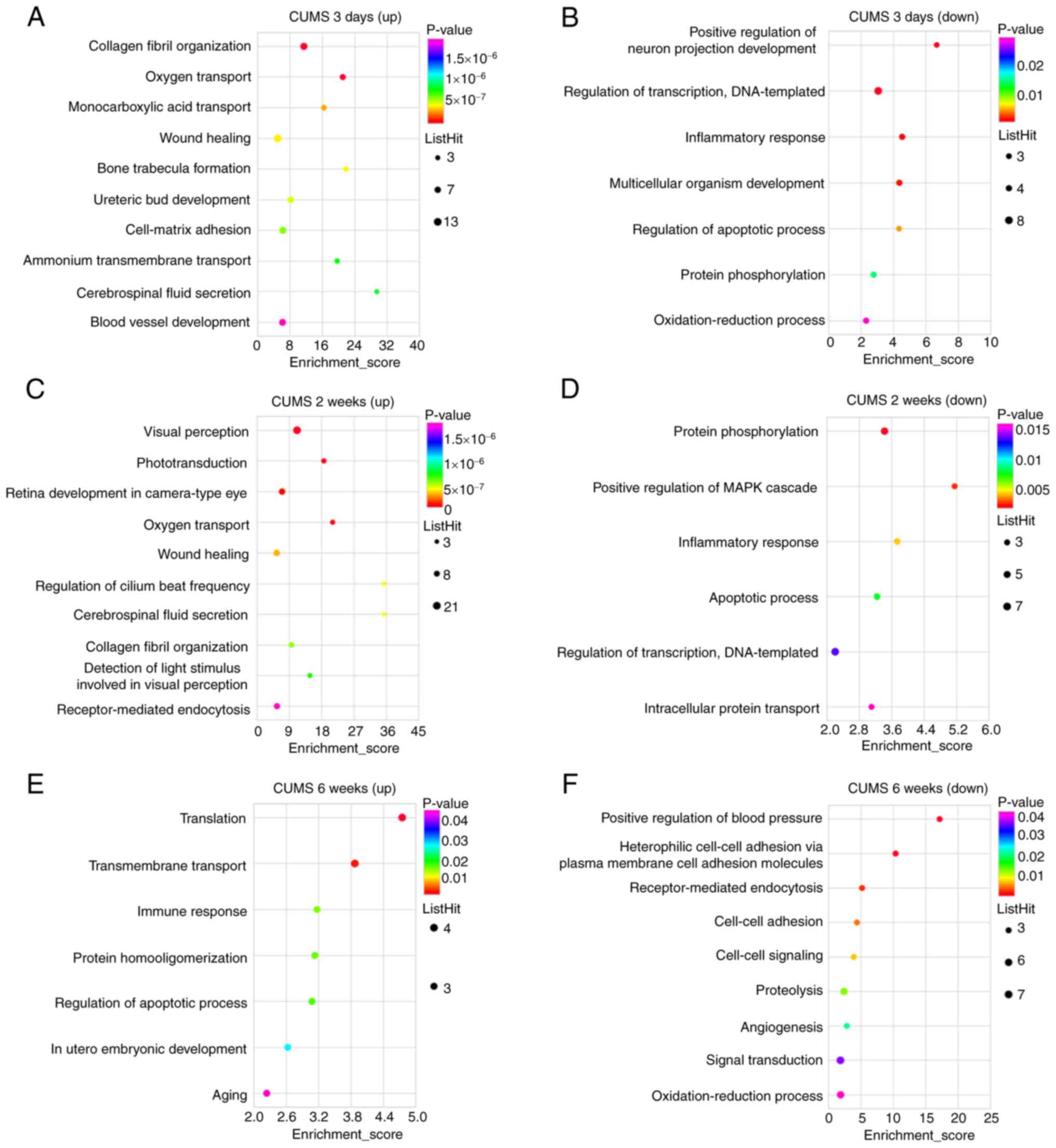
(<0.05) were selected from each category. Significant biological
processes of upregulated DEGs of the hippocampus in CUMS (3 days)
rats included ‘collagen fibril organization’, ‘oxygen transport’,
‘wound healing’, ‘cell-matrix adhesion’, ‘ammonium transmembrane
transport’, ‘cerebrospinal fluid secretion’ and ‘blood vessel
development’ (Fig. 3A), while
those of downregulated DEGs involved ‘positive regulation of neuron
projection development’, ‘inflammatory response’, ‘multicellular
organism development’, ‘regulation of apoptotic process’, ‘protein
phosphorylation’ and ‘oxidation-reduction process’ (Fig. 3B). Significant biological
processes of upregulated DEGs in CUMS (2 weeks) rats included
‘visual perception’, ‘phototransduction’, ‘retina development in
camera-type eye’, ‘oxygen transport’, ‘wound healing’, ‘regulation
of cilium beat frequency’, ‘cerebrospinal fluid secretion’,
‘collagen fibril organization’, ‘detection of light stimuli
involved in visual perception’ and ‘receptor-mediated endocytosis’
(Fig. 3C), while those of the
downregulated DEGs in this group involved ‘protein
phosphorylation’, ‘positive regulation of MAPK cascade’,
‘inflammatory response’, ‘apoptotic process’ and ‘intracellular
protein transport’ (Fig. 3D).
Certain biological processes were found after both 3 days and 2
weeks of CUMS. Significant biological processes of upregulated DEGs
in CUMS (6 weeks) rats involved ‘translation’, ‘transmembrane
transport’, ‘immune response’, ‘protein homooligomerization’,
‘regulation of apoptotic process’ and ‘aging’ (Fig. 3E), while those of the
