Introduction
Nephroblastoma, also known as Wilms' tumor, is the
fourth most common malignant abdominal cancer in children and
typically occurs between the ages of 3 and 5 years (1,2).
Over the past 50 years, therapeutic options for patients with
nephroblastoma have been primarily based on nephrectomy,
complemented by radiotherapy, systemic chemotherapy and autologous
stem cell transplantation (3).
The improvement of treatment methods has led to long-term survival
rate of >90% (4); thus
nephroblastoma has one of the best prognoses among types of
pediatric cancer. However, certain patients still experience poor
prognosis due to cancer recurrence and metastasis (5). Therefore, discovering targeted
biomarkers of nephroblastoma may contribute to understanding of the
pathogenesis and metastatic mechanism and development of novel
therapeutic strategies for management of patients with
nephroblastoma.
DEP domain containing 1 (DEPDC1) is a
tumor-associated gene (6,7) involved in regulation of cellular
processes such as transcription, mitosis and apoptosis (8). In addition, other studies have shown
that DEPDC1 participates in tumorigenesis and cancer progression in
lung adenocarcinoma (9), gastric
cancer (10), oral squamous cell
carcinoma (11) and colorectal
(12), liver (13) and breast cancer (14). By analyzing Therapeutically
Applicable Research to Generate Effective Treatments database, Su
et al (5) found that DEPDC1 is
highly expressed in nephroblastoma and is associated with poor
patient survival, indicating that DEPDC1 may serve as a potential
prognostic and diagnostic biomarker for nephroblastoma. To the best
of our knowledge, however, the role of DEPDC1 in the onset and
development of nephroblastoma have not been investigated.
Forkhead box transcription factor O (FOXO), a
subfamily of the forkhead transcription factor family, serves an
essential role in cell function, including apoptosis, energy
metabolism, DNA damage repair and oxidative stress (15–17). The FOXO subfamily includes four
primary subtypes (FOXO1, FOXO3/FOXO3a, FOXO4 and FOXO6) that
possess similar structure and function (18). The expression of FOXO4 is higher
in skeletal muscle, while FOXO3a is primarily expressed in the
brain, heart, kidney and spleen (19). FOXO6 is highly expressed in
nervous tissue (19). Studies
have demonstrated that downregulation of the functioning of FOXO
protein promotes tumorigenesis and cancer progression (20,21). In particular, FOXO3a, a tumor
suppressor, has been reported to be associated with metastasis of
malignant tumor, such as breast (22), pancreatic (23) and kidney cancer (24). A recent study found that FOXO3a
inhibits nephroblastoma cell proliferation, migration and invasion
and suppresses apoptosis by disrupting the Wnt/β-catenin signaling
pathway (25). Downregulation of
DEPDC1 also inhibits the Wnt/β-catenin signaling pathway (26). To the best of our knowledge,
however, no report has studied the association between FOXO3a and
DEPDC1 in onset and progression of nephroblastoma.
The aim of the present study was to clarify the
association between DEPDC1 expression, prognosis of nephroblastoma
and FOXO3a and associated signaling pathways to identify potential
therapeutic targets.
Materials and methods
Cell culture and treatment
Human normal renal (HK-2) and nephroblastoma cell
lines (WiT49, WILTU-1 and GHINK-1) were obtained from American Type
Culture Collection. All cell lines were cultured in DMEM
supplemented with 10% FBS (both HyClone; Cytiva), 1% penicillin and
1% streptomycin (both from Beyotime Institute of Biotechnology) at
37°C with 5% CO2.
Bioinformatics analysis
JASPAR database (jaspar.genereg.net/) was used to
predict the binding site of transcription factor FOXO3a to DEPDC1
promoter.
Cell proliferation
WiT49 cell (1×105 cells/well)
proliferation was assessed by 5-ethynyl-2′-deoxyuridine (EdU)
incorporation using a Click-iT™ EdU Imaging kit (cat.
no. C10337; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Subsequently, 1 µg/ml DAPI (Beyotime
Institute of Biotechnology) was applied to stain the cells for 10
min at room temperature. Images were captured in five randomized
fields of view using a confocal microscope.
Cell transfection
WiT49 cells (3×105 cells/well) were
inoculated onto 6-well plates and cultured for 24 h at 37°C with 5%
CO2. Following incubation, cells were transfected with
overexpression (Ov)-DEPDC1, Ov-FOXO3a vector, negative control
empty vector (Ov-NC), DEPDC1-targeting short hairpin (sh)RNA
(sh-DEPDC1#1, and sh-DEPDC1#2) and sh-NC at a concentration of 25
nM. shRNA sequences are presented in Table I. All plasmids were synthesized by
Shanghai GenePharma Co., Ltd. and transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Blank control group cells were untreated. Following transfection
for 48 h at 37°C, transfection efficiency was detected using
reverse transcription-quantitative (RT-q)PCR and western blotting
within 48 h.
 | Table I.shRNA and reverse
transcription-quantitative PCR primer sequences. |
Table I.
shRNA and reverse
transcription-quantitative PCR primer sequences.
| Name | Sequence |
|---|
| sh-DEPDC1#1 |
5′-GGAAGTTGTCATAATTTAA-3′ |
| sh-DEPDC1#2 |
5′-CGAGATGTATTCAGAACAA-3′ |
| sh-NC |
5′-TTCGGGTCATCCGATGGGCC-3′ |
| DEPDC1 | Forward:
5′-TCTGCCATGAAGTGCCTAGC-3′ |
|
| Reverse:
5′-TGATGTAGCCACAAACAACCAAA-3′ |
| FOXO3a | Forward:
5′-GAGGAGGACGATGAAGACGAC-3′ |
|
| Reverse:
5′-TGTGCCGGATGGAGTTCTTC-3′ |
| GAPDH | Forward:
5′-AGCCACATCGCTCAGACAC-3′ |
|
| Reverse:
5′-GCCCAATACGACCAAATCC-3′ |
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was performed to assess cell viability.
Briefly, WiT49 cells were seeded into a 96-well plate
(6×103 cells/well) and incubated for 24, 48 and 72 h at
37°C under 5% CO2. Following incubation, 10 µl CCK-8
solution (cat. no. P0037; Beyotime Institute of Biotechnology) was
added to each well and cells were cultured for another 2 h at 37°C
with 5% CO2. The optical density (OD) was measured at
450 nm using a microplate reader (BioTek Instruments, Inc.),
Relative cell viability (%) was calculated as follows: (Treated
ODA450-blank ODA450)/(control
ODA450-blank ODA450) ×100%.
Colony formation assay
WiT49 cells (5×102 cells/well) suspended
in DMEM were inoculated into 6-well plates and incubated at 37°C in
5% CO2 for 14 days. The cells were fixed with 70%
ethanol at room temperature for 15 min, stained with 0.05% crystal
violet at 37°C for 20 min and number of colonies was counted
manually (≥50 cells) using an Olympus GX53 light microscope
(Olympus Corporation; magnification, ×4).
