Introduction
In 1892, Ivanowski reported that extracts from
infected tobacco plants remained infectious following filtration
through a Chamberland filter candle (1). As bacteria did not pass through such
filters, a novel group of filterable pathogens were discovered.
Several years after this discovery, Beijerinck was the first to
refer to the causative agent of the tobacco mosaic as a ‘virus’. He
revealed that the ‘cause of illness’ was able to migrate in an agar
gel, which indicated that it was an infectious soluble agent, and
not fixed in place as would be the case for bacteria. Discoveries
made on the tobacco mosaic virus triggered the beginning of
virology (1), while the subsequent
discovery of bacteriophages by Felix d'Herelle and of numerous
other viruses by the early 20th century further advanced the field
(2). In 1940, the first electron
micrograph of a bacteriophage was published, which convinced
sceptics who had argued that bacteriophages were relatively simple
enzymes and not viruses (3). At
present, the current concept of a virus refers to an
ultramicroscopic (20–300 nm in diameter), metabolically inert,
infectious agent that replicates only within the cells of living
hosts, primarily bacteria, plants and animals, is composed of
either RNA or DNA as its genetic material, and is enclosed by a
protein coat or capsid and, in more complex types, by a surrounding
envelope (4,5). Viruses are common pathogens in humans,
causing a number of diseases such as hepatitis, measles,
poliomyelitis and smallpox (6).
However, during the last twenty years, viruses have emerged as
powerful tools in gene therapy, as they have been extensively used
as vehicles of therapeutic genes for the treatment of several
monogenic diseases such as immunodeficiencies (X-SCID, ADA-SCID),
β-thalassemia, sickle cell disease or hemophilia, but also for
complex diseases such as cancer (7). The advent of CAR-T cell gene therapy
by utilization of lentiviral vectors has further revolutionized
cancer treatment (8) while T-VEC,
an engineered herpes simplex virus-1, has shown great promise in
melanoma (9).
In 2003, La Scola et al (10) described the Mimivirus, which is a
parasite of amoebae and was long considered to be a bacterium due
to its size. Specifically, this particular virus has a diameter of
500 nm and a 1.2 Mb DNA genome. However, the Mimivirus is not the
only type of giant virus. Philippe et al (11) later discovered two Pandoraviruses
with 1.9 and 2.5 Mb DNA genomes, respectively.
The Mimivirus was observed for the first time in
1992, when a case of nosocomial pneumonia was investigated for
amoeba-associated microorganisms; however, at the time it was
considered to be an intracellular bacterium based on its appearance
under the light microscope. Furthermore, Acanthamoeba
polyphaga isolated from a water-cooling tower were reported to
contain this organism, which years later was characterized as a
giant virus and termed Acanthamoeba polyphaga Mimivirus
(APMV). This was the origin of the identification of the novel
family of Mimiviridae (10,12). Results from transmission electron
microscopy combined with relevant genomic data confirmed that the
organism was in fact a virus (10).
The Mimivirus has unique characteristics. Its genome
is larger than the majority of other viruses, and certain bacteria
and archaea, and it is comparable to the genome of certain
eukaryotes. The genome of the Mimivirus contains 1,262 genes, which
is three times higher than the number of genes contained in any
other virus (10). Furthermore,
genomic analysis has indicated that the Mimivirus may be a chimera,
due to their ability to exchange genetic material with their hosts
and also with other parasites that exist within the same host cell.
Additionally, these viruses exchange genes with other large DNA
viruses of amoeba, such as the Marseillevirus (13). The virions of the Marseilleviruses
enclose a genome ranging from 348 to 404 kb that encodes for
386–545 predicted proteins, while their overall genome organization
resembles that of the Mimivirus (13). These data prompted virologists to
establish Megavirales, a novel virus order composed of Mimivirus,
Marseillevirus and other similar viruses, as well as members of the
Poxviridae, Iridoviridae, Ascoviridae, Phycodnaviridae and
Asfarviridae families (14).
However, this order remains under discussion among virologists.
Evolution of the Mimivirus
A central characteristic of other large DNA viruses,
such as Poxviridae, is that their genome encompasses a stable
region that is unique to the virus and a variable region that is
frequently composed of, or is similar to, host genes. Specifically,
for the Mimivirus, the large diversity observed in the variable
regions prompted scientists to propose that these viruses are as
old as the three traditional domains on Earth, consisting of
Archaea, Bacteria and Eukarya, as proposed by Woese et al
(15). Notably, Raoult (16) have proposed that phylogenesis should
be performed not based on the classical ribosomal RNA genes
sequences, but on transfer RNA (tRNA) and RNA polymerase-encoding
genes. Such clustering resulted in four groups with different
genetic repertoires: Giant viruses, Archaea, Bacteria and Eukarya.