downregulated DEGs in this group included ‘positive regulation of
blood pressure’, ‘receptor-mediated endocytosis’, ‘cell-cell
adhesion’, ‘cell-cell signaling’, ‘proteolysis’, ‘angiogenesis’,
‘signal transduction’ and ‘oxidation-reduction process’ (Fig. 3F). Significant cellular components
and molecular functions of DEGs are shown in the supplementary
materials (Figs. S2 and S3). ‘Integral component of the plasma
membrane’ was involved in the cellular component of downregulated
DEGs in CUMS (3 days) rats and upregulated DEGs in CUMS (6 weeks)
rats. ‘Protein binding’, ‘identical protein binding’, ‘metal ion
binding’ and ‘ATP binding’ were involved in the molecular functions
of downregulated DEGs in CUMS (3 days) rats and upregulated DEGs in
CUMS (6 weeks) rats. Furthermore, significant cellular components
of downregulated DEGs in CUMS (6 weeks) rats involved ‘synapse’,
‘dendrite’ and ‘axon’.
Results of RT-qPCR coincide with the
changes in differential gene identified by mRNA-seq
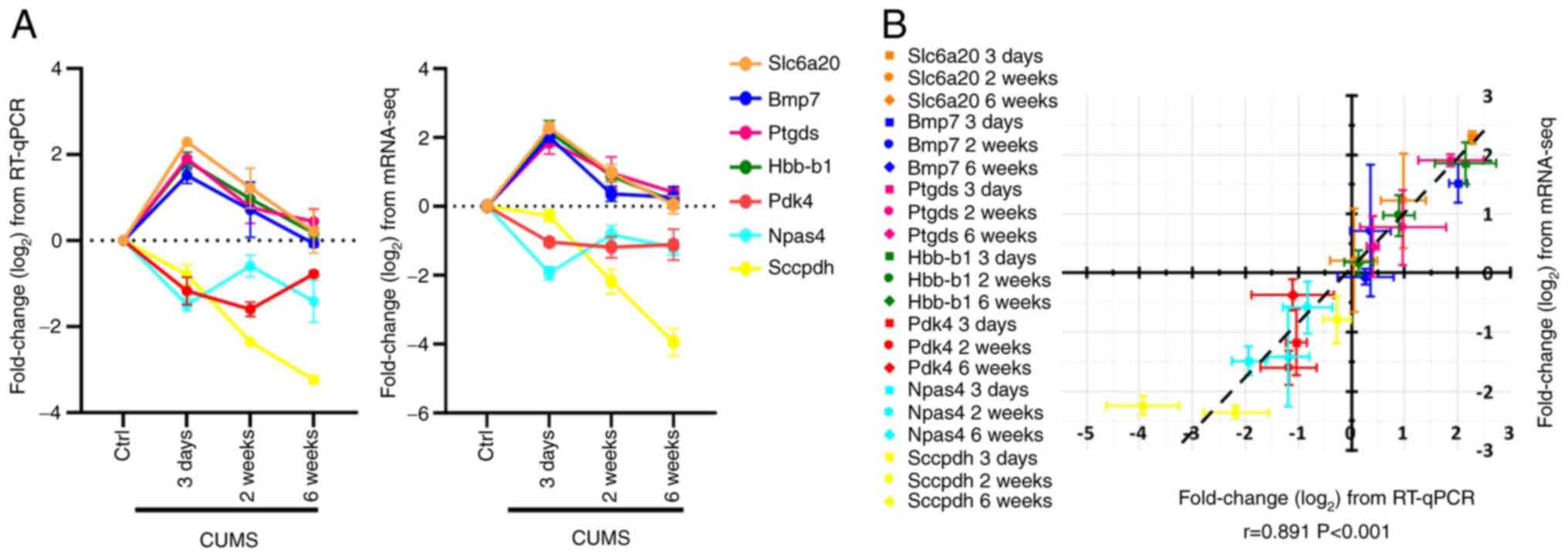
A total of seven differentially expressed
transcripts were verified by RT-qPCR. Slc6a20, Bmp7, Ptgds and
Hbb-b1 were upregulated during CUMS for 3 days and 2 weeks, and
gradually decreased to normal levels at 6 weeks of CUMS. These
changes in differential gene detected in mRNA-seq data were
confirmed by RT-qPCR. Similarly, Pdk4, Npas4 and Sccpdh were
downregulated in the whole CUMS process and their changes
identified by RT-qPCR were consistent with mRNA-seq data (Fig. 4A). A significant positive
correlation between the expression level data from mRNA-seq and
