Introduction
Ankylosing spondylitis (AS) is a type of immune
inflammatory disease that primarily affects the spine and
sacroiliac joints. The majority of patients develop AS between the
ages of 18–22 years, with a male to female ratio of 2:1 (1,2).
AS is a highly disabling disease, and numerous patients require
surgical correction of morphological deformities caused by joint
involvement as traditional symptomatic treatment is ineffective
(3). AS is associated with immune
disorders, cytokine imbalance and genetic factors, but the exact
cause of the disease is not completely understood (4). Therefore, in recent years, the
exploration of the mechanism underlying AS and the search for
targeted drugs for the treatment of AS have become novel research
hotspots.
Recent studies have reported that oxidative stress
and inflammation serve a crucial role in the occurrence and
development of AS (5,6). In the active stage of AS, there are
high levels of oxidative stress, and the biomarkers of oxidative
damage are significantly increased (7). In addition, the early stage of AS is
primarily characterized by inflammatory bone destruction. With the
development of the disease, inflammatory bone destruction and new
bone formation can occur simultaneously, indicating that the
inflammatory response serves a key role in the pathogenesis of AS
(8). Furthermore, there is a
close relationship between oxidative stress and inflammation. After
the activation of inflammatory cells, a variety of cytokines, such
as TNF-α, IL-1β and interferon-γ, can be produced, which then
activate the cells to produce a large number of oxidative active
substances. Solmaz et al (9) revealed that the level of oxidative
stress in patients with AS was significantly higher compared with
that in healthy subjects, and that the level of oxidative stress in
patients with active disease state was higher compared with that in
patients with inactive disease state and the control group.
Therefore, the occurrence and development of AS can be controlled
via the improvement of the oxidative stress and inflammatory
response in AS.
Fibroblast growth factor 21 (FGF21) is a polypeptide
hormone that regulates energy homeostasis and is synthesized by
numerous organs (10). The
mechanism of action underlying FGF21 is complex, and it can exert
different metabolic functions in multiple target organs in the form
of autocrine, paracrine and endocrine factors (11). Increasing evidence has suggested
that FGF21 is associated with a variety of chronic inflammatory
diseases. A previous study has shown that the expression level of
FGF21 was significantly reduced in the cartilage of mice with
rheumatoid arthritis (12). FGF21
also improves collagen-induced arthritis by regulating oxidative
stress and inhibiting the NF-κB signaling pathway (13). Furthermore, it has been revealed
that metformin improves experimental obesity-related autoimmune
arthritis by inducing FGF21 expression (14). However, to the best of our
knowledge, the effect of FGF21 on oxidative stress and inflammation
in AS has not been previously reported.
KLF, a zinc finger protein family with several
structurally similar members, serves an important role in gene
transcriptional regulation in eukaryotic cells (15). KLF4 is a nuclear transcription
factor with important biological functions. Currently, KLF4 is
widely studied in chronic inflammatory diseases. For instance, KLF4
expression is increased in the synovial tissues of patients with
rheumatoid arthritis, and KLF4 deficiency reduces the inflammatory
response induced by collagen antibodies (16). Therefore, we hypothesized that the
binding of the transcription factor KLF4 with the FGF21 promoter
serves an important role in AS.
A previous study reported that FGF21 could regulate
the expression of the sirtuin 1 (SIRT1) signaling pathway, serving
a role in the development of diseases, such as acute pancreatitis
and myocardial energy metabolism disease (17,18). The expression levels of anti-SIRT1
autoantibodies in patients with AS are abnormally high, indicating
that anti-SIRT1 autoantibodies could be used as a marker for early
diagnosis and prediction of hip involvement in AS (19). In addition, SIRT1 can improve the
inflammatory response in diseases by blocking the NF-κB/p53
signaling pathway (20,21).
Therefore, the present study aimed to investigate
the regulatory role and mechanism underlying KLF4 on FGF21 in AS to
provide a theoretical basis for the pathogenesis and targeted
therapy of AS.
Materials and methods
Cell culture and treatments
Mouse chondrocyte ATDC5 cells were obtained from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences (Chinese Academy of Sciences, Shanghai, China). Cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) with 5% CO2
at 37°C. Subsequently, 5 µg/ml lipopolysaccharide (LPS;
Sigma-Aldrich; Merck KGaA) was added to the cells for 12 h to
establish the AS inflammatory injury model.
Database
The interaction between KLF4 and FGF21 was predicted
by bioinformatics analysis using the JASPAR database
(jaspar.genereg.net).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and reverse transcribed into cDNA using the High Capacity
cDNA Reverse Transcription kit according to the manufacturer's
protocol and random hexamer primers (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The following thermocycling conditions
were used for qPCR: Initial denaturation at 95°C for 5 min;
followed by 40 cycles of denaturation at 95°C for 30 sec and
annealing at 60°C for 30 sec; and final extension at 72°C for 20
sec. qPCR was performed using a SYBR Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). mRNA expression levels
were quantified using the 2−∆∆Cq method (22). The sequences of the primers used
for qPCR were as follows: FGF21 forward, 5′-AGATCAGGGAGGACGGAACA-3′
and reverse, 5′-TCAGGATCAAAGTGAGGCGAT-3′; TNF-α forward,
5′-CCACCACGCTCTTCTGTCTA-3′ and reverse,
5′-ACTGATGAGAGGGAGCCCATT-3′; IL-6 forward,
5′-ACTGATGAGAGGGAGCCCATT-3′ and reverse,
5′-ACTGATGAGAGGGAGCCCATT-3′; IL-1β forward,
5′-TAGGGCTGGCAGAAAGGGAACA-3′ and reverse,
5′-GTGGGAGCGAATGACAGAGGGT-3′; KLF4 forward,
5′-CGGGCTGATGGGCAAGTT-3′ and reverse, 5′-GGGCAGGAAGGATGGGTAA-3′;
and GAPDH forward, 5′-TGAAGGTCGGAGTCAACGG-3′ and reverse,
5′-CCTGGAAGATGGTGATGGG-3′. GAPDH was used as the internal reference
gene.
Western blotting
Total protein was extracted from ATDC5 cells using
RIPA reagent (Protech Technology Enterprise Co., Ltd.). Protein
concentrations were determined using a BCA kit (Abcam). Proteins
(30 µg per lane) were separated via 10% SDS-PAGE and then
transferred to PVDF membranes. After blocking with 5% fat-free milk
was used for 1.5 h at 37°C, the membranes were incubated at 4°C
overnight with the following antibodies: Anti-FGF21 (1:1,000;
Abcam; cat. no. ab171941), anti-KLF4 (1:1,000; Abcam; cat. no.
ab214666), anti-Bcl-2 (1:1,000; Abcam; cat. no. ab182858), anti-Bax
(1:1,000; Abcam; cat. no. ab32503), anti-cleaved caspase 3
(1:1,000; Abcam; cat. no. ab32042), anti-SIRT1 (1:1,000; Abcam;
cat. no. ab110304), anti-phosphorylated (p)-NF-κB p65 (1:1,000;
Abcam; cat. no. ab239882), anti-NF-κB p65 (1:1,000; Abcam; cat. no.
ab207297), anti-Acetyl-p53 (1:1,000; Abcam; cat. no. ab17496) and
GAPDH (1:1,000; Abcam; cat. no. ab8245). Subsequently, the membrane
was incubated with a Goat Anti-Rabbit IgG H&L HRP-conjugated
secondary antibody (1:5,000; Abcam; cat. no. ab70902) a Goat
Anti-Mouse IgG H&L (Alexa Fluor® 488) secondary
antibody (1:5,000; Abcam; cat. no. ab150117) at room temperature
for 1 h. The bands were visualized using enhanced chemiluminescence
reagent (GE Healthcare). Protein expression levels were
semi-quantified using ImageJ software (version 1.46; National
Institutes of Health) with GAPDH as the loading control.
