Introduction
Colorectal cancer (CRC) is currently one of the most
common types of gastrointestinal cancer. Its incidence rate ranks
third among all types of gastrointestinal cancer (1), seriously affecting human life and
health. Oxaliplatin (OXA) was initially marketed in France in 1996
and can inhibit the DNA synthesis of tumor cells to prevent their
division and proliferation, thereby causing toxic effects (2). Currently, a combined chemotherapy
regimen of OXA, 5-fluorouracil and leucovorin is the main treatment
for stage III and metastatic CRC (2–6).
However, the wide use of OXA in cancer treatment has resulted in
adverse effects involving neurotoxicity, bone marrow suppression
and liver toxicity, seriously affecting chemotherapy and the
quality of life of the patients (7–9).
In 2004, it was reported that 79% of the patients
with metastatic colorectal cancer undergoing OXA-containing
chemotherapy had hepatic sinusoidal injuries (10). OXA-induced hepatic sinusoidal
obstruction syndrome (HSOS) has become a major concern for patients
with CRC receiving chemotherapy with OXA. OXA can cause edema and
continuous disintegration of liver sinusoidal endothelial cells
(LSECs). The damaged LSECs can secrete cytokines that induce
weakness of the mucosal barrier between the LSECs, thus resulting
in the escape of red blood cells, white blood cells and platelets
into the space of Disse between the hepatocytes and sinusoidal ECs
and leading to dissection of Disse. The cascade of actions, and the
activation of the healing mechanism resulting in fibrosis, lead to
normal blood flow obstruction and increased venous resistance, thus
resulting in the development of high portal blood pressure, further
liver dysfunction and ascites (11). Notably, the pathological
characteristics of OXA-induced HSOS mainly include sinusoidal
dilatation, sinusoidal obstruction and peripheral lobular vein
fibrosis (12,13). Patients with CRC undergoing
OXA-based chemotherapy who develop sinusoidal injury and do not
receive timely intervention may further develop liver fibrosis and
cirrhosis (14). Previous studies
have demonstrated that in patients with CRC that experience
OXA-induced HSOS following partial hepatectomy, their liver
function reserve is significantly reduced and some patients may
even develop liver failure, which greatly aggravates the course of
the disease after hepatectomy (9,15,16). In addition to liver damage,
OXA-induced HSOS is associated with higher rates of overall
morbidity, bleeding risk, decreased tumor response and longer
hospital stays (15–17).
Although a number of clinical reports have proven
the universality of OXA-induced HSOS (10,13,18,19), little is known about its
pathogenesis and therapeutic drugs to treat the disease are scarce.
An important reason for this is the lack of recognized animal
models, which limits the development of studies to investigate the
mechanisms involved. Therefore, in the present study, a mouse model
of OXA-induced HSOS was established and the possible pathological
mechanisms were preliminarily analyzed using mRNA microarray
analysis to provide a basis for future research on the intervention
of OXA-induced HSOS.
Materials and methods
Animals and treatments
A total of 45 male C57BL/6 mice (weight, 20±2 g;
age, 8–10 weeks) were obtained from Sipeifu (Beijing) Biotechnology
Co., Ltd. All animals were maintained at 23±2°C and 65±5% humidity,
under a 12-h light/dark cycle, and were provided with free access
to standard laboratory food and water. The present study was
approved by the Animal Experiment Ethics Committee of Tongji
Medical College, Huazhong University of Science and Technology
(Wuhan, China; approval no. 2646). The experimental animals were
provided with humane care in accordance with the institutional
animal care guidelines (20).
Humane endpoints were in place where animals would be sacrificed if
they had lost 20% weight or exhibited 10% weight loss alongside
hypotrichosis, anorexia or decreased vitality decreases.
The 45 C57BL/6 male mice were randomly divided into
the following five groups: i) Control, ii) 5 mg/kg OXA, iii) 10
mg/kg OXA, iv) 15 mg/kg OXA and v) 20 mg/kg OXA (n=9/group). Mice
in the different OXA groups were injected intraperitoneally with 5,
10, 15 or 20 mg/kg OXA solution once a week for 6 consecutive
weeks. Mice in the control group were given a corresponding volume
of 5% glucose solution (0.02 ml/g) each time. Each day, the
behaviors and health of mice were monitored and the body weight was
recorded. After the last administration, the mice were fasted but
had access to water overnight before sacrifice. All of the mice
were sacrificed via cervical dislocation under anesthesia with an
initial intraperitoneal injection of sodium pentobarbital (30 mg/kg
body weight) to minimize animal suffering. Mice whose breathing and
heartbeat had stopped were considered to have succumbed. Blood
samples was collected from orbital venous plexus before anesthesia.
