Introduction
Cell senescence is defined as the irreversible cell
cycle arrest state that occurs in response to various stress and
damage signals (1,2). The senescence phenotype is
characterized by cell cycle arrest, resistance to apoptotic stimuli
(3,4), the release of inflammatory cytokines
and chemokines (5), endoplasmic
reticulum stress (6), metabolism
dysregulation (7,8), genomic instability due to DNA damage
and chromatin changes affecting transcription (9,10).
The molecular mechanism of senescence involves
proteins such as p16INK4A and p21WAF1, which
serve as key cell cycle regulators via their function as
cyclin-dependent kinase inhibitors (CDKIs) (11). p16INK4A mediates
senescence via the retinoblastoma signaling pathway, inhibiting
CDKs and leading to G1 cell cycle arrest (12). Regulation of CDKI 2A
(CDKN2A)/p16 gene expression involves transcription factors and
epigenetic mechanisms such as histone post-translational
modification. Specifically, histone acetylation is performed by
histone acetyltransferases and deacetylases, which are responsible
for the addition and removal, respectively, of acetyl groups from
lysine residues on the N-terminal tails of histones (13). Acetylation at histone lysine
residues causes neutralization of its positive charge and a
decrease in DNA-nucleosome affinity associated with transcriptional
activation (13). Deacetylation
is associated with compacted chromatin and a transcriptionally
silent state (14).
Sirtuins (SIRTs) are a conserved family of
nicotinamide adenine dinucleotide (NAD+)-dependent
deacetylases. SIRTs act in different cell compartments. In the
nucleus, SIRTs deacetylate histones and regulate expression. In the
mitochondria, SIRTs are components of the metabolic machinery. In
the cytoplasm, SIRTs modulate cytoskeletal and signaling molecules
(15). Collectively, SIRTs
modulate metabolic processes such as energy availability, stress
response, protein aggregation, inflammation, and genome stability
(15). A total of seven sirtuins
have been identified in mammals (16). They share structural homology,
particularly in their highly conserved catalytic and
NAD+-binding domains (16). SIRT1 is a deacetylase that
contains both nuclear and exporting sequences and therefore serves
as a key regulator of certain proteins such as NF-κB, peroxisome
proliferators-activated receptor γ and its coactivator peroxisome
proliferator-activated receptor gamma coactivator 1-α, protein
tyrosine phosphatase, forkhead transcriptional factors, adenosine
monophosphate activated protein kinase, CRE-binding
protein-regulated transcription coactivator 2, endothelial nitric
oxide synthase, p53, myogenic differentiation, liver X receptor and
Transcription factor E2F1 (17).
SIRT2 is a deacetylase with cytosolic localization (18–20). SIRT3-5 are mitochondrial SIRTs
containing mitochondrial-targeting sequences and SIRT6-7 are
predominantly localized in the nucleus (18–20).
In the SIRT family, SIRT1 is the most studied in
cellular senescence and is reported as specifically catalyzing the
removal of acetylation at residues H3K9, H3K14, H3K56, H4K16, and
H1K26 and regulating transcription-associated genes, such as
CDKN2A/p16 and CDKN1A/p21 (21–24). In the present study, the
regulatory role of the histone deacetylase enzymes SIRT1, SIRT2,
SIRT6, and SIRT7 on gene expression of CDKN2A/p16 were
evaluated in human MRC5 cells, which were used as a model of
replicative cellular senescence.
Materials and methods
Cell culture
Primary human lung fibroblast MRC5 cells (derived
from 14-week-old male fetus normal lung tissue) were purchased from
American Type Culture Collection. MRC5 cells were cultured in
Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10%
fetal bovine serum (GIBCO), 100 U/ml penicillin and 100 mg/ml
streptomycin. Cells were maintained in a humidified atmosphere at
37°C and 5% CO2. Cells were cultured for 8 weeks and
harvested at different time points (2, 4, 6 and 8 weeks) to perform
the assays.
Senescence-associated β-galactosidase
(SA-β-gal) activity
The endogenous SA-β-gal activity in MRC5 culture was
assessed at 2, 4, 6 and 8 weeks using a Senescence Detection kit
(cat. no. ab65351; Abcam) according to the manufacturer's protocol.
Positive cells were imaged and counted in inverted white light
microscope (KERN OCM 161) at 40× magnification Percentage of
SA-β-galactosidase-positive tissue area was calculated and plotted
with GraphPad Prism.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from culture cells was extracted using
TRIzol® (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. An equal amount of each sample (2 µg)
was used for RT using a ProtoScript® First Strand cDNA
Synthesis kit (New England BioLabs, Inc.) following to the
manufacturer's protocol. qPCR was performed using a FastStart
Essential DNA green Master kit (Roche Diagnostics) using
LightCycler® Nano (Roche Diagnostics). The reaction
conditions were as follows: Initial denaturation for 10 min at
95°C, followed by 45 cycles of denaturation for 10 sec at 95°C;
annealing for 15 sec at 62°C for P16INK4α and Laminin
B primers, and 60°C for SIRT7 and GAPDH primers;
ending with 20 sec of elongation at 72°C, The results were
quantified using the 2−ΔΔCq method (25). Data are presented as relative mRNA
expression levels normalized to GAPDH mRNA expression
levels. The sequences of the primers used to amplify genes of
interest are presented in Table
SI.
Nuclear extract and western
blotting
Nuclear extracts were prepared from MRC5 cultures
with buffer containing 420.0 mM NaCl, 25.0% glycerol, 0.2 mM EDTA,
1.0 mM DTT, 20.0 mM HEPES (pH 7.9) and 1.5 mM MgCl2
using the Dignam method (26).
Total protein was quantified using the Bradford technique (27). A total of 25 µg protein/lane was
separated using 10% SDS-PAGE. Subsequently, the proteins were
transferred to nitrocellulose membranes. Membranes were blocked
with 5% milk solution in TBS-Tween (0.1%) for 1 h at room
temperature. Then, the membranes were incubated at 4°C overnight
with primary anti-SIRT7 (1:500; anti-rabbit; cat. no. D2K5A Cell
Signaling Technology, Inc.) and anti-transcription factor IIB
(TFIIB), dilution 1/100 anti-rabbit (C-18 sc-225 Santa Cruz
Biotechnology) were used as a control. Goat anti-Rabbit IgG
Poly-HRP was used as secondary antibody, dilution 1/5000,
incubation 2 h (32260 Thermo Fisher Scientific), at room
temperature. The immunoblots were visualized in CL-Xposure Film
using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo
Scientific, Inc.).
