Introduction
Colon cancer is a common malignant tumor of the
digestive system (1,2). Colon cancer has high morbidity and
mortality rates in developed western countries and is the third
commonest tumor in the world (3–5).
However, due to the rapid development of China's economy and
improvements to the living standard of its citizens, the increases
in life pressures and irregular work and rest patterns of the
evolving population, the incidence and mortality of colon cancer in
China are increasing. According to the latest data released by the
National Cancer Center, there were 387,600 new colorectal cancer
(CRC) cases in China in 2015, accounting for 9.87% of all malignant
tumor cases (6,7) and the mortality rate of colon cancer
in 2015 was ~8% (8). At present,
the primary treatment for colon cancer is surgery, supplemented by
chemoradiotherapy, gene therapy, targeted therapy and other
modalities; however, the strong invasive and migratory ability of
colon cancer lead to a poor prognosis and low survival rates
(9). Therefore, it is of great
significance to explore the factors and signaling pathways
associated with colon cancer cell proliferation and invasion in
order to improve the clinical treatment of colon cancer and patient
prognosis.
Nuclear receptor subfamily 3, group C, member 2
(NR3C2) is a nuclear transcription factor that encodes the MR
protein (also known as the halocorticoid receptor) (10). NR3C2 is downregulated in several
types of cancer and has been demonstrated to serve a tumor
suppressor role. In colon cancer, microRNA (miR)-4709 can promote
the proliferation, migration and invasion of colon adenocarcinoma
cells and can target the downregulation of NR3C2 (11), indirectly indicating that NR3C2
may exert a tumor suppressive effect. However, the mechanism of its
role in colon cancer remains to be elucidated. NR3C2 encodes a
corticosteroid receptor, which has been reported to promote
angiogenesis in colon cancer (12). In addition, the AKT/ERK signaling
pathway is an important signaling pathway affecting angiogenesis
(13). Research by Yang et
al (14) demonstrated that
NR3C2 can negatively regulate the AKT signaling pathway in
pancreatic cancer. Therefore, the aim of the present study was to
investigate whether NR3C2 can inhibit the proliferation, migration,
invasion and angiogenesis of colon cancer cells through inhibiting
the AKT/ERK signaling pathway, although the present study is not
the first to detect NR3C2 expression in colon cancer, it will
provide a reference for understanding the mechanism of NR3C2 in
colon cancer.
Materials and methods
Cell culture
Normal colonic mucosa cells (NCM460), colon cancer
cell lines (LoVo, CaCo2, SW1116, SW480 and HCT-116) and
immortalized cell line human umbilical vein endothelial cells
(HUVECs) were obtained from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. All cells were
cultured in the DMEM supplemented with 10% FBS (both from Gibco;
Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C
with 5% CO2.
Cell viability assay
LoVo cells were plated in a 96-well plate
(5×103 cells/well) and then pre-incubated for 24 h in a
humidified incubator at 37°C with 5% CO2. A total of 10
µl Cell Counting Kit-8 (CCK-8) solution (Beyotime Institute of
Biotechnology; cat. no. C0037) was added to each well of the plate.
Light absorbance at 450-nm was measured using a microplate reader
(BioTek Instruments, Inc.).
Cell transfection
The NR3C2 overexpression vector, pcDNA3.1-NR3C2
(Oe-NR3C2), and empty control vector, pcDNA3.1-NC (Oe-NC), were
synthesized by Shanghai GeneChem Co., Ltd. The cells were
inoculated on 12-well plates at a density of 3×105
cells/well and cultured in a 5% CO2 incubator at 37°C
for 24 h. Following incubation, cells were transfected with the
aforementioned plasmids (50 ng/ml) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. Following transfection for 48 h at 37°C
in a 5% CO2 humid incubator, the protein expression
level was evaluated by reverse transcription-quantitative (RT-q)
PCR.
Wound healing and Transwell
assays
For the wound healing assays, transfected LoVo cells
were cultured in 6-well plates to 70–80% confluency, after which
the monolayer was scratched with a 200-µl sterile pipette tip.
After washing with PBS, the cells were cultured in serum-free
medium and images were captured at 0 and 24 h under an optical
microscope (magnification, ×100).
For the Transwell assays, the invasive ability of
cells was evaluated using Transwell chambers coated with Matrigel
(BD Biosciences) as previously described (15). Briefly, LoVo cells
(3×104 cells) were plated in the plasma-free medium. The
upper chamber was pre-coated with Matrigel at 37°C for 30 min
(Sigma-Aldrich; Merck KGaA). A total of 0.1 ml cell suspension was
added to the upper chamber and the lower chamber was filled with
medium supplemented with 20% FBS. Cells were cultured for 24 h.
Subsequently, the upper chamber was collected and cleaned and the
cells that had invaded were stained with 0.5% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min. The
stained cells were counted under an optical microscope
(magnification, ×200).
Colony formation assay
The cells were inoculated in 6-well plates at the
density of 4×102 cells/well for 14 days. Subsequently,
the cells were fixed with 70% ethanol and then stained with 0.05%
crystal violet for 20 min at 37°C. The number of colonies formed
(>50 cells/colony) were counted by an Olympus BX40 light
microscope (Olympus Corporation).
ELISA
An ELISA kit (Nanjing Jiancheng Bioengineering
Institute; cat. no. H044-1) was used to analyze the levels of VEGF
according to the manufacturer's protocol. Each group was quantified
using an Automatic Microplate Reader (Syngene).
