Introduction
Breast cancer is the most common type of cancer
among women worldwide. It is estimated that ~1.4 million women are
diagnosed with breast cancer every year, and 458,000 women succumb
to the disease, making it the second most frequently diagnosed type
of cancer worldwide (1). The
incidence of early-onset breast cancer in America is 10.3% of all
new female breast cancer cases in 2012–2016 and has markedly
increased among young women aged 18–45 years (2). Breast cancer involves a multi-step
process associated with the abnormal expression of several
oncogenes and tumor suppressor genes (3). Therefore, the study of biomarkers
and active molecules in the initial stages of breast cancer can
provide a theoretical basis for the prevention and early detection
of breast cancer (4–7).
Cell division cycle-associated 5 (CDCA5), also known
as sororin, is the encoding gene of CDCA5, which is located on
chromosome 11. CDCA5 is a transcriptional protein with a molecular
weight of ~27 kDa (8,9) that is widely expressed in bone
marrow, testes, lymph nodes, bladder, lung, breast, stomach, and
other tissues and organs (10,11). CDCA5 has been reported to promote
cancer (10,12,13). The following genes have been shown
to be significantly upregulated in breast cancer tissues, which
lead to a decrease in the survival rates of patients: Kinetochore
scaffold 1 (CASC5), cytoskeleton-associated protein 2 like
(CKAP2L), family with sequence similarity 83 member D (FAM83D),
kinesin family member 18B (KIF18B), kinesin family member 23
(KIF23), spindle and kinetochore-associated complex subunit 1
(SKA1), GINS complex subunit 1 (GINS1), CDCA5 and minichromosome
maintenance complex component 6 (MCM6) (14). However, to the best of our
knowledge, the role of CDCA5 in breast cancer has not been studied
to date.
PDS5A, a sister chromatid cohesion protein PDS
homolog A, is responsible for unloading auxin from chromatin, while
CDCA5 (sororin) is involved in establishing sister chromatid
cohesion (SCC) (15). Alterations
in PDS5A expression levels have been observed in breast, kidney,
esophagus, stomach, liver and colon cancer (15,16). PDS5A has been found to be
significantly upregulated at both the mRNA and protein levels, and
this upregulation is positively correlated with World Health
Organization glioma grade (17).
PDS5A is upregulated in nasopharyngeal carcinoma tumor tissue, and
PDS5A overexpression in 293T cells promotes proliferation and clone
formation (18).
In the present study, the effect of CDCA5 and PDS5A
on breast cancer cell proliferation, migration and invasion was
determined. The present findings may provide a new target and
strategy for the clinical treatment of breast cancer.
Materials and methods
Cell culture and cell
transfection
Human normal mammary MCF-10A and human breast cancer
MCF-7, MDA-MB-231, SUM190PT and SK-BR-3 cell lines were purchased
from BeNa Culture Collection and cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) under constant conditions at 37°C in a
humidified atmosphere with 5% CO2.
Short hairpin RNAs (shRNAs) against CDCA5
(shRNA-CDCA5-1,
5′-CCGGCCAAAGTACCATAGCCAGTTTCTCGAGAAACTGGCTATGGTACTTTGGTTTTTG-3′;
and shRNA-CDCA5-2,
5′-CCGGGAGCAGTTTGATCTCCTGGTTCTCGAGAACCAGGAGATCAAACTGCTCTTTTTG-3′),
scrambled shRNA [transfected with empty vector; shRNA-negative
control (NC), 5′-TTCGGGTCATCCGATGGGCC-3′], adenovirus
overexpressing PDS5A (pcDNA3.1-PDS5A) and control adenovirus
(pcDNA3.1-NC) were constructed by Hanbio Biotechnology Co., Ltd. A
total of 0.4 µg shRNA/adenovirus was transfected into
1×105 cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) and incubated for 10
min at room temperature, according to the manufacturer's
instructions. At 48 h post-transfection, the transfected cells were
used for subsequent experiments.
Bioinformatics
STRING (https://string-db.org/) is a database that can be used
to predict interactions among proteins (accession date, September
2020, version. 11.0). The differences in PDS5A expression between
breast cancer and normal tissues were predicted using Encyclopaedia
of RNA Interactomes website (https://starbase.sysu.edu.cn/) and Gene Expression
Profiling Interactive Analysis (GEPIA) database
(gepia.cancer-pku.cn/), and Pearson was used as the statistical
tests.
Cell proliferation assays
For Cell Counting Kit-8 (CCK-8) assay, MDA-MB-231
cells (1×103 cells/well) were seeded in 96-well plates
and cultured for 24 h. At 24, 48 and 72 h after incubation, CCK-8
solution (Merck KGaA) was added to each well. The cells were
incubated for an additional 1 h to determine cell viability.
Optical density was determined at 450 nm using a microtiter plate
reader.
For colony formation assay, transfected MDA-MB-231
cells (1×105 cells/well) were seeded into 6-well plates,
and the MDA-MB-231 cells treated with sh-NC or pcDNA3.1 were used
as the negative controls. Following an incubation period of 14 days
at 37°C, the colonies were fixed with 100% methanol for 15 min at
room temperature and stained with 0.1% crystal violet at room
temperature in absolute ethanol for 15 min. Visible colonies of
>50 cells were counted and analyzed under an inverted light
microscope.
Wound healing and Transwell
assays
For the wound healing assay, transfected MDA-MB-231
cells (1×105 cells/well) were cultured in 6-well plates
until they reached 70–80% confluence, and then wounded using a
200-µl sterile pipette tip. Upon washing with PBS, the cells were
cultured in serum-free DMEM. Images were acquired using a light
microscope at each 0 and 24 h. The cell migration ratio was
calculated as the percentage of remaining cell-free area compared
to the area of the initially scratched area.