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was performed in cells at 4 and 8 weeks
to identify regulatory components that mediated epigenetic changes
associated with CDKN2A/p16 transcriptional control during
replicative senescence in MRC5 cells. Cross-linked chromatin
samples were prepared as described previously by Rojas et al
(28). Chromatin was sheared
using a Bioruptor® Pico sonication device (Diagenode SA)
to obtain ≤500 bp fragments and stored at −80°C; one aliquot was
used for quantification using A260 assessment. Chromatin size was
confirmed by electrophoretic analysis. Cross-linked extracts were
resuspended in sonication buffer to a final volume of 500 µl. The
samples were precleared by incubation with 2–4 µg normal IgG and 40
µl protein A/G PLUS-agarose beads (Santa Cruz Biotechnology
Inc.sc-2003) for 1.5 h at 4°C with agitation. Samples were
centrifuged at 4,000 × g for 5 min, at 4°C. The supernatant was
collected and immunoprecipitated with specific antibodies (Table SII) for 12–16 h at 4°C. The
immunocomplexes were recovered with the addition of 50 µl protein A
(for rabbit antibodies) or G agarose beads (for mouse antibodies),
followed by incubation for 1 h at 4°C with gentle agitation.
Immunoprecipitated complexes were washed once with sonication
buffer (50 mm HEPES, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton
X-100, 0.1% deoxycholate acid, 0.1% SDS, and a mixture of
proteinase inhibitors), twice with LiCl buffer (100 mM Tris-HCl; pH
8.0; 500 mM LiCl; 0.1% Nonidet P40 and 0.1% deoxycholic acid) and
once with Tris-EDTA (50 mM Tris-HCl, pH 8.0, and 2 mM EDTA) for 5
min each at 4°C, followed by centrifugation at 4,000 × g for 5 min
at 4°C. The protein-DNA complexes were eluted by incubation with
100 µl elution buffer (50 mM NaHCO3 and 1% SDS) for 15
min at 65°C. Extracts were centrifuged at 10,000 × g for 5 min at
room temperature. The supernatant was collected and incubated for
12–16 h at 65°C to reverse cross-linking. Proteins were digested
with 100 µg/ml proteinase K for 2 h at 50°C and the DNA was
recovered using a ChIP DNA Clean & Concentrator kit (cat. no.
D5201; Zymo Research Corp.). qPCR was performed as aforementioned.
The qPCR primers used to evaluate the human CDKN2A/p16
promoter region are presented in Table SI.
Small interfering RNA (siRNA)
knockdown
MRC5 cells cultured for 2 weeks (non-senescent
cells) were plated on 6-well plates at 50% confluence overnight and
transfected with 50 µM siRNA oligonucleotides targeting
SIRT7 (siSIRT7; cat. no. sc-63030; Santa Cruz Biotechnology,
Inc.). siSIRT7 is a mix of three target-specific 19–25 nt siRNAs
designed to knock down SIRT7 gene expression. Control siRNA
(siCtrl), a non-targeting 20–25 nt siRNA (cat. no. sc-37007; Santa
Cruz Biotechnology, Inc.), was used as a negative control. siRNA
sequences are presented in Table
SIII. Transfection was performed using transfection reagent
according to the manufacturer's protocol (sc-29528 Santa Cruz
Biotechnology, Inc.). Briefly, 1ug of siRNA duplex was diluted into
100 µl siRNA transfection Medium (sc-36868; Santa Cruz
Biotechnology, Inc.). For each transfection, 4 µl of siRNA
transfection reagent were used. This transfection reagent mixture
was overlayed onto the washed cells and incubated for 6 h at 37°C
in CO2 incubator. Finally, transfection medium was
replaced with fresh 1X normal growth medium. Subsequent experiments
were performed 48 h later.
Statistical analysis
ChIP assay results and mRNA expression levels were
analyzed by one-way ANOVA followed by Dunnett's post hoc test to
assess significant changes between three or more samples and the
control, while the Mann-Whitney assay was applied to establish
differences between two nonparametric data. Data are presented as
mean ± SD. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyzes were performed
with GraphPad Prism version 8 (GraphPad Software, Inc.). All
experiments were performed in triplicate.
Results
CDKN2A/p16 expression in MRC5 human
fibroblast cells and during activation of senescence
Replicative senescence limits somatic cell
proliferation in culture and may reflect cellular aging in
vivo (26). The most widely
used biomarker for senescent and aging cells is SA-β-gal, assessed
as the level of β-gal activity at pH 6.0 in senescent cells
(29). mRNA expression levels of
CDKN2A/p16, a cell cycle regulator, and lamin B, a
nuclear morphology factor, were analyzed as senescent cells
demonstrate an increase and decrease in these genes, respectively
(30). A time course experiment
was used to assess changes during senescence. MRC5 cells were
cultured for 2, 4, 6 and 8 weeks and used in SA-β-gal and RT-qPCR
assays to evaluate the in vitro senescence model.
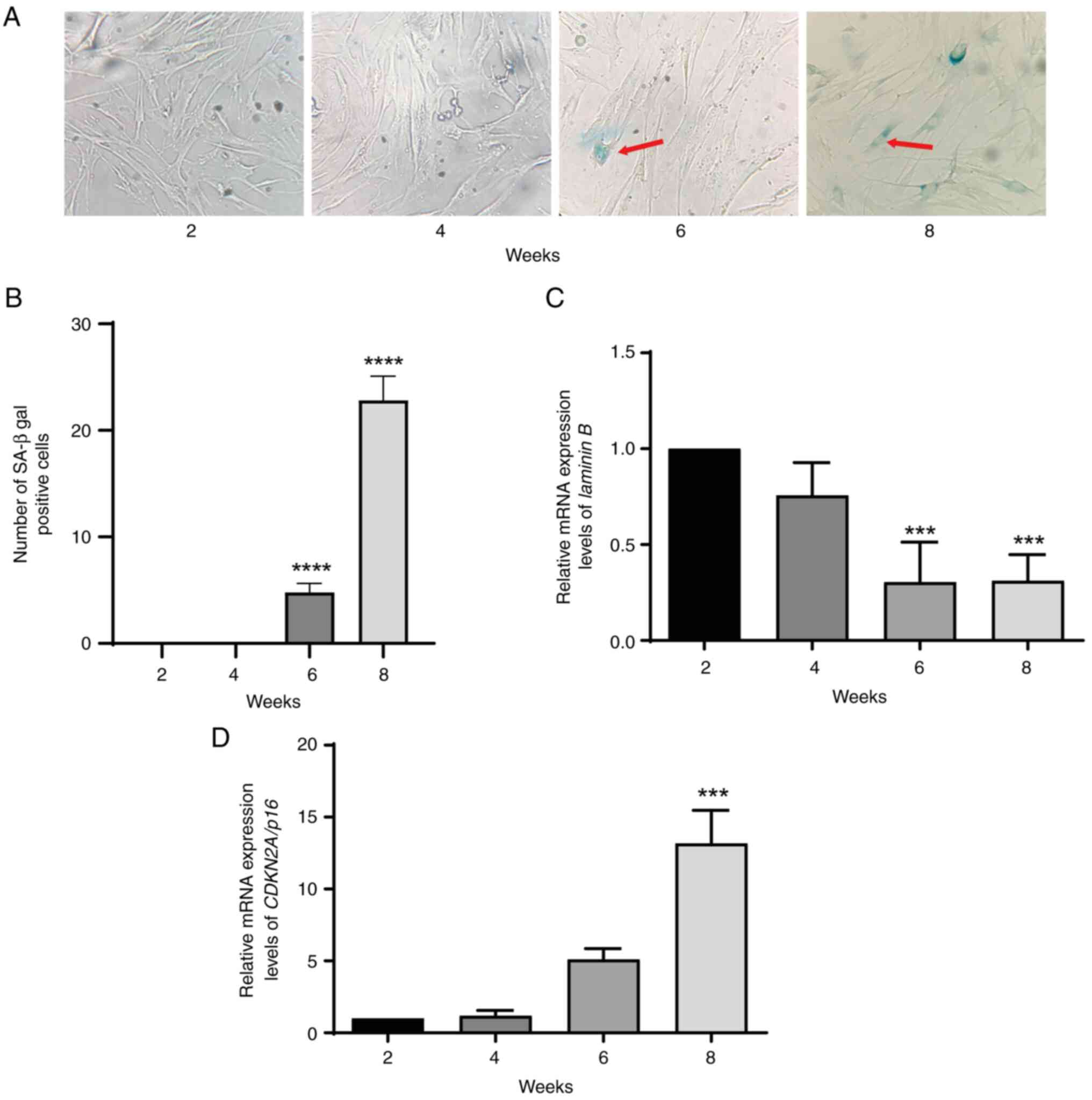
MRC5 cells were assessed for SA-β-gal activity using
X-gal at pH 6.0. β-gal activity was demonstrated at 6 and 8 weeks
of cell culture (Fig. 1A and B).