Cell transfection
ATDC5 cells were seeded (1×106 cells/ml)
into a 6-well plate and cultured for 12 h until the cells grew to
~80% confluence. Cells were transfected with 20 nM FGF21
overexpression (Ov) plasmid (Ov-FGF21; Shanghai GenePharma Co.,
Ltd.) or negative control (NC) plasmid (Ov-NC; empty vector) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
The eukaryotic expression vector pcDNA3.1-KLF4 was
constructed by Tiandz, Inc.. Cells were transfected with 20 nM
pcDNA3.1-NC (an empty vector) or pcDNA3.1-KLF4 using Lipofectamine
2000. After transfection for 48 h at 37°C with 5% CO2,
transfection efficiencies were assessed via RT-qPCR and western
blotting.
Cell Counting Kit-8 (CCK-8) assay
ATDC5 cells were seeded (8×103
cells/ml/well) into 96-well plates and cultured for 12 h. After
transfection, cell viability was assessed using a CCK-8 assay
(Vazyme Biotech Co., Ltd.). Briefly, 10 µl CCK-8 solution was added
to each well and incubated for 2 h at 37°C. The absorbance was
measured at a wavelength of 450 nm using a VersaMax microplate
reader (Molecular Devices, LLC).
TUNEL assay
ATDC5 cells were seeded (1×106
cells/well) into 6-well plates. After transfection, cells were
fixed with 4% paraformaldehyde for 15 min at 37°C. After washing
with PBS, TUNEL solution was added for 1 h at 37°C. Biotin labeling
and subsequent DAB color development were performed for 10 min at
15°C using reagents provided in the kit according to the
manufacturer's instructions. The cells were stained with 0.1 µg/ml
DAPI for 10 min at room temperature, prior to detecting nuclear DNA
fragmentation using the DeadEnd™ Fluorometric TUNEL system (Promega
Corporation). Finally, after the cells were washed with PBS, five
random fields of views were selected for analysis, in which cell
apoptosis was observed using glass coverslips with PBS as mounting
medium. Stained cells were visualized using an IX71 fluorescent
microscope (Olympus Corporation).
ELISAs
The levels of IL-6 (cat. no. ab222503), IL-1β (cat.
no. ab197742) and TNF-α (cat. no. ab208348) in the cell medium were
measured using commercially available ELISA kits (all Abcam)
according to the manufacturer's protocols. The levels of
inflammatory cytokines from the cell supernatant were calculated
from standard curves. Lactate dehydrogenase (LDH) levels in cell
medium were measured using an LDH Cytotoxicity Assay kit (cat. no.
A020-2-2, Nanjing Jiancheng Bioengineering Institute) according to
the manufacturer's protocol.
Oxidative stress markers
Malondialdehyde (MDA; cat. no. A003-1-2), reduced
glutathione (GSH; cat. no. A005-1-2) and superoxide dismutase (SOD;
cat. no. A001-3-2) levels were determined using commercially
available ELISA kits (all Nanjing Jiancheng Bioengineering
Institute) according to the manufacturer's protocol.
Dual-luciferase reporter assay
The FGF21 reporter, which contains 1 kb of the FGF21
promoter, was created using RT-qPCR as aforementioned and cloned
into the pGL3 plasmid (Shanghai GenePharma Co., Ltd). Then, 20 nM
pcDNA3.1-KLF4 plasmid or 20 nM pcDNA3.1 plasmid and pGL3-FGF21
plasmid (WT and MUT version) were co-transfected into ATDC5 cells
using Lipofectamine 2000 at 37°C for 48 h. At 48 h
post-transfection, cells were harvested and luciferase activities
were analyzed using a dual-luciferase assay kit (Promega
Corporation) according to the manufacturer's protocol.
Renilla luciferase activity was used to normalize the
firefly luciferase activity.
Chromatin immunoprecipitation (ChIP)
assay
Cells were collected and fixed with 1% formaldehyde
for 15 min at 37°C, after which the cells were scraped and lysed in
100 µl SDS lysis buffer (Beyotime Institute of Biotechnology).
After purified by soluble material, extracted proteins were
immunoprecipitated using 5 µl KLF4 antibody (1:1,000; Abcam; cat.
no. ab214666) or control rabbit IgG (1:1,000; Abcam; cat. no.
ab133470)for 12 h at 4°C. Subsequently, 20 µl protein A+G agarose
beads (Beyotime Institute of Biotechnology) was added and incubated
for another 4 h at 4°C. After the supernatant was removed, the
collected agarose beads were washed five times with 100 µl PBS
(0.01 M; pH 7.4) and denatured by boiling for 5 min. The relative
expression levels were determined via RT-qPCR according to the
aforementioned protocol.
Statistical analysis
Comparisons among multiple groups were analyzed
using one-way ANOVA followed by Tukey's post hoc test. Comparisons
between two groups were analyzed using an unpaired Student's
t-test. Statistical analyses were performed using GraphPad Prism
software (version 22.0; GraphPad Software, Inc.). Data are
presented as the mean ± standard deviation of three independent
repeats. P<0.05 was considered to indicate a statistically
significant difference.
Results
FGF21 overexpression increases
viability and inhibits the apoptosis of LPS-induced ATDC5
cells
Western blotting and RT-qPCR were used to detect the
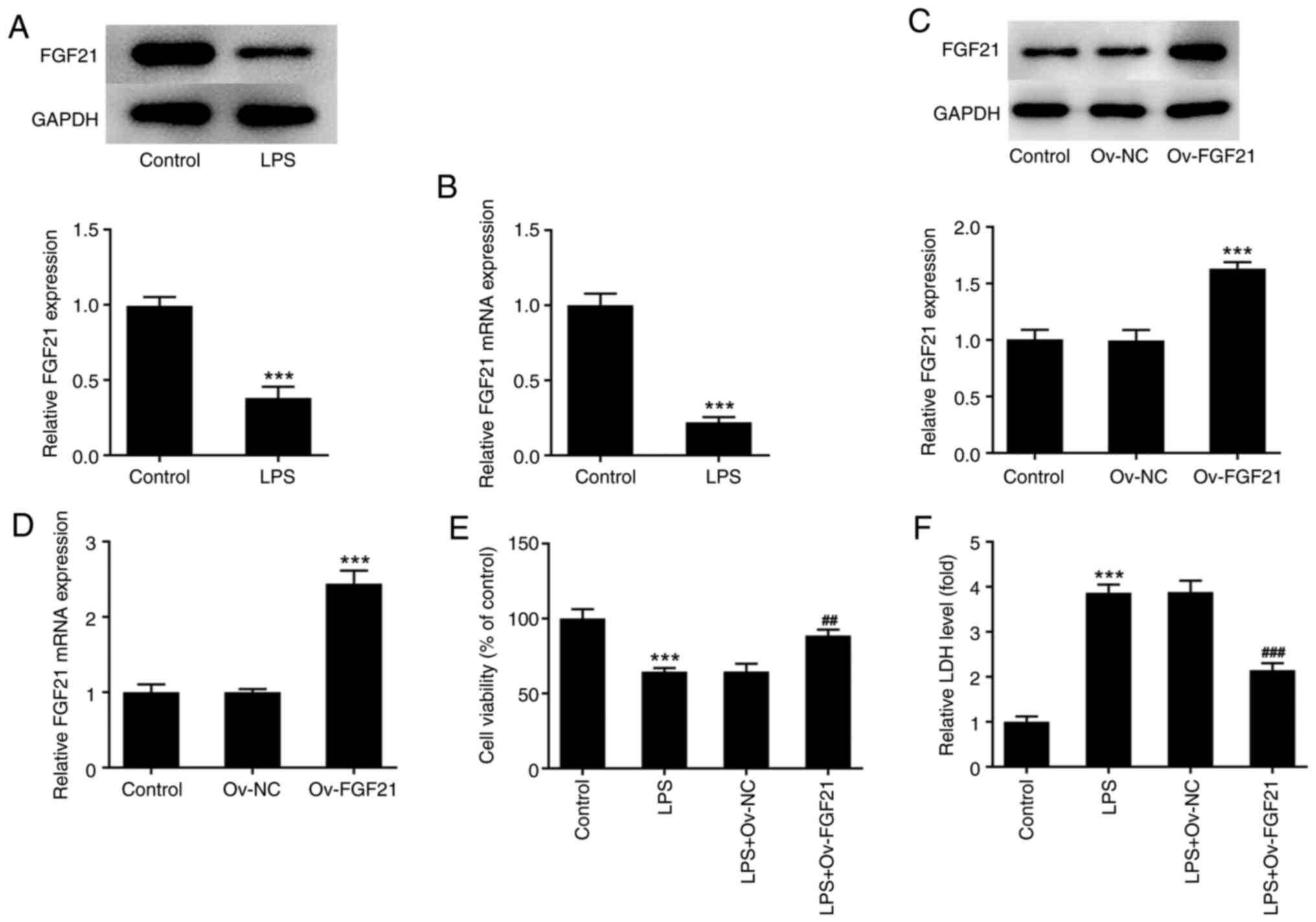
expression levels of FGF21 in LPS-induced ATDC5 cells. The results
demonstrated that the expression level of FGF21 was significantly
decreased in the LPS-induced group compared with that in the
control group (Fig. 1A and B).