Wound healing assay
WiT49 cells were inoculated in 6-well plates
(1×105 cells/well). When they reached 70–80% confluence,
medium was replaced with serum-free DMEM (Shanghai Yimiao Chemical
Technology Co., Ltd.) and cells were incubated overnight at 37°C at
5% CO2. Subsequently, a 200-µl sterile pipette tip was
used to scratch the cell monolayer. Following washing, with PBS
three times, plates were maintained at 37°C with 5% CO2.
Images were captured at 0 and 24 h using a BX51 inverted microscope
(Olympus Corporation; magnification, ×100) Cell migration (%) was
calculated as follows: Final wound width/original wound width
×100.
Cell invasion assay
Cell invasion ability was assessed using
Matrigel-coated Transwell chambers (BD Biosciences). Briefly,
24-well Transwell plates (Corning, Inc.) with 8-µm pore inserts
were coated with Matrigel (BD Biosciences) at 37°C for 30 min.
Next, 4×104 WiT49 cells were plated in serum-free DMEM
(Shanghai Yimiao Chemical Technology Co., Ltd.). The upper chamber
was precoated with Matrigel and cells were cultured in the upper
chamber (0.1 ml cell suspension/well); the lower chamber was filled
with DMEM (Shenzhen Baienwei Biotechnology Co., Ltd.) supplemented
with 20% FBS. Following incubation at 37°C with 5% CO2
for 24 h, the upper chamber was collected and cleaned. The invading
cells were fixed with 4% formaldehyde at 25°C for 15 min and
stained with 0.3% crystal violet solution (Sigma-Aldrich; Merck
KGaA) at room temperature for 30 min. The invading cells were
observed under a light microscope in five randomly selected fields
of view (Olympus Corporation; magnification, ×100).
RT-qPCR
Total RNA was extracted from WiT49 cells using
RNAzol RT (Sigma-Aldrich; Merck KGaA), according to the
manufacturer's instructions. RNA concentration and quantification
were assessed using a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.). Following DNase I digestion, total RNA was
reverse-transcribed into cDNA using a QuantiTect Reverse
Transcription kit (Qiagen GmbH), according to the manufacturer's
instructions. Subsequently, RT-qPCR was performed using a
QuantiTect SYBR Green PCR kit (Qiagen GmbH), according to the
manufacturer's instructions. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles of 95°C for 10 sec and 60°C for 60 sec. Primers (GenScript)
are listed in Table I. mRNA
expression levels were quantified using the 2-ΔΔCq
method (27) and normalized to
the internal reference gene GAPDH.
Western blotting
WiT49 cells were washed twice with PBS, lysed with
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
incubated for 30 min on ice. Cell lysate was centrifuged at 300 × g
at 4°C for 20 min and protein supernatant was transferred into
Eppendorf tubes. Total protein was quantified using a BCA protein
assay kit (Bio-Rad Laboratories, Inc.). Following denaturation in a
95°C metal bath, Protein (20 µg/lane) was separated using 10%
SDS-PAGE and transferred onto PVDF membranes (GE Healthcare), which
were blocked with 10% skimmed milk for 1 h at room temperature.
Subsequently, membranes were incubated overnight at 4°C with the
primary antibodies (all 1:1,000; all Abcam) as follows: Anti-DEPDC1
(cat. no. ab197246), anti-FOXO3a (cat. no. ab109629),
anti-phosphorylated (p-)glycogen synthase kinase-3β (GSK-3β; cat.
no. ab75814), anti-GSK-3β (cat. no. ab93926), anti-Wnt3a (cat. no.
ab219412), anti-β-catenin (cat. no. ab32572) and anti-GAPDH (cat.
no. ab181602). Following incubation with primary antibodies, the
membranes were incubated with goat anti-rabbit horseradish
peroxidase-conjugated IgG secondary antibody (1:5,000; cat. no.
ab150077; Abcam) at room temperature for 1 h. Protein bands were
visualized using enhanced chemiluminescence reagent (GE
Healthcare). Protein expression levels were semi-quantified using
ImageJ software (version 1.46; National Institutes of Health) with
GAPDH as the loading control.
Dual-luciferase reporter assay
WiT49 cells were inoculated in a 24-well plate
(1×105 cells/well). When cells reached 80% confluence,
luciferase activity was detected using the Promega Double
Fluorescence Detection kit (Promega Corporation), according to the
manufacturer's instructions. The DEPDC1 sequence including the
putative binding sites of FOXO3a was sub-cloned and inserted into
the pmirGLO vector (Promega Corporation) to construct
DEPDC1-wild-type (WT) and mutant (MUT) reporter plasmids. 0.1 µg
DEPDC1-WT or DEPDC1-MUT reporter plasmids containing Ov-FOXO3a or
Ov-NC were constructed (Shanghai GenePharma) to be co-transfected
into WiT49 cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). After 24 h, culture supernatant was
discarded and collected cells were rinsed with PBS three times.
Subsequently, 120 µl cell lysate was added to each well and shaken
on a horizontal oscillator for 45 min at 50 × g. Next, 10 µl lysed
cell mixture and 50 µl firefly luciferase reagent was added to a
1.5 ml tube and mixed. A total of 50 µl Stop/Glo Sealing Luciferase
Reagent was added. Firefly/Renilla luciferase values were recorded
and DEPDC1 promoter transfer activity was analyzed. The luciferase
activity was also normalized to that of Renilla. Each group was set
up in three wells and each experiment was repeated three times.
Chromatin immunoprecipitation
(ChIP)-PCR assay
Total genomic DNA was isolated by 1 ml SDS
(Beyotime)and sonicated using the EZChip™ kit (EMD
Millipore). Cells (1×107/ml were sonicated for 10s on
ice for 5 times and fragmented DNA was visualized on an agarose
gel. For chromatin isolation, the sample was centrifuged at 15,000
× g for 10 min at 4°C to remove insoluble material and 10X ChIP
dilution buffer was added to the collected supernatant. The sample
was pre-cleared with protein G-agarose beads at 4°C for 1 h and
pre-cleared chromatin was incubated with antibodies against FOXO3a
(cat. no. ab70315;3 µg/mg; Abcam) at 4°C overnight according to the
manufacture's protocol. Finally, PBS was adopted for the rinse of
the sample. Immunochromatin was amplified using RT-qPCR as
aforementioned.