RT-qPCR was identified (r=0.891; P<0.001) and the slope of the
correlation line was 0.915 (Fig.
4B).
Discussion
In the present study, gene expression profiling of
the hippocampus in rats exposed to stress was investigated using
mRNA-seq. The GO analysis of DEGs in CUMS for 3 days and 6 weeks
shows an inversion of some biological functions. The downregulated
genes in CUMS (3 days) rats shared some GO terms with upregulated
genes in CUMS (6 weeks) rats, including ‘regulation of the
apoptotic process’, ‘integral component of the plasma membrane’,
‘protein binding’, ‘identical protein binding’, ‘metal ion binding’
and ‘ATP binding’. These results suggested a functional change from
adaptation to damage during the early and late stages of chronic
stress. Other studies have also reported similar inverted changes
in hormones and proteins during stress (22–25). At the onset of stress,
glucocorticoids (GCs) exert anti-apoptotic effects targeting
protein Bcl-2, while prolonged stress increases free radicals and
facilitates apoptosis (22).
Moreover, as a key neurotrophic factor, brain-derived neurotrophic
factor (BDNF) exhibits a transient increase in the hippocampus
following acute stress, whereas chronic stress leads to
downregulation of BDNF in the CA3 region of the hippocampus
(23–25). The mechanism underlying inverted
changes during CUMS is still unknown. Differences between GC
receptors [mineralocorticoid receptor (MR) and glucocorticoid
receptor (GR)] may account for the inverted changes. GCs at low
concentrations in the early stage of stress act via high-affinity
MR, which will promote the increase of neuronal excitability.