Subsequently, cell transfection was performed to overexpress FGF21
and cells were divided into control, Ov-NC and Ov-FGF21 groups.
RT-qPCR and western blotting were used to assess cell transfection
efficiency. Compared with that in the Ov-NC group, the expression
level of FGF21 in Ov-FGF21 group was significantly increased,
indicating successful transfection (Fig. 1C and D).
Subsequently, cells were divided into control, LPS,
LPS + Ov-NC and LPS + Ov-FGF21 groups. Cell viability was detected
by performing a CCK-8 assay. The results showed that the viability
of cells in the LPS group was significantly decreased compared with
that in the control group. Moreover, compared with that in the LPS
+ Ov-NC group, the cell viability of LPS-treated,
FGF21-overexpression cells was significantly increased (Fig. 1E). LDH release by cells was
detected using an LDH kit. Compared with that in the control group,
the release of LDH by cells in the LPS group was significantly
increased. However, LDH release by LPS-induced cells was
significantly decreased by FGF21 overexpression compared with that
in the LPS + Ov-NC group (Fig.
1F).
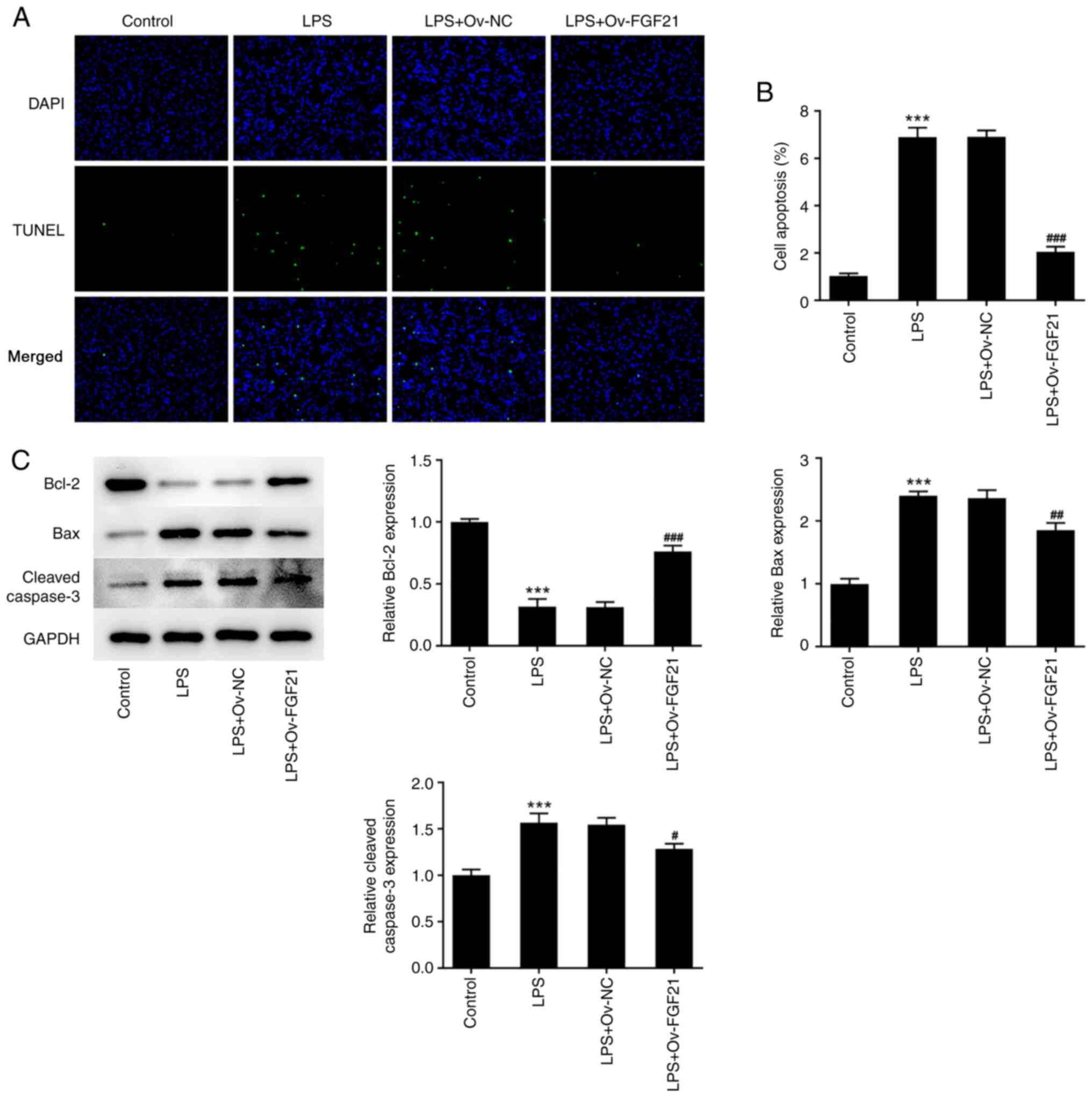
A TUNEL assay was conducted to detect apoptosis and
western blotting was performed to detect the expression levels of
apoptosis-related proteins. The apoptotic rate was significantly
increased after LPS induction compared with that in the control
group (Fig. 2A and B), which was
accompanied by significantly decreased expression levels of Bcl-2
and significantly increased expression levels of Bax and cleaved
caspase 3 (Fig. 2C). Compared
with LPS + Ov-NC, the apoptotic rate was significantly decreased in
the LPS + Ov-FGF21 group; this was accompanied by significantly
increased expression levels of Bcl-2 and significantly decreased
expression levels of Bax and cleaved caspase 3. These results
indicated that FGF21 overexpression increased the activity and
inhibited the apoptosis of LPS-induced ATDC5 cells.