Statistical analysis
Data are presented as the mean ± SD of ≥3
independent experiments. GraphPad Prism 8.0.2 software (GraphPad
Software, Inc.) was used for data analysis. Statistical differences
were determined using one-way ANOVA followed by Tukey's post hoc
test for group comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
DEPDC1 is highly expressed in
nephroblastoma cell lines and sh-DEPDC1 inhibits WiT49 cell
proliferation
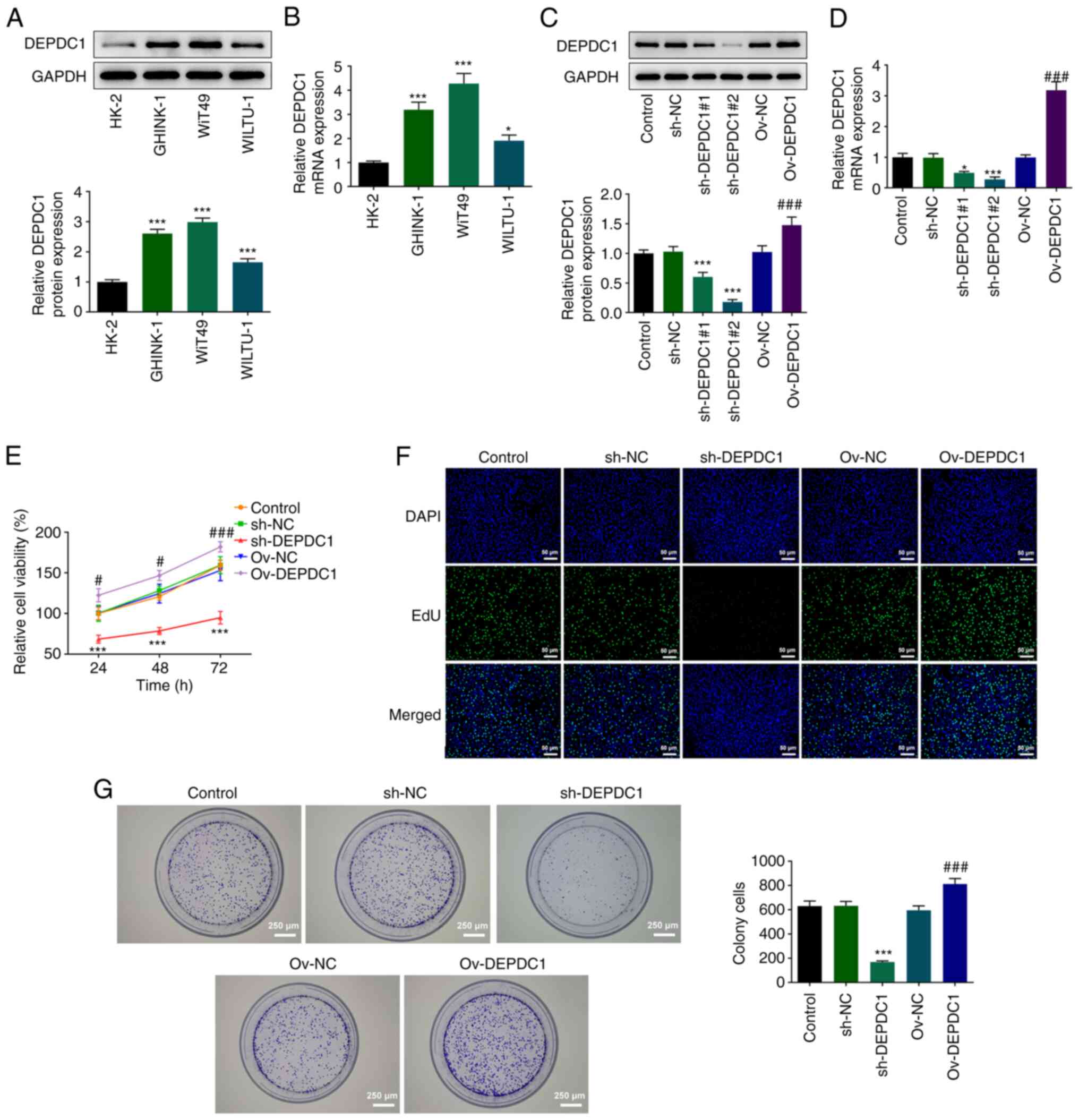
Western blotting (Fig.
1A) and RT-qPCR (Fig. 1B)
were used to detect DEPDC1 expression in nephroblastoma cell lines.
Compared with normal renal cell line (HK-2), DEPDC1 expression was
significantly increased in nephroblastoma cell lines (WiT49,
WILTU-1 and GHINK-1), particularly WiT49 cells. Therefore, WiT49
cells were selected for subsequent experiments. To investigate the
effects of DEPDC1 on nephroblastoma cell proliferation, DEPDC1
interference and overexpression plasmids were constructed and
transfected into WiT49 cells. Compared with the control group,
protein and mRNA expression levels of DEPDC1 in the Ov-DEPDC1 group
significantly increased (Fig. 1C and
D). In addition, transfection of interference plasmids
significantly inhibited expression of DEPDC1, with sh-DEPDC1#2
exhibiting a greater interference efficiency than sh-DEPDC1#1.
Therefore, sh-DEPDC1#2 was selected for subsequent experiments.
CCK-8 assay results showed that sh-DEPDC1 significantly inhibited
the viability of WiT49 cells. By contrast, DEPDC1 overexpression
significantly increased cell viability (Fig. 1E). Proliferation of WiT49 cells
was assessed using EdU incorporation (Fig. 1F) and colony formation experiments
(Fig. 1G). sh-DEPDC1 inhibited
cell proliferation compared with the control group, with the
opposite results observed following DEPDC1 overexpression,
evidenced by increased cell proliferation and tumorigenic ability
in Ov-DEPDC1 group. These results indicated that DEPDC1 may promote
nephroblastoma progression.