For the Transwell assay, MDA-MB-231 cells
(3×104 cells/well) were cultured in serum-free DMEM. The
upper chamber of 24-well Transwell was pre-coated with Matrigel
(Merck KGaA) at 37°C for 30 min. The cells were then cultured in
the upper chamber with 0.1 ml cell suspension in each well, while
the lower chamber was filled with cell culture DMEM containing 20%
FBS. The cells were cultured for 24 h at 37°C. Subsequently, the
membrane in the lower chamber was collected and cleaned, followed
by staining with 0.5% crystal violet dye (Merck KGaA) at room
temperature for 10 min. The stained cells were counted using an
optical microscope.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from 1×104
MDA-MB-231 cells using TRIzol® reagent (Invitrogen,
cat.no.15596018). The total RNA concentration was assessed with
NanoDrop 2000™ (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.) before being reverse transcribed to cDNA using Titan One Tube
RT-PCR reagent (Merck KGaA; cat. No. 11939823001), according to the
manufacturer's instructions. Subsequently, RT-qPCR was performed
using a QuantiTect SYBR Green PCR kit (Qiagen GmbH), according to
the manufacturer's instructions. The following thermocycling
conditions were used for qPCR: Initial denaturation at 95°C for 5
min, followed by 30 cycles of 95°C for 10 sec and 60°C for 45 sec.
The following primers (GenScript) were used for RT-qPCR: CDCA5
forward, 5′-GGCCAGAGACTTGGAAATGT-3′ and reverse,
5′-GGCCAGAGACTTGGAAATGT-3′; PDS5A forward,
5′-GATCACCACGGACGAGATGA-3′ and reverse,
5′-AAGGCTAGTGGGAGATACTGCTGTT-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. mRNA expression levels were
quantified using the 2−ΔΔCq method (19) and normalized to the internal
reference gene GAPDH.
Western blotting
Total protein was extracted from 1×106
MDA-MB-231 cells using RIPA lysis buffer (Elabscience, cat. No.
E-BC-R327) and quantified using a Pierce™ BCA Protein Assay Kit
(Pierce; Thermo Fisher Scientific, Inc.). Following denaturation,
electrophoresis of 30 µg protein/lane was performed using 12%
SDS-PAGE. Following gel transfer onto PVDF membranes, the membranes
were blocked in 5% fat-free milk for 2 h at room temperature.
Subsequently, the membranes were incubated overnight at 4°C with
the following primary antibodies: Anti-MMP2 (1:1,000; cat. no.
ab215986; Abcam), anti-MMP9 (1:1,000; cat. no. ab219372; Abcam),
anti-GAPDH (1:1,000; cat. no. ab181602; Abcam), anti-Ki67 (1:1,000;
cat. no. ab92742; Abcam), anti-proliferating cell nuclear antigen
(PCNA; 1:1,000; cat. no. ab92552; Abcam), anti-CDCA5 (1:1,000; cat.
no. ab192237; Abcam) and anti-PDS5A (1:500; cat. no. 203627-T34;
Sino Biological Inc.). Next, the membranes were incubated with a
goat anti-rabbit HRP-conjugated IgG secondary antibody (1:2,000;
cat. no. ab6721; Abcam) at room temperature for 4 h. The protein
bands were visualized using an enhanced chemiluminescence reagent
(cat. no. abs920, Absin). The protein expression levels were
semi-quantified using ImageJ software (v1.46; National Institutes
of Health) with GAPDH as the loading control.
Co-immunoprecipitation (co-IP)
assay
Total protein was isolated from 1×107
MDA-MB-231 cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology), followed by centrifugation at 14,000 × g for 15 min
at 4°C and quantified using a BCA kit (Beyotime Institute of
Biotechnology). For the co-IP assay, 400 µg protein was incubated
with 2 µg the appropriate antibodies including anti-PDS5A (1:500,
203627-T34, Sino Biological), anti-CDCA5 (1:500; FineTest,
FNab01542), Goat Anti-Rabbit IgG (1:2000, ab6721, Abcam) overnight
at 4°C. Subsequently, 30 µl Protein G/A agarose beads (Invitrogen;
Thermo Fisher Scientific, Inc.) were added to the cell lysates for
a 3-h incubation at 4°C. Following three washes with PBS, the
precipitated proteins were resuspended in 5X SDS-PAGE loading
buffer (cat. no. ZY81204, Shanghai Zeye Biological Inc.), boiled
for 5 min at 99°C and eluted from the beads with 1 ml lysis buffer
for 3 times. Finally, western blotting was used to detect the co-IP
products in the way as aforementioned.
Statistical analysis
All experiments were repeated at least three
independent times, and the results are expressed as the mean ± SD.