Liver tissues and spleen tissues were collected and weighed for
subsequent analyses.
Serum biochemistry analysis
The blood samples was placed in EP tubes at room
temperature for 1 h and then centrifuged at 855 × g for 10 min, and
the supernatant was taken to obtain the serum. Commercial kits
(Nanjing Jiancheng Institute of Biological Engineering) were used
to determine the serum levels of aspartate aminotransferase (AST,
cat. no. C009-3-1) and alanine aminotransferase (ALT, cat. no.
C010-3-1) in the mice from each experimental group in strict
accordance with the manufacturer's instructions.
Liver histological analyses
The mice livers were removed, washed with sterile
saline and fixed in 10% neutral formaldehyde at room temperature
for 48 h. The tissue samples were then dehydrated and embedded in
paraffin. The paraffin blocks were sectioned at a thickness of 4 µm
and each section was stained with hematoxylin and eosin (H&E)
at room temperature for 5 mins. Pathological changes in the livers
were observed under an optical microscope (Thermo Fisher
Scientific, Inc.).
For Sirius red staining, at room temperature,
sections were first stained with celestine blue solution for 5–10
min, then with Sirius red-saturated picric acid for 15–30 min.
After sealing with neutral glue, the sections were observed under
an optical microscope (Thermo Fisher Scientific, Inc.).
For scanning electron microscopy (SEM), liver tissue
was harvested within 1–3 min after the mice were sacrificed. Liver
tissue was fixed in 2.5% glutaraldehyde (Structure Probe, Inc.) at
room temperature for 2 h and then transferred to 4°C for storage.
After repeated washing with 0.1 M phosphate buffer, fixed samples
were prepared with 1% osmic acid at room temperature and protected
from light for 1–2 h. Subsequently, the samples were sequentially
dehydrated in alcohol and isoamyl acetate of different
concentrations. After drying, each sample was coated by ion
sputtering and observed by SEM (Hitachi, Ltd.).
mRNA microarray sample processing
A total of four liver tissue samples/group was
randomly selected from mice in the control group and the 10 mg/kg
OXA group. Total RNA was extracted from the tissues using Takara
RNAiso Plus (cat. no. 9109; Takara Bio, Inc.). After passing an
electrophoretic quality inspection (Agilent Technologies, Inc.),
the extracted total RNA was purified using RNeasy Mini kit and an
RNase-free DNase kit (Qiagen GmbH). The total RNA of the samples
was amplified and labeled using the Agilent Expression Profile Chip
kit (Agilent Technologies, Inc.) and then the labeled cRNA was
purified by RNeasy Mini kit (Qiagen GmbH). Each slide was
hybridized with 1.65 µg Cy3-labeled cRNA using Gene Expression
Hybridization Kit (Agilent Technologies, Inc.) in Hybridization
Oven (Agilent Technologies, Inc.), according to the manufacturer's
instructions After 17 h hybridization, slides were washed in
staining dishes (Thermo Shandon, Waltham) with Gene Expression Wash
Buffer Kit (Agilent technologies, Inc.), followed the
manufacturer's instructions. The slides that completed
hybridization were scanned by an Agilent microarray scanner
(Agilent Technologies, Inc.). The data were extracted using Feature
Extraction software 10.7 (Agilent Technologies, Inc.). The ‘limma’
package in R (https://www.rdocumentation.org/packages/limma/versions/3.28.14)
was used to normalize the raw data by the quantile algorithm.
Normalized data were screened by a fold-change statistical method
using the following selection conditions: Fold-change (linear) ≤0.5
or fold-change (linear) ≥2. In addition, the normalized data were
processed by a boxplot, sample cluster map, sample correlation
analysis, principal component analysis (PCA), scatter plot, volcano
plot and cluster heatmap using ‘ggplot2’ package in R (https://www.rdocumentation.org/packages/ggplot2/versions/3.3.6).
Technical support was provided by Shanghai Bohao Biotechnology Co.,
Ltd.