However, higher GC levels in the late stage activate low-affinity
GR to exert different effects, such as inhabitation of
proliferation and integration (26). Norepinephrine is secreted rapidly
in the early stage of stress, while GC levels increase during
long-term stress. In the hippocampal dentate gyrus, norepinephrine
enhances excitability and synaptic plasticity, but the high levels
of corticosterone suppress noradrenergic activity (27). The duration and antagonism of
hormones during stress may contribute to the inversion of
biological functions. Further studies are needed to investigate the
mechanism underlying altered biological processes during chronic
stress.
GO analysis of upregulated genes in CUMS (2 weeks)
rats revealed vision-associated functional alterations, such as
‘visual perception’, ‘phototransduction’, ‘retina development in
the camera-type eye’, ‘detection of light stimuli involved in
visual perception’, ‘photoreceptor outer segment’, ‘photoreceptor
inner segment’ and ‘retinal binding’. This indicated that visual
function was significantly altered after 2 weeks of stress, when
cognitive impairment had not yet developed. The association between
changes in visual function and stress is still unclear. Certain
studies have found that stress leads to attentional narrowing and
enhanced ability for detection and observation (28,29). However, other studies have found
decreased attention and accuracy of visual perceptual processing in
stressful states (30,31). The effects of acute stress on
visual function are controversial. Chronic stress is hypothesized
to be the most common cause of depression. In addition to symptoms
such as anxiety, depression is often accompanied by perceptual
abnormality, including visual changes; such changes may comprise
notable diminished visual acuity or increased sensitivity to light
stimuli (32,33). Certain studies have reported
visual impairment in patients with depression, including impaired
ability to distinguish intensity of light in a room and the ability
to read (32,34). Golomb et al (33) used a classical visual stimulus,
the moving grating test, to test patients with major depressive
disorder who had normal visual acuity and found that motion
perception was enhanced in patients with depression compared with
healthy individuals. The present findings suggested that visual
function may be an early indicator of impairment due to stress.
Further studies are needed to determine changes in visual
perception during stress.
GO analysis of upregulated genes in CUMS (3 days and
2 weeks) rats showed functional changes in ‘extracellular matrix’
(ECM), ‘extracellular space’ and ‘extracellular region’. GO
analysis of downregulated genes in these groups also showed
functional changes in ‘extracellular space’ and ‘extracellular
region’, suggesting an imbalance in synthesis and degradation of
extracellular components of hippocampal tissue during the early and
middle stages of CUMS. This imbalance might cause excessive
deposition of ECM components in the hippocampal tissue (35), such as collagen,
metalloproteinase, fibronectin, bone morphogenetic protein 7,
nidogen 1 and elastin, which contributes to development of organ
fibrosis (36). Collagen is a
primary structural component of ECM and serves a key role in the
cell cycle stimulated by mitogen (37). However, certain reports have shown
that collagen inhibits cell proliferation (38,39). Excessive deposition of fibrous
collagen, such as type I collagen, occurs during organ fibrosis,
which leads to tissue and organ sclerosis and loss of function.
Activation of type I collagen COL1A1 in the hippocampus contributes
to the development of hippocampal sclerosis (40). The primary pathological
manifestations of hippocampal sclerosis include hippocampal
atrophy, neuronal loss, granule cell reorganization, alteration of
interneuronal populations and chronic fibrillary gliosis centered
on the pyramidal cell layer (41). GO analysis of upregulated DEGs in
CUMS (3 days and 2 weeks) groups also indicated a wound healing
function. Increased levels of collagen fibers serve an important
role in tissue injury healing (42,43). Activation of ECM in the
hippocampus during the early and middle stages of CUMS is
hypothesized to contribute to self-repair of damaged hippocampal
cells but also promotes hippocampal sclerosis, which leads to
partial neuronal loss (35). ECM
changes are often secondary to inflammatory reactions (36), which indicates that hippocampal
inflammation occurs during the early and middle stages of CUMS.
Excessive ECM deposition is a damage repair response to hippocampal
cell damage and inflammation caused by stress, which contributes to
the development of hippocampal sclerosis while promoting
self-repair (44).