Statistical analysis was performed using SPSS 19.0 software (IBM
Corp.). Unpaired Student's t-test and one-way ANOVA followed by
Tukey's post-hoc test were used to determine statistical
significance. Correlation between CDCA5 and PDS5A was evaluated
using Pearson's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
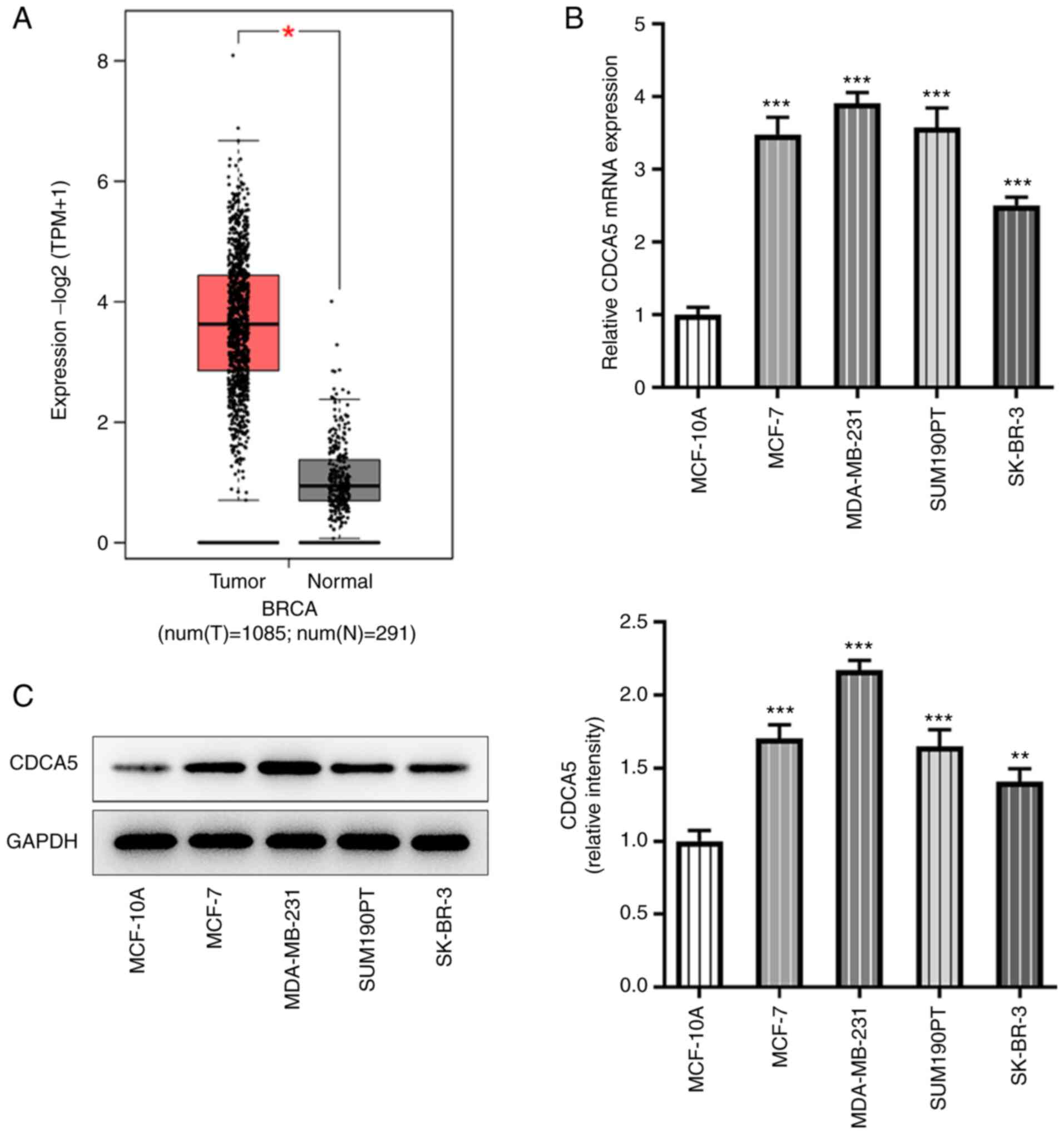
CDCA5 is upregulated in human breast
cancer tissues and cell lines
According to GEPIA database (gepia.cancer-pku.cn/)
prediction, CDCA5 expression was increased in breast cancer tissues
(Fig. 1A). Further RT-qPCR
(Fig. 1B) and western blotting
(Fig. 1C) were performed to
detect the expression levels of CDCA5 in breast cancer cells. The
results indicated that CDCA5 was upregulated in breast cancer
cells, particularly in MDA-MB-231 cells, compared with the
expression levels in the breast cancer lines MCF-7, SUM190T and
SK-BR-3. The MDA-MB-231 cell line was therefore selected for
subsequent experiments.
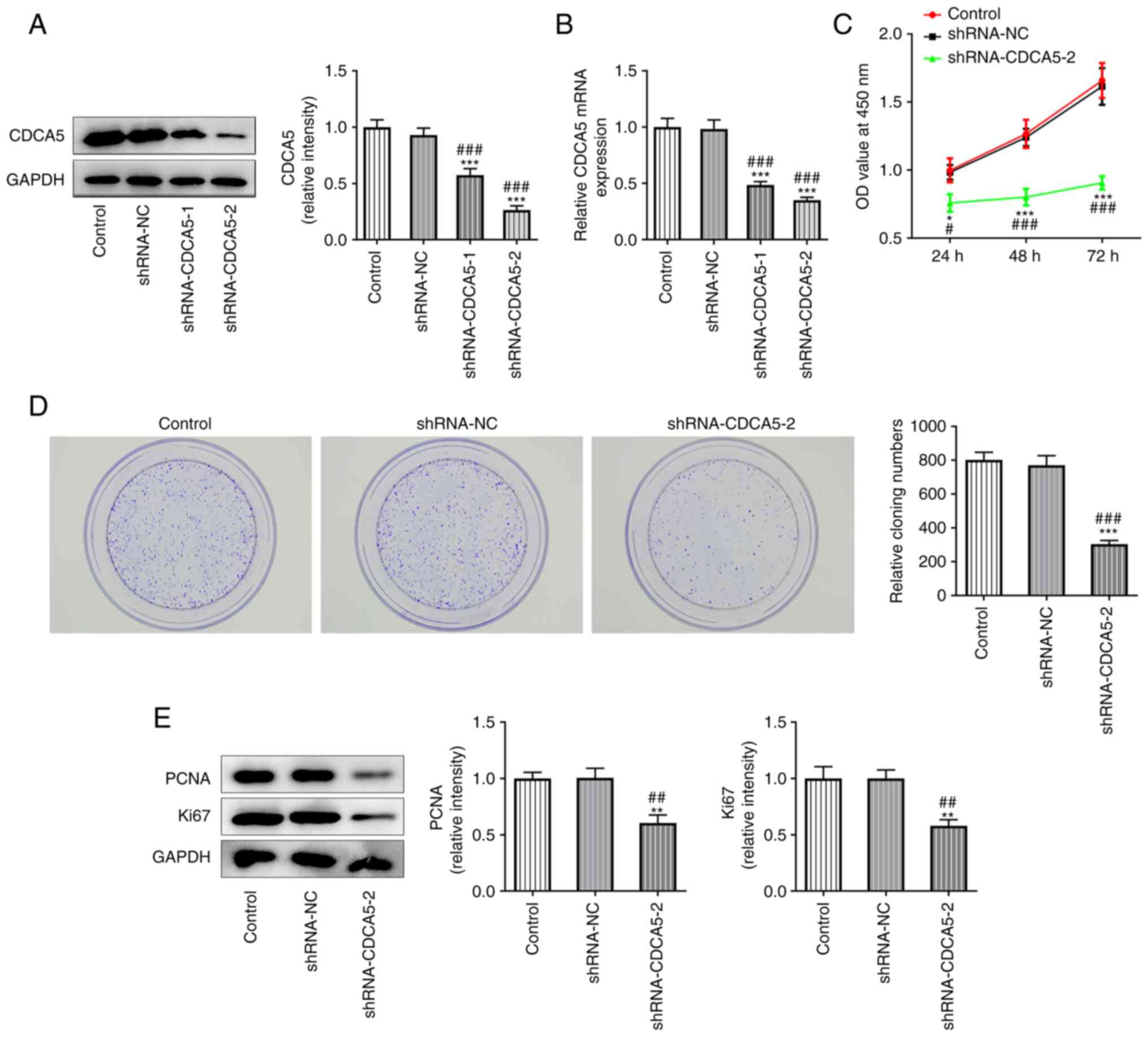
CDCA5 knockdown inhibits
proliferation, invasion and migration in MDA-MB-231 cell
To further explore the role of CDCA5 in breast
cancer, CDCA5 knockdown plasmids were constructed. The inhibition
level of CDCA5 was subsequently detected by RT-qPCR and western
blotting. As presented in Fig. 2A and
B, CDCA5 was significantly inhibited in the shRNA-CDCA5-1/2
groups compared with the shRNA-NC group, indicating that the
plasmid-mediated inhibition of CDCA5 had been successful.