The β-gal activity level in cells cultured for eight weeks was 4–5
times higher than that in cells cultured for 6 weeks. RT-qPCR was
used to assess lamin B and CDKN2A/p16 mRNA expression
levels in MRC5 culture cells at 2, 4, 6 and 8 weeks. The results
demonstrated that lamin B was significantly downregulated in
cultured cells at 6 and 8 weeks compared with 2 weeks (Fig. 1C). Lamin B downregulation
was associated with CDKN2A/p16 mRNA expression levels, which
were significantly increased at 8 weeks compared with 2 weeks
(Fig. 1D). These results
demonstrated that MRC5 cells entered senescence by 6 weeks.
Epigenetic changes in histone
modifications are associated with CDKN2A/p16 gene expression in
senescent cells
Histone acetylation is associated with
transcriptional activation (13).
To determine whether epigenetic modifications participated in
CDKN2A/p16 transcriptional control in senescence, changes in
histone modification were assessed using ChIP assay using 4
(non-senescent) and 8-week cultured cells (senescent).
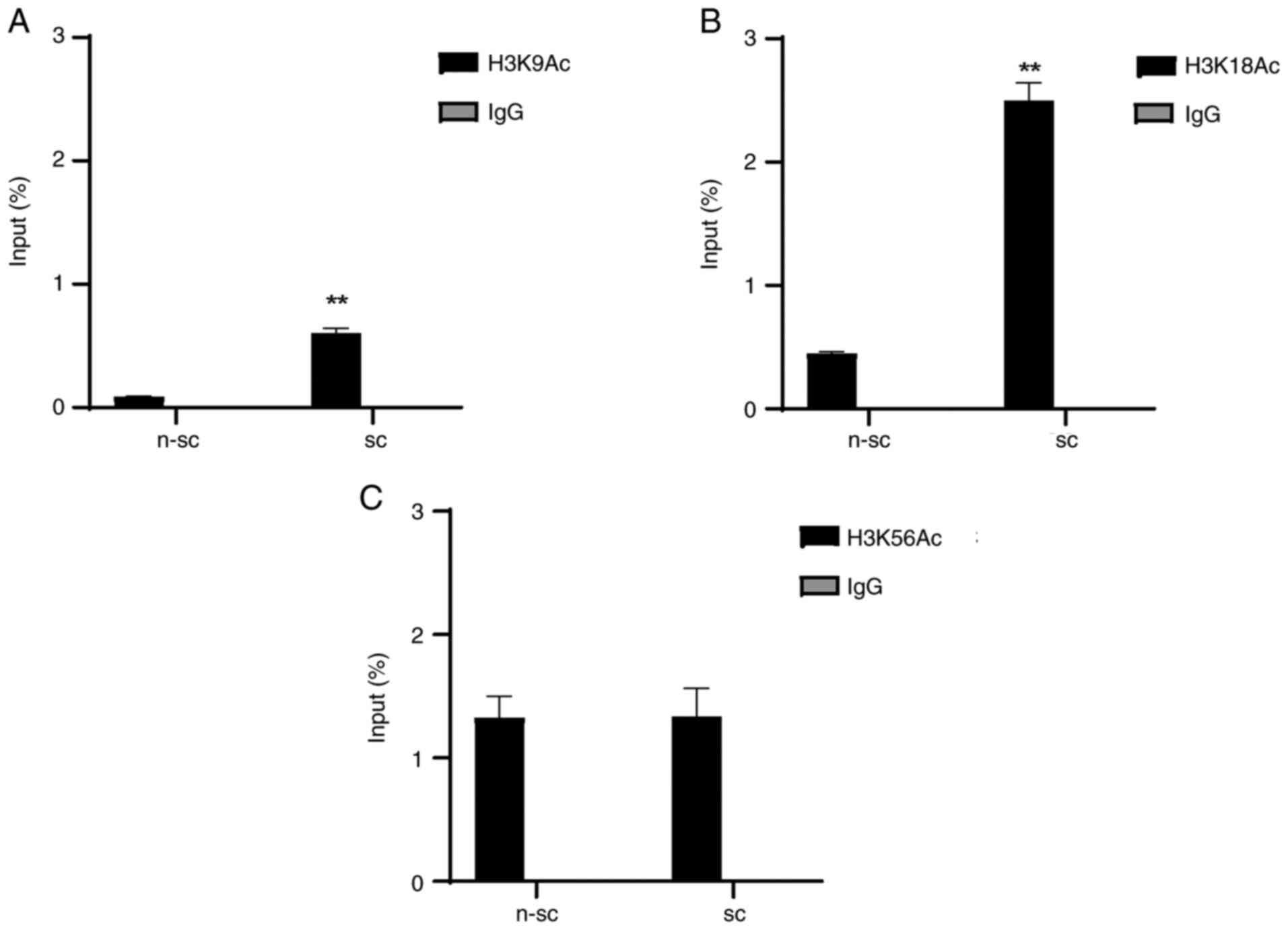
The results demonstrated that senescent cells
exhibited histone H3 post-translational modifications
characteristic of transcriptionally active genes. Specifically,
8-week-old cultured cells presented enrichment levels of H3K9Ac
(0.8%) and H3K18Ac (2.5%) in the CDKN2A/p16 gene promoter
region (Fig. 2A-C). These
enrichment percentages were significantly increased in senescence
cells compared with non-senescence cells.