sh-DEPDC1 inhibits nephroblastoma cell
invasion and migration
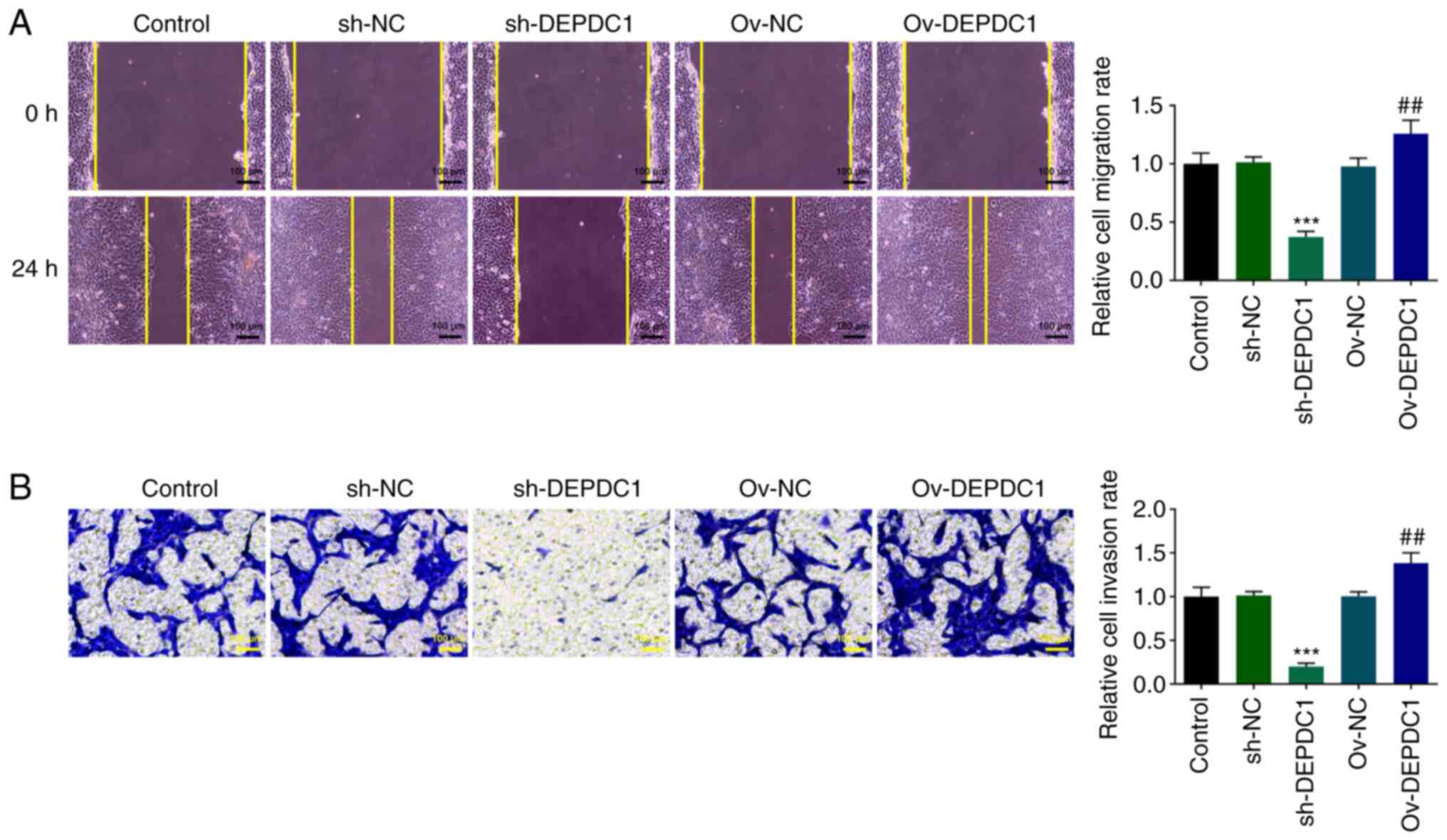
Wound healing (Fig.
2A) and Transwell (Fig. 2B)
assay were used to detect cell invasion and migration. Compared
with the Ov-NC group, cell migration and invasion in the Ov-DEPDC1
group significantly increased. In addition, sh-DEPDC1 significantly
inhibited migration and invasion of WiT49 cells. These results
suggested that DEPDC1 may promote invasion and migration of
nephroblastoma cells.
FOXO3a inhibits transcription of
DEPDC1
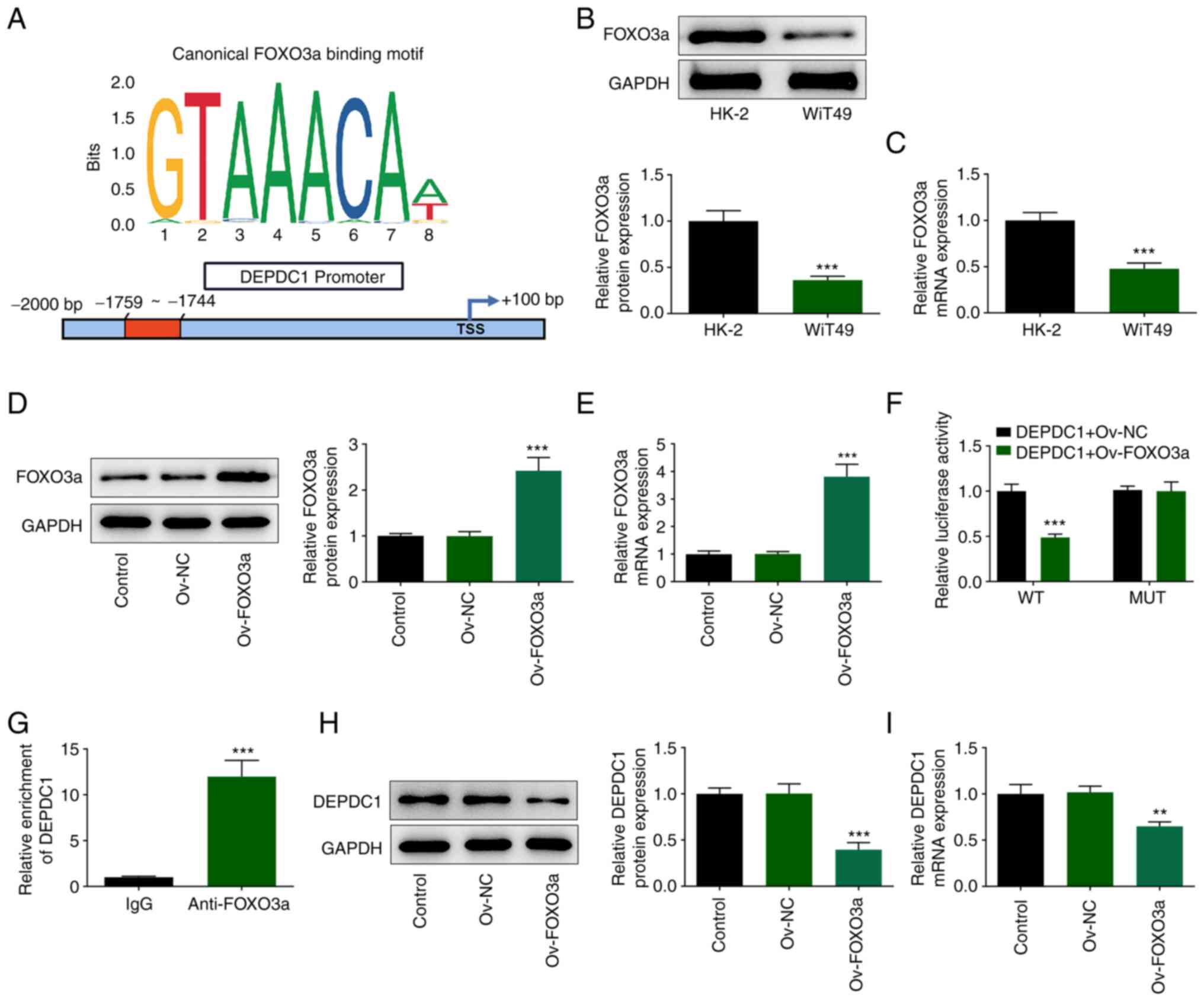
The binding sites of transcription factors FOXO3a
and DEPDC1 promoter were predicted using JASPAR (Fig. 3A). Next, western blotting
(Fig. 3B) and RT-qPCR (Fig. 3C) were performed to detect FOXO3a
expression in WiT49 cells. FOXO3a mRNA and protein expression were
significantly downregulated in WiT49 compared with HK-2 cells.