HUVEC tube formation assay
Matrigel matrix glue, 24-well culture plates and
pipette tips were placed at 4°C overnight. The culture medium of
HUVECs in each group was changed to serum-free culture medium for
24 h. Next, the cells were digested with 0.25% trypsin EDTA
solution and were aspirated into a single-cell suspension following
termination of digestion. After counting the number of cells, the
cell density was adjusted to 7.5×106 ml. A total of 250
µl Matrigel was added to each well of the 24-well plate and the
plate was placed in the incubator at 37°C with 5% CO2
with saturated humidity and allowed to cure for 30 min. The culture
medium containing 300 µl serum was mixed with 10 µl single-cell
suspension and then added to the 24-well plates. After culturing
for 10 h, five fields were randomly selected for observation under
an inverted microscope (Olympus Corporation; magnification,
×40).
RT-qPCR
Total RNA from LoVo cells (1×106
cells/well) was extracted using TRIzol® reagent (Thermo
Fisher Scientific, Inc.). Subsequently, according to the
manufacturer's protocol. RNA concentration and quantification were
assessed using a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.). Following DNase I digestion, total RNA was
reverse transcribed into cDNA using a QuantiTect Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Subsequently, qPCR
was performed using a QuantiTect SYBR Green PCR kit (Qiagen GmbH),
according to the manufacturer's protocol. The following
thermocycling conditions were used for qPCR: 95°C for 10 min;
followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The
following primers (GenScript) were used for qPCR: NR3C2 forward,
5′-GATTGACAGTTGGTCGGC-3′ and reverse, 5′-TTAGTCAGCTCAGGCTTGC-3′ and
GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′ and reverse,
5′-GCCCAATACGACCAAATCC-3′. mRNA expression levels were quantified
using the 2−ΔΔCq method (16) and normalized to the internal
reference gene GAPDH. The experiments were performed in
triplicate.
Total protein extraction and western
blotting
Total protein was extracted from LoVo cells using
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology; cat. no. P0013C) and quantified using a
Pierce BCA Protein assay kit (Thermo Fisher Scientific, Inc.).
Following denaturing, electrophoresis was performed using SDS-PAGE
with 12% SDS gels. Following protein transfer to PVDF membranes,
the membranes were blocked in 5% fat-free milk for 2 h at room
temperature. The membranes were incubated with primary antibodies
(all purchased from Abcam) against, NR3C2 (1:5,000; cat. no.
ab64457), MMP2 (1:1,000; cat. no. ab92536), MMP9 (1:1,000; cat. no.
ab76003), AKT (1,000; cat. no. ab18785), phosphorylated (p)-AKT
(1:1,000; cat. no. ab38449), VEGFR2 (1:5,000; cat. no. ab134191),
p-VEGFR2 (1:1,000; cat. no. ab5473), ERK (1:1,000; cat. no.
ab32537), p-ERK (1,000; cat. no. ab194776) and GAPDH (1:10,000;
cat. no. ab181602) overnight at 4°C. This was followed by
incubation with IgG-horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (1:3,000; cat. no. ab6721) for 1 h
at room temperature. Signals were visualized using ECL reagent
(MilliporeSigma). Densitometry analysis was performed using ImageJ
software (version 1.46; National Institutes of Health) with GAPDH
as the loading control.
Statistical analysis
All experiments were repeated at least three times
independently and results are expressed as the mean ± standard
deviation. Statistical analyses were performed using SPSS version
19.0 (IBM Corp.). One-way ANOVA followed by Tukey's post hoc test
was used to compare differences between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
NR3C2 expression is downregulated in
colon cancer cells
The expression levels of NR3C2 in normal intestinal
mucosa cells and colon cancer cell lines (CaCo2, SW1116, SW480,
HCT-116 and LoVo) was detected by western blotting and RT-qPCR. As
demonstrated in Fig. 1A, NR3C2
expression was downregulated in a variety of colon cancer cells
(CaCo2, SW1116, SW480, HCT-116 and LoVo) and the expression of
NR3C2 was lowest in LoVo cells. Therefore, it was hypothesized that
LoVo cells are representative to study the role of NR3C2 in colon
cancer and the LoVo cell line was selected for all subsequent
experiments, which will be conducive to the significance of
experimental results (11,14).