However, enrichment of 1,2% were detected in H3K56Ac
in non-senescent cells. These results suggested that H3K9Ac and
H3K18Ac may be involved in CDKN2A/p16 gene overexpression in
senescent cells.
SIRT7 weakly binds to the CDKN2A/p16
promoter in senescent MRC5 cells
To identify the regulatory components that mediated
the epigenetic changes associated with transcriptional control of
the CDKN2A/p16 promoter during senescence, ChIP assay was
performed for epigenetic suppressors (SIRT1, SIRT2, SIRT6, and
SIRT7) that modulate post-translational histone modification.
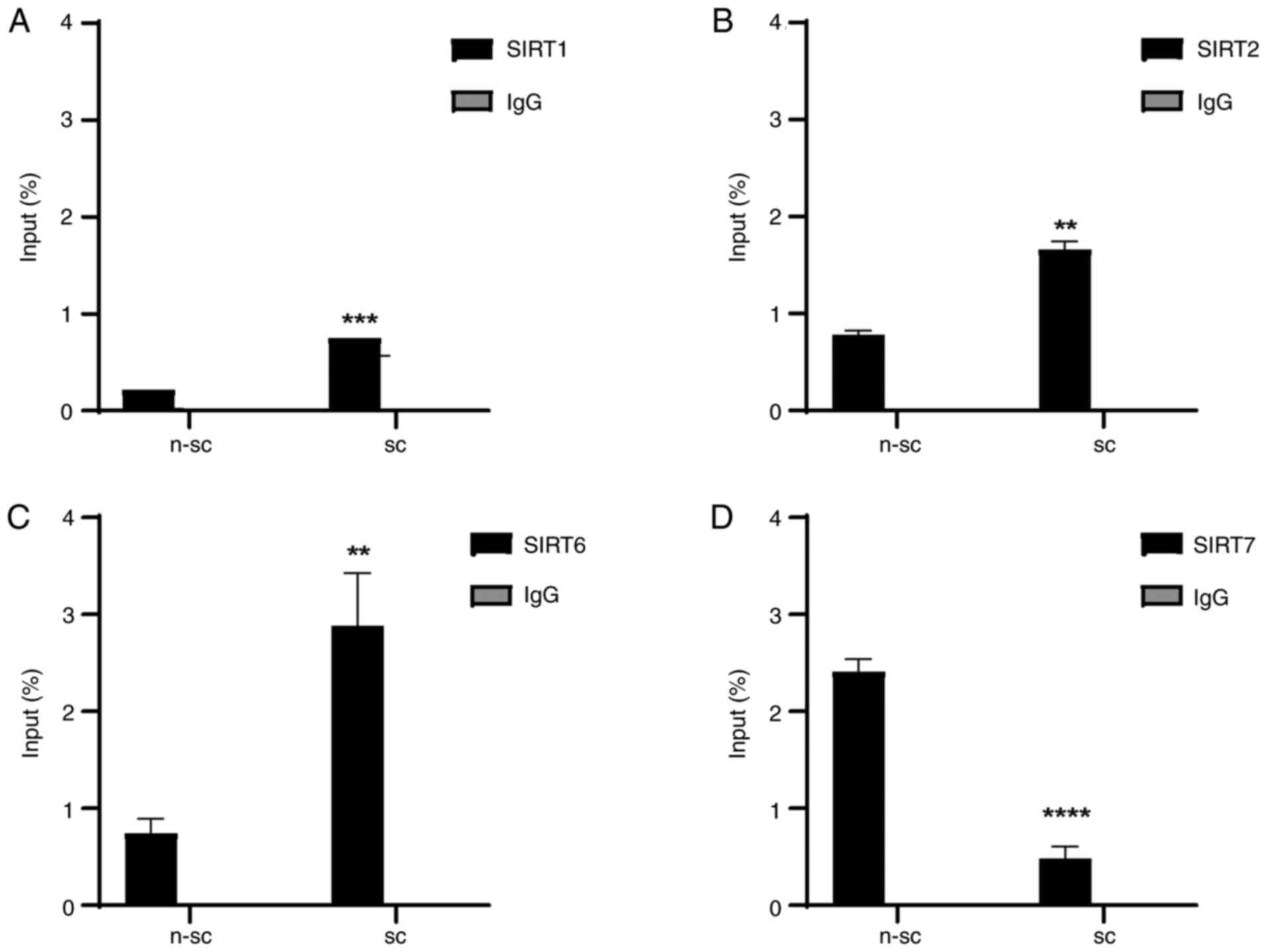
ChIP assay with senescent cells demonstrated that
SIRT1, SIRT2 and SIRT6 were significantly enriched in the
CDKN2A/p16 promoter (Fig.
3A-C) compared with non-senescent cells. SIRT7 association with
the CDKN2A/p16 promoter was significantly decreased in
senescent compared with non-senescent cells (Fig. 3D). This may indicate
CDKN2A/p16 transcriptional activation.
mRNA expression levels of SIRT1, SIRT2,
SIRT6, and SIRT7 were assessed using RT-qPCR at 2, 4, 6
and 8 weeks. Low mRNA expression levels of SIRT1 and
SIRT2 were demonstrated and mRNA expression levels of
SIRT6 and SIRT7 were markedly higher at all times.
(Fig. S1). SIRT1 expression
levels decreased significantly compared with non-senescent cells.
Additionally, SIRT7 protein expression levels were analyzed by
western blot at different times evaluated (2,4,6 and 8 weeks).
Fig. S2) show that the levels of
protein expression in the different times didn't change.