Thus, giant viruses may be considered as a fourth branch in the
tree of life (16–18). Interestingly, in 2017 Marcelino
et al (19) demonstrated
that Mimiviruses are a sister group of Eukarya using analysis of
evolutionary relationship of proteins involved in the translation
system.
Furthermore, Forterre (20) argues that giant viruses lie at the
origin of the eukaryotic nucleus and, according to the theory of
viral eukaryogenesis, the large DNA viruses were important in the
formation of the nucleus. Forterre (20) and others (21–23)
also propose that DNA may have been ‘devised’ by viruses in order
to convert a world of RNA-based organisms to one where DNA is the
major hereditary material. According to the ‘RNA world’ hypothesis,
RNA is considered to be the molecular basis of the origin of life
on Earth (24), primarily due to
its catalytic potential. Therefore, the theory proposed by Forterre
hypothesizes that early RNA cells and ancient RNA viruses
coexisted, and that early RNA cells were parasitized by these
viruses. However, the introduction of a DNA virus into such
primitive cells may have been facilitated by the fact that these
hosts may have begun to develop RNA-specific defense mechanisms to
protect them against an RNA viral infection. During that process,
viruses may have shared or exchanged gene sequences with the host
cells, leading to the introduction of DNA membrane-surrounded
vacuoles that later evolved into nuclei, which contained DNA
molecules that are steadier as genetic material compared to RNA
molecules in terms of resistance to degradation, and thus were
favored by natural selection (17).
Genome organization and replication cycle of
the Mimivirus
The Mimivirus is considered to be a unique type of
virus. Although their genome does not contain any ribosomal
RNA-encoding genes, it does include genes responsible for cellular
processes, such as protein translation and metabolism, with genes
including amino-acyl tRNA synthase, tRNA and translation factors.
The presence of these genes within the Mimivirus genome confers a
degree of independence in terms of viral replication, as the
protein machinery of the host is not an absolute necessity. The
Mimiviruses and Marseilleviruses possess numerous chimeric genes
with sequences that are derived from other viruses, bacteria or
eukaryotic organisms, suggesting the occurrence of lateral gene
transfer (25). The genome of the
Mimivirus comprises four primary groups of open reading frames
(ORFs), including Megavirales core genes, genes involved in lateral
gene transfer, duplicated genes and ORFans, which represents genes
with limited or no homology with any other characterized or mapped
nucleotide sequence (26). Their
genome also contains a component that is termed transpoviron, which
is equivalent to a transposon and is a mobile genetic element of ~7
kb that encompasses 6–8 protein-encoding genes (27,28).
Transpovirons encode a superfamily 1 helicase, which includes an
inactivated family B DNA polymerase domain. Based on the
phylogenetic analysis of the helicase domain, it has been concluded
that transpovirons evolved from polinton-like viruses via the
deletion of several genes (27).