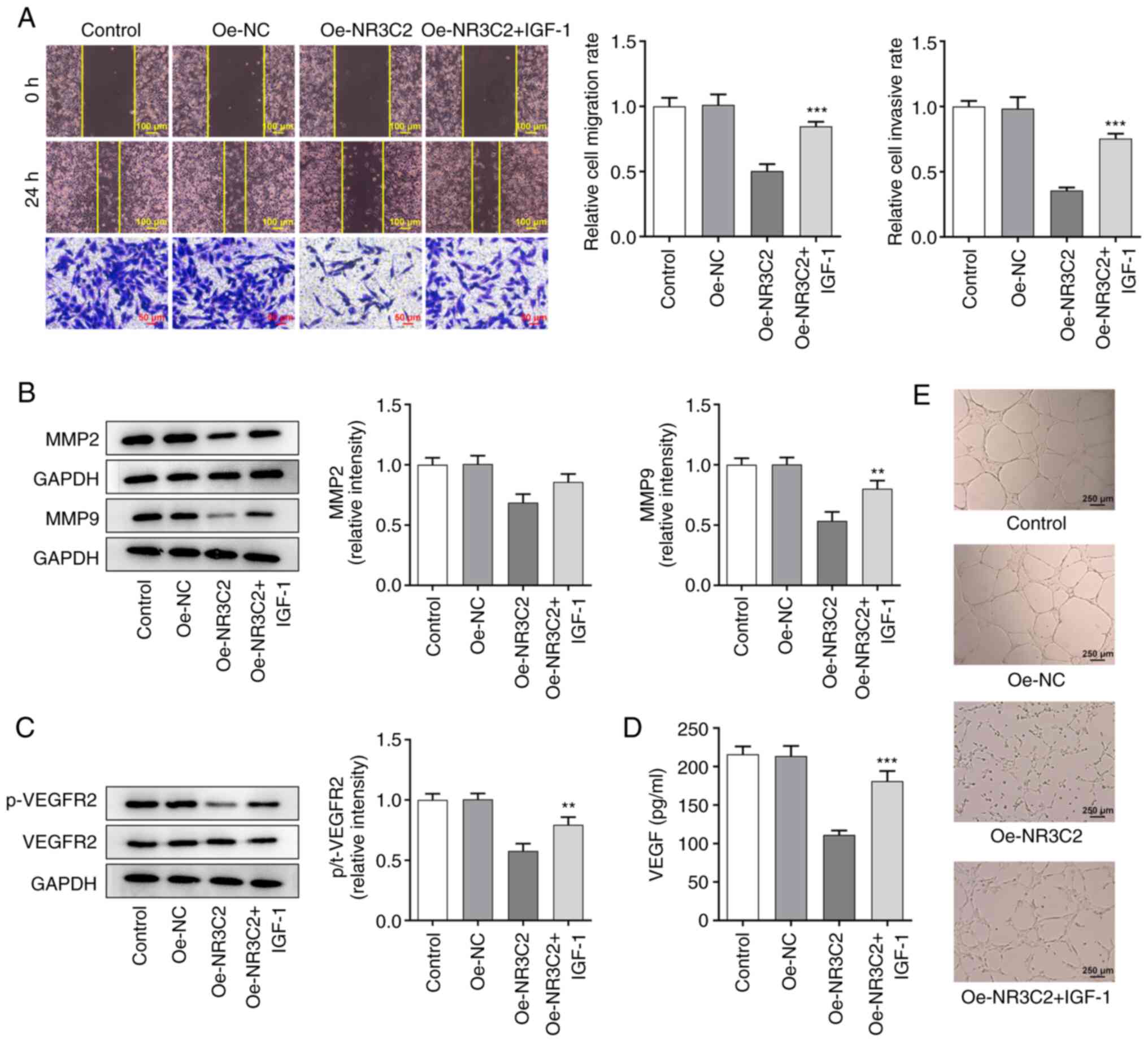
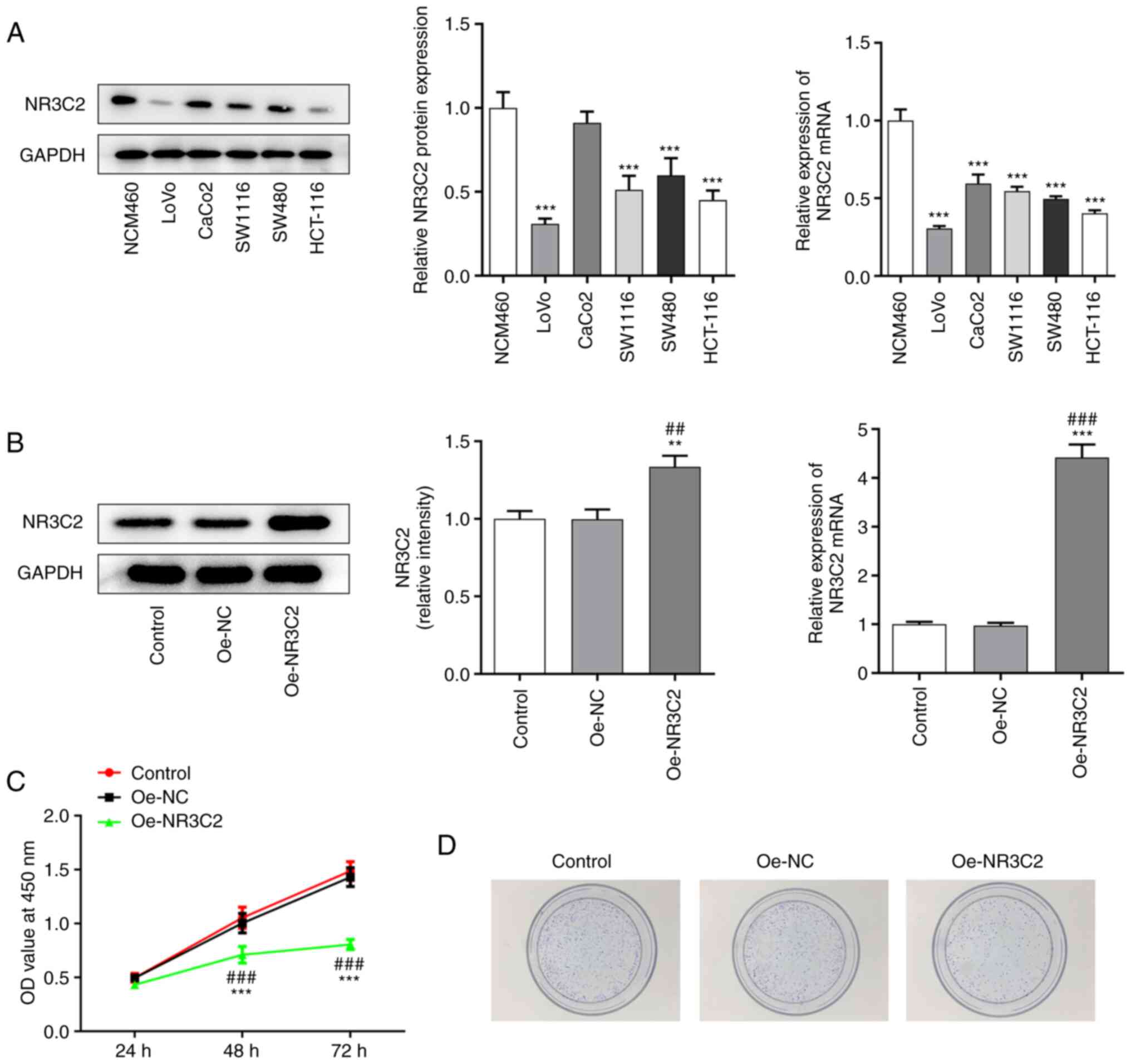 | Figure 1.Overexpression of NR3C2 inhibited the
proliferation of LoVo cells. (A) The expressions of NR3C2 in normal
intestinal mucosa cells and colon cancer cell lines (CaCo2, SW1116,
SW480, HCT-116 and LoVo) were detected by western blot and RT-qPCR.
