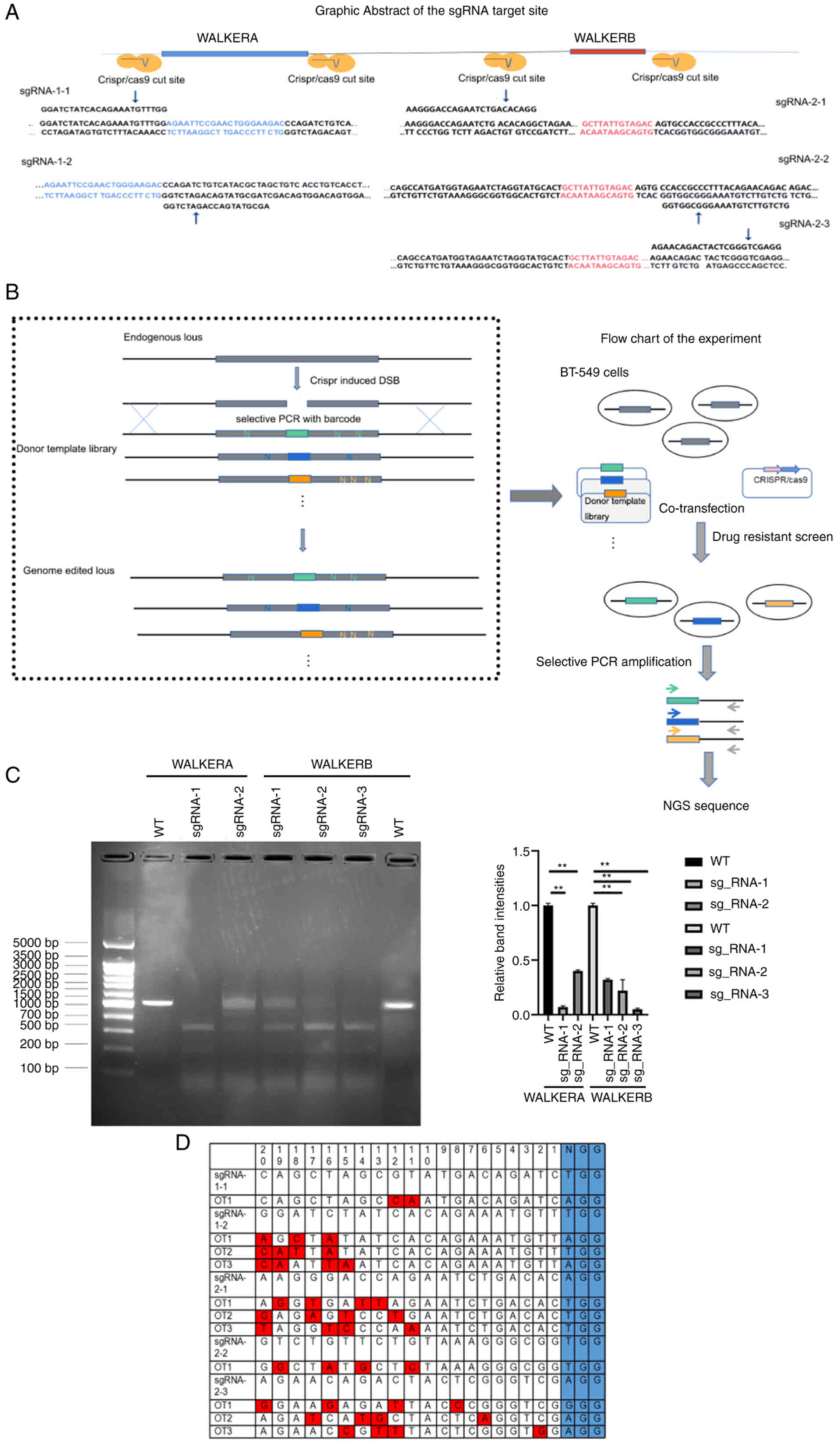
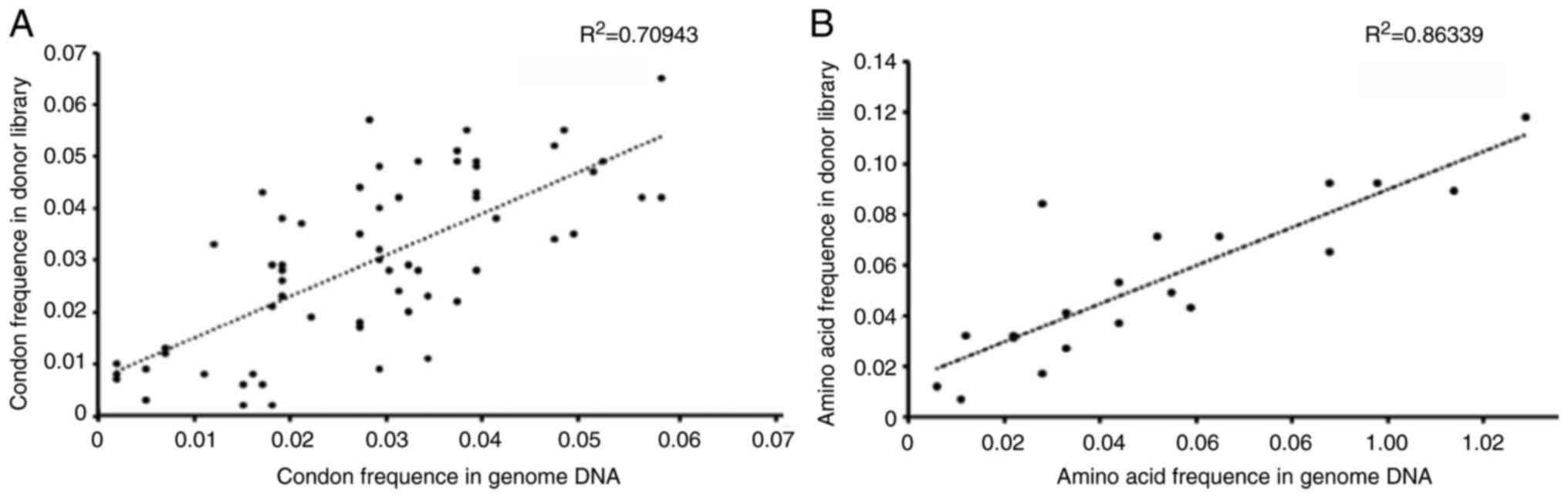
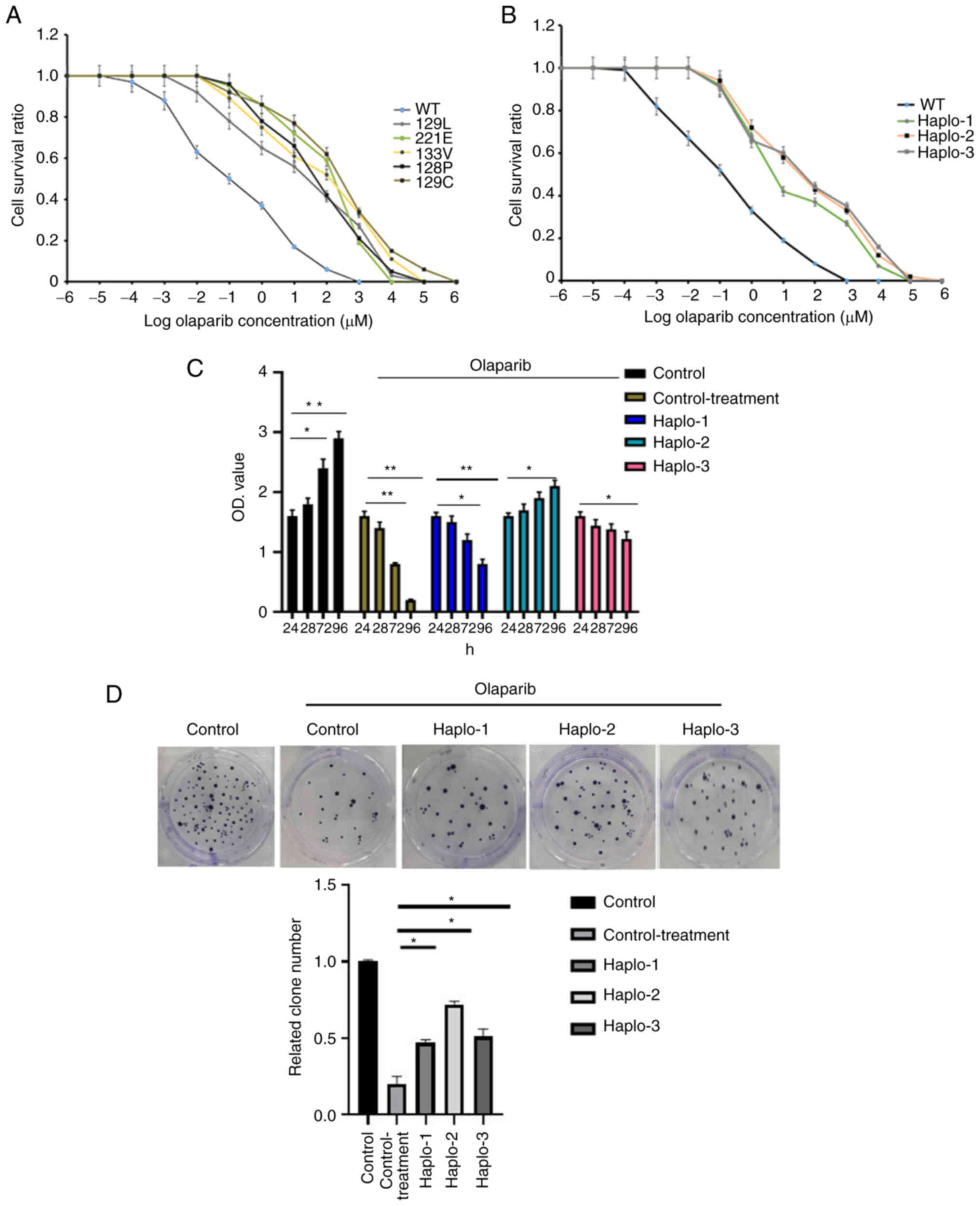
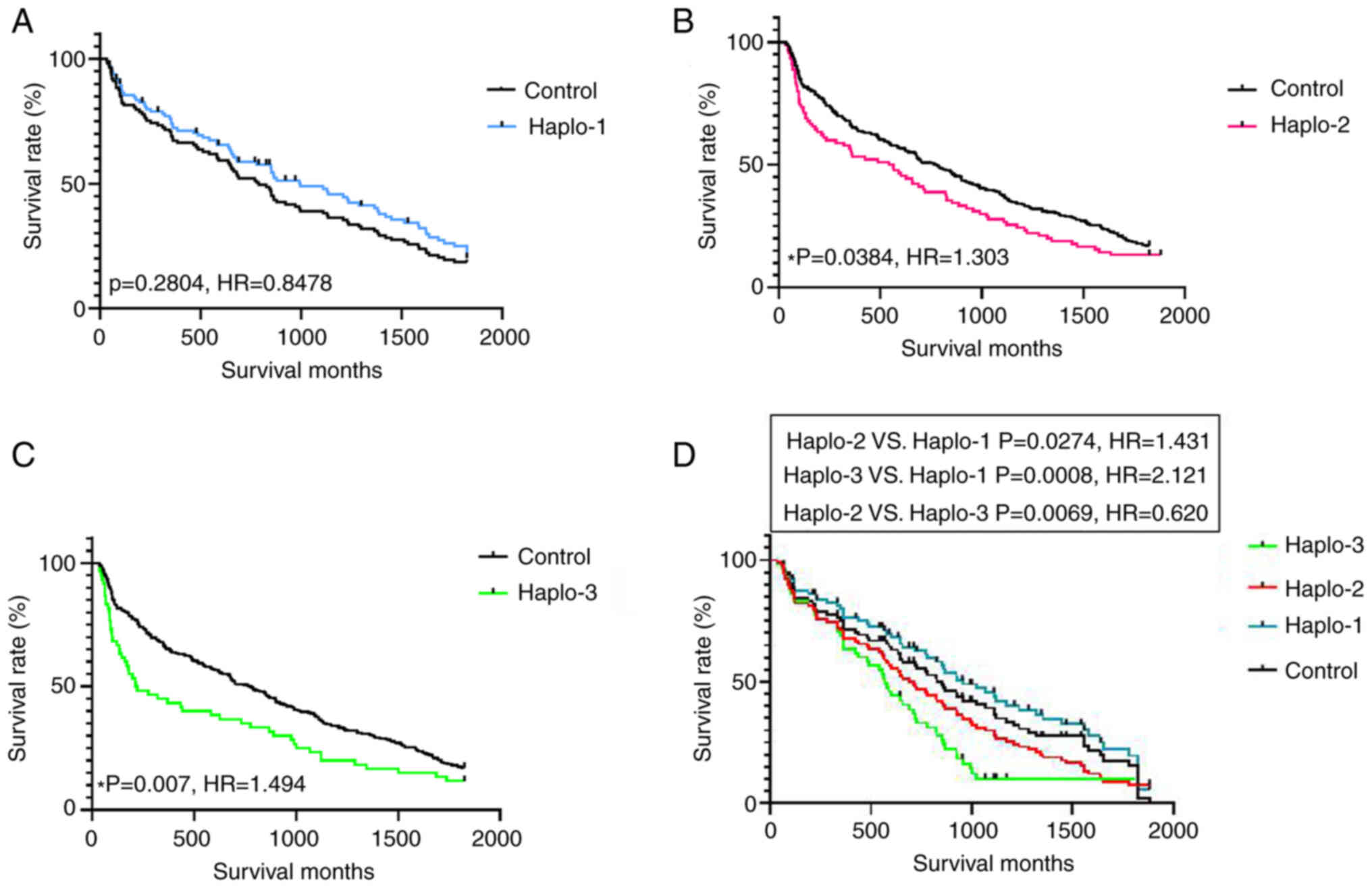
|
1
|
Mavaddat N, Antoniou AC, Easton DF and
Garcia-Closas M: Genetic susceptibility to breast cancer. Mol
Oncol. 4:174–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lord CJ and Ashworth A: BRCAness
revisited. Nat Rev Cancer. 16:110–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Apostolou P and Fostira F: Hereditary
breast cancer: The era of new susceptibility genes. Biomed Res Int.
2013:7473182013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Noordermeer SM and van Attikum H: PARP
Inhibitor Resistance: A tug-of-war in BRCA-mutated cells. Trends
Cell Biol. 29:820–834. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Risdon EN, Chau CH, Price DK, Sartor O and
Figg WD: PARP Inhibitors and prostate cancer: To infinity and
beyond BRCA. Oncologist. 26:e115–e129. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bajrami I, Frankum JR, Konde A, Miller RE,
Rehman FL, Brough R, Campbell J, Sims D, Rafiq R, Hooper S, et al:
Genome-wide profiling of genetic synthetic lethality identifies
CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity.
Cancer Res. 74:287–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grundy MK, Buckanovich RJ and Bernstein
KA: Regulation and pharmacological targeting of RAD51 in cancer.