GO analysis of the upregulated DEGs in CUMS (6
weeks) rats involved changes in ‘aging’, suggesting that chronic
stress shares certain pathways with aging or that long-term chronic
stress promotes aging. Aging is often accompanied by pathological
signs such as memory loss, Alzheimer's disease, dementia and
depression, and chronic stress is one of the contributing factors
to these pathological signs (45,46). The results of numerous animal
stress experiments and clinical trials show that stress leads to
changes in brain structure and function, cognitive decline and
accelerated cellular aging (46,47). Sapolsky et al (48) proposed a GC cascade hypothesis of
aging, suggesting that stress and GC accelerate the aging process
in rats. The hippocampus is a target and a highly vulnerable area
of GC in the brain. Elevated GC levels in the blood of elderly
people leads to hippocampal atrophy, the degree of which is
positively correlated with degree of GC elevation, resulting in
hippocampal dysfunction and deficits in memory and learning and
cognitive ability (49,50). In addition, one study found that
chronic stress disrupts the normal processing of Aβ precursor
protein, thus leading to the accumulation of Aβ in the brain
(14). The development of
physiological aging may interact with certain genetic background or
negative environmental conditions (such as chronic stressors and
stress in infancy and early childhood) to trigger dysregulation of
the hypothalamus-pituitary-adrenal (HPA) axis (51) and changes in the neurotransmitter
system and brain structures such as the hippocampus, prefrontal
cortex and amygdala (1), making
the brain more vulnerable to increased levels of GCs and metabolic
changes and less resilient to damage from exposure to stressors
(52,53); these alterations lead to formation
of an abnormal aging brain with decreased function and neuronal
death and stress serves a key role in brain aging.
GO analysis of upregulated DEGs in CUMS (6 weeks)
rats showed functional alterations in ‘regulation of the apoptotic
process’ and ‘negative regulation of cell proliferation’ and
downregulated DEGs showed functional alterations in
‘receptor-mediated endocytosis’, ‘angiogenesis’, ‘synapse’,
‘dendrite’ and ‘axon’. This suggested that under chronic stress,
neuronal apoptosis increased, multiple functions, such as cell
proliferation and endocytosis, were inhibited and expression of
genes associated with neuronal structures, such as synapses, axons
and dendrites, decreased, resulting in damaging effects, such as
impaired neuronal function and synaptic plasticity (46,49). Research has shown that under
chronic stress, hippocampal dentate gyrus and CA3 area pyramidal
cells atrophy and die (54),
mossy fiber-CA3 synaptic transmission (55) dentate gyrus granule cell
production decrease, dentate gyrus neurogenesis is inhibited
(56) and CA3 area dendritic
spine length and number are significantly decreased (57). Chronic stress causes functional
and structural damage to hippocampal neurons, which exacerbates
neuroendocrine abnormality and leads to mood changes and cognitive
impairment (1). Parul et
al (58) found that chronic
unpredictable stress inhibits hippocampal neurogenesis in adult
rats and increases glial cell and neuronal apoptosis in the
cerebral cortex and hippocampus, which may be associated with
reduced AKT and increased ERK signaling. Dagyte et al
(59) concluded that strong
activation of the HPA axis during acute stress is not sufficient to
decrease hippocampal cell proliferation, but prolonged and repeated
exposure to stressful stimuli inhibits hippocampal cell
proliferation and leads to neurodevelopmental dysplasia. The
present study supported the hypothesis that chronic stress may lead
to cumulative changes in the brain that are not observed following
acute stress. Such changes may indicate compromised brain
plasticity and increased vulnerability to neuropathology (60).