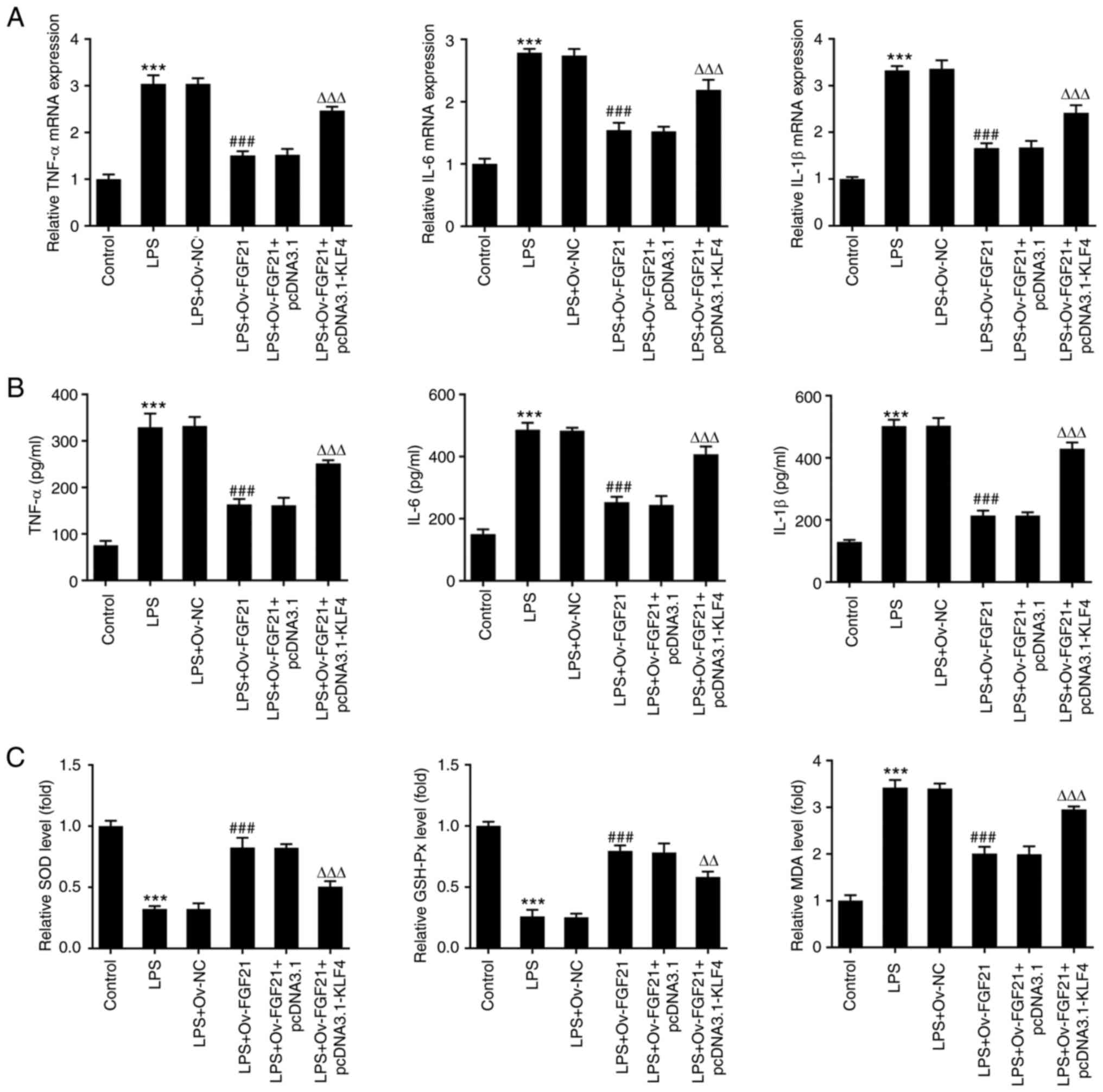
FGF21 overexpression attenuates the
inflammatory response and oxidative stress of LPS-induced ATDC5
cells
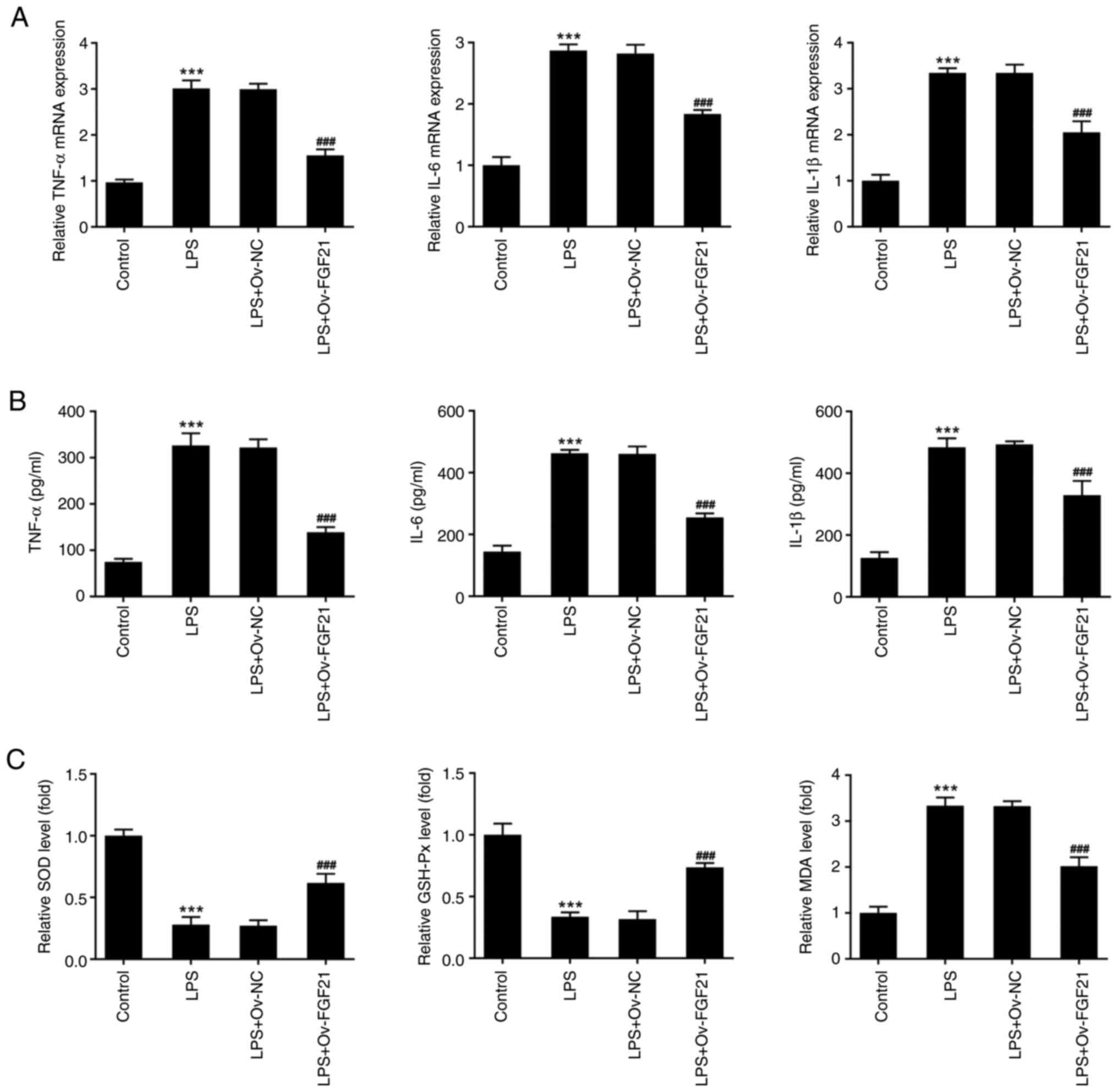
To verify the effect of FGF21 overexpression on the
inflammatory response of cells, the expression levels of
inflammation-related factors, including TNF-α, IL-6 and IL-1β, were
detected via RT-qPCR and ELISAs. The expression levels of TNF-α,
IL-6 and IL-1β in LPS-induced ATDC5 cells were significantly
increased compared with those in the control group (Fig. 3A and B). Compared with those in
the LPS + Ov-NC group, the expression levels of these inflammatory
cytokines were significantly decreased in the LPS + Ov-FGF21
group.