Functional enrichment analysis of
differentially expressed mRNA
Through Gene Ontology (GO; geneontology.org) and
Kyoto Encyclopedia of Genes and Genomes (KEGG; genome.jp/kegg)
enrichment analyses, differentially expressed genes (DEGs) were
classified based on different functions. The adopted method is
fisher exact test, and the data packet is cluster Profiler from
R/bioconductor (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html);
The selection criterion is that the number of DEGs on a certain GO
term or KEGG pathway is ≥2 and the P-value is <0.05. The GO
terms and KEGG pathways were obtained, presented in descending
order according to the enrichment factor values, and the top 30
results were assessed in the present study. Enrichment factor was
calculated as follows: Enrichment factor=(the number of DEGs in a
term/the total number of DEGs)/(the total number of genes in the
database term/the total number of genes in the database).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from liver samples obtained
from the mice in 10 mg/kg OXA group and control group using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA samples were quantified and the purity was assessed
using a NanoDrop BioChrom apparatus (Harvard Bioscience). A total
of 1 µg RNA was isolated and reverse transcribed to cDNA using
PrimeScript™ RT Master Mix (Takara Biotechnology Co.,
Ltd.) at 37°C for 15 mins and 95°C for 5 sec. qPCR was performed
using SYBR Green (Takara, Dalian, China) on an Applied Biosystems
StepOnePlus Real-time PCR system (Applied Biosystems). qPCR cycle
parameters were as follows: 95°C for 5 min, followed by 40 cycles
at 95°C for 10 sec and 60°C for 30 sec, and extension at 72°C for
20 sec. A final extension step at 72°C for 10 min was conducted.
Results were obtained with Bio-Rad CFX Manager (Bio-Rad
Laboratories, Inc.) and were analyzed using the 2-ΔΔCq
method (21) with the β-actin
gene used as the endogenous control. The PCR primers are listed in
Table I.
 | Table I.Sequence of primers used for
quantitative polymerase chain reaction. |
Table I.
Sequence of primers used for
quantitative polymerase chain reaction.
| Gene name | Sequence
(5′-3′) |
|---|
| Fmo3 | F:
GGCCTGTGGAAATTCTCAGAC |
|
| R:
AAGTCATCGGGATAGGGGAAG |
| Sult1e1 | F:
ATGGAGACTTCTATGCCTGAGT |
|
| R:
ACACAACTTCACTAATCCAGGTG |
| 9530077C05Rik | F:
TCTATTCGCGTAACGGAAAAGC |
|
| R:
TGAGAATCCCAGAGGACAAACTC |
| Cyp2b9 | F:
GCTCATTCTCTGGTCAGATGTTT |
|
| R:
CGCTTGTGGTCTCAGTTCCA |
| Sult3a2 | F:
GACCCACGAGCAAACAATGAA |
|
| R:
TCCAGTCTCCAACGATACCTT |
| Hsd3b5 | F:
GCTCTTGGAAACAAAAGGAAC |
|
| ACT |
|
| R:
TTCGACCGAAGGTCCTGAAC |
| Hsd3b4 | F:
GAGGTTTCTCATAAGCACAGG |
|
| AGT |
|
| R:
TCCTCCTGCACCAACATTCG |
| Elovl3 | F:
TTCTCACGCGGGTTAAAAATGG |
|
| R:
GAGCAACAGATAGACGACCAC |
| Slco1a1 | F:
GTGCATACCTAGCCAAATCACT |
|
| R:
CCAGGCCCATAACCACACA |
| Ugt2b38 | F:
TGCGCCACAAAAGGGCTAA |
|
| R:
ACACAAGAGAGTAGGAAGCCG |
| β-actin | F:
GGCTGTATTCCCCTCCATCG |
|
| R:
CCAGTTGGTAACAATGCCATGT |
Analysis of oxidative stress
indexes
At 4°C, mouse liver tissue homogenates (10%) were
prepared by adding nine volumes (ml) of normal saline to the liver
mass (g)=1/9 and centrifuging for 10 min at 627-1,164 × g; the
supernatant was used for subsequent analyses. The malondialdehyde
(MDA, cat. no. A003-1-2), superoxide dismutase (SOD, cat. no.
A001-3-2), reduced glutathione (GSH, cat. no. A006-2-1) and
catalase (CAT, cat. no. A007-1-1) levels in the mice livers of each
experimental group were measured according to the kit instructions
(Nanjing Jiancheng Bioengineering Institute), in order to evaluate
the oxidative stress-related damage in the mice livers.