In summary, genome-wide expression data and
functional analysis reported here revealed dynamic functional
changes during the different stages of CUMS. During the early and
middle stages of chronic stress, the hippocampal ECM appears
significantly altered and damage repair capacity is enhanced. In
the late stage of CUMS, the balance of hippocampal apoptosis and
proliferation is disrupted and chronic stress induces increased
hippocampal apoptosis, aging, and the inhibition of multiple
neuronal structures and functions. Further experiments are required
to analyzed differential gene expression profiles and provide novel
directions for gene therapy to treat chronic stress.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 31771290 and 81702454), National
Natural Science Foundation of Beijing (grant no. 5222033) and
Military Logistics Scientific Research Foundation of China (grant
no. BWS17J027).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LJQ, FX and FL contributed to the conception and
design of the study. FL, FX, XW and YZ collected and interpreted
the data. FL and FX confirm the authenticity of all the raw data.
FL, YW and FX performed statistical analysis and wrote the
manuscript. All authors contributed to manuscript revision. All
authors have read and approved the manuscript.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Animal Care and Use Committee of the Beijing
Institute of Basic Medical Sciences (approval no.
IACUC-DWZX-2020670) and were performed in accordance with the NIH
Guide for the Care and Use of Laboratory Animals (NIH Publications
no. 8023) and the ARRIVE guidelines from NC3Rs. During treatment,
efforts were made to minimize the potential pain and distress of
animals. Animals were monitored at least four times a week.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McEwen BS, Bowles NP, Gray JD, Hill MN,
Hunter RG, Karatsoreos IN and Nasca C: Mechanisms of stress in the
brain. Nat Neurosci. 18:1353–1363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weinberg A and Creed F: Stress and
psychiatric disorder in healthcare professionals and hospital
staff. Lancet. 355:533–537. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sinha R: Chronic stress, drug use, and
vulnerability to addiction. Ann NY Acad Sci. 1141:105–130. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rozanski A, Blumenthal JA and Kaplan J:
Impact of psychological factors on the pathogenesis of
cardiovascular disease and implications for therapy. Circulation.
99:2192–2217. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhatia V and Tandon RK: Stress and the
gastrointestinal tract. J Gastroenterol Hepatol. 20:332–339. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reiche EM, Nunes SO and Morimoto HK:
Stress, depression, the immune system, and cancer. Lancet Oncol.
5:617–625. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sapolsky RM: Stress and the brain:
Individual variability and the inverted-U. Nat Neurosci.
18:1344–1346. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JJ and Diamond DM: The stressed
hippocampus, synaptic plasticity and lost memories. Nat Rev
Neurosci. 3:453–462. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kempermann G and Kronenberg G: Depressed
new neurons-adult hippocampal neurogenesis and a cellular
plasticity hypothesis of major depression. Biol Psychiatry.
54:499–503. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chao HM, Ma LY, McEwen BS and Sakai RR:
Regulation of glucocorticoid receptor and mineralocorticoid
receptor messenger ribonucleic acids by selective agonists in the
rat hippocampus. Endocrinology. 139:1810–1814. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li XH, Chen JX, Yue GX, Liu YY, Zhao X,
Guo XL, Liu Q, Jiang YM and Bai MH: Gene expression profile of the
hippocampus of rats subjected to chronic immobilization stress.
PLoS One. 8:e576212013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ubaldi M, Ricciardelli E, Pasqualini L,
Sannino G, Soverchia L, Ruggeri B, Falcinelli S, Renzi A, Ludka C,
Ciccocioppo R and Hardiman G: Biomarkers of hippocampal gene
expression in a mouse restraint chronic stress model.
Pharmacogenomics. 16:471–482. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Antoniuk S, Bijata M, Ponimaskin E and
Wlodarczyk J: Chronic unpredictable mild stress for modeling
depression in rodents: Meta-analysis of model reliability. Neurosci
Biobehav Rev. 99:101–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie F, Zhao Y, Ma J, Gong JB, Wang SD,
Zhang L, Gao XJ and Qian LJ: The involvement of homocysteine in
stress-induced Aβ precursor protein misprocessing and related
cognitive decline in rats. Cell Stress Chaperones. 21:915–926.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du J, Wang Y, Hunter R, Wei Y, Blumenthal
R, Falke C, Khairova R, Zhou R, Yuan P, Machado-Vieira R, et al:
Dynamic regulation of mitochondrial function by glucocorticoids.