SIRT7 is an epigenetic regulator of
CDKN2A/p16 gene in replicative senescence
ChIP and RT-qPCR assay in non-senescent cells
demonstrated SIRT7 binding in the promoter region (Fig. 3D) and low CDKN2A/p16 mRNA
expression levels (Fig. 1D). The
role of SIRT7 in epigenetic control of CDKN2A/p16 gene was
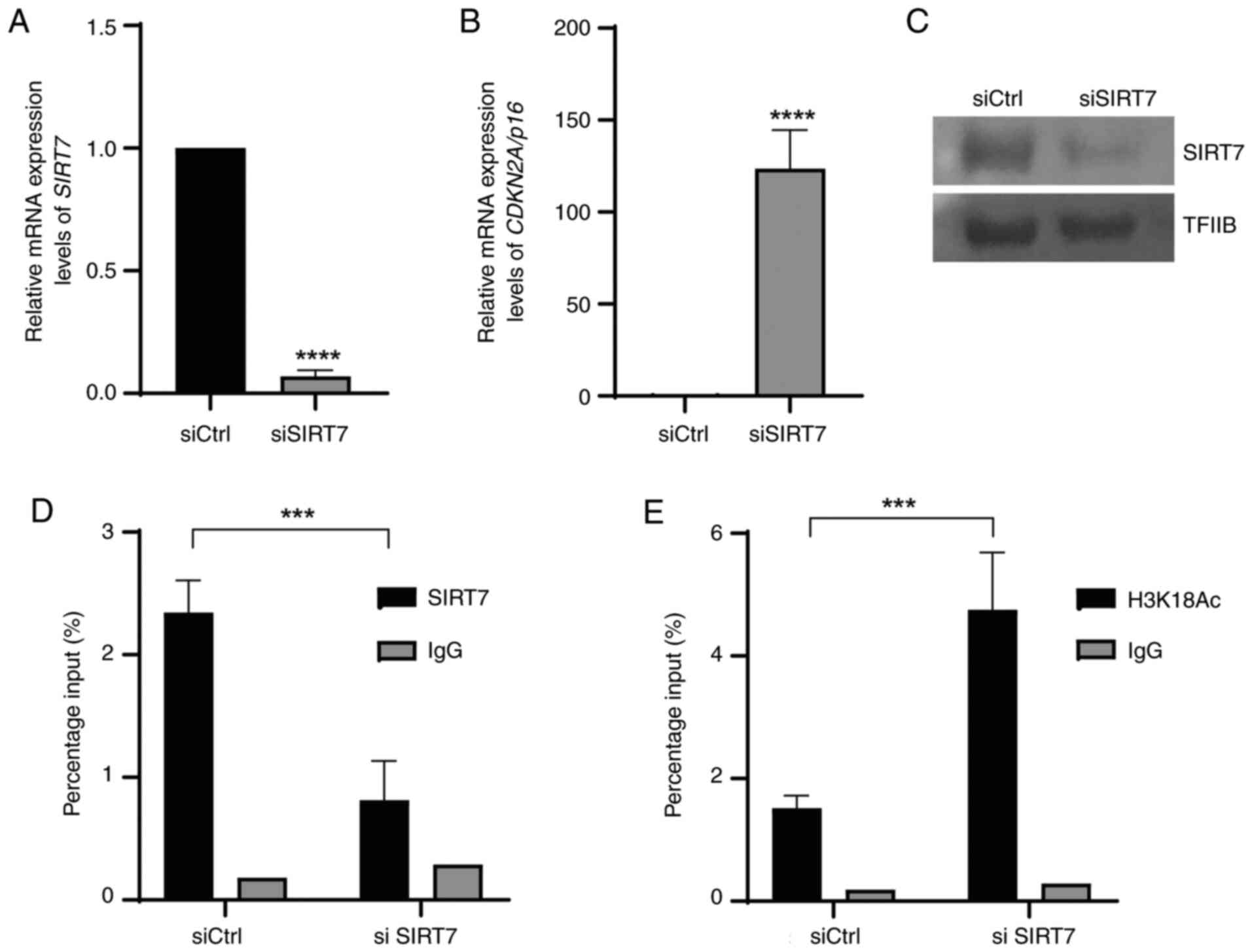
assessed. siRNA-mediated knockdown of SIRT7 in non-senescent cells
(2 weeks) was performed. SIRT7 mRNA expression levels were
significantly downregulated and protein expression levels were
markedly downregulated in cells transfected with siSIRT7 at 48 h
compared with cells transfected with siCtrl when assessed using
qPCR (Fig. 4A) and western
blotting (Fig. 4C). Furthermore,
this decrease was associated with a significant increase in
CDKN2A/p16 expression in cells transfected with siSIRT7 at
48 h compared with cells transfected with siCtrl (Fig. 4B). However, SIRT7 knockdown
demonstrated a significant decrease in SIRT7 binding to the
CDKN2A/p16 promoter sequence (Fig. 4D) and significant enrichment of
H3K18Ac compared with siCtrl (Fig.
4E).
Discussion
Histone modification is an epigenetic mechanism that
regulates gene expression. These modifications are catalyzed by
enzyme complexes that act on the N-terminal ends of histones that
form nucleosomes, mediating the removal or aggregation of chemical
marks such as acetylation, methylation and phosphorylation
(16). SIRTs are enzymes that
regulate gene expression and biological activities, such as cell
senescence (16). This is
mediated by removing acetyl groups from histones, favoring
compaction of chromatin and therefore mediating gene repression
(31). A total of seven SIRTs has
been reported in mammals, SIRT1-7 (16). SIRT1 and SIRT2 participate in cell
senescence and SIRT2 ortholog overexpression extends lifespan in a
range of lower eukaryotes (32).
Previous reports showed that extra copies of SIR2, a member of
Sirtuin in budding yeast Saccharomyces cerevisiae, extended the
lifespan by 30% by preventing the formation of extrachromosomal DNA
circles. Caenorhabditis elegans has four Sirtuins (sir-2.1,
sir-2.2, sir-2.3, and sir-2.4), where sir-2.1 is the most similar
to the S. cerevisiae SIR2. On the other hand, Drosophila
melanogaster has five Sirtuins (dSirt1, dSirt2, dSirt4, dSirt6, and
dSirt7), of which Sirt1 (better known as dSir2) is most similar to
S. cerevisiae SIR2, and high levels are found in the nuclei and/or
cytoplasm of neurons and fat bodies (33).
However, Huang et al (32) demonstrated that under stress,
SIRT1 overexpression contributes to cell proliferation and prevents
senescence in human diploid fibroblasts. The SIRT1-mediated delay
of senescence is associated with P16INK4A/Rb pathway
downregulation and ERK/S6K1 signaling activation (32). However, the role of SIRT7 in cell
senescence is unknown.
To elucidate the role of SIRT7 in senescence, a
senescent cell model was developed in vitro. Pulmonary
fibroblast MRC5 cells were cultured for 2, 4, 6 and 8 weeks. β-gal
activity was assessed at 6 and 8 weeks, allowing acquisition of the
senescent phenotype (3). In
general, induction of the senescent phenotype induces expression of
transcription factor EB, which increases lysosomal biogenesis,
leading to overproduction of lysosomes and a decrease in their
elimination (31). These lipid
vesicles contain the enzyme SA-β-gal and when a chromogenic
substrate is added to senescent cells, SA-β gal releases the
chromogen from galactose, which resulted in formation of a blue
coloration that demonstrates the increase in lysosomal content, is
therefore an indicator that cells exhibit the senescent phenotype
(3).