Staining of dihydroethidium
(DHE)-reactive oxygen species (ROS)
Frozen liver sections (−26°C; 6 µm thickness) were
warmed at room temperature and mounted with an anti-fluorescence
quenching solution for 5 min. ROS dye solution (cat. no. D7008;
MilliporeSigma) was added dropwise and sections were incubated for
30 min at 37°C in the dark. Subsequently, the sections were washed
three times with phosphate-buffered saline (PBS, pH 7.4) and DAPI
staining solution (Beijing Solarbio Science & Technology Co.,
Ltd.) was added dropwise and incubated for 10 min at room
temperature in the dark. After washing three times with PBS and
drying, the sections were mounted with an anti-fluorescence
quenching solution. The sections were observed under a fluorescence
microscope (Nikon Corporation) and images were captured.
Statistical analysis
GraphPad Prism 8.0 software (GraphPad Software,
Inc.) was used for the statistical analyses. The data are presented
as the mean ± standard deviation. Data between groups were analyzed
by one-way ANOVA followed by Dunnett's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
OXA-induces HSOS in mice
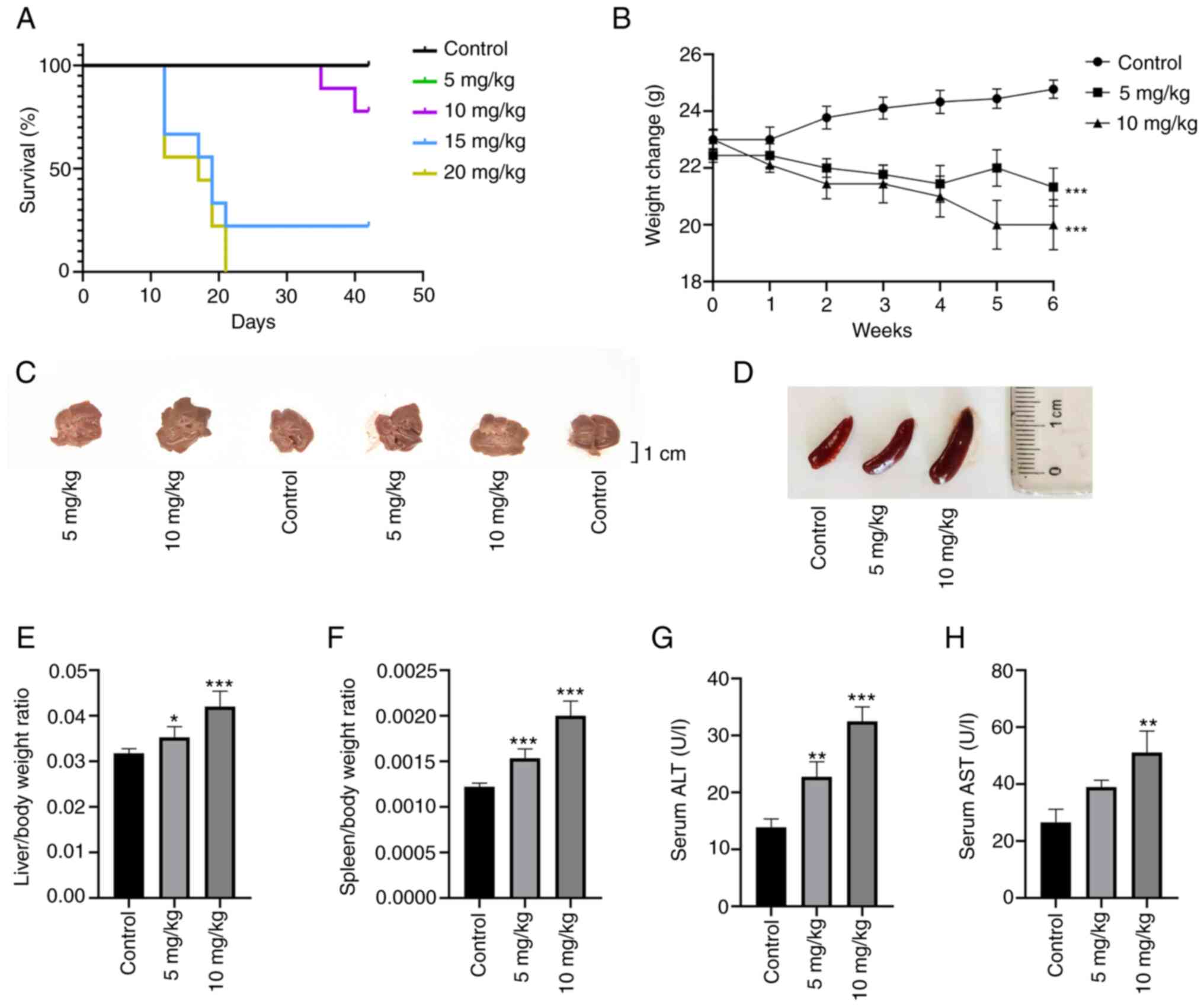
To explore the optimal dose of OXA in the mouse
model, the present study selected 5, 10, 15 and 20 mg/kg OXA based
on previous reports and the clinical dose (22–26). It was found that 3 weeks after the