NAR Cancer. 2:zcaa0242020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kolinjivadi AM, Sannino V, de Antoni A,
Técher H, Baldi G and Costanzo V: Moonlighting at replication
forks-a new life for homologous recombination proteins BRCA1, BRCA2
and RAD51. FEBS Lett. 591:1083–1100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cruz C, Castroviejo-Bermejo M,
Gutiérrez-Enríquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver A,
Bonache S, Morancho B, Bruna A, Rueda OM, et al: RAD51 foci as a
functional biomarker of homologous recombination repair and PARP
inhibitor resistance in germline BRCA-mutated breast cancer. Ann
Oncol. 29:1203–1210. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Castroviejo-Bermejo M, Cruz C,
Llop-Guevara A, Gutiérrez-Enríquez S, Ducy M, Ibrahim YH,
Gris-Oliver A, Pellegrino B, Bruna A, Guzmán M, et al: A RAD51
assay feasible in routine tumor samples calls PARP inhibitor
response beyond BRCA mutation. EMBO Mol Med. 10:e91722018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malka MM, Eberle J, Niedermayer K, Zlotos
DP and Wiesmüller L: Dual PARP and RAD51 inhibitory drug conjugates
show synergistic and selective effects on breast cancer cells.
Biomolecules. 11:9812021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moudry P, Watanabe K, Wolanin KM, Bartkova
J, Wassing IE, Watanabe S, Strauss R, Troelsgaard Pedersen R,
Oestergaard VH, Lisby M, et al: TOPBP1 regulates RAD51
phosphorylation and chromatin loading and determines PARP inhibitor
sensitivity. J Cell Biol. 212:281–288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garcin EB, Gon S, Sullivan MR, Brunette
GJ, Cian A, Concordet JP, Giovannangeli C, Dirks WG, Eberth S,
Bernstein KA, et al: Differential requirements for the Rad51
paralogs in genome repair and maintenance in human cells. PLoS
Genet. 15:e10083552019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abreu CM, Prakash R, Romanienko PJ, Roig
I, Keeney S and Jasin M: Shu complex SWS1-SWSAP1 promotes early
steps in mouse meiotic recombination. Nat Commun. 9:39612018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang P, De Souza C, Minn K and Chien J:
Genome-scale CRISPR knockout screen identifies TIGAR as a modifier
of PARP inhibitor sensitivity. Commun Biol. 2:3352019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma L, Boucher JI, Paulsen J, Matuszewski
S, Eide CA, Ou J, Eickelberg G, Press RD, Zhu LJ, Druker BJ, et al:
CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical
drug resistance with improved accuracy. Proc Natl Acad Sci USA.
114:11751–11756. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Findlay GM, Boyle EA, Hause RJ, Klein JC
and Shendure J: Saturation editing of genomic regions by multiplex
homology-directed repair. Nature. 513:120–123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Burger A, Lindsay H, Felker A, Hess C,
Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, et al:
Maximizing mutagenesis with solubilized CRISPR-Cas9
ribonucleoprotein complexes. Development. 143:2025–2037.
2016.PubMed/NCBI
|
|
19
|
Pelttari LM, Kiiski JI, Ranta S, Vilske S,
Blomqvist C, Aittomäki K and Nevanlinna H: RAD51, XRCC3, and XRCC2
mutation screening in Finnish breast cancer families. Springerplus.
4:922015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Short JM, Liu Y, Chen S, Soni N,
Madhusudhan MS, Shivji MK and Venkitaraman AR: High-resolution
structure of the presynaptic RAD51 filament on single-stranded DNA
by electron cryo-microscopy. Nucleic Acids Res. 44:9017–9030.
2016.PubMed/NCBI
|
|
21
|
Xu J, Zhao L, Xu Y, Zhao W, Sung P and
Wang HW: Cryo-EM structures of human RAD51 recombinase filaments
during catalysis of DNA-strand exchange. Nat Struct Mol Biol.
24:40–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen J, Villanueva N, Rould MA and
Morrical SW: Insights into the mechanism of RAD51 recombinase from
the structure and properties of a filament interface mutant.