Ov-FOXO3a plasmid was constructed to detect DEPDC1 promoter
activity using luciferase assay. Protein and mRNA expression of
FOXO3a were increased by overexpressing FOXO3a compared with that
in Ov-NC (Fig. 3D and E).
Ov-FOXO3a induced a significant decrease in luciferase activity in
the presence of DEPDC1-WT promoter, while there was no significant
change in the presence of MUT promoter (Fig. 3F). Subsequently, binding of FOXO3a
to DEPDC1 promoter was verified using ChIP. DEPDC1 has abundant
enrichment in anti-FOXO3a in comparison with the IgG (Fig. 3G). Furthermore, DEPDC1 expression
was significantly downregulated in Ov-FOXO3a nephroblastoma cells
compared with Ov-NC group (Fig. 3H
and I).
FOXO3a mediates DEPDC1 promotion of
malignant progression of nephroblastoma via Wnt/β-catenin
signaling
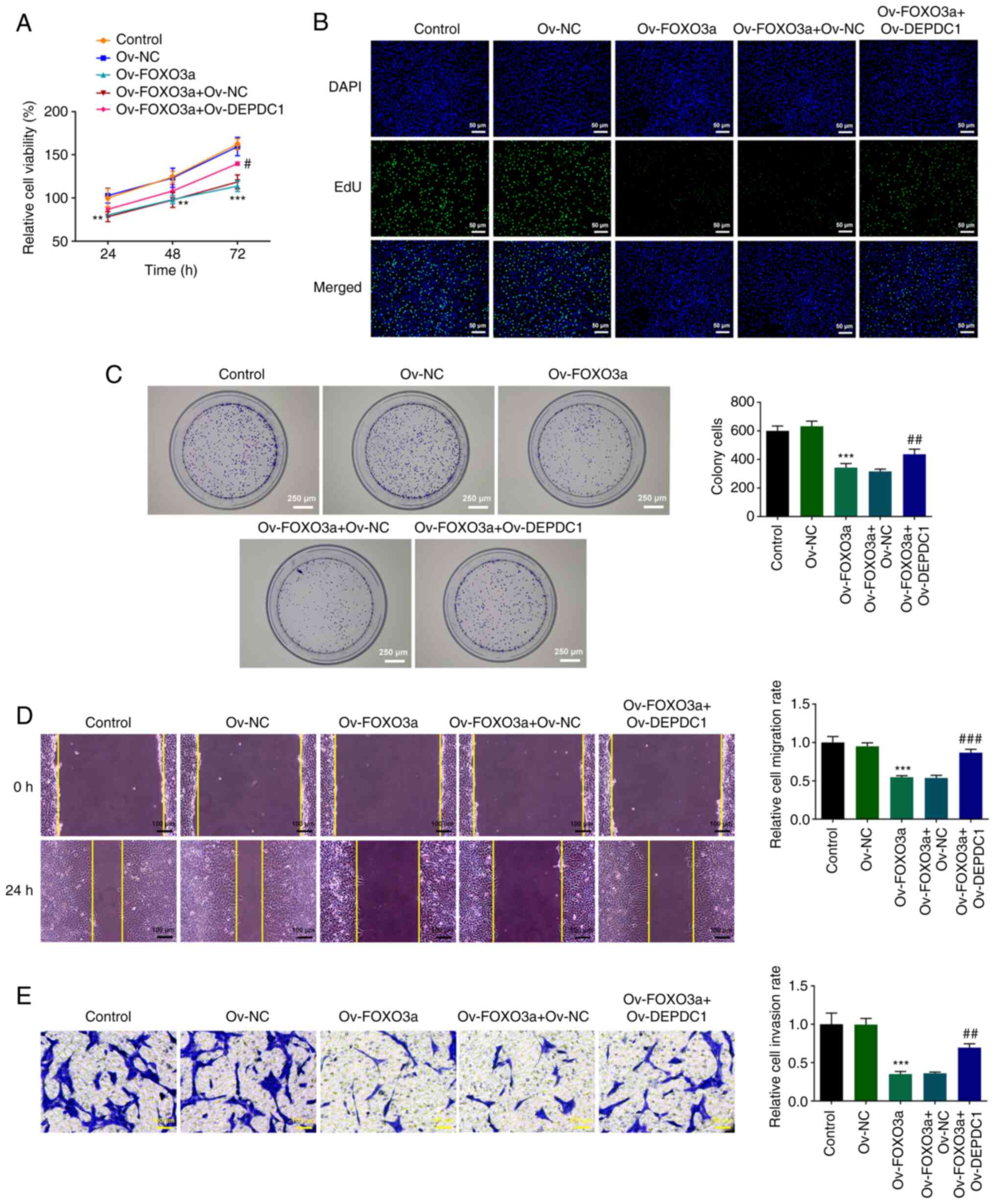
Considering the negative association between FOXO3a
and DEPDC1 expression (Fig. 3H and
I), DEPDC1 was overexpressed using Ov-FOXO3a to assess the
effect of DEPDC1 on proliferation, migration and invasion of
nephroblastoma cells and the potential mechanism. Compared with
Ov-NC, Ov-FOXO3a significantly inhibited WiT49 cell viability,
which was reversed by DEPDC1 overexpression, evidenced by the
increased WiT49 cell viability in Ov-FOXO3a + Ov-DEPDC1 when
compared with Ov-FOXO3a + Ov-NC group (Fig. 4A). EdU (Fig. 4B) and colony formation assay
(Fig. 4C), which detected cell
proliferation, showed that overexpression of DEPDC1 reversed
inhibition of cell proliferation induced by Ov-FOXO3a in comparison
with the Ov-FOXO3a + Ov-NC group. Wound healing (Fig. 4D) and Transwell assay (Fig. 4E) also showed that DEPDC1
overexpression reversed the inhibitory effect of Ov-FOXO3a on cell
migration and invasion compared with Ov-FOXO3a + Ov-NC. Assessment
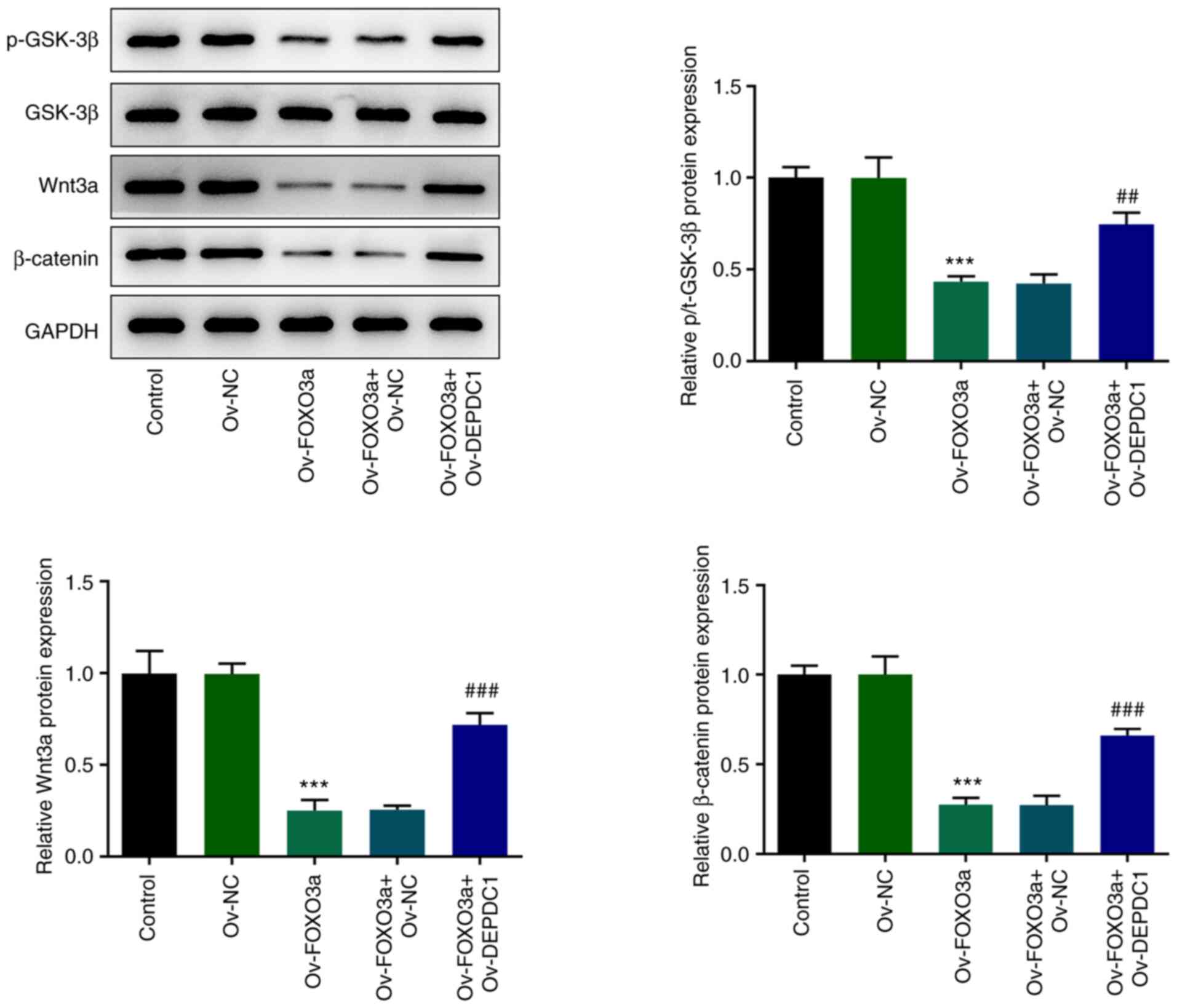
of Wnt/β-catenin signaling pathway-associated protein expression
levels (Fig. 5) showed that
FOXO3a overexpression inhibited protein expression of p-GSK-3β,
Wnt3a and β-catenin, while overexpression of DEPDC1 increased
expression of the aforementioned proteins, suggesting that FOXO3a
mediates DEPDC1-induced promotion of malignant progression of
nephroblastoma via the Wnt/β-catenin signaling pathway.
Discussion
Nephroblastoma typically occurs in children aged
<5 years and is one of the most common types of abdominal
malignancy, accounting for 8% of all pediatric malignancy (28). Although the prognosis for patients
with nephroblastoma has markedly improved over past decades, the
mortality rate in 1969 to 1995 remained high in the USA (29). It is therefore important to
determine the biological and molecular mechanisms underlying
nephroblastoma progression.
The aberrant expression of DEPDC1, a recently
identified tumor-associated gene, is associated with tumorigenesis
in multiple types of cancer, such as lung adenocarcinoma (30), hepatocellular carcinoma (31) and gastric cancer (10) and contributes to cancer
progression (7). The aberrant
upregulation of DEPDC1 has been observed in bladder (32) and breast cancer (33), multiple myeloma (34) and hepatocellular carcinoma
(35), indicating its potential