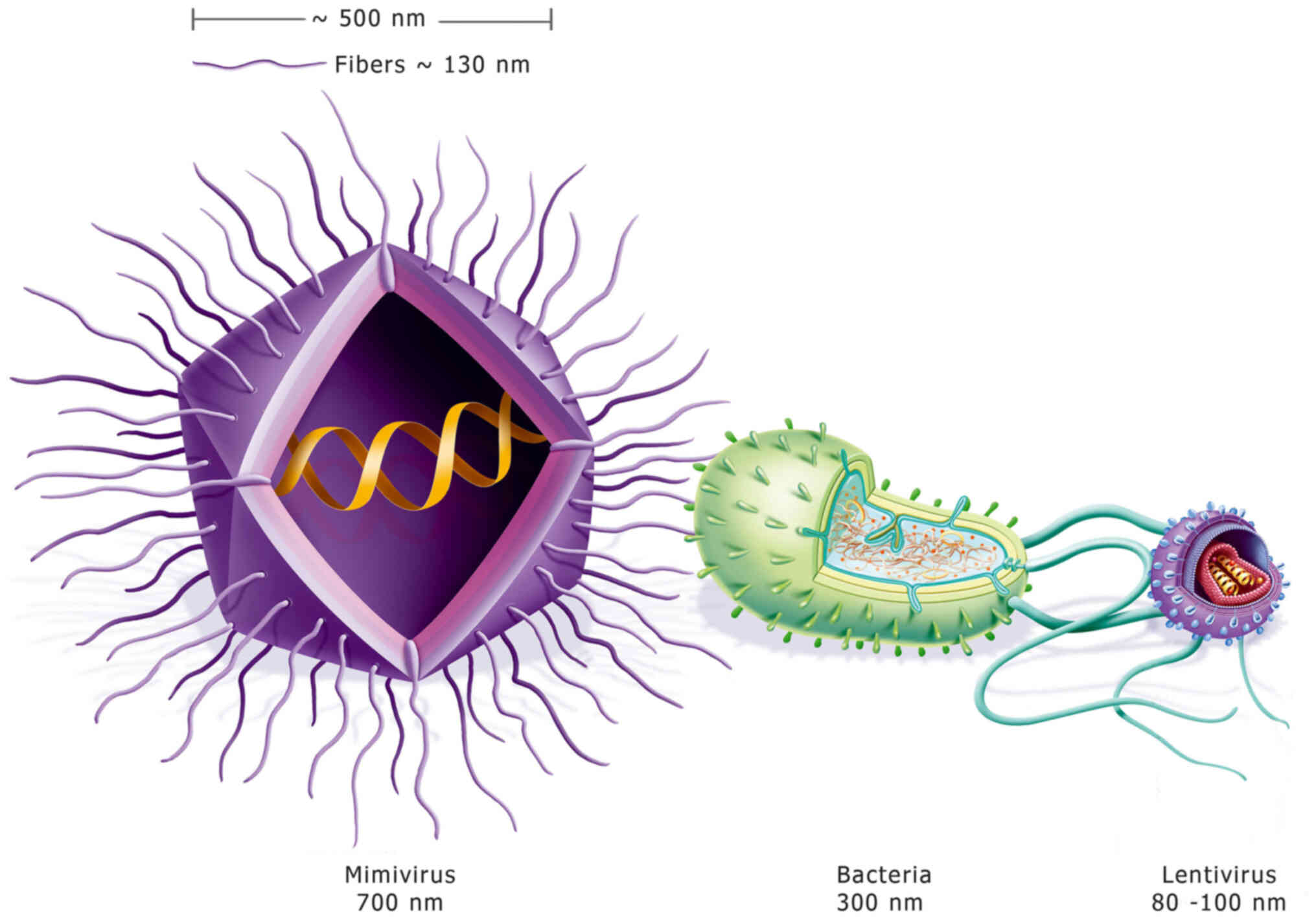
Mimivirus viral particles are composed of a protein
capsid of ~500 nm in diameter, which is enclosed within a
polysaccharide layer in which multiple fibers are embedded. These
fibers are ~120–140 nm long and 1.4 nm thick, which contributes to
an overall diameter of ~700 nm, as shown in Fig. 1. Another important characteristic of
the Mimivirus is their capacity to propagate exclusively within the
cytoplasm of the infected cell. Notably, although they are DNA
viruses that infect eukaryotic cells, Mimiviruses never enter the
nucleus of the host cell. Initially, Mimivirus particles are
internalized by endocytosis and are surrounded by a membrane
vesicle until the viral ‘star-gate’ channels open, which leads to
membrane fusion and results in the release of the genome-containing
capsids into the cytoplasm of the host cell (26). However, recent data using electron
microscopy, indicate that all giant viruses studied so far
including Mimivirus enter cells by phagocytosis. Fusion of
lysosomes surrounding the virion-containing phagosome has been
observed and suggested to trigger the virus uncoating (29). Recent studies attempt to
characterize proteins released from Giant viruses including Samba
virus, a member of Mimivirus lineage A, during infection
elucidating the molecular forces that trigger it. Remarkably, not
only the expected protein types are released, such as those
involved in genome translocation, blocking host replication and
hijack cell machinery, but also proteins that play a role in virus
protection from oxidative stress and chemotaxis (30). Mimivirus DNA replication occurs
exclusively in the cytoplasm and it is dependent on the host
nucleus. A vast number of nuclear factors that originate from the
endoplasmic reticulum or the outer nuclear membrane, which are
necessary for replication, are transferred through vesicle
transportation from the nucleus to the cytoplasm. The vesicles fuse
together in the cytoplasm to create what is termed a ‘viral
factory’, which is a transcriptional and translational mechanism
that copies the viral genome and ensures viral replication by
utilizing nucleotides of the host cell. The host cell dies within
~16 h following infection, a process which leads to the production
of ~10,000 new viruses (26).