Oxidative stress levels in the cells were also
measured. Compared with those in the control group, the levels of
SOD and GSH-Px were significantly decreased in the LPS group,
whereas the level of MDA was significantly increased. Compared with
those in the LPS + Ov-NC group, the levels of SOD and GSH-Px were
significantly increased in the LPS + Ov-FGF21 group, whereas the
level of MDA was significantly decreased (Fig. 3C). These results revealed that
FGF21 overexpression decreased the inflammatory response and
oxidative stress of LPS-induced ATDC5 cells.
Transcription factor KLF4 inhibits
FGF21 expression
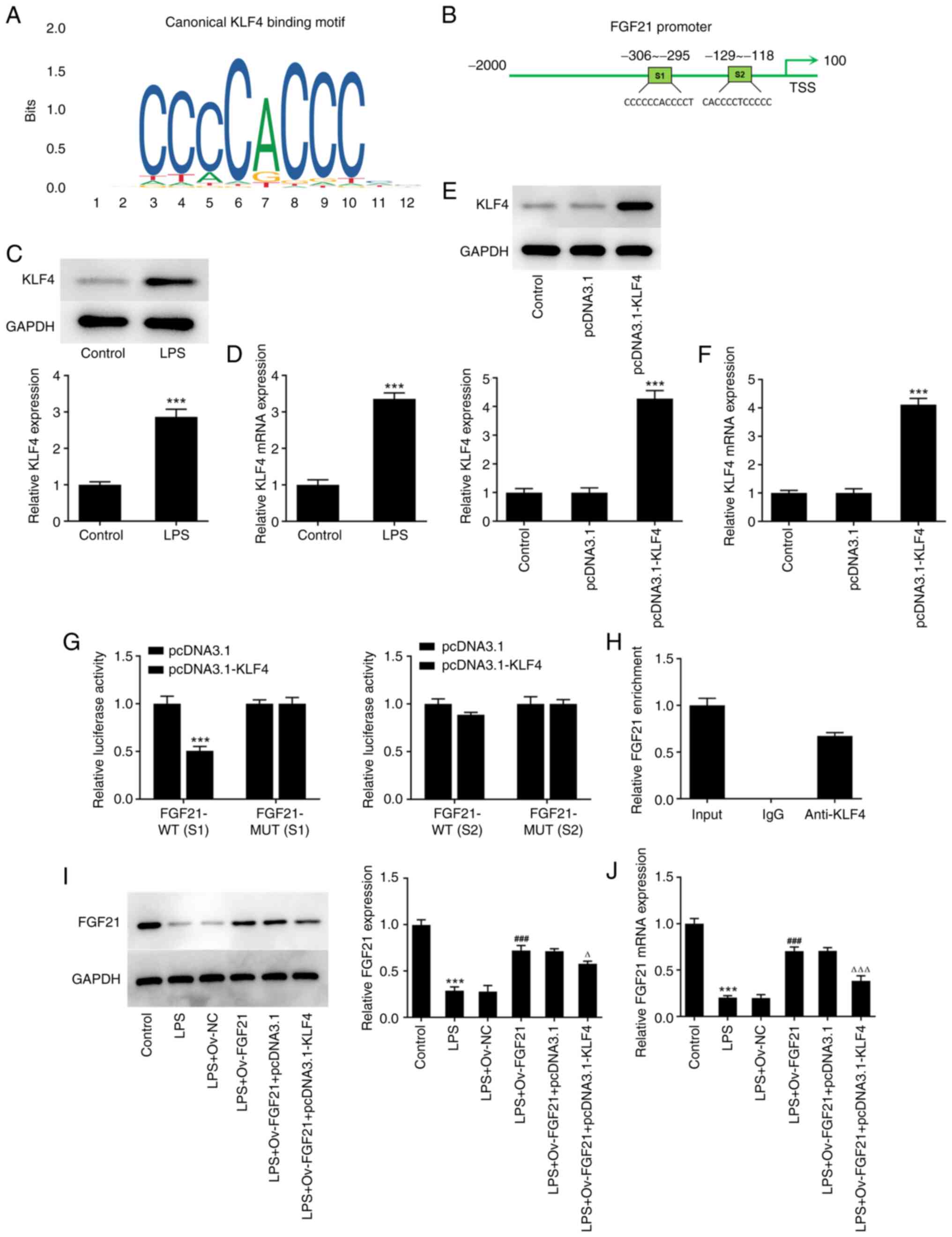
The JASPAR database was used to predict the binding
sites between KLF4 and FGF21 (Fig. 4A
and B). Subsequently, the expression level of KLF4 in cells was
detected via RT-qPCR and western blotting. KLF4 expression was
significantly increased in the LPS group compared with that in the
control group (Fig. 4C and D).
Subsequently, the KLF4 overexpression plasmid was constructed, and
the transfection efficiency was determined via RT-qPCR and western
blotting. The expression level of KLF4 in the pcDNA3.1-KLF4 group
was significantly increased compared with that in the pcDNA3.1
group (Fig. 4E and F).
Subsequently, the binding sites between FGF21 and
KLF4 were verified using luciferase reporter gene assays. The S1
and S2 sequences of FGF21 were mutated (MUT), namely FGF21-MUT (S1)
and FGF21-MUT (S2) (Fig. 4B).
Compared with that in the pcDNA3.1 group, the luciferase activity
in the pcDNA3.1-KLF4 group was significantly decreased in FGF21-WT
(S1), whereas the activity level was not significantly altered in
FGF21-MUT (S1). Similarly, there was no significant change in the
luciferase activity of FGF21-WT (S2) and FGF21-MUT (S2) between the
pcDNA3.1 and pcDNA3.1-KLF4 groups, indicating that the binding site
of KLF4 and FGF21 was at the S1 site of FGF21 (Fig. 4G). The ChIP assay results also
confirmed the association between FGF21 and KLF4 (Fig. 4F and H). Cells were divided into
control, LPS, LPS + Ov-NC, LPS + Ov-FGF21, LPS + Ov-FGF21 +
pcDNA3.1 and LPS + Ov-FGF21 + pcDNA3.1-KLF4 groups. The expression
level of FGF21 was detected via RT-qPCR and western blotting. KLF4
overexpression significantly downregulated the expression level of
FGF21 in cells compared with LPS + Ov-FGF21 + pcDNA3.1 (Fig. 4I and J). Collectively, these
results suggested that KLF4 inhibited FGF21 expression.