(B) Expression levels of NR3C2 in the LoVo were detected by western
blot and RT-qPCR. (C) CCK-8 and (D) clone formation assays were
performed to determine cell proliferation. **P<0.01,
***P<0.001 vs. NCM460 or control; ##P<0.01,
###P<0.001 vs. Oe -NC. NR3C2, nuclear receptor
subfamily 3, group C, member 2; RT-qPCR, reverse
transcription-quantitative PCR; Oe, overexpression; NC, negative
control. |
Overexpression of NR3C2 inhibits the
proliferation of LoVo cells
Next, the expression of NR3C2 was increased by
transfection of an Oe-NR3C2 vector into LoVo cells. Compared with
the Oe-NC group, the expression of NR3C2 in the Oe-NR3C2 group was
significantly higher (Fig. 1B).
CCK-8 and colony formation assays were performed to assess cell
proliferation. As demonstrated in Fig. 1C and D, overexpression of NR3C2
significantly reduced cell viability compared with the Oe-NC
group.
Overexpression of NR3C2 inhibits
migration and invasion of LoVo cells
Wound healing and Transwell assays were used to
assess cell migration and invasion. As demonstrated in Fig. 2A, overexpression of NR3C2
significantly reduced the migration and invasion of LoVo cells.
Western blotting was used to detect the expression of the
invasion-related proteins, MMP2 and MMP9. As demonstrated in
Fig. 2B, overexpression of NR3C2
significantly decreased the expression of MMP2 and MMP9 compared
with the Oe-NC group.
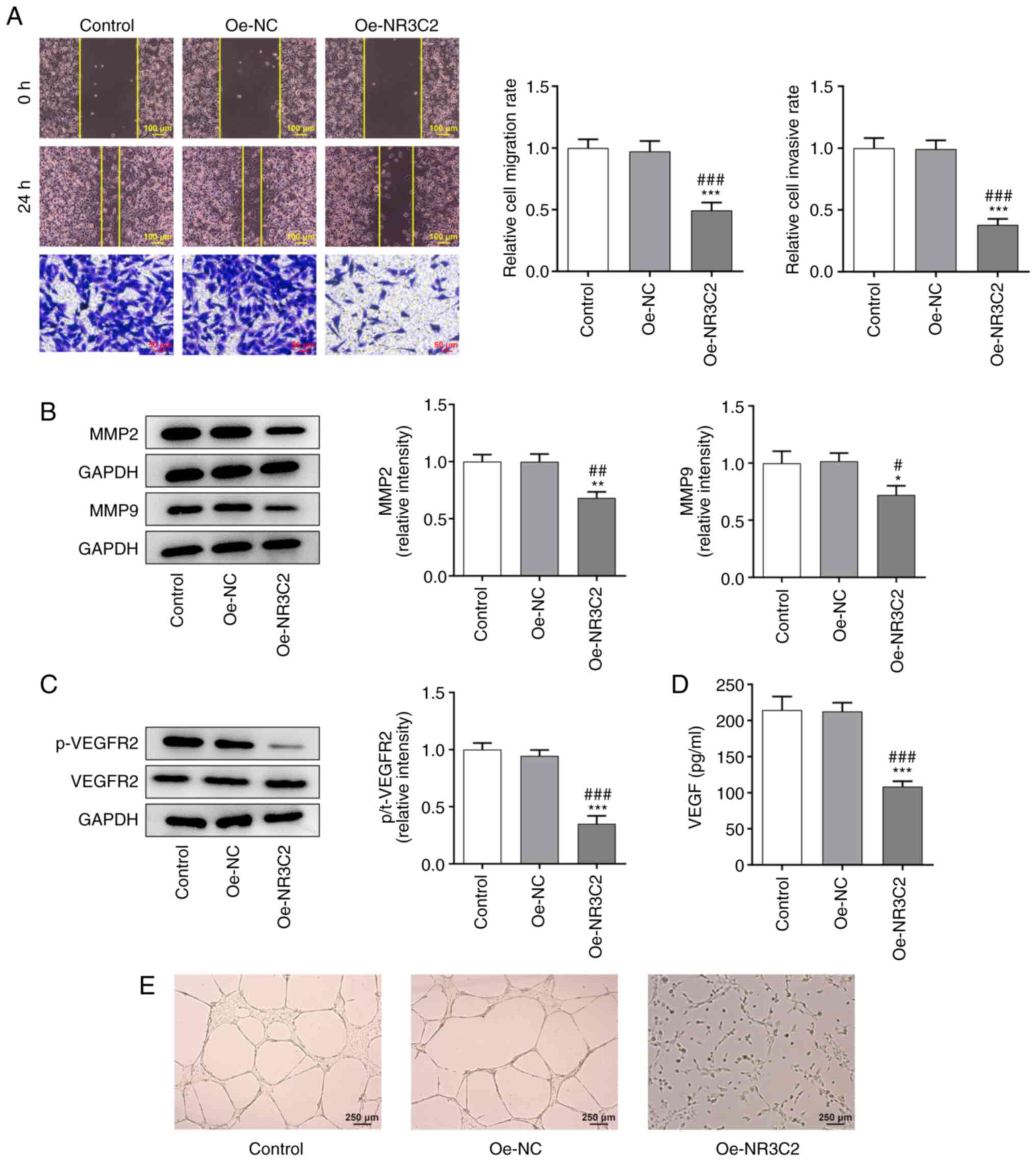 | Figure 2.Overexpression of NR3C2 inhibitions
migration, invasion and angiogenesis of LoVo cells. (A)
Wound-healing and Transwell assays were used to detect cell
migration and invasion. (B) Western blotting was used to detect the
expression of invasion-related proteins MMP2 and MMP9. (C) Western
blotting was used to detect the expression of invasion-related
proteins p-VEGFR2 and VEGFR2. (D) VEGF levels were assessed using
ELISA. (E) Tubule formation experiment was used to detect the
angiogenesis of LoVo cells (magnification, ×40). *P<0.05,
**P<0.01, ***P<0.001 vs. control; #P<0.05,
##P<0.01, ###P<0.001 vs. Oe-NC. NR3C2,
nuclear receptor subfamily 3, group C, member 2; p-,