value as a therapeutic target. A recent study revealed that
increased expression of DEPDC1 leads to poor survival in patients
with nephroblastoma (36). Su et
al (5) found that DEPDC1 is
highly expressed in nephroblastoma and associated with poor
survival using database network analysis. The present study
investigated the role and underlying mechanism of DEPDC1 in
nephroblastoma progression. DEPDC1 was highly expressed in
nephroblastoma cells at both the protein and mRNA level. These
results supported the hypothesis that high expression of DEPDC1
serves a carcinogenic role in nephroblastoma progression. To verify
this, shRNA interference vectors were constructed to knock down
DEPDC1 expression. Silencing expression of DEPDC1 significantly
inhibited proliferation, migration and invasion of nephroblastoma
cells. This is similar to results of Shen and Xi (37), who reported that DEPDC1 is highly
expressed in lung adenocarcinoma and promotes tumor cell
proliferation.
FOXO proteins are considered to be tumor suppressors
due to their pro-apoptotic and anti-proliferative activity
(17). Accumulating evidence
indicates that suppressing the function of FOXO proteins promotes
cancer progression and tumorigenesis (20,38). FOXO3a, also known as FOXO3 or
forehead in rhabdomyosarcoma-like 1, is a key transcription factor
regulating biological processes, including apoptosis, inflammation
and DNA damage response (39–41) and is associated with extreme human
longevity (18). As a tumor
suppressor, FOXO3a exerts an inhibitory effect on cancer
progression and is frequently inactivated in breast cancer cell
lines (42). In the present
study, expression of FOXO3a was downregulated in WiT49 cells
compared with normal HK-2 cells; this is consistent with results of
Qian and Liu (25).
JASPAR predicted that FOXO3a bound to and regulated
DEPDC1, indicating an association between FOXO3a and DEPDC1
expression in nephroblastoma. In addition, co-immunoprecipitation
results demonstrated that FOXO3a bound to DEPDC1 and Ov-FOXO3a
decreased levels of DEPDC1. Ov-FOXO3a also prevented the promoting
effect of Ov-DEPDC1 on proliferation, invasion and migration of
nephroblastoma cells, indicating an association between FOXO3a and
DEPDC1. Increasing evidence indicates that aberrant Wnt/β-catenin
cascade leads to onset and progression of certain types of solid
tumor and hematological malignancy (43,44). The Wnt signaling pathway is a key
signal transduction pathway involved in renal development (45). The transcription factor β-catenin
is a key component of the Wnt signaling pathway and its
upregulation leads to occurrence of acute myeloid leukemia
(46). Therefore, it may be
important to study the effect of abnormal Wnt/β-catenin signaling
on the occurrence of nephroblastoma. In addition, DEPDC1 has been
reported to activate the Wnt/β-catenin signaling pathway to
regulate proliferation and metastasis of hepatocellular carcinoma
cells (26,47). In the present study, FOXO3a
overexpression decreased expression levels of p-GSK-3β, β-catenin
and Wnt3a protein, which are involved in the Wnt/β-catenin
signaling pathway, and inhibited the DEPDC1-dependent Wnt/β-catenin
pathway.