Nucleic Acids Res. 38:4889–4906. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bonilla B, Hengel SR, Grundy MK and
Bernstein KA: Rad51 gene family structure and function. Annu Rev
Genet. 54:25–46. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baldock RA, Pressimone CA, Baird JM,
Khodakov A, Luong TT, Grundy MK, Smith CM, Karpenshif Y,
Bratton-Palmer DS, Prakash R, et al: Rad51D splice variants and
cancer-associated mutations reveal XRCC2 interaction to be critical
for homologous recombination. DNA Repair (Amst). 76:99–107. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Corrales-Sánchez V, Noblejas-López MDM,
Nieto-Jiménez C, Pérez-Peña J, Montero JC, Burgos M, Galán-Moya EM,
Pandiella A and Ocaña A: Pharmacological screening and
transcriptomic functional analyses identify a synergistic
interaction between dasatinib and olaparib in triple-negative
breast cancer. J Cell Mol Med. 24:3117–3127. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stemmer M, Thumberger T, Del Sol Keyer M,
Wittbrodt J and Mateo JL: CCTop: An intuitive, flexible and
reliable CRISPR/Cas9 target prediction tool. PloS One.
10:e01246332015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamel D, Gray C, Walia JS and Kumar V:
PARP inhibitor drugs in the treatment of breast, ovarian, prostate
and pancreatic cancers: An update of clinical trials. Curr Drug
Targets. 19:21–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chu CY, Lee YC, Hsieh CH, Yeh CT, Chao TY,
Chen PH, Lin IH, Hsieh TH, Shih JW, Cheng CH, et al: Genome-wide
CRISPR/Cas9 knockout screening uncovers a novel inflammatory
pathway critical for resistance to arginine-deprivation therapy.
Theranostics. 11:3624–3641. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang B, Wang M, Zhang W, Xiao T, Chen CH,
Wu A, Wu F, Traugh N, Wang X, Li Z, et al: Integrative analysis of
pooled CRISPR genetic screens using MAGeCKFlute. Nat Prot.
14:756–780. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giuliano AE, Edge SB and Hortobagyi GN:
Eighth edition of the AJCC cancer staging manual: Breast cancer.
Ann Surg Oncol. 25:1783–1785. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z,
Zhang X, Zhang P and Huang X: Generation of gene modified mice via
Cas9/RNA-mediated gene targeting. Cell Res. 23:720–723. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stephens M and Scheet P: Accounting for
decay of linkage disequilibrium in haplotype inference and
missing-data imputation. Am J Hum Genet. 76:449–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li P, Guo M, Wang C, Liu X and Zou Q: An
overview of SNP interactions in genome-wide association studies.
Brief Funct Genomics. 2:143–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tulbah S, Alabdulkarim H, Alanazi M,
Parine NR, Shaik J, Pathan AA, Al-Amri A, Khan W and Warsy A:
Polymorphisms in Rad51 and their relation with breast cancer in
Saudi females. Onco Targets. 9:269–277. 2016.PubMed/NCBI
|
|
35
|
Vral A, Willems P, Claes K, Poppe B,
Perletti G and Thierens H: Combined effect of polymorphisms in
Rad51 and Xrcc3 on breast cancer risk and chromosomal
radiosensitivity. Mol Med Rep. 4:901–912. 2011.PubMed/NCBI
|
|
36
|
Brooks J, Shore RE, Zeleniuch-Jacquotte A,
Currie D, Afanasyeva Y, Koenig KL, Arslan AA, Toniolo P and Wirgin
I: Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to
breast cancer risk. Cancer Epidemiol Biomark Prev. 17:1016–1019.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim JH and Hong YC: Modification of PARP4,
XRCC3, and RAD51 gene polymorphisms on the relation between
Bisphenol A exposure and liver abnormality. Int J Environ Res
Public Health. 17:27942020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Marchenko YV, Carroll RJ, Lin DY, Amos CI
and Gutierrez RG: Semiparametric analysis of case-control genetic
data in the presence of environmental factors. Stata J. 8:305–333.