shRNA-CDCA5-2 showed a better knockdown than shRNA-CDCA5-1 and was
selected for subsequent experiments.
CCK-8 assay was performed to detect the level of
cell proliferation (Fig. 2C) and
a colony formation experiment was performed to detect the colony
formation potential of MDA-MB-231 cells (Fig. 2D). The results indicated that
CDCA5 knockdown inhibited the proliferation of MDA-MB-231 cells.
The expression of the proliferation-related proteins Ki67 and PCNA
was also decreased, as shown by western blot analysis, which
confirmed the aforementioned observations (Fig. 2E).
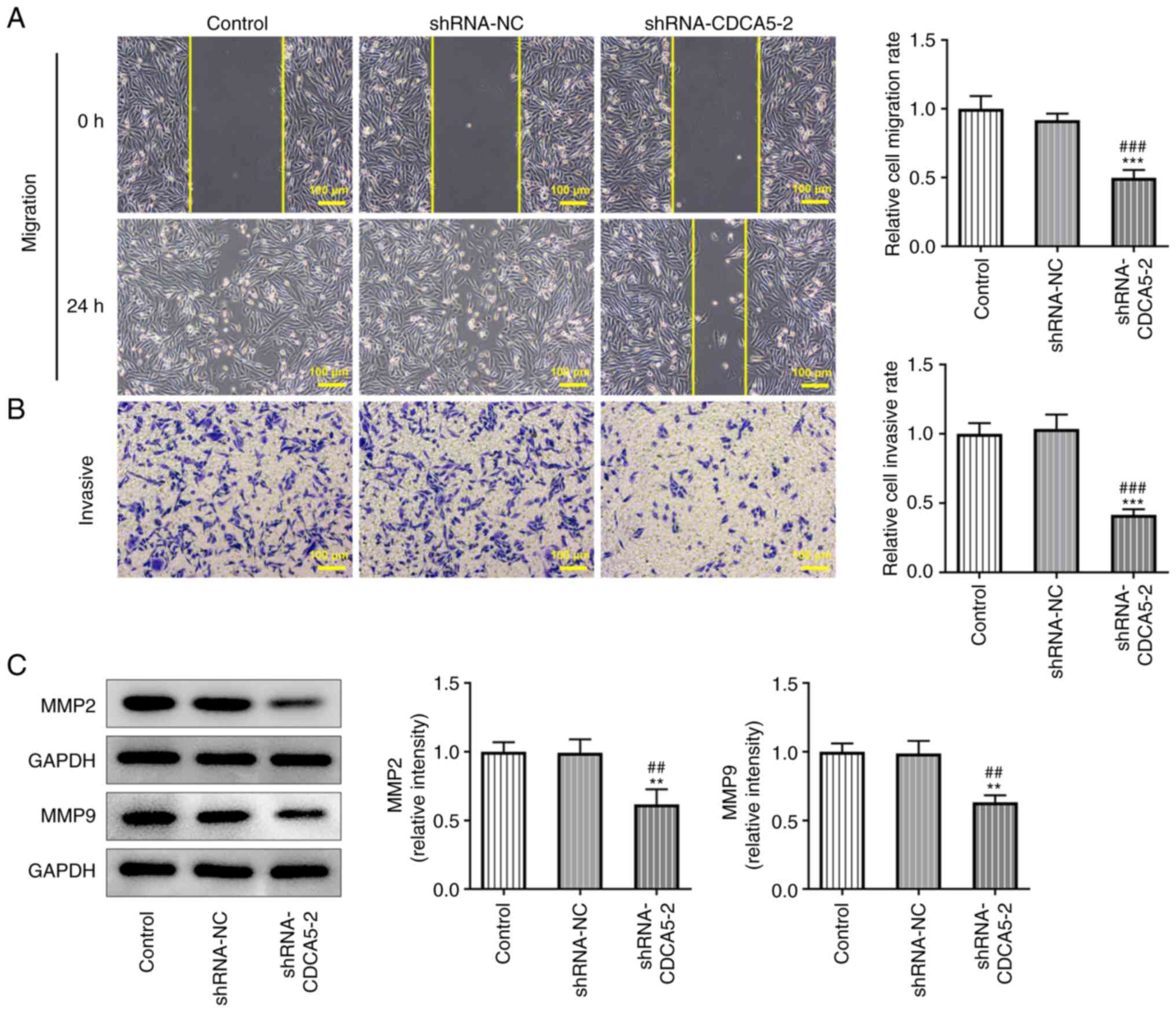
Furthermore, wound healing and Transwell assays
detected a marked decrease in the migration (Fig. 3A) and invasion (Fig. 3B) of MDA-MB-231 cells. In
addition, the expressions of the migration-related proteins MMP2
and MMP9 were reduced in MDA-MB-231 cells (Fig. 3C). The results indicated that
CDCA5 knockdown inhibited the invasion and migration of breast
cancer cells.