The Mimivirus immune system
While examining the Mamavirus, a strain of
Mimivirus, La Scola et al (31) identified a virus that infects other
giant viruses, which was termed Sputnik. These small viruses were
detected using transmission electron microscopy and were later
termed virophages, in accordance with the name given to
bacteriophages. Virophage replication utilizes the viral mechanism
that the Mamavirus manufactures within its amoeba host. Sputnik,
which is not the only characterized virophage, has a genome size of
18 kb and carries genes from various host types. Another virus that
is similar to the Mimivirus, termed Cafeteria roenbergensis
virus (CroV), is parasitized by a virophage that is termed
‘Mavirus’ (32). CroV is a giant
virus that infects the marine bicosoecid flagellate, Cafeteria
roenbergensis, and has one of the largest genomes of all
established marine viruses (32,33).
In 2014, another virophage was characterized and was termed
Zamilon, originating from the Arabic word ‘xamilon’, which means
‘the neighbor’. Surprisingly, Zamilon was not able to infect all
strains of Mimivirus (34). In
2019, a novel virophage named Guarani was described. Guarani has a
19 kb double-stranded DNA genome encoding 22 genes, quite similar
to Sputnik genes, despite the fact that Guarani seems to be more
related to Zamilon. As all Sputnik strains, Guarani is capable of
infecting the three lineages of the Mimiviridae family (35).
Zamilon was used to infect different Mimiviruses
strains that were derived from the three characterized lineages, A,
B and C, of the Mimiviridae family (36). The results demonstrated that Zamilon
was able to infect B and C lineages, but strains from the A lineage
were resistant to Zamilon infection. All resistant strains,
including all strains from the A lineage and one strain from the C
lineage (Megavirus chilensis), were demonstrated to have a
28 bp Zamilon sequence incorporated in their genome. This sequence
is encoded by the ORF4 in the genome of Zamilon and results in the
expression of a protein similar to transposase A; the sequence is
integrated within the Mimivirus gene R349 and the corresponding
orthologous genes in APMV A Mimiviridae strains. Notably, all APMV
A genomes contained a 15 nucleotide-long sequence in four copies,
which was derived from the Zamilon 28 bp sequence. However, the
respective sequence was not detectable in group B and C genomes.
Furthermore, a significant association between Zamilon resistance
and the presence of the repeated Zamilon sequence in Mimiviruses
was observed (36). A recently
isolated Mimivirus that belongs to lineage A, lacking three of four
repeats of R349 gene, exhibited susceptibility to Zamilon (37). A more detailed analysis of this 15
bp repeat sequence and its vicinity within the Mimivirus genome
revealed that, downstream of the 15 bp repeats, there is a putative
phage-type endonuclease, which is encoded by the APMV A ORF R354
and is associated with a lamda exonuclease protein belonging to the
Cas4 nuclease family. Adjacent to the R349 gene, there is a
putative helicase domain associated with a SNF2 domain, encoded by
the APMV A ORF R350, which contains motifs characteristic of the
Cas3 protein. In addition, a putative RNase III gene, encoded by
the APMV A ORF R343, is localized upstream of the 15 bp sequence
repeats (36). In order to
determine the role of the aforementioned system in Zamilon
infection, Levasseur et al (36) employed RNA interference technology
to silence all associated gene sequences, 27 genes in total, in the
Mimivirus genome. The results demonstrated that the silencing of
R354, R350 and R349 significantly inhibited Zamilon infection.
Therefore, this network of ORFs was termed the ‘Mimivirus virophage
resistance element’ or MIMIVIRE (36).