2008. View Article : Google Scholar
|
|
39
|
Hurley RM, Wahner Hendrickson AE, Visscher
DW, Ansell P, Harrell MI, Wagner JM, Negron V, Goergen KM, Maurer
MJ, Oberg AL, et al: 53BP1 as a potential predictor of response in
PARP inhibitor-treated homologous recombination-deficient ovarian
cancer. Gynecol Oncol. 153:127–134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nacson J, Krais JJ, Bernhardy AJ, Clausen
E, Feng W, Wang Y, Nicolas E, Cai KQ, Tricarico R, Hua X, et al:
Brca1 mutation-specific responses to 53bp1 loss-induced homologous
recombination and PARP inhibitor resistance. Cell Rep.
24:3513–3527. e72018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jaspers JE, Kersbergen A, Boon U, Sol W,
van Deemter L, Zander SA, Drost R, Wientjens E, Ji J, Aly A, et al:
Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated
mouse mammary tumors. Cancer Discov. 3:68–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Symington LS and Gautier J: Double-strand
break end resection and repair pathway choice. Annu Rev Genet.
45:247–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shalem O, Sanjana NE, Hartenian E, Shi X,
Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG and
Zhang F: Genome-scale CRISPR-Cas9 knockout screening in human
cells. Science. 343:84–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Koike-Yusa H, Li Y, Tan EP,
Velasco-Herrera Mdel C and Yusa K: Genome-wide recessive genetic
screening in mammalian cells with a lentiviral CRISPR-guide RNA
library. Nat Biotechnol. 32:267–273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamauchi T, Masuda T, Canver MC, Seiler M,
Semba Y, Shboul M, Al-Raqad M, Maeda M, Schoonenberg VAC, Cole MA,
et al: Genome-wide CRISPR-Cas9 screen identifies leukemia-specific
dependence on a Pre-mRNA metabolic pathway regulated by DCPS.
Cancer Cell. 33:386–400. e52018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ali M, Kaltenbrun E, Anderson GR, Stephens
SJ, Arena S, Bardelli A, Counter CM and Wood KC: Codon bias imposes
a targetable limitation on KRAS-driven therapeutic resistance. Nat
Commun. 8:156172017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Morio F, Lombardi L and Butler G: The
CRISPR toolbox in medical mycology: State of the art and
perspectives. PLoS Pathog. 16:e10082012020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gout J, Perkhofer L, Morawe M, Arnold F,
Ihle M, Biber S, Lange S, Roger E, Kraus JM, Stifter K, et al:
Synergistic targeting and resistance to PARP inhibition in DNA
damage repair-deficient pancreatic cancer. Gut. 70:743–760. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dallavalle S, Dobričić V, Lazzarato L,
Gazzano E, Machuqueiro M, Pajeva I, Tsakovska I, Zidar N and
Fruttero R: Improvement of conventional anti-cancer drugs as new
tools against multidrug resistant tumors. Drug Resist Update.
50:1006822020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Laucht M, Skowronek MH, Becker K, Schmidt
MH, Esser G, Schulze TG and Rietschel M: Interacting effects of the
dopamine transporter gene and psychosocial adversity on
attention-deficit/hyperactivity disorder symptoms among
15-year-olds from a high-risk community sample. Arch Gen
Psychiatry. 64:585–590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Caspi A, Sugden K, Moffitt TE, Taylor A,
Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A
and Poulton R: Influence of life stress on depression: Moderation
by a polymorphism in the 5-HTT gene. Science. 301:386–389. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aguiar D, Wong WS and Istrail S: Tumor
haplotype assembly algorithms for cancer genomics. Pac Symp
Biocomput. 3–14. 2014.PubMed/NCBI
|
|
53
|
Azam M, Latek RR and Daley GQ: Mechanisms
of autoinhibition and STI-571/imatinib resistance revealed by
mutagenesis of BCR-ABL. Cell. 112:831–843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma Y, Zhang J, Yin W, Zhang Z, Song Y and
Chang X: Targeted AID-mediated mutagenesis (TAM) enables efficient
genomic diversification in mammalian cells. Nat Methods.
13:1029–1035. 2016. View Article : Google Scholar : PubMed/NCBI
|