To the best of our knowledge, the present study is
the first to investigate the role and mechanism of DEPDC1 in
nephroblastoma. The present study only performed in vitro
experiments; therefore, in vivo studies are needed to verify the
results. DNMT1-mediated hypermethylation of the FOXO3a promoter
downregulates FOXO3a expression in breast cancer (48). However, the mechanism underlying
downregulation of FOXO3a expression and the effect of FOXO3a
depletion on cell proliferation and DNA damage in nephroblastoma
need further investigation. In addition, the association between
signaling pathways other than Wnt/β-catenin (for example, NF-κB)
and the mechanism of DEPDC1 in nephroblastoma need to be
verified.
In conclusion, the present study demonstrated that
DEPDC1 promoted malignant progression of nephroblastoma via the
Wnt/β-catenin signaling pathway; this may be regulated by FOXO3a.
The present findings suggested that DEPDC1 may serve as a valuable
biomarker for understanding the mechanism of nephroblastoma
progression.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GG, MM, QHL and XQG designed the study. QBN, YTX,
ZLM and YJW performed the experiments. GG, MM, ZLM and YJW revised
the manuscript. QHL, YTX and XQG collected and analyzed the data.
ZLM, YJW and QBN confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xie W, Wei L, Guo J, Guo H, Song X and
Sheng X: Physiological functions of Wilms' tumor 1-associating
protein and its role in tumourigenesis. J Cell Biochem. Feb
12–2019.(Epub ahead of print). doi: 10.1002/jcb.28402.
|
|
2
|
Cone EB, Dalton SS, Van Noord M, Tracy ET,
Rice HE and Routh JC: Biomarkers for Wilms tumor: A systematic
review. J Urol. 196:1530–1535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chakumatha E, Weijers J, Banda K, Bailey
S, Molyneux E, Chagaluka G and Israels T: Outcome at the end of
treatment of patients with common and curable childhood cancer
types in Blantyre, Malawi. Pediatr Blood Cancer. 67:e283222020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fuchs J: Surgical concepts in the
treatment of Wilms tumor: An update. Urologe A. 54:1784–1791.
2015.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su C, Huang R, Yu Z, Zheng J, Liu F, Liang
H and Mo Z: Myelin and lymphocyte protein serves as a prognostic
biomarker and is closely associated with the tumor microenvironment
in the nephroblastoma. Cancer Med. 11:1427–1438. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanehira M, Harada Y, Takata R, Shuin T,
Miki T, Fujioka T, Nakamura Y and Katagiri T: Involvement of
upregulation of DEPDC1 (DEP domain containing 1) in bladder
carcinogenesis. Oncogene. 26:6448–6455. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu Y, Sun L, Yu J, Xiang Y, Shen M, Wasan
HS, Ruan S and Qiu S: Identification of biomarkers in colon cancer
based on bioinformatic analysis. Transl Cancer Res. 9:4879–4895.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harada Y, Kanehira M, Fujisawa Y, Takata
R, Shuin T, Miki T, Fujioka T, Nakamura Y and Katagiri T:
Cell-permeable peptide DEPDC1-ZNF224 interferes with
transcriptional repression and oncogenicity in bladder cancer
cells. Cancer Res. 70:5829–5839. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li YY, Li W, Chang GZ and Li YM: Long
noncoding RNA KTN1 antisense RNA 1exerts an oncogenic function in
lung adenocarcinoma by regulating DEP domain containing 1
expression via activating epithelial-mesenchymal transition.
Anticancer Drugs. 32:614–625. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gong Z, Chu H, Chen J, Jiang L, Gong B,
Zhu P, Zhang C, Wang Z, Zhang W, Wang J, et al: DEPDC1 upregulation
promotes cell proliferation and predicts poor prognosis in patients
with gastric cancer. Cancer Biomark. 30:299–307. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo J, Zhou S, Huang P, Xu S, Zhang G, He
H, Zeng Y, Xu CX, Kim H and Tan Y: NNK-mediated upregulation of
DEPDC1 stimulates the progression of oral squamous cell carcinoma
by inhibiting CYP27B1 expression. Am J Cancer Res. 10:1745–1760.
2020.PubMed/NCBI
|
|
12
|
Wang Q, Jiang S, Liu J, Ma G, Zheng J and
Zhang Y: DEP domain containing 1 promotes proliferation, invasion,
and epithelial-mesenchymal transition in colorectal cancer by
enhancing expression of suppressor of zest 12. Cancer Biother
Radiopharm. 36:36–44. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo W, Li H, Liu H, Ma X, Yang S and Wang
Z: DEPDC1 drives hepatocellular carcinoma cell proliferation,
invasion and angiogenesis by regulating the CCL20/CCR6 signaling
pathway. Oncol Rep. 42:1075–1089. 2019.PubMed/NCBI
|
|
14
|
Zhao H, Yu M, Sui L, Gong B, Zhou B, Chen
J, Gong Z and Hao C: High expression of DEPDC1 promotes malignant
phenotypes of breast cancer cells and predicts poor prognosis in
patients with breast cancer. Front Oncol. 9:2622019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gomes AR, Brosens JJ and Lam EW: Resist or
die: FOXO transcription factors determine the cellular response to
chemotherapy. Cell Cycle. 7:3133–3136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calissi G, Lam EW and Link W: Therapeutic
strategies targeting FOXO transcription factors. Nat Rev Drug
Discov. 20:21–38. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen YH, Li CL, Chen WJ, Liu J and Wu HT:
Diverse roles of FOXO family members in gastric cancer. World J
Gastrointest Oncol. 13:1367–1382. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiramongkol Y and Lam EW: FOXO
transcription factor family in cancer and metastasis. Cancer
Metastasis Rev. 39:681–709. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yadav RK, Chauhan AS, Zhuang L and Gan B:
FoxO transcription factors in cancer metabolism. Semin Cancer Biol.
50:65–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin L, Zhang J, Fu HQ, Zhang X and Pan YL:
FOXO3a inhibits the EMT and metastasis of breast cancer by
regulating TWIST-1 mediated miR-10b/CADM2 axis. Transl Oncol.