MIMIVIRE: The Mimivirus CRISPR-Cas
system
The above findings prompted scientists to associate
MIMIVIRE with the recently characterized CRISPR-Cas system
(38,39). In prokaryotes, the CRISPR-Cas system
has a key role in defense and is detected in ~48% of bacteria and
80% of archaea (38). The bacterial
type II clustered, regularly interspaced, short palindromic repeats
(CRISPR) system, comprises three minimal components (40): A CRISPR-associated effector nuclease
Cas9, a specificity-determining CRISPR RNA (crRNA), and an
auxiliary trans-activating crRNA (tracrRNA). The hybridization of
crRNA and tracrRNA leads Cas9 to target genomic loci that match a
20-nucleotide guide sequence (gRNA) contained at the 5′ end of
crRNA, and residing upstream of an essential 5′-NGG protospacer
adjacent motif (PAM). In addition, crRNA and tracrRNA duplexes may
also be fused to form chimeric single guide RNA (sgRNA), which
mimics the natural crRNA-tracrRNA hybrid. Thus, crRNA-tracrRNA
duplexes and sgRNAs may be used to bind to Cas9 and target specific
loci. Furthermore, this system allows bacteria to retain a memory
of viruses that have infected them previously, and allows them to
respond more efficiently during subsequent infections.
Specifically, during the first viral infection, a fragment of the
viral DNA is incorporated into the CRISPR locus. When bacteria
encounter the same virus strain again, they recognize the viral DNA
and digest it via the Cas9 nuclease, which is directed to cleave
the exogenous DNA via the crRNA-tracrRNA duplexes transcribed from
the respective region of the CRISPR locus. As crRNA-tracrRNA
duplexes and sgRNAs may be used to target Cas9 for multiplexed
genome editing in eukaryotic cells, the CRISPR system has
revolutionized the field of genome editing (41).
Comparison of the MIMIVIRE system to the CRISPR-Cas
system revealed that nuclease R354 may cleave the invading nucleic
acid and lead to unspecific cleavage and partial degradation of the
double-stranded DNA, being more efficient on low content (28–38%)
GC templates. Therefore, Mimivirus and virophage genes with ~29% GC
content are readily degraded, while the Acanthamoeba
polyphaga genome, with 59% GC-rich content, is not. Thus, the
components of the MIMIVIRE system that correspond to those of the
CRISPR-Cas system are the R354 protein, which cleaves the DNA, the
R350 gene with the proposed helicase activity and the 15 bp
sequence in the R349 ORF (36,42).
However, it should be noted that the similarity between the
MIMIVIRE and the CRISPR system remains controversial (43).
Mimiviruses and human health
At present, the importance of Mimiviruses in human
health is an area that has not been investigated extensively.
However, antibodies against Mimiviruses have been detected in a
technician who developed pneumonia after working with Mimiviruses,
while in a small number of reported cases, strains of Mimivirus
were detected in the lung of patients who developed pneumonia
(44–46), compatible with features fulfilling
several of the criteria for viral disease causation (47). Furthermore, antibodies against
Mimivirus-encoded collagen, have been implicated in rheumatoid
arthritis (48). Interestingly,
immunofluorescence studies of young asymptomatic adults revealed
that humans are frequently exposed to Marseillevirus (49). Marseillevirus has also been detected
in a lymph node of a patient with Hodgkin's lymphoma, associated
with IgG antibodies against the virus (50). However, the hypothesis suggesting
Marseillevirus as a potential additional viral causative agent of
Hodgkin's lymphoma requires further investigation. Another case
report documented the detection of Marseillevirus in the lymph node
of a child with adenitis of unknown etiology, suggesting that this
virus can cause symptomatic infection on a background of a
defective immune system (49).
Additionally, APMV has been reported to grow in human peripheral
blood mononuclear cells and induce immune reaction via the
production of type I interferons (IFNs) (51). This observation is the first
convincing evidence of a host-pathogen interaction between APMV and
humans in terms of immunity. Moreover, APMV is able to replicate on
IFN-α pretreated cells, but not on IFN-β pretreated cells, as they
are sensitive to the antiviral action of IFN-β in a dose-dependent
manner (52). However, APMV
infection is not able to induce the expression of IFN-stimulated
genes, and infected peripheral blood mononuclear cells do not
express viroceptors for IFN-α2 and IFN-β (52).
The IFN-α and β receptor subunit 1 complex controls
an exclusive group of genes by a signaling pathway that remains
unknown and is thought to be mediated by IFN regulatory factor 1.
One of these genes is the immune responsive gene 1 (IRG1) that
codes an enzyme responsible for producing itaconic acid, which is
an organic compound that inhibits isocitrate lyase. Isocitrate
lyase is an enzyme that has an important function in the glyoxylate
shunt, a process that is required for bacterial growth (53). IRG1 is expressed in macrophages thus
leading to an association between metabolism with immune defense.