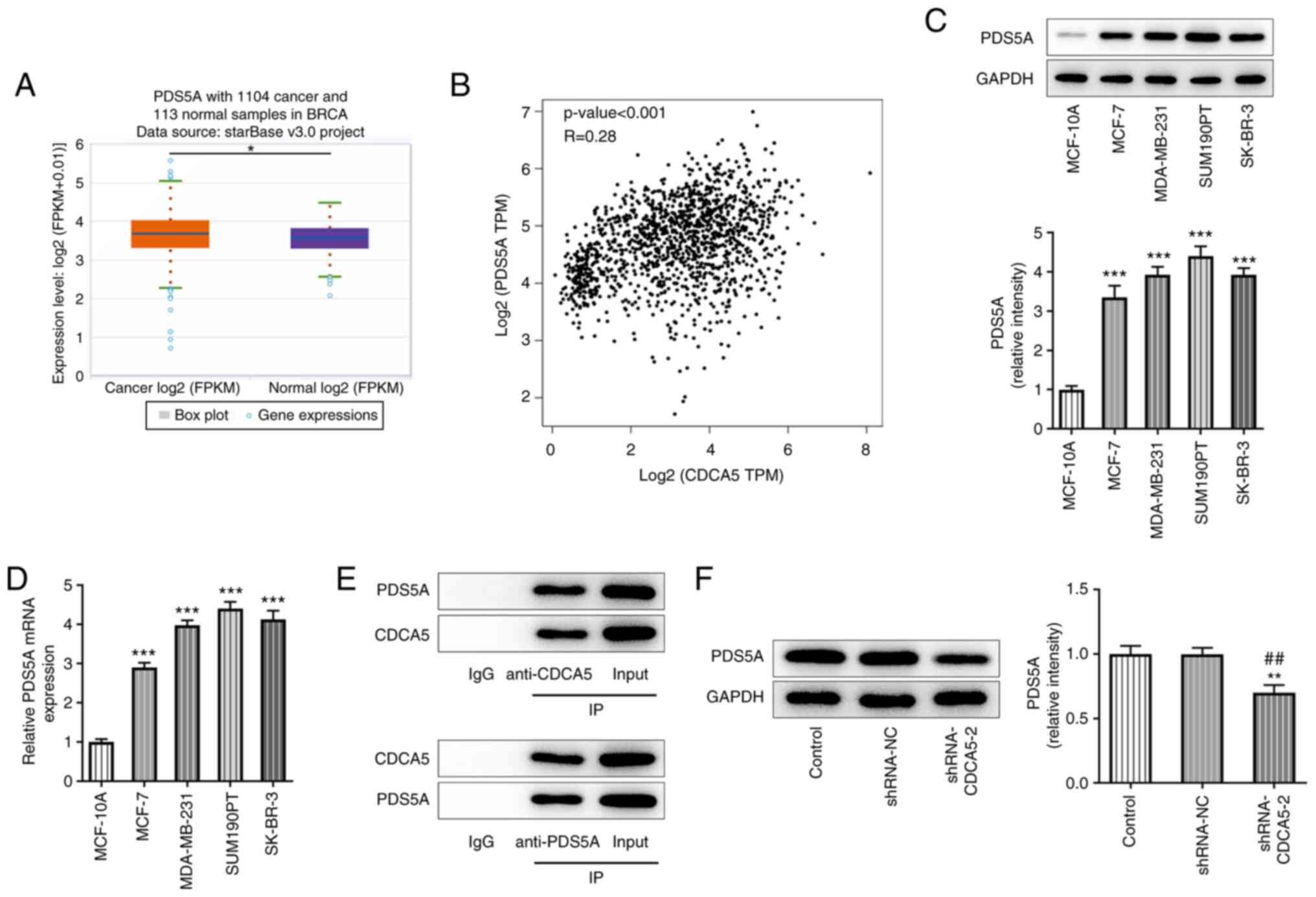
CDCA5 knockdown downregulates PDS5A
expression in breast cancer cells
starBase database analysis demonstrated that PDS5A
expression was upregulated in breast cancer tissues (Fig. 4A), while the prediction results
from the GEPIA website revealed a positive correlation between
CDCA5 and PDS5A expression in patients with breast cancer (Fig. 4B). Furthermore, RT-qPCR and
western blot analyses were used to detect PDS5A expression in
breast cancer cell lines, and PDS5A was found to be upregulated in
MDA-MB-231 cells (Fig. 4C and D).
Co-IP assay further indicated that CDCA5 could bind to PDS5A
(Fig. 4E). In addition, western
blot analysis showed that CDCA5 knockdown inhibited PDS5A
expression (Fig. 4F).