phosphorylated; Oe, overexpression; NC, negative control. |
Overexpression of NR3C2 inhibits
angiogenesis in LoVo cells
The expression levels of angiogenesis-related
proteins were detected by western blotting and the results
demonstrated that the expression of phosphorylated- (p-)VEGF
receptor 2 was significantly decreased in the Oe-NR3C2 group
(Fig. 2C). VEGF levels detected
by ELISA kit demonstrated that overexpression of NR3C2 inhibited
the expression of VEGF (Fig. 2D).
The results of the tube formation assays further demonstrated that
NR3C2 overexpression inhibited angiogenesis in colon cancer cells
(Fig. 2E).
Overexpression of NR3C2 inhibits the
AKT/ERK signaling pathway in colon cancer cells
Yang et al (14) demonstrated that NR3C2 can
negatively regulate the AKT signaling pathway; thus, western
blotting was used to detect the expression levels of proteins
associated with the AKT/ERK signaling pathway. Compared with the
Oe-NC group, the overexpression of NR3C2 significantly reduced the
expression of p-AKT and p-ERK (Fig.
3A).
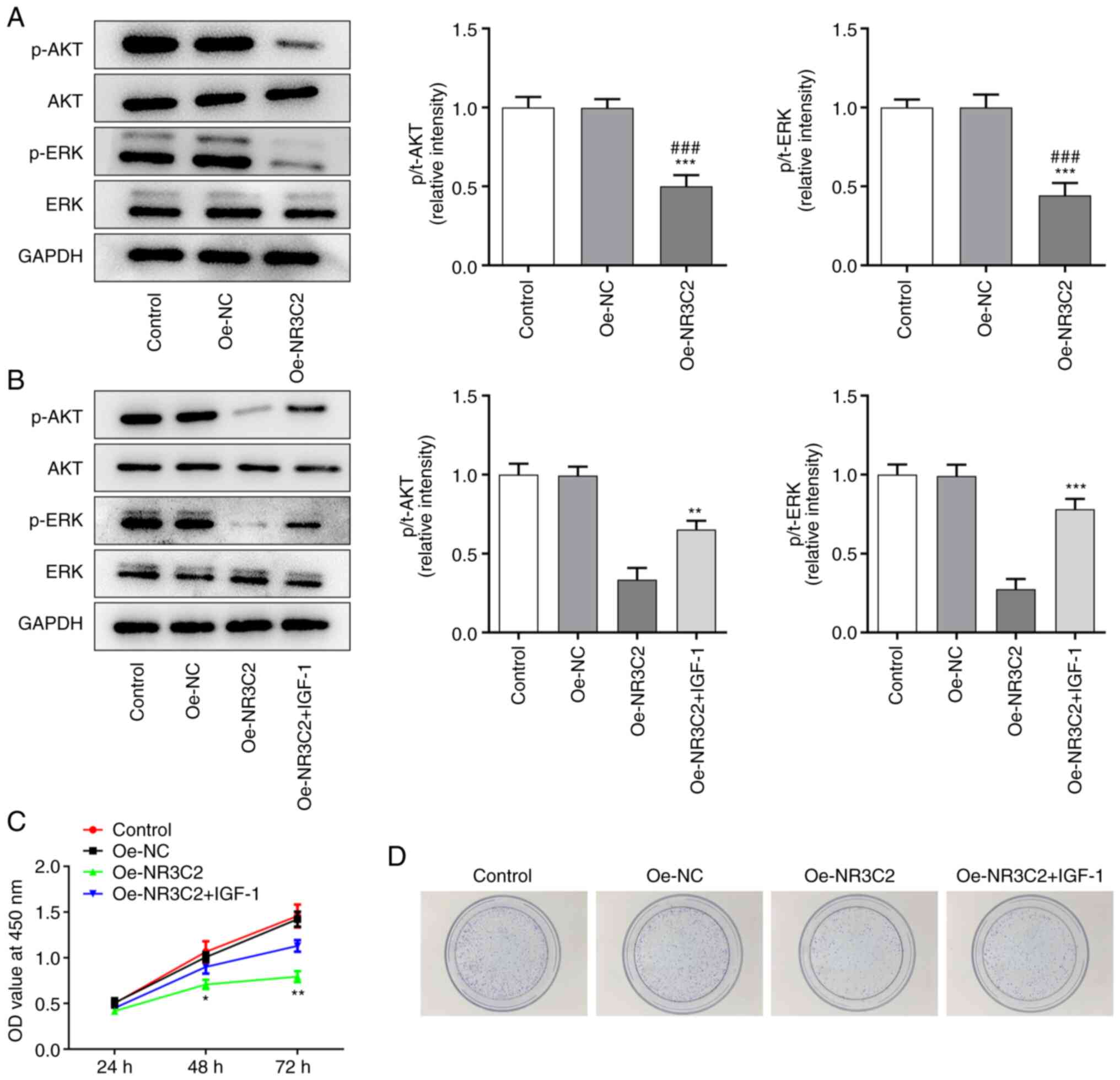 | Figure 3.NR3C2 inhibits the proliferation and
cloning of colon cancer cells through the AKT/ERK signaling
pathway. (A) The expressions of p-AKT, AKT, ERK and p-ERK in LoVo
cells were detected by western blotting. (B) The expressions of
p-AKT, AKT, ERK and p-ERK in LoVo cells were detected by western
blotting. (C) CCK-8 and assays (D) clone formation were performed
to determine the cell proliferation. *P<0.05, **P<0.01,
***P<0.001 vs. Oe-NR3C2 or control; ###P<0.001 vs.
Oe-NC. NR3C2, nuclear receptor subfamily 3, group C, member 2; p-,
phosphorylated; Oe, overexpression; NC, negative control; IGF,
insulin growth factor-1. |
NR3C2 inhibits the proliferation and
colony formation of colon cancer cells through the AKT/ERK
signaling pathway
To further investigate the role of NR3C2 in colon
cancer, AKT agonist insulin growth factor-1 (IGF-1; 10 nM) was
added to the medium. Western blotting demonstrated that
overexpression of NR3C2 inhibited the expression of p-AKT and p-ERK
and this was reversed by IGF-1 (Fig.