Therefore, itaconic acid functions as an immune-supportive
metabolite in mammalian immune cells by exhibiting antibacterial
action (53). Previous studies
(51,54) have investigated itaconic acid as a
virucidal agent and reported its ability to inactivate APMV in
vitro in a dose-dependent manner. However, experimental data
clarifying whether the action of itaconic acid against APMV is
direct or indirect are still missing. Thus, as the production of
itaconic acid is mediated by IFN, this novel mechanism may also be
implicated in viral infections, and IFN-β antiviral activity may be
responsible for inhibiting APMV infections in human cells. Such
studies for the experimental validation of this working hypothesis
need to be performed.
Conclusions
Undoubtedly, Mimiviruses represent a novel concept
in biology and medicine, and constitute a novel and intriguing
field for future research, since their discovery has challenged our
traditional perception about viruses and the definition of life in
general. Their existence is undeniable; however, as with any novel
discovery, uncertainty exists concerning their pathogenicity in
humans and the importance of the MIMIVIRE system, which comprises
an immune network of ORFs that resembles the CRISPR-Cas system.
Thus, although numerous scientific issues are yet to
be addressed, research concerning giant viruses may provide
important opportunities for future experimentation and clinical
investigations in all aspects of biomedicine, including immunology,
molecular biology, oncology and internal medicine.
Acknowledgements
Not applicable.
Funding
The present review was co-funded by the European Union's
European Social Fund and Greek National Funds through the Program
THALIS, under the Operational Program Education and Lifelong
Learning of the National Strategic Reference Framework (project no.
383418; grant no. 70-3-11830) to Professor Kalliopi I. Pappa.
Availability of data and materials
Not applicable.
Authors' contributions
EK searched the literature and wrote the manuscript.
EP and EM reviewed the manuscript. KIP and NPA conceived and
designed the study, and edited the manuscript. Data authentication
is not applicable. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lecoq H: Discovery of the first virus, the
tobacco mosaic virus: 1892 or 1898? C R Acad Sci III. 324:929–933.
2001.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duckworth DH: Who discovered
bacteriophage? Bacteriol Rev. 40:793–802. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ackermann HW: Phage classification and
characterization. Methods Mol Biol. 501:127–140. 2009. View Article : Google Scholar
|
|
4
|
Wagner R: Chemical and biologic approaches
to the therapy of viral diseases. Am Rev Respir Dis. 88:404–419.
1963.
|
|
5
|
Gillen AL: Viruses: fallen genes coated
with protein. The Genesis of Germs: The Origin of Diseases and the
Coming Plagues. Alan G: New Leaf Publishing Group Inc.; Green
Forest, AR: pp. 92–105. 2007
|
|
6
|
Baron S, Fons M and Albrecht T: Viral
pathogens. Viral Pathogenesis Medical Microbiology. Baron S: 4th
edition. University of Texas Medical Branch at Galveston;
Galveston, TX: Chapter 45. 1996
|
|
7
|
Morgan RA, Gray D, Lomova A and Kohn DB:
Hematopoietic stem cell gene therapy: Progress and lessons learned.
Cell Stem Cell. 21:574–590. 2017. View Article : Google Scholar
|
|
8
|
Lee DW, Kochenderfer JN, Stetler-Stevenson
M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M,
Shah NN, et al: T cells expressing CD19 chimeric antigen receptors
for acute lymphoblastic leukaemia in children and young adults: A
phase 1 dose-escalation trial. Lancet. 385:517–528. 2015.
View Article : Google Scholar
|
|
9
|
Rehman H, Silk AW, Kane MP and Kaufman HL:
Into the clinic: Talimogene laherparepvec (T-VEC), a first-in-class
intratumoral oncolytic viral therapy. J Immunother Cancer.
4:532016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
La Scola B, Audic S, Robert C, Jungang L,
de Lamballerie X, Drancourt M, Birtles R, Claverie JM and Raoult D:
A giant virus in amoebae. Science. 299:20332003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Philippe N, Legendre M, Doutre G, Couté Y,
Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, et
al: Amoeba viruses with genomes up to 2.5 Mb reaching that of
parasitic eukaryotes. Science. 341:281–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raoult D, Audic S, Robert C, Abergel C,
Renesto P, Ogata H, La Scola B, Suzan M and Claverie JM: The
1.2-megabase genome sequence of Mimivirus. Science. 306:1344–1350.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boyer M, Yutin N, Pagnier I, Barrassi L,
Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, et
al: Giant Marseillevirus highlights the role of amoebae as a
melting pot in emergence of chimeric microorganisms. Proc Natl Acad
Sci USA. 106:21848–21853. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colson P, De Lamballerie X, Yutin N,
Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL,
Koonin EV, et al: ‘Megavirales’, a proposed new order for
eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol.