start of the experiment, all mice in the 20 mg/kg OXA group and
seven mice in the 15 mg/kg OXA group met the humane endpoints
before the end of the experiment. No mice succumbed in the control
group or the 5 mg/kg OXA group and only two mice met the humane
endpoints and were sacrificed in the 10 mg/kg OXA group at the
fifth week (Fig. 1A). Therefore,
subsequent experiments were mainly performed in these three groups.
Compared with in the control group, the body weight of the mice in
the 5 and 10 mg/kg OXA groups were decreased following the
administration of OXA (Fig. 1B)
(P<0.01) and body weight in the 10 mg/kg OXA group decreased
more than that in 5 mg/kg OXA group, but the difference was not
statistically significant.
Liver changes and basic biochemical indexes showed
that the liver weight ratio, and serum levels of ALT and AST, which
are common indicators of liver injury (Fig. 1C, E, G and H), were increased
after OXA treatment and the changes were more significant in the 10
mg/kg OXA group (P<0.01). Splenomegaly is closely related to
OXA-induced HSOS (17,27) and can be used as a reliable
predictor of the occurrence and severity of OXA-induced HSOS
(28). In the present study, the
ratio of spleen weight to body weight in the different OXA groups
showed a dose-dependent increase compared with that in the control
group (Fig. 1D and F).