3B). CCK-8 and colony formation assays were used to assess cell
proliferation and the results demonstrated that both were reduced
by NR3C2 overexpression and increased by IGF-1 treatment (Fig. 3C-D).
NR3C2 inhibits the migration and
invasion of colon cancer cells via the AKT/ERK signaling
pathway
Transwell and wound healing assays were used to
detect cell invasion and migration and it was found that NR3C2
overexpression could inhibit the metastasis of LoVo cells, whereas
IGF-1 treatment enhanced invasion and migration (Fig. 4A). AKT is located upstream of ERK,
so AKT/ERK can act as a cascade reaction to regulate the expression
of MMPs (17), as demonstrated in
Fig. 4B, NR3C2 overexpression
inhibited the expression of metastasis-related proteins MMP2 and
MMP9 in LoVo cells and IGF-1 treatment increased the expression of
MMP2 and MMP9 (Fig. 4B). As
demonstrated in Fig. 4C, IGF-1
markedly increased the expressions of p-VEGFR2 compared with the
Oe-NR3C2 group by western blot. in addition, VEGF levels detected
by ELISA kit demonstrated that overexpression of NR3C2 inhibited
the expression of VEGF, which reversed by IGF-1 (Fig. 4D). Tubule formation test was used
to detect angiogenesis. Compared with the control group, tubule
formation rate of HUVEC in Oe-NR3C2 group was significantly
decreased and tubule formation rate was significantly increased
after IGF-1 was added (Fig. 4E).
Taken together, NR3C2 inhibits the migration, invasion and of colon
cancer cells through the AKT/ERK signaling pathway.
Discussion
The present study demonstrated that NR3C2 expression
was downregulated in colon cancer cells. It has been demonstrated
that the expression of NR3C2 is higher in normal tissues compared
with cancerous tissues (14).
Patients with CRC with upregulated expression of NR3C2 have a
longer 5-year overall survival (OS) rate, earlier-stage disease and
lower rate of lymphatic and distant metastasis (11). Colon cancer is more common than
rectal cancer and, consistent with the findings of the present
study, the 5-year OS rate of colon cancer has been found to be
higher than that of rectal cancer (6,18).