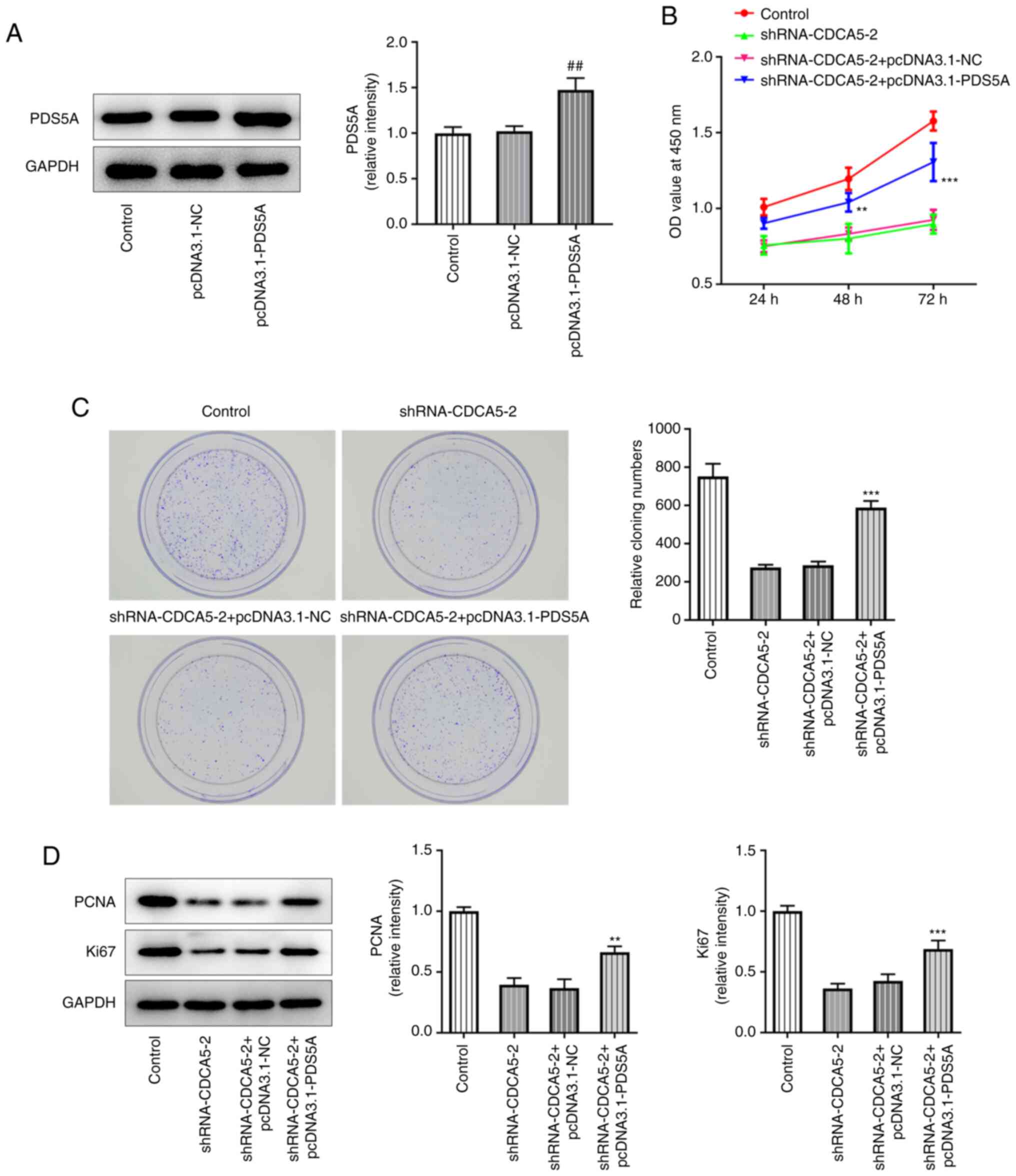
PDS5A overexpression reverses the
consequences of CDCA5 inhibition on breast cancer cell
proliferation and migration
The overexpression of PDS5A in MDA-MB-231 cells by
using a pcDNA3.1-PDS5A vector was confirmed by western blotting,
which revealed that PDS5A expression was upregulated in the
pcDNA3.1-PDS5A group compared with the pcDNA3.1-NC group (Fig. 5A). Furthermore, in the
shRNA-CDCA5-2+pcDNA3.1-PDS5A group, CCK-8 assay revealed a marked
increase in cell viability (Fig.
5B), increased levels of colony formation (Fig. 5C) and also elevated expression of
the proliferation-related proteins Ki67 and PCNA (Fig. 5D), compared to
shRNA-CDCA5-2+pcDNA3.1-NCgroup. The results showed that PDS5A
overexpression reversed the consequences of CDCA5 inhibition on
breast cancer cell proliferation.
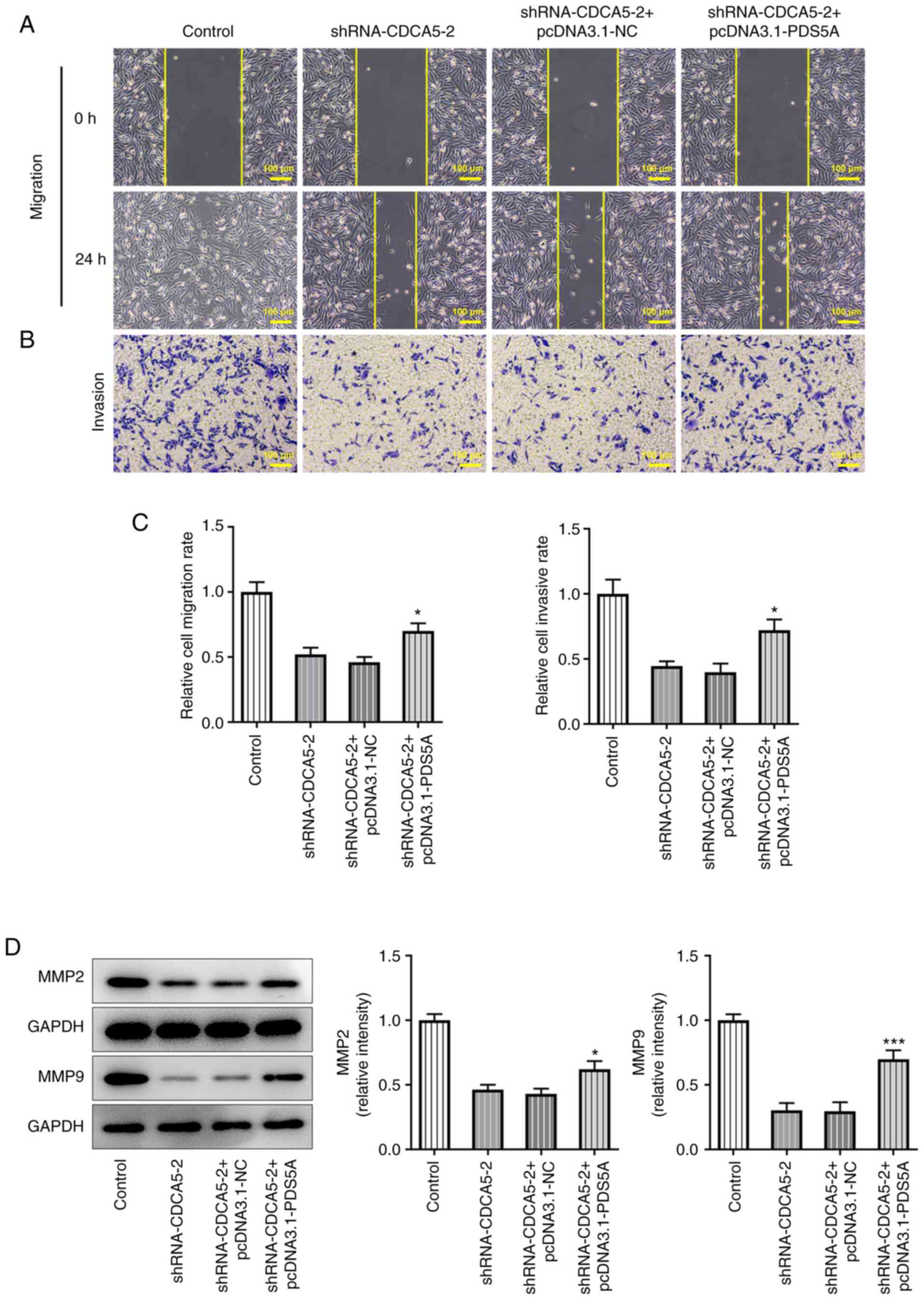
Subsequently, wound healing (Fig. 6A) and Transwell (Fig. 6B) assays were performed.