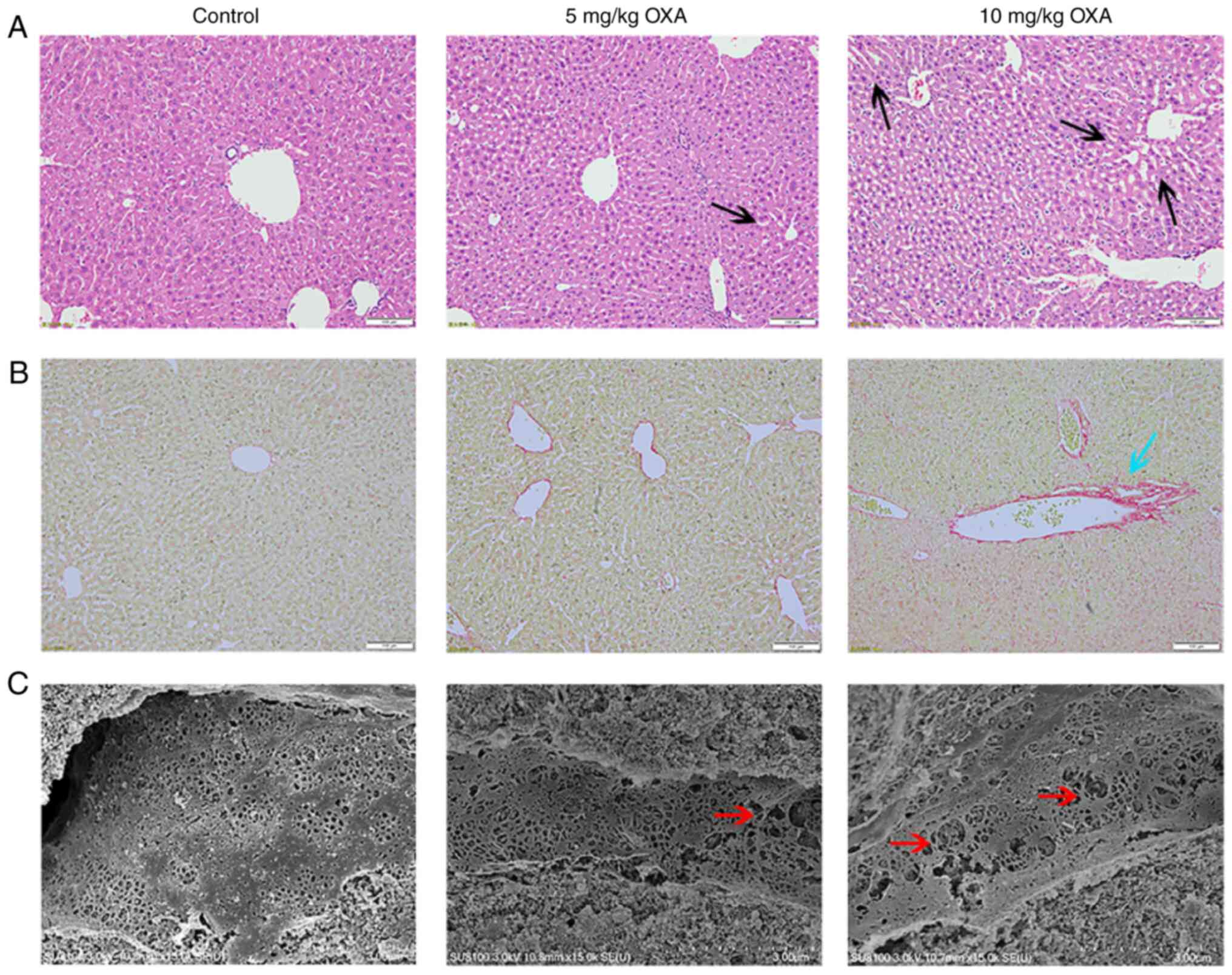
Next, the pathological changes in the livers were
analyzed. H&E staining of the liver tissue of mice in the
control group did not show evident abnormalities. The livers of the
mice in the 5 mg/kg OXA group showed mild dilatation of hepatic
sinusoids. Furthermore, the mice livers in the 10 mg/kg OXA group
showed more serious injuries and there was marked dilatation of
hepatic sinusoids around the central vein and increased hepatocyte
spaces (Fig. 2). Next, liver
fibrosis was determined using Sirius red staining, which showed
that liver fibrosis was very mild in mice administered 5 mg/kg OXA.
In addition, the staining of the mice livers was more intense and
the collagen deposition increased after the administration of 10
mg/kg OXA, thereby indicating that 10 mg/kg OXA exacerbated
fibrotic changes in the mice livers (Fig. 2). Moreover, SEM showed that OXA
resulted in obvious damage to the mice liver sinusoids in a
dose-dependent manner, resulting in the enlargement of hepatic
sinusoidal endothelial spaces and expansion of the fenestra
(Fig. 2).
Taken together, these results indicated that 10
mg/kg OXA could cause obvious liver damage, fibrosis and hepatic
sinusoidal dilation without excessive death. Therefore, it was
decided that 10 mg/kg OXA could be used for the modeling of
OXA-induced HSOS.