Several studies have demonstrated that NR3C2 is a tumor suppressor
gene and NR3C2 expression of the MR protein can inhibit
angiogenesis and, thus, inhibit the progression of colorectal
tumors (12). In other tumors,
such as pancreatic cancer, NR3C2 also acts as a tumor suppressor
gene, which can inhibit the proliferation and EMT of pancreatic
cancer cells and increase sensitivity to certain chemotherapeutic
agents (14,19). In hepatocellular carcinoma, NR3C2
can regulate the β-catenin signaling pathway to reduce the
proliferation- and invasion-promoting effects of miR-766 on
hepatocellular carcinoma cells (20). In renal cancer, the overexpression
of NR3C2 is demonstrated to inhibit the proliferation, migration
and other activities of renal cancer cells (21). Thus, it may be hypothesized that
NR3C2 may act as a tumor suppressor gene in CRC and inhibit the
occurrence and development of CRC to some extent. The present study
also confirmed that overexpression of NR3C2 inhibited
proliferation, colony formation, migration and invasion of colon
cancer cells. Overexpression of NR3C2 inhibited angiogenesis in
colon cancer cells.
During tumor progression, pathological vascular
growth is in a state of activation, which promotes the distal
metastasis of tumor cells (22).
VEGFA can effectively promote cell division and proliferation of
tumor cells during tumor growth, invasion and metastasis (23). Clinical studies have demonstrated
that the expression levels of VEGFA in the serum of patients with
CRC is closely associated with the state of angiogenesis and is an
independent predictor of prognosis (23,24). The MAPK/ERK and PI3K/AKT signaling
pathways serve important roles in vascular endothelial cell growth
and angiogenesis of tumor cells (25). Studies have demonstrated that
decreased VEGFA expression can inhibit the activation of the
MAPK/ERK and PI3K/AKT signaling pathways, thus serving an
anti-angiogenic role (26,27).
Angiogenesis is important for tumor growth and metastasis. Studies
have demonstrated that the expression levels of VEGF in the tissues
of patients with cancer is significantly higher compared with that
in normal tissues (28). VEGF
promotes angiogenesis by activating proliferation and invasion of
vascular endothelial cells through AKT/PI3K/MAPK signaling
(29). In the present study,
overexpression of NR3C2 significantly reduced the expression levels
of migration and invasion related proteins MMP2 and MMP9, which
might be due to the fact that AKT was located upstream of ERK and
AKT/ERK could regulate the expression of MMPs as a cascade reaction
(17). Therefore, NR3C2
overexpression might inhibit the malignant process of colon cancer
cells through AKT/ERK signaling pathway, which provided a reference
for studying the mechanism of NR3C2 in colon cancer.
Although the present study obtained some meaningful
results, there were some shortcomings, which could serve as the
direction of future research. First, the expression level of NR3C2
was only detected in colon cancer cells, while the expression level
in colon cancer tissues needs further investigation. Second,
another colon cancer cell with low NR3C2 expression should be
selected to further confirm the results of the present study.
Third, it is necessary to fully discuss the relationship between
NR3C2 and AKT/ERK signaling pathway through another agonist.
Fourth, the results of the present study demonstrated that NR3C2
was downregulated in colon cancer cells, but the reason for its
differential expression in different types of colon cancer cells
needs further investigation, Finally, it should be emphasized that
in future research it will be worthwhile to further detect cell
activation and proliferation under NR3C2 inhibition on the basis of
NR3C2 overexpression colon cancer cell.
In conclusion, the present study demonstrated that
NR3C2 expression was downregulated in colon cancer tissues and that
overexpression of NR3C2 inhibited the proliferation, migration,
invasion and angiogenesis of colon cancer cells. In addition, NR3C2
affected the proliferation, migration, invasion and angiogenesis of
colon cancer cells through the AKT/ERK signaling pathway. These
findings may highlight novel targets for the treatment of colon
cancer.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of
Jiangxi province (grant no. 20202ACB216003).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JL conceptualized and designed the current study. JL
and ZX acquired, analyzed and interpreted data. JL and ZX drafted
the manuscript and revised it critically for important intellectual
content. JL and ZX confirm the authenticity of all the raw data.
Both authors agreed to be held accountable for the current study in
ensuring questions related to the integrity of any part of the work
are appropriately investigated and resolved. Both authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu J, Yi J, Zhang Z, Cao D, Li L and Yao
Y: Deoxyribonuclease 1-like 3 may be a potential prognostic
biomarker associated with immune infiltration in colon cancer.
Aging (Albany NY). 13:16513–16526. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Troncone E, Marafini I, Stolfi C and
Monteleone G: Involvement of Smad7 in inflammatory diseases of the
gut and colon cancer. Int J Mol Sci. 22:39222021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Finotti M, Vitale A, Gringeri E, D'Amico
FE, Boetto R, Bertacco A, Lonardi S, Bergamo F, Feltracco P and
Cillo U: Colon rectal liver metastases: The role of the liver
transplantation in the Era of the transplant oncology and precision
medicine. Front Surg. 8:6933872021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tauriello DVF, Palomo-Ponce S, Stork D,
Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M,
Ibiza S, Cañellas A, Hernando-Momblona X, et al: TGFβ drives immune
evasion in genetically reconstituted colon cancer metastasis.
Nature. 554:538–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Araghi M, Arnold M, Rutherford MJ, Guren
MG, Cabasag CJ, Bardot A, Ferlay J, Tervonen H, Shack L, Woods RR,
et al: Colon and rectal cancer survival in seven high-income
countries 2010-2014: Variation by age and stage at diagnosis (the
ICBP SURVMARK-2 project). Gut. 70:114–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qin Q, Yang L, Sun YK, Ying JM, Song Y,
Zhang W, Wang JW and Zhou AP: Comparison of 627 patients with
right- and left-sided colon cancer in China: Differences in
clinicopathology, recurrence, and survival. Chronic Dis Transl Med.