Quantitative analysis of the experimental results (Fig. 6C) indicated that CDCA5 knockdown
inhibited the invasion and migration of breast cancer cells, and
PDS5A overexpression could reverse this trend. Consistently,
migration-associated proteins also showed an upregulation (Fig. 6D). Overall, the present findings
demonstrated that PDS5A overexpression reversed the inhibitory
effect of CDCA5 interference on breast cancer cell proliferation
and migration.
Discussion
It has been reported that the CASC5, CKAP2L, FAM83D,
KIF18B, KIF23, SKA1, GINS1, CDCA5 and MCM6 genes are significantly
upregulated in breast cancer tissues, and that this upregulation
leads to a decrease in patient survival rate (14). CDCA5 expression was found to be
higher in bladder cancer tissues compared with that in adjacent
healthy tissues, and high CDCA5 expression was found to be
associated with poor survival in patients with breast cancer
(13,20). A previous study indicated that
CDCA5 knockout in T24 and 5637 cells reduced cell proliferation and
induced cell apoptosis (13).
Compared with that in adjacent tissues, CDCA5 expression was
upregulated in hepatocellular carcinoma (HCC) tissue and was
negatively correlated with HCC patient survival (21). CDCA5 knockdown can inhibit cell
proliferation and tumorigenesis, and induce cell apoptosis,
suggesting that CDCA5 plays a carcinogenic role in liver cancer
(22). The present study also
indicated that CDCA5 was upregulated in breast cancer tissue, as
queried through the GEPIA website. The present findings also
demonstrated that CDCA5 was upregulated in breast cancer cells.
In recent years, the role of CDCA5 in cancer has
been gradually revealed (11,13). In a subcutaneous tumorigenesis
experiment on nude mice, CDCA5 knockdown could effectively inhibit
tumor formation and growth in tumor-bearing mice (10). In a mechanistic study, CDCA5
knockdown was able to block the cell cycle of triple-negative
breast cancer cells (14). CDCA5
enhanced the proliferation and inhibited the apoptosis of bladder
cancer cells (10,13). CDCA5 promoted bladder cancer cell
proliferation by activating the Akt signaling pathway in bladder
tumors (9). In the present study,
CDCA5 knockdown inhibited breast cancer cell proliferation, and the
expression level of proliferation-related proteins was
downregulated following CDCA5 knockdown.
A previous study has indicated that PDS5A
overexpression in breast cancer cells could increase the number of
cells in G1 phase and enhance the activation of caspase
3, leading to tumor cell apoptosis (23). In addition, a decrease in PDS5A
mRNA expression was observed in breast and kidney tumor samples
compared with that in the corresponding normal tissues (24). Another study reported an increase
in PDS5A mRNA expression in esophageal, stomach, liver and
transverse colon tumors (25). In
conclusion, these data suggested that PDS5A may be highly
tissue-specific in the process of tumorigenesis and development,
and that, depending on the tissue or cell type, it could inhibit
the proliferation or induce the over-proliferation of tumor cells
18,226). In the present study, CDCA5 knockdown downregulated PDS5A
expression in breast cancer cells, and PDS5A overexpression
reversed the inhibitory effects of CDCA5 knockdown on breast cancer
cell proliferation and migration.
In conclusion, CDCA5 could be a valuable oncogene,
and it should be further investigated in breast cancer cell
progression. However, the present study has certain limitations.
Firstly, the drug sensitivity of shRNA-CDCA5-transfected cells was
not analyzed. Secondly, the mRNA and protein expression levels of
CDCA5 and PDS5A in breast cancer and normal tissue biopsy specimens
were not analyzed. In addition, further analysis on whether CDCA5
knockdown causes the inhibition of the Akt signaling pathway in
breast cancer cell lines would be helpful for confirming the
accuracy of the present experimental results. Therefore, future
research should focus on the aforementioned issues.
To conclude, CDCA5 knockdown was found to suppress
the malignant progression of breast cancer by regulating PDS5A in
the current study. The present findings may provide new potential
targets for breast cancer treatment, since CDCA5 was found to be a
potential molecular target for breast cancer therapy and
diagnosis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and JY designed the study and performed the
experiments. YW, XJZ and YLZ drafted and revised the manuscript.
XJZ, JL, YJ, FS and YLZ analyzed the data. JL and FS performed the
literature search. JL and YW confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smaili F, Boudjella A, Dib A, Braikia S,
Zidane H, Reggad R, Bendib A, Abdelouahab A, Bereksi-Reguig F,
Yekrou D, et al: Epidemiology of breast cancer in women based on
diagnosis data from oncologists and senologists in Algeria. Cancer
Treat Res Commun. 25:1002202020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chelmow D, Pearlman MD, Young A, Bozzuto
L, Dayaratna S, Jeudy M, Kremer ME, Scott DM and O'Hara JS:
Executive summary of the early-onset breast cancer evidence review
conference. Obstet Gynecol. 135:1457–1478. 2020. View Article : Google Scholar
|
|
3
|
Kontomanolis EN, Koutras A, Syllaios A,
Schizas D, Mastoraki A, Garmpis N, Diakosavvas M, Angelou K,
Tsatsaris G, Pagkalos A, et al: Role of oncogenes and
tumor-suppressor genes in carcinogenesis: A Review. Anticancer Res.