Proc Natl Acad Sci USA. 106:3543–3548. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daskalakis NP, Cohen H, Cai G, Buxbaum JD
and Yehuda R: Expression profiling associates blood and brain
glucocorticoid receptor signaling with trauma-related individual
differences in both sexes. Proc Natl Acad Sci USA. 111:13529–13534.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Govindarajan A, Rao BS, Nair D, Trinh M,
Mawjee N, Tonegawa S and Chattarji S: Transgenic brain-derived
neurotrophic factor expression causes both anxiogenic and
antidepressant effects. Proc Natl Acad Sci USA. 103:13208–13213.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lakshminarasimhan H and Chattarji S:
Stress leads to contrasting effects on the levels of brain derived
neurotrophic factor in the hippocampus and amygdala. PLoS One.
7:e304812012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Joëls M and Baram TZ: The neuro-symphony
of stress. Nat Rev Neurosci. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pu Z, Krugers HJ and Joëls M:
Corticosterone time-dependently modulates beta-adrenergic effects
on long-term potentiation in the hippocampal dentate gyrus. Learn
Mem. 14:359–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clark WC, Yang JC and Janal MN: Altered
pain and visual sensitivity in humans: The effects of acute and
chronic stress. Ann NY Acad Sci. 467:116–129. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tijerina L, Garrott WR, Stoltzfus D and
Parmer E: Eye glance behavior of van and passenger car drivers
during lane change decision phase. Transp Res Rec. 1937:37–43.
2005. View Article : Google Scholar
|
|
30
|
Paul M, Lech RK, Scheil J, Dierolf AM,
Suchan B and Wolf OT: Acute stress influences the discrimination of
complex scenes and complex faces in young healthy men.
Psychoneuroendocrinology. 66:125–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smith KE, Leitzke BT and Pollak SD:
Youths' processing of emotion information: Responses to chronic and
video-based laboratory stress. Psychoneuroendocrinology.
122:1048732020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Norton DJ, McBain RK, Pizzagalli DA,
Cronin-Golomb A and Chen Y: Dysregulation of visual motion
inhibition in major depression. Psychiatry Res. 240:214–221. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Golomb JD, McDavitt JR, Ruf BM, Chen JI,
Saricicek A, Maloney KH, Hu J, Chun MM and Bhagwagar Z: Enhanced
visual motion perception in major depressive disorder. J Neurosci.
29:9072–9077. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Friberg TR and Borrero G: Diminished
perception of ambient light: A symptom of clinical depression? J
Affect Disord. 61:113–118. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anwar MM, Özkan E and Gürsoy-Özdemir Y:
The role of extracellular matrix alterations in mediating astrocyte
damage and pericyte dysfunction in Alzheimer's disease: A
comprehensive review. Eur J Neurosci. Jun 28–2021.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rosenberg GA: Extracellular matrix
inflammation in vascular cognitive impairment and dementia. Clin
Sci (Lond). 131:425–437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hornberger LK, Singhroy S, Cavalle-Garrido
T, Tsang W, Keeley F and Rabinovitch M: Synthesis of extracellular
matrix and adhesion through beta(1) integrins are critical for
fetal ventricular myocyte proliferation. Circ Res. 87:508–515.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Henriet P, Zhong ZD, Brooks PC, Weinberg
KI and DeClerck YA: Contact with fibrillar collagen inhibits
melanoma cell proliferation by up-regulating p27KIP1. Proc Natl
Acad Sci USA. 97:10026–10031. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kelly KK, MacPherson AM, Grewal H, Strnad
F, Jones JW, Yu J, Pierzchalski K, Kane MA, Herson PS and
Siegenthaler JA: Col1a1+ perivascular cells in the brain
are a source of retinoic acid following stroke. BMC Neurosci.
17:492016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Falconer MA, Serafetinides EA and
Corsellis JA: Etiology and pathogenesis of temporal lobe epilepsy.
Arch Neurol. 10:233–248. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thom M: Review: Hippocampal sclerosis in
epilepsy: A neuropathology review. Neuropathol Appl Neurobiol.