MRC5 cells cultured for 6 or 8 weeks demonstrated a
significant decrease in lamin B mRNA expression, a biomarker
of senescence, compared with 2-week cells. Senescent cells have a
distinct gene expression profile, often accompanied by spatial
redistribution of heterochromatin into senescence-associated
heterochromatic foci (SAHFs) (34). Previously, a genome-wide mapping
study reported that lamin B1 is depleted during senescence,
preferentially at central regions of lamin-associated domains, and
Lys9 trimethylation on histone H3 (H3K9me3) is enriched (30). Lamin B1 knockdown
facilitates the spatial re-localization of perinuclear
H3K9me3-positive heterochromatin, thus promoting SAHF formation,
which is inhibited by ectopic lamin B expression (34). In the present study, MRC5 cells at
6 and 8 weeks demonstrated a marked increase in CDKN2A/p16
mRNA expression levels. It has been previously reported that
expression of CDKN2A/p16 is crucial for CDKI
activation-dependent senescence (3). Induction of senescence phenotype
causes the cell to express other typical senescence
characteristics, such as proinflammatory protein synthesis,
resistance to apoptosis, active metabolism, endoplasmic reticulum
stress, p53 overexpression, lamin B1 downregulation
and increased lysosomal content (3). The present study demonstrated that
senescence started at 6 weeks in cultured cells, which validated
this senescent cellular model. Several studies have reported that
MRC5 cells are an ideal biological model to study senescence and
analyze molecular changes as such as nuclear laminin-associated
protein, lamin B1 and loss of the epigenetic suppressive marker
tri-methylation of Lys 27 on histone H3 in chromatin (35–38).
The epigenetic mechanisms involved in
CDKN2A/p16 transcriptional activation, were evaluated using
ChIP assay, which demonstrated significant enrichment of H3K9Ac and
H3K18Ac accompanied by a significant decrease in SIRT7 in the
promoter region in cells with a senescent compared with
non-senescent phenotype. A decrease in SIRT7 expression has been
reported in aging tissue (39)
and SIRT7 enzyme loss in mice leads to a decrease in embryonic
viability and lifespan, aging-associated pathology and the loss of
regenerative potential hematopoietic stem cells (40,41). Previous studies have reported
increased levels of the senescence marker CDKN2A/p16 in
SIRT7-deficient cell cultures and increased CDKN2A/p16 mRNA
expression levels in splenocytes and fibroblasts obtained from
SIRT7-negative mice (40,41).
The present study demonstrated that SIRT7 knockdown
markedly increased CDKN2A/p16 mRNA and protein expression
levels compared with siCtrl in cells cultured for 2 weeks. This
enhanced CDKN2A/p16 expression was associated with
CDKN2A/p16 gene changes, which demonstrated a 2-fold H3K18Ac
enrichment on the promoter. These results demonstrated that the
SIRT7 deacetylase enzyme participated in cell senescence via
transcriptional regulation of CDKN2A/p16. Previous studies
reported that SIRT7 serves key roles in cell senescence and aging,
SIRT7-deficient mice demonstrate a shortened lifespan and
aging-associated phenotypes (39,40,42–45) and overexpression of SIRT7 in
senescent-induced cells suppresses expression of senescence markers
such as p53 and p21 (39,46).
In terms of the epigenetic mechanisms that control
CDKN2A/p16 expression, DNA methylation of its promoter
region has been reported in pathology, such as cancer (47). However, in normal cells of young
mammals, the INK4/ARF locus remains silenced (embryonic and
fetal stem cells) due to suppressive Polycomb complexes PRC1 and
PRC2 action (48). However, the
INK4/ARF locus responds to oncogenic stress signals when
stem cells lose self-renewal and differentiation capacity. In these
cases, alterations in PRC1 and PRC2 complex member proteins
(Chromobox 7, BMI1 proto-oncogene polycomb ring finger and enhancer
of zeste 2 Polycomb repressive complex 2 subunit) that produce loss
of suppressor markers in trimethylated lysine 27 of histone H3
(H3K27). This activates CDKN2A/p16 expression to induce
senescence (48). The aim of the
present study was to assess the role of the epigenetic enzyme SIRT7
in transcriptional control of CDKN2A/P16. In future, the protein
expression profiles of P16 and lamin B1 should be assessed in
biological models of cellular senescence such as fibroblasts or
culture of neurons.
The present study demonstrated that in non-senescent
cells, transcriptional suppression of CDKN2A/p16 gene was
mediated by binding of SIRT7 and low levels of H3K18Ac in its
promoter region. In this context, there were low levels of cellular
CDKN2A/p16 RNA messenger. If this mRNA low levels translate
into low protein levels, it could be proposed that CDK and the CycD
complex to phosphorylate the retinoblastoma protein, favoring
transcription factor E2F release and inducing the protein synthesis
necessary for DNA replication and cell cycle progression (Fig. 5A). However, in the senescent cell
model, there was no evidence of the repressor enzyme SIRT7 binding
in the CDKN2A/p16 promoter region, which allowed its
transcriptional activation. It was hypothesized that
p16INK4A protein in senescent cells would therefore
inhibit CDK4/CycD binding, avoiding retinoblastoma protein
phosphorylation and inhibiting the release of the transcription
factor E2F, thus leading to cell cycle arrest (Fig. 5B). However, the present study did
not perform protein expression assay; this is required in future
works.