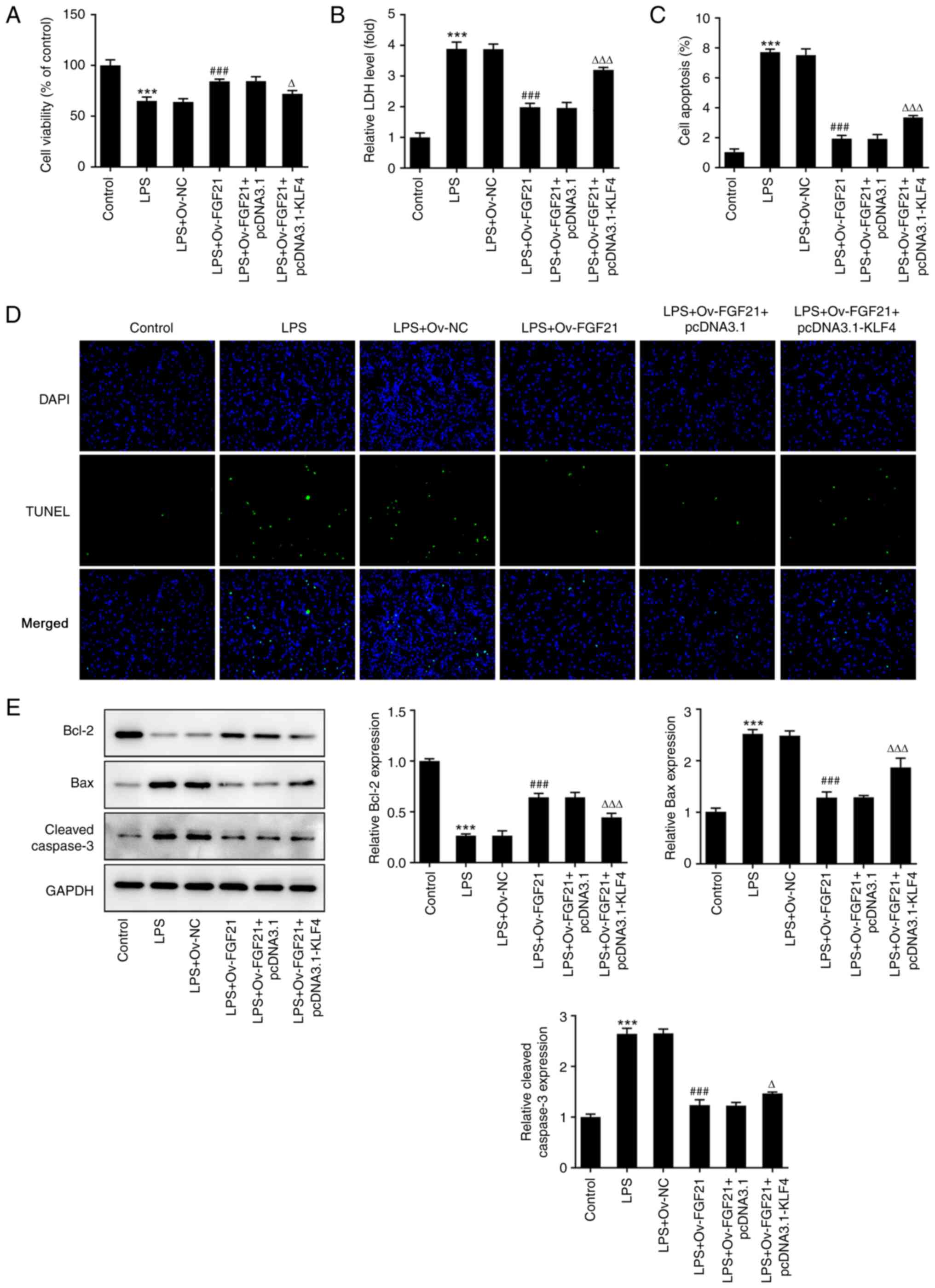
KLF4 overexpression reverses the
protective effect of FGF21 overexpression on the inflammatory
injury of LPS-induced ATDC5 cells
To further investigate the effects of KLF4 and FGF21
on LPS-induced inflammatory injury in ATDC5 cells, cells were
divided into the control, LPS, LPS + Ov-NC, LPS + Ov-FGF21 +
pcDNA3.1 and LPS + Ov-FGF21 + pcDNA3.1-KLF4 groups. Cell viability
was detected using a CCK-8 assay. The results demonstrated that
cell viability was significantly decreased in the LPS + Ov-FGF21 +
pcDNA3.1-KLF4 group compared with that in the LPS + Ov-FGF21 +
pcDNA3.1 group (Fig. 5A). LDH
release was detected using the LDH kit. Compared with that in the
LPS + Ov-FGF21 + pcDNA3.1 group, LDH release was increased in the
LPS + Ov-FGF21 + pcDNA3.1-KLF4 group (Fig. 5B). Cell apoptosis was detected by
performing a TUNEL assay and western blotting. Compared with the
LPS + Ov-FGF21 + pcDNA3.1 group, the apoptotic rate was
significantly increased in the LPS + Ov-FGF21 + pcDNA3.1-KLF4
group, which was accompanied by significantly decreased Bcl-2
protein expression, and significantly increased Bax and cleaved
caspase 3 protein expression (Fig.
5C-E). In addition, the levels of inflammatory factors and
oxidative stress were detected. Compared with those in the LPS +
Ov-FGF21 + pcDNA3.1 group, the levels of TNF-α, IL-6, IL-1β and MDA
in the LPS + Ov-FGF21 + pcDNA3.1-KLF4 group were significantly
increased, whereas the levels of SOD and GSH-Px were significantly
decreased (Fig. 6A-C). These
results suggested that KLF4 overexpression reversed the inhibitory
effect of FGF21 overexpression on LPS-induced inflammation and
oxidative stress in ATDC5 cells.
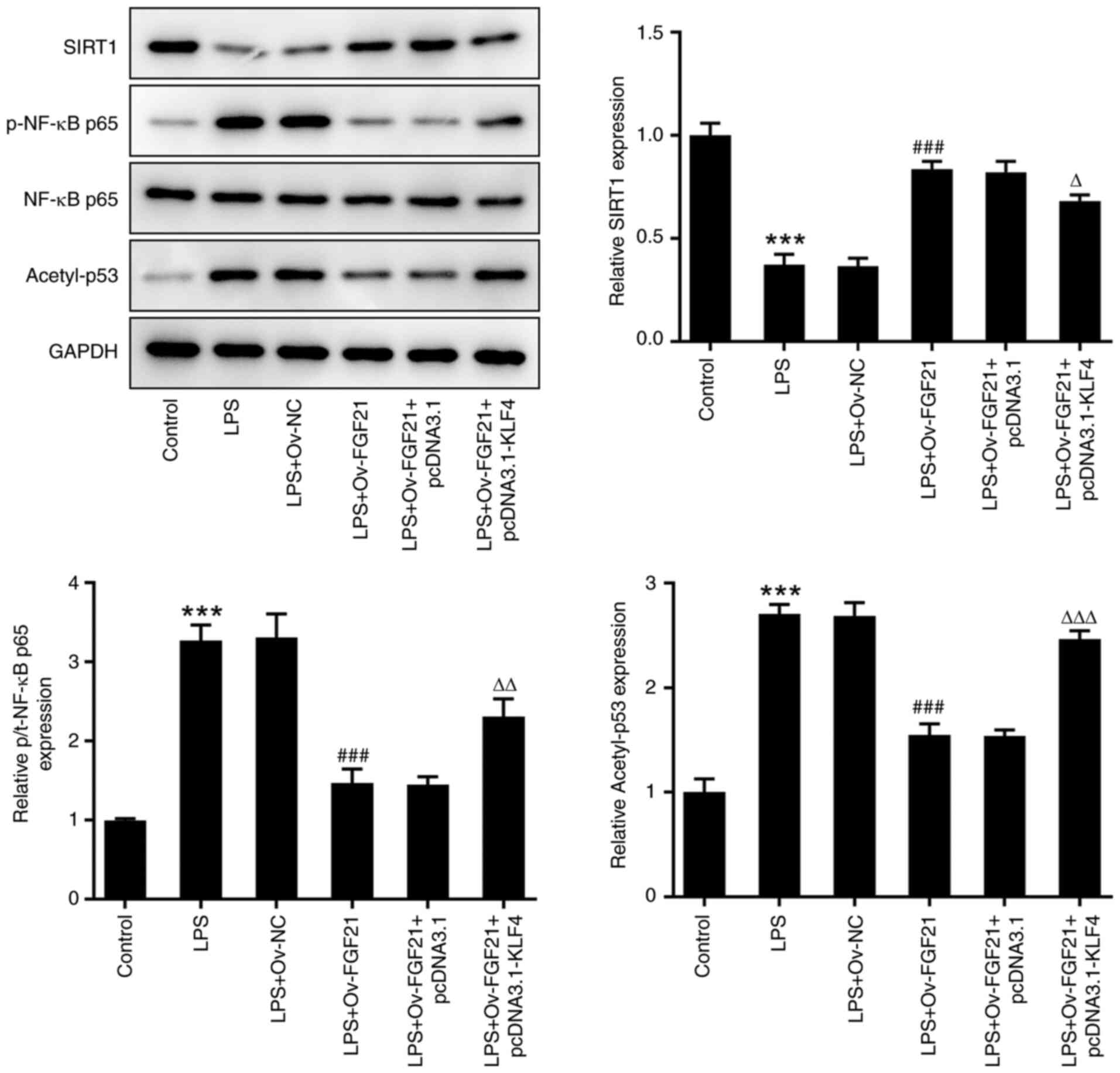
FGF21 overexpression protects
LPS-induced ATDC5 cells via the SIRT1/NF-κB/p53 signaling
pathway
Finally, the expression levels of SIRT1/NF-κB/p53
pathway-related proteins, including SIRT1, p-NF-κB p65, NF-κB p65
and acetyl-p53, were detected via western blotting. Compared with
those in the control group, the expression levels of SIRT1 were
significantly decreased, whereas the expression levels of p-NF-κB
p65 and acetyl-p53 were significantly increased in the LPS group.