3:51–59. 2017.PubMed/NCBI
|
|
7
|
Gao J, Yang M, Liu L, Guo S, Li Y and
Cheng C: The aggressive surgical treatment and outcome of a colon
cancer patient with COVID-19 in Wuhan, China. BMC Gastroenterol.
20:2692020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao M, Li H, Sun D and Chen W: Cancer
burden of major cancers in China: A need for sustainable actions.
Cancer Commun (Lond). 40:205–210. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Villalba M, Evans SR, Vidal-Vanaclocha F
and Calvo A: Role of TGF-β in metastatic colon cancer: It is
finally time for targeted therapy. Cell Tissue Res. 370:29–39.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Kloet ER, Van Acker SA, Sibug RM, Oitzl
MS, Meijer OC, Rahmouni K and de Jong W: Brain mineralocorticoid
receptors and centrally regulated functions. Kidney Int.
57:1329–1336. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu M, Yu HL, Li QH, Zhang L and Chen YX:
miR-4709 overexpression facilitates cancer proliferation and
invasion via downregulating NR3C2 and is an unfavorable prognosis
factor in colon adenocarcinoma. J Biochem Mol Toxicol.
33:e224112019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tiberio L, Nascimbeni R, Villanacci V,
Casella C, Fra A, Vezzoli V, Furlan L, Meyer G, Parrinello G,
Baroni MD, et al: The decrease of mineralcorticoid receptor drives
angiogenic pathways in colorectal cancer. PLoS One. 8:e594102013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Wang X, Li Z, Zhang Z and Zhang Y:
Chemokine receptor 7 targets the vascular endothelial growth factor
via the AKT/ERK pathway to regulate angiogenesis in colon cancer.
Cancer Med. 8:5327–5340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Wu X and She W: lncRNA POU3F3
promotes cancer cell migration and invasion in nasopharyngeal
carcinoma by up-regulating TGF-β1. Biosci Rep. 39:BSR201816322019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agraval H and Yadav UCS: MMP-2 and MMP-9
mediate cigarette smoke extract-induced epithelial-mesenchymal
transition in airway epithelial cells via EGFR/Akt/GSK3β/β-catenin
pathway: Amelioration by fisetin. Chem Biol Interact.
314:1088462019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Han F, Liu W and Shi X: PTBP1
promotes tumorigenesis by regulating apoptosis and cell cycle in
colon cancer. Bull Cancer. 105:1193–1201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao
L and Pei H: miR-135b-5p Promotes migration, invasion and EMT of
pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother.
96:1341–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang C, Ma X, Guan G, Liu H, Yang Y, Niu
Q, Wu Z, Jiang Y, Bian C, Zang Y and Zhuang L: MicroRNA-766
promotes cancer progression by targeting NR3C2 in hepatocellular
carcinoma. FASEB J. 33:1456–1467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Z, Zhang M, Duan X, Deng T, Qiu H and
Zeng G: Low NR3C2 levels correlate with aggressive features and
poor prognosis in non-distant metastatic clear-cell renal cell
carcinoma. J Cell Physiol. 233:6825–6838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muche A, Bigl M, Arendt T and Schliebs R:
Expression of vascular endothelial growth factor (VEGF) mRNA, VEGF
receptor 2 (Flk-1) mRNA, and of VEGF co-receptor neuropilin (Nrp)-1
mRNA in brain tissue of aging Tg2576 mice by in situ hybridization.
Int J Dev Neurosci. 43:25–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mizuno R, Kawada K, Itatani Y, Ogawa R,
Kiyasu Y and Sakai Y: The role of tumor-associated neutrophils in
colorectal cancer. Int J Mol Sci. 20:5292019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boeckx N, Janssens K, Van Camp G,
Rasschaert M, Papadimitriou K, Peeters M and Op de Beeck K: The
predictive value of primary tumor location in patients with
metastatic colorectal cancer: A systematic review. Crit Rev Oncol
Hematol. 121:1–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng JQ, Ruggeri B, Klein WM, Sonoda G,
Altomare DA, Watson DK and Testa JR: Amplification of AKT2 in human
pancreatic cells and inhibition of AKT2 expression and
tumorigenicity by antisense RNA. Proc Natl Acad Sci USA.
93:3636–3641. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu Y, Wang Q, Guo K, Qin W, Liao W, Wang
S, Ding Y and Lin J: TUSC3 promotes colorectal cancer progression
and epithelial-mesenchymal transition (EMT) through WNT/β-catenin
and MAPK signalling. J Pathol. 239:60–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen H, Feng J, Zhang Y, Shen A, Chen Y,
Lin J, Lin W, Sferra TJ and Peng J: Pien Tze Huang inhibits
hypoxia-induced angiogenesis via HIF-1 α/VEGF-a pathway in
colorectal cancer. Evid Based Complement Alternat Med.
2015:4542792015.PubMed/NCBI
|
|
29
|
Wang R, Luo Z, Zhang H and Wang T:
Tanshinone IIA reverses gefitinib-resistance in human
non-small-cell lung cancer via regulation of VEGFR/Akt pathway.
Onco Targets Ther. 12:9355–9365. 2019. View Article : Google Scholar : PubMed/NCBI
|