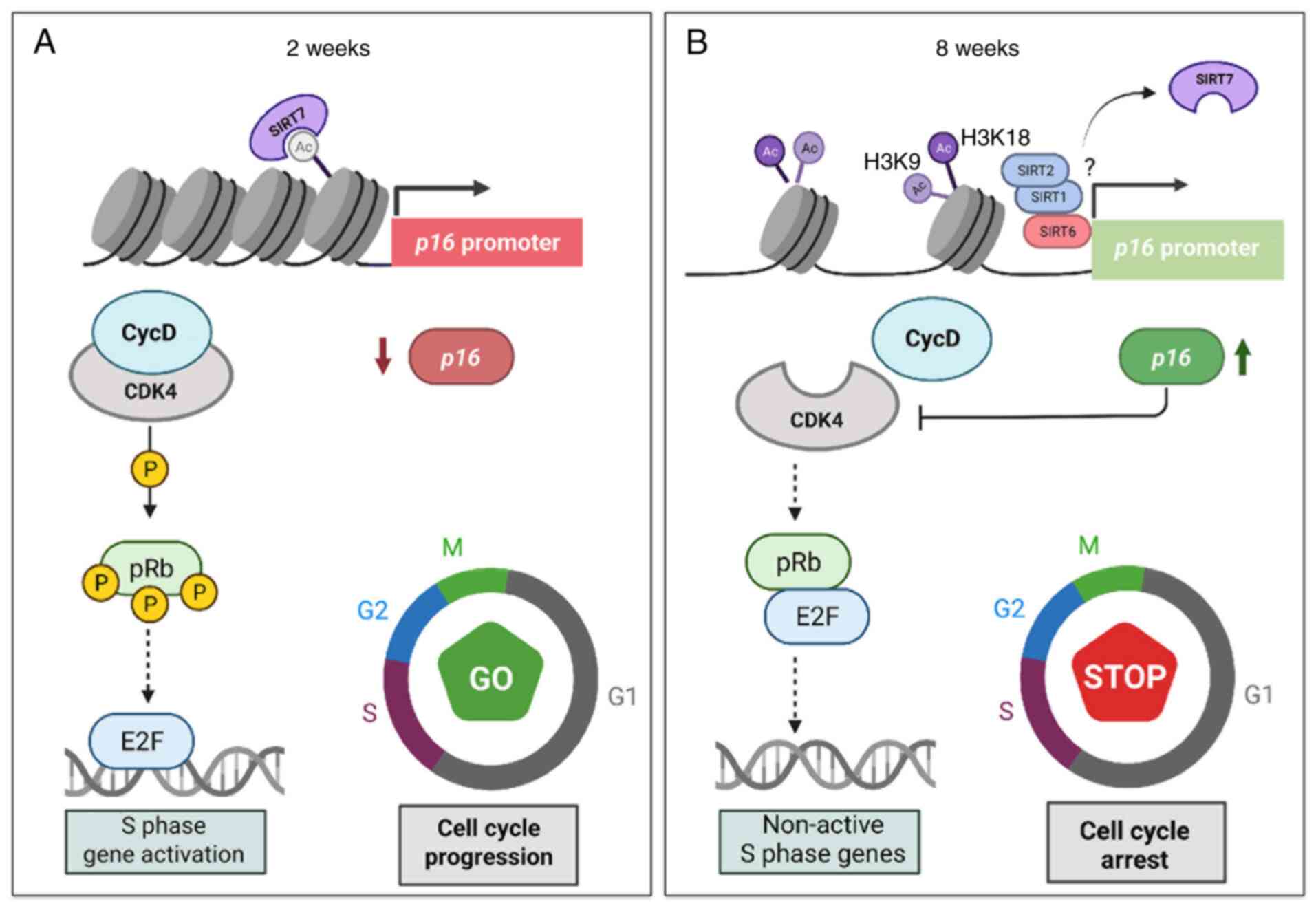 | Figure 5.Proposed model of CDKN2A/p16
gene epigenetic regulation in cellular senescence. (A)
Non-senescent cell state. Cells demonstrated SIRT7 enrichment in
the CDKN2A/p16 promoter region. SIRT7 binding was associated
with activity as a suppressor enzyme mediating H3K18Ac removal and
inhibiting CDKN2A/p16 expression. As there were low cellular
p16INK4A levels, CDK and the CycD complex phosphorylate
Rb, favoring release of transcription factor E2F and inducing
protein synthesis necessary for DNA replication and cell cycle
progression. (B) Senescent cell model. Lack of suppressor enzyme
SIRT7 binding in the CDKN2A/p16 promoter region
allows its transcriptional activation. The presence of
P-16INK4A protein in senescent cells inhibits CDK4/CycD
binding, avoiding Rb phosphorylation and inhibiting release of
transcription factor E2F, thus leading to cell cycle arrest. SIRT;
sirtuin; CDK, cyclin-dependent kinase; CDKN2A, CDK inhibitor 2A; p,
phosphorylated; Rb, retinoblastoma protein; E2F, E2 factor; CycD,
cyclin D. |
The present study assessed epigenetic parameters
regulating CDKN2A/p16 transcription during senescence. These
results validated the MRC5 cell line as a model of senescence.
Furthermore, it was demonstrated that SIRT7 decreased H3K18Ac in
the CDKN2A/p6 promoter region and was directly associated
with suppression of this gene.
In the present study, the regulatory effect of the
histone deacetylase enzyme SIRT7 on the gene expression of
CDKN2A/p16 was evaluated in human MRC5 cells used as a model
of replicative cell senescence. The results demonstrated that
CDKN2A/p16 transcriptional repression was regulated by SIRT7
via direct binding of the promoter region and deacetylation of the
activating epigenetic marker H3K18Ac.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Fundación
para la promoción de la Investigación y la tecnología of the Banco
de la República de Colombia (grant number: P.T.I 4171 and
Pontificia Universidad Javeriana, Grant no. PUJ ID 6659.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BH and AR conceived and designed the experiments.
SR, LGB, CB, and DG performed the experiments, analyzed the data
and wrote the manuscript. AC and TMR analyzed data and designed
experiments. AR and BH supervised the study. AR and BH confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kuilman T, Michaloglou C, Mooi WJ and
Peeper DS: The essence of senescence. Genes Dev. 24:2463–2479.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muñoz-Espín D and Serrano M: Cellular
senescence: From physiology to pathology. Nat Rev Mol Cell Biol.
15:482–496. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hernandez-Segura A, Nehme J and Demaria M:
Hallmarks of Cellular Senescence. Trends Cell Biol. 28:436–453.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Childs BG, Baker DJ, Kirkland JL, Campisi
J and van Deursen JM: Senescence and apoptosis: Dueling or
complementary cell fates? EMBO Rep. 15:1139–1153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coppé JP, Desprez PY, Krtolica A and
Campisi J: The senescence-associated secretory phenotype: The dark
side of tumor suppression. Annu Rev Pathol. 5:99–118. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pluquet O, Pourtier A and Abbadie C: The
unfolded protein response and cellular senescence. A review in the
theme: Cellular mechanisms of endoplasmic reticulum stress
signaling in health and disease. Am J Physiol Cell Physiol.
308:C415–C425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gorgoulis V, Adams PD, Alimonti A, Bennett
DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K,
Ferbeyre G, et al: Cellular senescence: Defining a path forward.
Cell. 179:813–827. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
James EL, Michalek RD, Pitiyage GN, de
Castro AM, Vignola KS, Jones J, Mohney RP, Karoly ED, Prime SS and
Parkinson EK: Senescent human fibroblasts show increased glycolysis
and redox homeostasis with extracellular metabolomes that overlap
with those of irreparable DNA damage, aging, and disease. J
Proteome Res. 14:1854–1871. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmeer C, Kretz A, Wengerodt D,
Stojiljkovic M and Witte OW: Dissecting aging and
senescence-current concepts and open lessons. Cells. 8:14462019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferrari S and Pesce M: Stiffness and aging
in cardiovascular diseases: The dangerous relationship between
force and senescence. Int J Mol Sci. 22:34042021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rayess H, Wang MB and Srivatsan ES:
Cellular senescence and tumor suppressor gene p16. Int J Cancer.
130:1715–1725. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sterner DE and Berger SL: Acetylation of
histones and transcription-related factors. Microbiol Mol Biol Rev.
64:435–459. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen HP, Zhao YT and Zhao TC: Histone
deacetylases and mechanisms of regulation of gene expression. Crit
Rev Oncog. 20:35–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Langley B and Sauve A: Sirtuin
deacetylases as therapeutic targets in the nervous system.