158:2517–25121. 2013. View Article : Google Scholar
|
|
15
|
Woese CR, Kandler O and Wheelis M: Towards
a natural system of organisms: Proposal for the domains archaea,
bacteria, and eucarya. Proc Natl Acad Sci USA. 87:4576–4579. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raoult D: TRUC or the need for a new
microbial classification. Intervirology. 56:349–353. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raoult D: Viruses reconsidered: The
discovery of more and more viruses of record-breaking size calls
for a reclassification of life on Earth. Scientist. 28:32014.
|
|
18
|
Boyer M, Madoui MA, Gimenez G, La Scola B
and Raoult D: Phylogenetic and phyletic studies of informational
genes in genomes highlight existence of a 4th domain of life
including giant viruses. PLoS One. 5:e155302010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marcelino VM, Espinola MVPC, Serrano-Solis
V and Farias ST: Evolution of the genus Mimivirus based on
translation protein homology and its implication in the tree of
life. Genet Mol Res. 16:gmr160397842017. View Article : Google Scholar
|
|
20
|
Forterre P: Giant viruses: Conflicts in
revisiting the virus concept. Intervirology. 53:362–378. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bell PJ: Viral eukaryogenesis: Was the
ancestor of the nucleus a complex DNA virus? J Mol Evol.
53:251–256. 2001. View Article : Google Scholar
|
|
22
|
Bell PJ: Sex and the eukaryotic cell cycle
is consistent with a viral ancestry for the eukaryotic nucleus. J
Theor Biol. 243:54–63. 2006. View Article : Google Scholar
|
|
23
|
Takemura M: Poxviruses and the origin of
the eukaryotic nucleus. J Mol Evol. 52:419–425. 2001. View Article : Google Scholar
|
|
24
|
Joyce GF: RNA evolution and the origins of
life. Nature. 338:217–224. 1989. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Colson P and Raoult D: Gene repertoire of
amoeba-associated giant viruses. Intervirology. 53:330–343. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abrahão JS, Dornas FP, Silva LC, Almeida
GM, Boratto PV, Colson P, La Scola B and Kroon EG: Acanthamoeba
polyphaga mimivirus and other giant viruses: An open field to
outstanding discoveries. Virol J. 11:1202014. View Article : Google Scholar
|
|
27
|
Krupovic M and Koonin EV:
Self-synthesizing transposons: Unexpected key players in the
evolution of viruses and defense systems. Curr Opin Microbiol.
31:25–33. 2016. View Article : Google Scholar
|
|
28
|
Desnues C, La Scola B, Yutin N, Fournous
G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV
and Raoult D: Provirophages and transpovirons as the diverse
mobilome of giant viruses. Proc Natl Acad Sci USA. 109:18078–18083.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Quemin ER, Corroyer-Dulmont S and
Krijnse-Locker J: Entry and disassembly of large DNA viruses:
Electron microscopy leads the way. J Mol Biol. 430:1714–1724. 2018.
View Article : Google Scholar
|
|
30
|
Schrad JR, Abrahão JS, Cortines JR and
Parent KN: Structural and proteomic characterization of the
initiation of giant virus infection. Cell. 181:1046–1061. 2020.
View Article : Google Scholar
|
|
31
|
La Scola B, Desnues C, Pagnier I, Robert
C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P,
Koonin E and Raoult D: The virophage as a unique parasite of the
giant mimivirus. Nature. 455:100–104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fischer MG and Hackl T: Host genome
integration and giant virus-induced reactivation of the virophage
mavirus. Nature. 540:288–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fischer MG, Allen MJ, Wilson WH and Suttle
CA: Giant virus with a remarkable complement of genes infects
marine zooplankton. Proc Natl Acad Sci USA. 107:19508–19513. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gaia M, Benamar S, Boughalmi M, Pagnier I,
Croce O, Colson P, Raoult D and La Scola B: Zamilon, a novel
virophage with Mimiviridae host specificity. PLoS One.
9:e949232014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mougari S, Bekliz M, Abrahao J, Di Pinto
F, Levasseur A and La Scola B: Guarani virophage, a new
Sputnik-like isolate from a Brazilian lake. Front Microbiol.