After FGF21 overexpression, the expression level of SIRT1 was
increased, and the expression levels of p-NF-κB p65 and acetyl-p53
were decreased in LPS-treated cells. After KLF4 overexpression, the
effects of LPS and FGF21 overexpression on the expression levels of
SIRT1, p-NF-κB p65 and acetyl-p53 were partially reversed (Fig. 7). These results suggested that
FGF21 overexpression may protect LPS-induced ATDC5 cells via the
SIRT1/NF-κB/p53 signaling pathway.
Discussion
ATDC5 cells can differentiate into chondrocytes. In
addition, ATDC5 cells are widely used in the study of AS (23,24). LPS is commonly used as an inducer
of inflammatory responses, stimulating cells to induce inflammatory
responses and produce proinflammatory cytokines (25). Therefore, in the present study,
ATDC5 cells were selected to establish an LPS-induced joint injury
model. The results demonstrated that LPS induced decreased cell
viability, and increased LDH release, apoptosis, inflammatory
cytokine levels and oxidative stress levels, indicating successful
model induction.
In the late stage of AS, progressive joint
destruction and bony fusion lead to a reduced range of motion and
even disability of the joint, severely affecting the quality of
life of the patient (26). At
present, the pathogenesis of AS is not completely understood; thus,
it is of great significance to further study the molecular
mechanism underlying its pathogenesis.
FGF21, a member of the FGF family, is an endocrine
factor that primarily mediates glucose and lipid metabolism, and
serves a key role in maintaining glucose homeostasis and protecting
the liver, heart, kidney and skin from damage and cancer cell
proliferation (27). Studies have
shown that FGF21 served an important role in chronic inflammatory
diseases. For instance, FGF21 improved collagen-induced arthritis
by regulating oxidative stress and inhibiting the NF-κB signaling
pathway (13). FGF21 and
adalimumab were found to exert similar pharmacological effects on
collagen-induced rheumatoid arthritis by regulating systemic
inflammation (28). In addition,
metformin was shown to improve experimental obesity-related
autoimmune arthritis by inducing FGF21 expression and brown
adipocyte differentiation (14).
These findings suggested that FGF21 serves an important role in the
regulation of articular inflammation. However, to the best of our
knowledge, the role of FGF21 in AS has not been reported so far. In
the present study, the expression level of FGF21 was significantly
decreased in LPS-induced ATDC5 cells, which was consistent with the
study conducted by Chen et al (12) that demonstrated a decrease in
FGF21 expression in the cartilage of rheumatoid arthritis model
mice. The present study further verified the role of FGF21 in AS.
FGF21 overexpression promoted LPS-induced viability of ATDC5 cells
and decreased cell apoptosis, as well as inhibiting the cellular
inflammatory response by downregulating the expression of TNF-α,
IL-6 and IL-1β in cells. In addition, FGF21 overexpression
inhibited cellular oxidative stress in LPS-induced ATDC5 cells. The
levels of SOD and GSH-Px were increased, and the level of MDA was
decreased after FGF21 overexpression in LPS-induced ATDC5
cells.
The transcription factor KLF4 was able to
transcriptionally activate the expression of FGF21, as determined
via bioinformatics analysis. Moreover, the relationship between the
two factors was confirmed by performing dual-luciferase reporter
gene and ChIP assays. KLF4 also serves an important role in
inflammatory joint diseases (16,29,30). In the present study, KLF4
overexpression reversed the protective effect of FGF21
overexpression on LPS-induced inflammatory injury in ATDC5
cells.
In addition, the SIRT1/NF-κB/p53 signaling pathway
was activated in LPS-induced ATDC5 cells. After FGF21
overexpression, the SIRT1/NF-κB/p53 signaling pathway was
inhibited. KLF4 overexpression reversed the inhibitory effect of
FGF21 overexpression on the SIRT1/NF-κB/p53 signaling pathway in
LPS-induced ATDC5 cells. SIRT1, as a histone III deacetylase widely
present in human cells, regulates the tumor suppressor factor p53,
NF-κB and other factors via deacetylation to exert a variety of
biological activities (31). In
addition, a previous study reported that dipeptidyl peptidase-4
inhibitors induce FGF21 expression via SIRT1 signaling, thereby
improving myocardial energy metabolism (17). FGF21 alleviates acute pancreatitis
by activating the SIRT1 signaling pathway (18). Therefore, it may be preliminarily
concluded that FGF21 overexpression may protect LPS-induced ATDC5
cells via the SIRT1/NF-κB/p53 signaling pathway. The mechanism
underlying the regulatory effects of KLF4 and FGF21 on the
SIRT1/NF-κB/p53 signaling pathway requires further
investigation.
In conclusion, the present study demonstrated that
KLF4 downregulates FGF21 to activate inflammatory injury and
oxidative stress of LPS-induced ATDC5 cells via SIRT1/NF-κB/p53
signaling. (Fig. S1). Therefore,
the present study provided a solid theoretical basis for the
pathogenesis of AS and the drug targeted therapy of AS.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ningxia Natural Science
Foundation (grant no. NZ14164).