Changes in the mRNA expression profile
in the livers of a mouse model of OXA-induced HSOS
To observe genetic changes in the pathological
process of OXA-induced HSOS, microarray technology was used to
determine the mRNA expression profiles in the mice livers in the
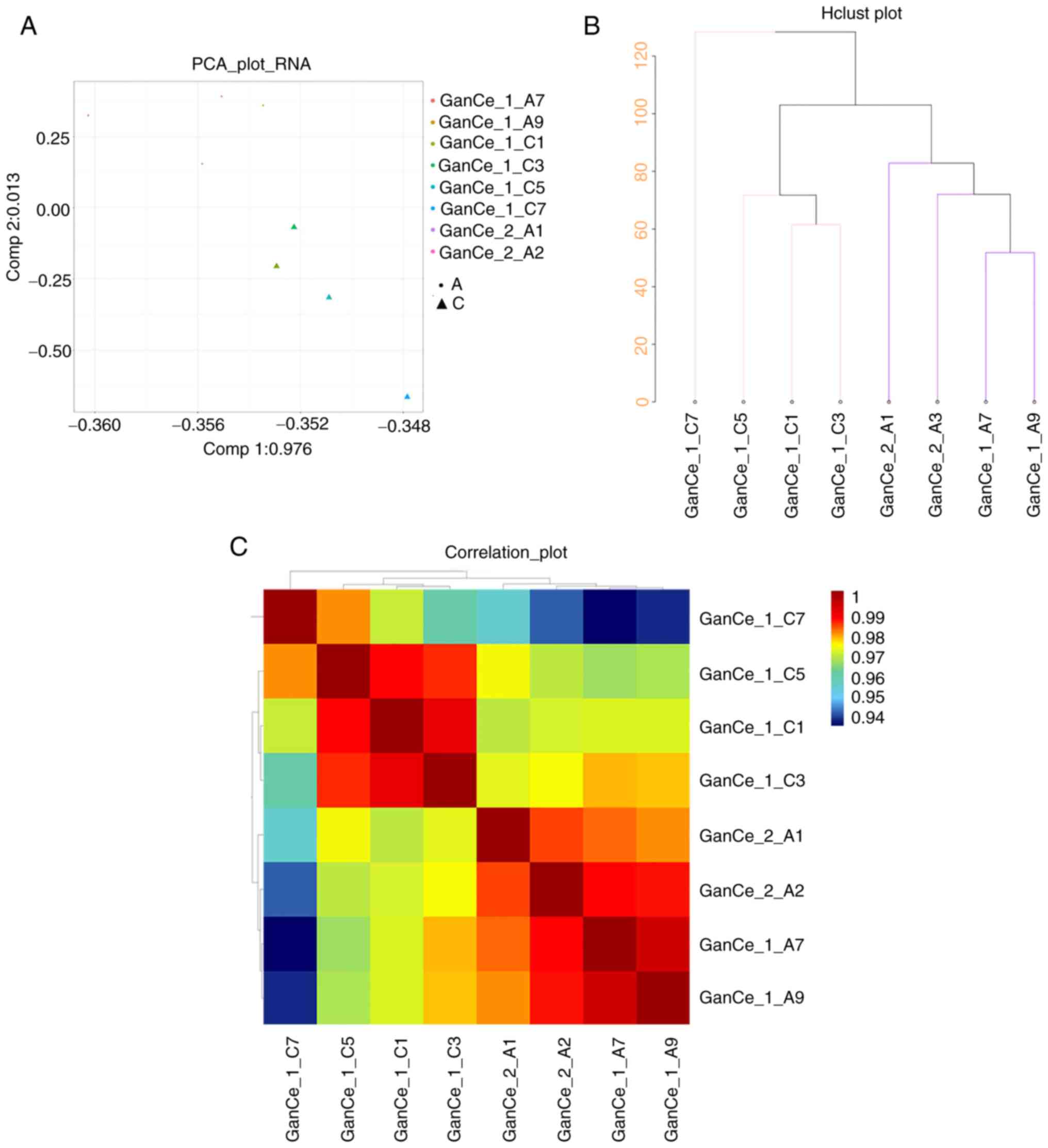
OXA (10 mg/kg) group and the control group. First, the differences
in the characteristics of the two groups were analyzed and
presented in a PCA diagram (Fig.
3A), cluster diagram (Fig.
3B) and pearson correlation analysis diagram (Fig. 3C). In the comparison of data
between the control group and the 10 mg/kg OXA group, the genes in
the livers of the mice in the two groups were not well correlated,
whereas those within the same group were correlated. Thus, the mRNA
expression in the liver samples from OXA-induced HSOS mice was
significantly different from that in the livers of mice in the
control group.