10:10032019. View Article : Google Scholar
|
|
36
|
Levasseur A, Bekliz M, Chabrière E,
Pontarotti P, La Scola B and Raoult D: MIMIVIRE is a defense system
in mimivirus that confers resistance to virophage. Nature.
531:249–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mougari S, Abrahão J, Oliveira GP, Bou
Khalil JY and La Scola B: Role of the R349 gene and its repeats in
the MIMIVIRE defense system. Front Microbiol. 10:11472019.
View Article : Google Scholar
|
|
38
|
Barrangou R, Fremaux C, Deveau H, Richards
M, Boyaval P, Moineau S, Romero DA and Horvath P: CRISPR provides
acquired resistance against viruses in prokaryotes. Science.
315:1709–17012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang F and Doudna JA: CRISPR-Cas9
structures and mechanisms. Ann Rev Biophys. 46:505–529. 2017.
View Article : Google Scholar
|
|
40
|
Jiang W, Bikar D, Co D, Zhan F and
Marraffini LA: RNA-guided editing of bacterial genomes using
CRISPR-Cas systems. Nat Biotechnol. 31:233–239. 2013. View Article : Google Scholar
|
|
41
|
Carroll M and Zhou X: Panacea in progress:
CRISPR and the future of its biological research introduction.
Microbiol Res. 201:63–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Colson P, La Scola B, Levasseur A,
Caetano-Anollés G and Raoult D: Mimivirus: Leading the way in the
discovery of giant viruses of amoebae. Nat Rev Microbiol.
15:243–254. 2017. View Article : Google Scholar
|
|
43
|
Levasseur A, Bekliz M, Chabrière E,
Pontarotti P, La Scola B and Raoult D: MIMIVIRE is a defence system
in mimivirus that confers resistance to virophage. Nature.
531:249–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raoult D, Renesto P and Brouqui P:
Laboratory infection of a technician by Mimivirus. Ann Intern Med.
144:702–703. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
La Scola B, Marrie TJ, Auffray JP and
Raoult D: Mimivirus in pneumonia patients. Emerg Infect Dis.
11:449–452. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sakhaee F, Vaziri F, Bahramali G, Siadat
SD and Fateh A: Pulmonary infection related to Mimivirus in patient
with primary ciliary dyskinesia. Emerg Infect Dis. 26:2524–2526.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Colson P, Aherfi S, La Scola B and Rault
D: The role of giant viruses of amoebas in humans. Curr Opin
Microbiol. 31:199–208. 2016. View Article : Google Scholar
|
|
48
|
Shah N, Hülsmeier AJ, Hochhold N, Neidhart
M, Gay S and Hennet T: Exposure to Mimivirus collagen promotes
arthritis. J Virol. 88:838–845. 2014. View Article : Google Scholar
|
|
49
|
Sahmi-Bounsiar D, Rolland C, Aherfi S,
Boudjemaa H, Levasseur A, La Scola B and Colson P:
Marseilleviruses: An update in 2021. Front Microbiol.
12:6487312021. View Article : Google Scholar
|
|
50
|
Aherfi S, Colson P, Audoly G, Nappez C,
Xerri L, Valensi A, Million M, Lepidi H, Costello R and Rault D:
Marseillevirus in lymphoma: A giant in the lymph node. Lancet
Infect Dis. 16:e255–e234. 2016. View Article : Google Scholar
|
|
51
|
Almeida GM, Silva LC, Colson P and Abrahão
JS: Mimiviruses and the human interferon system: Viral evasion of
classical antiviral activities, but inhibition by a novel
interferon-β regulated immunomodulatory pathway. J Interferon
Cytokine Res. 37:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Silva LC, Almeida GM, Oliveira DB, Dornas
FP, Campos RK, La Scola B, Ferreira PC, Kroon EG and Abrahão JS: A
resourceful giant: APMV is able to interfere with the human type I
interferon system. Microbes Infect. 16:187–195. 2014. View Article : Google Scholar
|
|
53
|
Michelucci A, Cordes T, Ghelfi J, Pailot
A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et
al: Immune-responsive gene 1 protein links metabolism to immunity
by catalyzing itaconic acid production. Proc Natl Acad Sci USA.
110:7820–7825. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Strelko CL, Lu W, Dufort FJ, Seyfried TN,
Chiles TC, Rabinowitz JD and Roberts MF: Itaconic acid is a
mammalian metabolite induced during macrophage activation. J Am
Chem Soc. 133:16386–16389. 2011. View Article : Google Scholar
|