40:520–543. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rosensteel SM, Wilson RP, White SL and
Ehrlich HP: COL1A1 oligodeoxynucleotides decoy: Biochemical and
morphologic effects in an acute wound repair model. Exp Mol Pathol.
89:307–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shi-Wen X, Leask A and Abraham D:
Regulation and function of connective tissue growth factor/CCN2 in
tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev.
19:133–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Krishnaswamy VR, Benbenishty A, Blinder P
and Sagi I: Demystifying the extracellular matrix and its
proteolytic remodeling in the brain: Structural and functional
insights. Cell Mol Life Sci. 76:3229–3248. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moreno-Fernandez RD, Tabbai S,
Castilla-Ortega E, Perez-Martin M, Estivill-Torrus G, Rodriguez de
Fonseca F, Santin LJ and Pedraza C: Stress, depression, resilience
and ageing: A role for the LPA-LPA1 pathway. Curr Neuropharmacol.
16:271–283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
de Magalhães JP and Passos JF: Stress,
cell senescence and organismal ageing. Mech Ageing Dev. 170:2–9.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Segar TM, Kasckow JW, Welge JA and Herman
JP: Heterogeneity of neuroendocrine stress responses in aging rat
strains. Physiol Behav. 96:6–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sapolsky RM, Krey LC and McEwen BS: The
neuroendocrinology of stress and aging: The glucocorticoid cascade
hypothesis. Endocr Rev. 7:284–301. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Raber J: Detrimental effects of chronic
hypothalamic-pituitary-adrenal axis activation. From obesity to
memory deficits. Mol Neurobiol. 18:1–22. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Elgh E, Lindqvist Astot A, Fagerlund M,
Eriksson S, Olsson T and Näsman B: Cognitive dysfunction,
hippocampal atrophy and glucocorticoid feedback in Alzheimer's
disease. Biol Psychiatry. 59:155–161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Buwembo A, Long H and Walker CD:
Participation of endocannabinoids in rapid suppression of stress
responses by glucocorticoids in neonates. Neuroscience.
249:154–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Conrad CD: Chronic stress-induced
hippocampal vulnerability: The glucocorticoid vulnerability
hypothesis. Rev Neurosci. 19:395–411. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bloss EB, Janssen WG, McEwen BS and
Morrison JH: Interactive effects of stress and aging on structural
plasticity in the prefrontal cortex. J Neurosci. 30:6726–6731.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Magariños AM, McEwen BS, Flügge G and
Fuchs E: Chronic psychosocial stress causes apical dendritic
atrophy of hippocampal CA3 pyramidal neurons in subordinate tree
shrews. J Neurosci. 16:3534–3540. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Magariños AM, Verdugo JM and McEwen BS:
Chronic stress alters synaptic terminal structure in hippocampus.
Proc Natl Acad Sci USA. 94:14002–14008. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gould E and Tanapat P: Stress and
hippocampal neurogenesis. Biol Psychiatry. 46:1472–1479. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
McEwen BS: Stress and hippocampal
plasticity. Annu Rev Neurosci. 22:105–122. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Parul Mishra A, Singh S, Singh S, Tiwari
V, Chaturvedi S, Wahajuddin M, Palit G and Shukla S: Chronic
unpredictable stress negatively regulates hippocampal neurogenesis
and promote anxious depression-like behavior via upregulating
apoptosis and inflammatory signals in adult rats. Brain Res Bull.
172:164–179. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dagyte G, Van der Zee EA, Postema F,
Luiten PG, Den Boer JA, Trentani A and Meerlo P: Chronic but not
acute foot-shock stress leads to temporary suppression of cell
proliferation in rat hippocampus. Neuroscience. 162:904–913. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
McEwen BS and Morrison JH: The brain on
stress: Vulnerability and plasticity of the prefrontal cortex over
the life course. Neuron. 79:16–29. 2013. View Article : Google Scholar : PubMed/NCBI
|