40:6009–6015. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Falzone L, Grimaldi M, Celentano E,
Augustin LSA and Libra M: Identification of modulated micrornas
associated with breast cancer, diet, and physical activity. Cancers
(Basel). 12:25552020. View Article : Google Scholar
|
|
5
|
Cocco S, Piezzo M, Calabrese A, Cianniello
D, Caputo R, Lauro VD, Fusco G, Gioia GD, Licenziato M and De
Laurentiis M: Biomarkers in Triple-negative breast cancer:
State-of-the-Art and future perspectives. Int J Mol Sci.
21:45792020. View Article : Google Scholar
|
|
6
|
Han Y, Wang J and Xu B: Novel biomarkers
and prediction model for the pathological complete response to
neoadjuvant treatment of triple-negative breast cancer. J Cancer.
12:936–945. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sporikova Z, Koudelakova V, Trojanec R and
Hajduch M: Genetic markers in triple-negative breast cancer. Clin
Breast Cancer. 18:e841–e850. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang N and Pati D: Sororin is a master
regulator of sister chromatid cohesion and separation. Cell Cycle.
11:2073–2083. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jordan PW, Eyster C, Chen J, Pezza RJ and
Rankin S: Sororin is enriched at the central region of synapsed
meiotic chromosomes. Chromosome Res. 25:115–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang IW, Lin VC, He HL, Hsu CT, Li CC, Wu
WJ, Huang CN, Wu TF and Li CF: CDCA5 overexpression is an indicator
of poor prognosis in patients with urothelial carcinomas of the
upper urinary tract and urinary bladder. Am J Transl Res.
7:710–722. 2015.PubMed/NCBI
|
|
11
|
Xu T, Ma M, Dai J, Yu S, Wu X, Tang H, Yu
J, Yan J, Yu H, Chi Z, et al: Gene expression screening identifies
CDCA5 as a potential therapeutic target in acral melanoma. Hum
Pathol. 75:137–145. 2018. View Article : Google Scholar
|
|
12
|
Zhang Z, Shen M and Zhou G: Upregulation
of CDCA5 promotes gastric cancer malignant progression via
influencing cyclin E1. Biochem Biophys Res Commun. 496:482–489.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu G, Xu Z, Chen X, Pan H, Wang Y and Jin
B: CDCA5 functions as a tumor promoter in bladder cancer by
dysregulating mitochondria-mediated apoptosis, cell cycle
regulation and PI3k/AKT/mTOR pathway activation. J Cancer.
11:2408–2420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu Y, Zhou QZ, Zhang XL, Wang ZZ and Wang
P: Identification of hub genes using Co-Expression network analysis
in breast cancer as a tool to predict different stages. Med Sci
Monit. 25:8873–8890. 2019. View Article : Google Scholar
|
|
15
|
Zhang N, Coutinho LE and Pati D: PDS5A and
PDS5B in cohesin function and human disease. Int J Mol Sci.
22:58682021. View Article : Google Scholar
|
|
16
|
Morales C, Ruiz-Torres M, Rodríguez-Acebes
S, Lafarga V, Rodríguez-Corsino M, Megías D, Cisneros DA, Peters
JM, Méndez J and Losada A: PDS5 proteins are required for proper
cohesin dynamics and participate in replication fork protection. J
Biol Chem. 295:146–157. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hagemann C, Weigelin B, Schommer S,
Schulze M, Al-Jomah N, Anacker J, Gerngras S, Kühnel S, Kessler AF,
Polat B, et al: The cohesin-interacting protein, precocious
dissociation of Sisters 5A/sister chromatid cohesion protein 112,
is up-regulated in human astrocytic tumors. Int J Mol Med.
27:39–51. 2011.PubMed/NCBI
|
|
18
|
Zheng MZ, Zheng LM and Zeng YX: SCC-112
gene is involved in tumor progression and promotes the cell
proliferation in G2/M phase. J Cancer Res Clin Oncol. 134:453–462.
2008. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data suing real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Phan NN, Wang CY, Li KL, Chen CF, Chiao
CC, Yu HG, Huang PL and Lin YC: Distinct expression of CDCA3,
CDCA5, and CDCA8 leads to shorter relapse free survival in breast
cancer patient. Oncotarget. 9:6977–6992. 2018. View Article : Google Scholar
|
|
21
|
Tian Y, Wu J, Chagas C, Du Y, Lyu H, He Y,
Qi S, Peng Y and Hu J: CDCA5 overexpression is an Indicator of poor
prognosis in patients with hepatocellular carcinoma (HCC). BMC
Cancer. 18:11872018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen H, Chen J, Zhao L, Song W, Xuan Z,
Chen J, Li Z, Song G, Hong L, Song P and Zheng S: CDCA5,
Transcribed by E2F1, promotes oncogenesis by enhancing cell
proliferation and inhibiting apoptosis via the AKT pathway in
hepatocellular carcinoma. J Cancer. 10:1846–1854. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kumar D, Sakabe I, Patel S, Zhang Y, Ahmad
I, Gehan EA, Whiteside TL and Kasid U: SCC-112, a novel cell
cycle-regulated molecule, exhibits reduced expression in human
renal carcinomas. Gene. 328:187–196. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hill VK, Kim JS and Waldman T: Cohesin
mutations in human cancer. Biochim Biophys Acta. 1866:1–11.
2016.
|
|
25
|
Put N, Van Roosbroeck K, Vande Broek I,
Michaux L and Vandenberghe P: PDS5A, a novel translocation partner
of MLL in acute myeloid leukemia. Leuk Res. 36:e87–e89. 2012.
View Article : Google Scholar : PubMed/NCBI
|