Neurotherapeutics. 10:605–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allis D, Caparros ML, Jenuwein T, Reinberg
D and Lachner M: Epigenetics. 2nd edition. Cold Spring Harbor
Laboratory Press; New York, NY: 2015
|
|
17
|
Elibol B and Kilic U: High levels of SIRT1
expression as a protective mechanism against disease-related
conditions. Front Endocrinol (Lausanne). 9:6142018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakagawa T and Guarente L: SnapShot:
Sirtuins, NAD, and aging. Cell Metab. 20:192–192.e1. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wątroba M, Dudek I, Skoda M, Stangret A,
Rzodkiewicz P and Szukiewicz D: Sirtuins, epigenetics and
longevity. Ageing Res Rev. 40:11–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Criscione SW, Teo YV and Neretti N: The
chromatin landscape of cellular senescence. Trends Genet.
32:751–761. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen C, Zhou M, Ge Y and Wang X: SIRT1 and
aging related signaling pathways. Mech Ageing Dev. 187:1112152020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nacarelli T and Sell C: Targeting
metabolism in cellular senescence, a role for intervention. Mol
Cell Endocrinol. 455:83–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jing H and Lin H: Sirtuins in epigenetic
regulation. Chem Rev. 115:2350–2375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang N and Sen P: The senescent cell
epigenome. Aging (Albany NY). 10:3590–3609. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carey MF, Peterson CL and Smale ST: Dignam
and roeder nuclear extract preparation. Cold Spring Harb Protoc.
2009.pdb.prot5330, 2009. View Article : Google Scholar
|
|
27
|
He F: Bradford Protein Assay. Bio.
101:e452011.
|
|
28
|
Rojas A, Aguilar R, Henriquez B, Lian JB,
Stein JL, Stein GS, van Wijnen AJ, van Zundert B, Allende ML and
Montecino M: Epigenetic control of the bone-master Runx2 gene
during osteoblast-lineage commitment by the histone demethylase
JARID1B/KDM5B. J Biol Chem. 290:28329–28342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee BY, Han JA, Im JS, Morrone A, Johung
K, Goodwin EC, Kleijer WJ, DiMaio D and Hwang ES:
Senescence-associated beta-galactosidase is lysosomal
beta-galactosidase. Aging Cell. 5:187–195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davan-Wetton CSA, Pessolano E, Perretti M
and Montero-Melendez T: Senescence under appraisal: Hopes and
challenges revisited. Cell Mol Life Sci. 78:3333–3354. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ropero S and Esteller M: The role of
histone deacetylases (HDACs) in human cancer. Mol Oncol. 1:19–25.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang J, Gan Q, Han L, Li J, Zhang H, Sun
Y, Zhang Z and Tong T: SIRT1 overexpression antagonizes cellular
senescence with activated ERK/S6k1 signaling in human diploid
fibroblasts. PLoS One. 3:e17102008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee SH, Lee JH, Lee HY and Min KJ: Sirtuin
signaling in cellular senescence and aging. BMB Rep. 52:24–34.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sadaie M, Salama R, Carroll T, Tomimatsu
K, Chandra T, Young AR, Narita M, Pérez-Mancera PA, Bennett DC,
Chong H, et al: Redistribution of the Lamin B1 genomic binding
profile affects rearrangement of heterochromatic domains and SAHF
formation during senescence. Genes Dev. 27:1800–1808. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Perrigue PM, Rakoczy M, Pawlicka KP,
Belter A, Giel-Pietraszuk M, Naskręt-Barciszewska M, Barciszewski J
and Figlerowicz M: Cancer stem cell-inducing media activates
senescence reprogramming in fibroblasts. Cancers (Basel).
12:17452020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dalle Pezze P, Nelson G, Otten EG,
Korolchuk VI, Kirkwood TB, von Zglinicki T and Shanley DP: Dynamic
modelling of pathways to cellular senescence reveals strategies for
targeted interventions. PLoS Comput Biol. 10:e10037282014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen P, Zhang Q, Zhang H, Gao Y, Zhou Y,
Chen Y, Guan L, Jiao T, Zhao Y, Huang M and Bi H: Carnitine
palmitoyltransferase 1C reverses cellular senescence of MRC-5
fibroblasts via regulating lipid accumulation and mitochondrial
function. J Cell Physiol. 236:958–970. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen B, Chai Q, Xu S, Li Q, Wu T, Chen S
and Wu L: Silver nanoparticle-activated COX2/PGE2 axis involves
alteration of lung cellular senescence in vitro and in vivo.
Ecotoxicol Environ Saf. 204:1110702020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun L and Dang W: SIRT7 slows down stem
cell aging by preserving heterochromatin: A perspective on the new
discovery. Protein Cell. 11:469–471. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vazquez BN, Thackray JK, Simonet NG,
Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero
A, Tischfield JA and Serrano L: SIRT 7 promotes genome integrity
and modulates non-homologous end joining DNA repair. EMBO J.
35:1488–1503. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Paredes S, Angulo-Ibanez M, Tasselli L,
Carlson SM, Zheng W, Li TM and Chua KF: The epigenetic regulator
SIRT7 guards against mammalian cellular senescence induced by
ribosomal DNA instability. J Biol Chem. 293:11242–11250. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vakhrusheva O, Smolka C, Gajawada P,
Kostin S, Boettger T, Kubin T, Braun T and Bober E: Sirt7 increases
stress resistance of cardiomyocytes and prevents apoptosis and
inflammatory cardiomyopathy in mice. Circ Res. 102:703–710. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shin J, He M, Liu Y, Paredes S, Villanova
L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, et al: SIRT7
represses Myc activity to suppress ER stress and prevent fatty
liver disease. Cell Rep. 5:654–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang
H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, et al: A
SIRT7-Dependent Acetylation Switch of GABPβ1 controls mitochondrial
function. Cell Metab. 20:856–869. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Adrados I, Larrasa-Alonso J, Galarreta A,
López-Antona I, Menéndez C, Abad M, Gil J, Moreno-Bueno G and
Palmero I: The homeoprotein SIX1 controls cellular senescence
through the regulation of p16INK4A and differentiation-related
genes. Oncogene. 35:3485–3494. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wronska A, Lawniczak A, Wierzbicki PM and
Kmiec Z: Age-Related Changes in Sirtuin 7 expression in
calorie-restricted and refed rats. Gerontology. 62:304–310. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhao R, Choi BY, Lee MH, Bode AM and Dong
Z: Implications of genetic and epigenetic alterations of CDKN2A
(p16INK4a) in cancer. EBioMedicine. 8:30–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sherr CJ: Ink4-Arf locus in cancer and
aging. Wiley Interdiscip Rev Dev Biol. 1:731–741. 2012. View Article : Google Scholar : PubMed/NCBI
|