14:1010962021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao H, Chen W, Zhu Y and Lou J: Hypoxia
promotes pancreatic cancer cell migration, invasion, and
epithelial-mesenchymal transition via modulating the
FOXO3a/DUSP6/ERK axis. J Gastrointest Oncol. 12:1691–1703. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding D, Ao X, Li M, Miao S, Liu Y, Lin Z,
Wang M, He Y and Wang J: FOXO3a-dependent Parkin regulates the
development of gastric cancer by targeting ATP-binding cassette
transporter E1. J Cell Physiol. 236:2740–2755. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qian C and Liu Q: FOXO3a inhibits
nephroblastoma cell proliferation, migration and invasion, and
induces apoptosis through downregulating the Wnt/β-catenin
signaling pathway. Mol Med Rep. 24:7962021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Tian Y, Zhong W, Wang N, Wang Y,
Zhang Y, Zhang Z, Li J, Ma F, Zhao Z and Peng Y: Artemisia argyi
essential oil inhibits hepatocellular carcinoma metastasis via
suppression of DEPDC1 dependent Wnt/β-catenin signaling pathway.
Front Cell Dev Biol. 9:6647912021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sharifi A, Vahedi H, Honarvar MR, Amiriani
T, Nikniaz Z, Rad EY and Hosseinzadeh-Attar MJ: Vitamin D decreases
CD40L gene expression in ulcerative colitis patients: A randomized,
double-blinded, placebo-controlled trial. Turk J Gastroenterol.
31:99–104. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Al-Hussain T, Ali A and Akhtar M: Wilms
tumor: An update. Adv Anat Pathol. 21:166–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cotton CA, Peterson S, Norkool PA,
Takashima J, Grigoriev Y, Green DM and Breslow NE: Early and late
mortality after diagnosis of wilms tumor. J Clin Oncol.
27:1304–1309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang W, Li A, Han X, Wang Q, Guo J, Wu Y,
Wang C and Huang G: DEPDC1 up-regulates RAS expression to inhibit
autophagy in lung adenocarcinoma cells. J Cell Mol Med.
24:13303–13313. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou C, Wang P, Tu M, Huang Y, Xiong F and
Wu Y: DEPDC1 promotes cell proliferation and suppresses sensitivity
to chemotherapy in human hepatocellular carcinoma. Biosci Rep.
39:BSR201909462019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Obara W, Ohsawa R, Kanehira M, Takata R,
Tsunoda T, Yoshida K, Takeda K, Katagiri T, Nakamura Y and Fujioka
T: Cancer peptide vaccine therapy developed from oncoantigens
identified through genome-wide expression profile analysis for
bladder cancer. Jpn J Clin Oncol. 42:591–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hao S, Tian W, Chen Y, Wang L, Jiang Y,
Gao B and Luo D: MicroRNA-374c-5p inhibits the development of
breast cancer through TATA-box binding protein associated factor
7-mediated transcriptional regulation of DEP domain containing 1. J
Cell Biochem. 120:15360–15368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kassambara A, Schoenhals M, Moreaux J,
Veyrune JL, Rème T, Goldschmidt H, Hose D and Klein B: Inhibition
of DEPDC1A, a bad prognostic marker in multiple myeloma, delays
growth and induces mature plasma cell markers in malignant plasma
cells. PLoS One. 8:e627522013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian C, Abudoureyimu M, Lin X, Chu X and
Wang R: Linc-ROR facilitates progression and angiogenesis of
hepatocellular carcinoma by modulating DEPDC1 expression. Cell
Death Dis. 12:10472021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng H, Li BH, Liu C, Jia L and Liu FT:
Comprehensive analysis of lncRNA-mediated ceRNA crosstalk and
identification of prognostic biomarkers in Wilms' tumor. Biomed Res
Int. 2020:49516922020.PubMed/NCBI
|
|
37
|
Shen J and Xi M: DEPDC1 is highly
expressed in lung adenocarcinoma and promotes tumor cell
proliferation. Zhongguo Fei Ai Za Zhi. 24:453–460. 2021.(In
Chinese). PubMed/NCBI
|
|
38
|
Liu Y, Ao X, Ding W, Ponnusamy M, Wu W,
Hao X, Yu W, Wang Y, Li P and Wang J: Critical role of FOXO3a in
carcinogenesis. Mol Cancer. 17:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen YF, Pandey S, Day CH, Chen YF, Jiang
AZ, Ho TJ, Chen RJ, Padma VV, Kuo WW and Huang CY: Synergistic
effect of HIF-1α and FoxO3a trigger cardiomyocyte apoptosis under
hyperglycemic ischemia condition. J Cell Physiol. 233:3660–3671.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Joseph J, Ametepe ES, Haribabu N, Agbayani
G, Krishnan L, Blais A and Sad S: Inhibition of ROS and
upregulation of inflammatory cytokines by FoxO3a promotes survival
against Salmonella typhimurium. Nat Commun. 7:127482016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fluteau A, Ince PG, Minett T, Matthews FE,
Brayne C, Garwood CJ, Ratcliffe LE, Morgan S, Heath PR, Shaw PJ, et
al: The nuclear retention of transcription factor FOXO3a correlates
with a DNA damage response and increased glutamine synthetase
expression by astrocytes suggesting a neuroprotective role in the
ageing brain. Neurosci Lett. 609:11–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zou Y, Tsai WB, Cheng CJ, Hsu C, Chung YM,
Li PC, Lin SH and Hu MC: Forkhead box transcription factor FOXO3a
suppresses estrogen-dependent breast cancer cell proliferation and
tumorigenesis. Breast Cancer Res. 10:R212018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y and Wang X: Targeting the
Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol.
13:1652020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ashihara E, Takada T and Maekawa T:
Targeting the canonical Wnt/β-catenin pathway in hematological
malignancies. Cancer Sci. 106:665–671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu Z, Li L, Cheng P, Liu Q, Zheng X, Peng
F and Zhang Q: lncRNA MSC-AS1 activates Wnt/β-catenin signaling
pathway to modulate cell proliferation and migration in kidney
renal clear cell carcinoma via miR-3924/WNT5A. J Cell Biochem.
121:4085–4093. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou J, Toh SH, Chan ZL, Quah JY, Chooi
JY, Tan TZ, Chong PSY, Zeng Q and Chng WJ: A loss-of-function
genetic screening reveals synergistic targeting of AKT/mTOR and
WTN/β-catenin pathways for treatment of AML with high PRL-3
phosphatase. J Hematol Oncol. 11:362018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qu D, Cui F, Lu D, Yang Y and Xu Y: DEP
domain containing 1 predicts prognosis of hepatocellular carcinoma
patients and regulates tumor proliferation and metastasis. Cancer
Sci. 110:157–165. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y,
Jiang G, Lu M, Zhang Z, Yin J, et al: Downregulation of FOXO3a by
DNMT1 promotes breast cancer stem cell properties and
tumorigenesis. Cell Death Differ. 27:966–983. 2020. View Article : Google Scholar : PubMed/NCBI
|