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
XC and DG wrote the manuscript and analyzed the
data. JW and CL carried out the experiments, supervised the present
study, searched the literature and revised the manuscript. All
authors read and approved the final manuscript. XC and DG confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Feldtkeller E, Khan MA, van der Heijde D,
van der Linden S and Braun J: Age at disease onset and diagnosis
delay in HLA-B27 negative vs. positive patients with ankylosing
spondylitis. Rheumatol Int. 23:61–66. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zvyagin IV, Mamedov IZ, Britanova OV,
Staroverov DB, Nasonov EL, Bochkova AG, Chkalina AV, Kotlobay AA,
Korostin DO, Rebrikov DV, et al: Contribution of functional KIR3DL1
to ankylosing spondylitis. Cell Mol Immunol. 7:471–476. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fiorillo MT, Haroon N, Ciccia F and Breban
M: Editorial: Ankylosing spondylitis and related immune-mediated
disorders. Front Immunol. 10:12322019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith JA: Update on ankylosing
spondylitis: Current concepts in pathogenesis. Curr Allergy Asthma
Rep. 15:4892015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye G, Xie Z, Zeng H, Wang P, Li J, Zheng
G, Wang S, Cao Q, Li M, Liu W, et al: Oxidative stress-mediated
mitochondrial dysfunction facilitates mesenchymal stem cell
senescence in ankylosing spondylitis. Cell Death Dis. 11:7752020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pishgahi A, Abolhasan R, Danaii S,
Amanifar B, Soltani-Zangbar MS, Zamani M, Kamrani A, Ghorbani F,
Mehdizadeh A, Kafil HS, et al: Immunological and oxidative stress
biomarkers in Ankylosing Spondylitis patients with or without
metabolic syndrome. Cytokine. 128:1550022020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feijóo M, Túnez I, Ruiz A, Tasset I, Muñoz
E and Collantes E: Oxidative stress biomarkers as indicator of
chronic inflammatory joint diseases stage. Reumatol Clin. 6:91–94.
2010.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maksymowych WP, Chiowchanwisawakit P,
Clare T, Pedersen SJ, Østergaard M and Lambert RG: Inflammatory
lesions of the spine on magnetic resonance imaging predict the
development of new syndesmophytes in ankylosing spondylitis:
Evidence of a relationship between inflammation and new bone
formation. Arthritis Rheum. 60:93–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Solmaz D, Kozaci D, Sari I, Taylan A, Önen
F, Akkoç N and Akar S: Oxidative stress and related factors in
patients with ankylosing spondylitis. Eur J Rheumatol. 3:20–24.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fisher FM and Maratos-Flier E:
Understanding the physiology of FGF21. Annu Rev Physiol.
78:223–241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Staiger H, Keuper M, Berti L, Hrabe de
Angelis M and Häring HU: Fibroblast growth factor 21-metabolic role
in mice and men. Endocr Rev. 38:468–488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen H, He C, Liu Y, Li X, Zhang C, Qin Q
and Pang Q: LncRNA-GAS5 inhibits expression of miR 103 and
ameliorates the articular cartilage in adjuvant-induced arthritis
in obese mice. Dose Response. 18:15593258209427182020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Y, Li S, Liu Y, Tian G, Yuan Q, Bai F,
Wang W, Zhang Z, Ren G, Zhang Y and Li D: Fibroblast growth factor
21 (FGF21) ameliorates collagen-induced arthritis through
modulating oxidative stress and suppressing nuclear factor-kappa B
pathway. Int Immunopharmacol. 25:74–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim EK, Lee SH, Lee SY, Kim JK, Jhun JY,
Na HS, Kim SY, Choi JY, Yang CW, Park SH and Cho ML: Metformin
ameliorates experimental-obesity-associated autoimmune arthritis by
inducing FGF21 expression and brown adipocyte differentiation. Exp
Mol Med. 50:e4322018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghaleb AM and Yang VW: Kruppel-like factor
4 (KLF4): What we currently know. Gene. 611:27–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi S, Lee K, Jung H, Park N, Kang J, Nam
KH, Kim EK, Ju JH and Kang KY: Kruppel-like factor 4 positively
regulates autoimmune arthritis in mouse models and rheumatoid
arthritis in patients via modulating cell survival and inflammation
factors of fibroblast-like synoviocyte. Front Immunol. 9:13392018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Furukawa N, Koitabashi N, Matsui H, Sunaga
H, Umbarawan Y, Syamsunarno MRAA, Yamaguchi A, Obokata M, Hanaoka
H, Yokoyama T and Kurabayashi M: DPP-4 inhibitor induces FGF21
expression via sirtuin 1 signaling and improves myocardial energy
metabolism. Heart Vessels. 36:136–146. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Q, Li J, Ma J, Yang X, Ni M, Zhang Y,
Li X, Lin Z and Gong F: Fibroblast growth factor 21 alleviates
acute pancreatitis via activation of the Sirt1-autophagy signalling
pathway. J Cell Mol Med. 24:5341–5351. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu Q, Sun Y, Li Y, Shi H, Teng J, Liu H,
Cheng X, Ye J, Su Y, Yin Y, et al: Anti-SIRT1 autoantibody is
elevated in ankylosing spondylitis: A potential disease biomarker.
BMC Immunol. 19:382018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen M, Chen C, Gao Y, Li D, Huang D, Chen
Z, Zhao X, Huang Q, Wu D, Lai T, et al: Bergenin-activated SIRT1
inhibits TNF-α-induced proinflammatory response by blocking the
NF-κB signaling pathway. Pulm Pharmacol Ther. 62:1019212020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lv C, He Y, Wei M, Xu G, Chen C, Xu Z and
Ding Z: CTRP3 ameliorates cerulein-induced severe acute
pancreatitis in mice via SIRT1/NF-κB/p53 axis. Biosci Rep.
40:BSR202000922020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Neerinckx B, Kollnberger S, Shaw J and
Lories R: No evidence for a direct role of HLA-B27 in pathological
bone formation in axial SpA. RMD Open. 3:e0004512017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong Y, Yan X, Yang X, Yu C, Deng Y, Song
X and Zhang L: Notoginsenoside R1 suppresses miR-301a via NF-κB
pathway in lipopolysaccharide-treated ATDC5 cells. Exp Mol Pathol.
112:1043552020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JW, Bae CJ, Choi YJ, Kim SI, Kwon YS,
Lee HJ, Kim SS and Chun W: 3,4,5-trihydroxycinnamic acid inhibits
lipopolysaccharide (LPS)-induced inflammation by Nrf2 activation in
vitro and improves survival of mice in LPS-induced endotoxemia
model in vivo. Mol Cell Biochem. 390:143–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Golder V and Schachna L: Ankylosing
spondylitis: An update. Aust Fam Physician. 42:780–784.
2013.PubMed/NCBI
|
|
27
|
Geng L, Lam KSL and Xu A: The therapeutic
potential of FGF21 in metabolic diseases: From bench to clinic. Nat
Rev Endocrinol. 16:654–667. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu D, Ye X, Che R, Wu Q, Qi J, Song L, Guo
X, Zhang S, Wu H, Ren G and Li D: FGF21 exerts comparable
pharmacological efficacy with Adalimumab in ameliorating
collagen-induced rheumatoid arthritis by regulating systematic
inflammatory response. Biomed Pharmacother. 89:751–760. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mo X, Chen J, Wang X, Pan Z, Ke Y, Zhou Z,
Xie J, Lv G and Luo X: Kruppel-like factor 4 regulates the
expression of inducible nitric oxide synthase induced by TNF-α in
human fibroblast-like synoviocyte MH7A cells. Mol Cell Biochem.
438:77–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Q, Gong L, Qi R, Qing W, Zou M, Ke Q,
Zhang L, Tang X, Nie Q, Yang Y, et al: Oxidative stress-induced
KLF4 activates inflammatory response through IL17RA and its
downstream targets in retinal pigment epithelial cells. Free Radic
Biol Med. 147:271–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Karbasforooshan H and Karimi G: The role
of SIRT1 in diabetic cardiomyopathy. Biomed Pharmacother.
90:386–392. 2017. View Article : Google Scholar : PubMed/NCBI
|