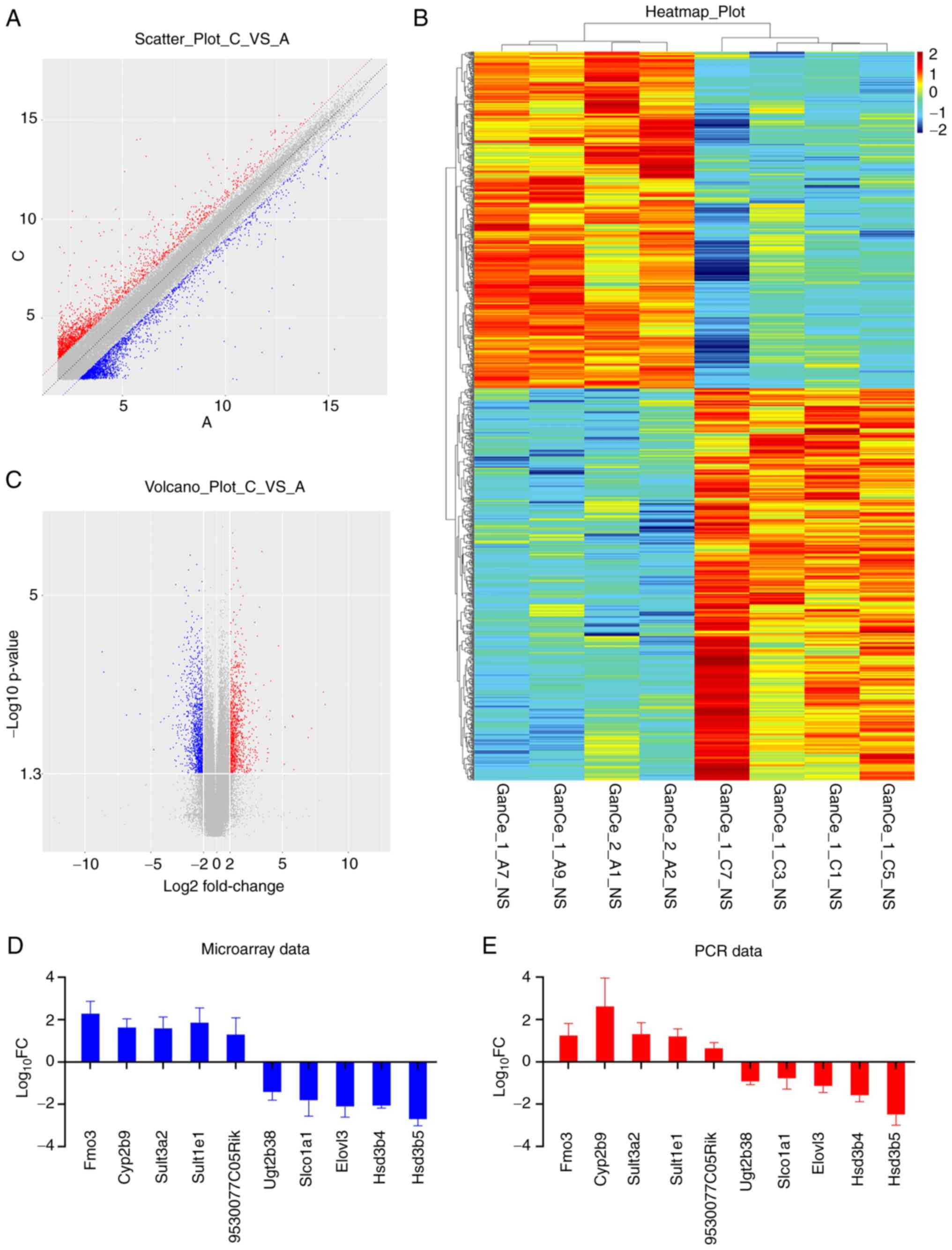
Next, the distribution of DEGs between the samples
of the two groups was analyzed and presented in a scatter plot,
volcano plot and heat map. As shown in Fig. 4, the distribution concentration
trend of data in the two groups was significantly different. Genes
with a similar expression clustered together with the samples and
DEGs between the two groups showed different expression levels,
thereby further indicating that the changes in gene expression in
the mice liver samples after OXA administration were significantly
different from those in the livers of mice in the control group
(Fig. 4A-C). A total of 1,109
DEGs were identified, including 512 downregulated genes and 597
upregulated genes. The top five downregulated and top five
upregulated DEGs are listed in Table
II. To confirm the reliability of the microarray data, the mRNA
expression levels of these candidate genes were investigated by
RT-qPCR. The 2−ΔΔCq levels and log10 fold
change of each of the 10 genes were listed in Table III. The results were consistent
with those from the microarray data (Fig. 4D and E).
 | Table II.Microarray analysis of top five
downregulated and upregulated DEGs. |
Table II.
Microarray analysis of top five
downregulated and upregulated DEGs.
| A, Upregulated
genes |
|---|
|
|---|
| Gene symbol | Log10
FC | P-value |
|---|
| Fmo3 | 2.4939 | 0.0018 |
| Sult1e1 | 2.1863 | 0.0057 |
| 9530077C05Rik | 1.7790 | 0.0411 |
| Cyp2b9 | 1.7591 | 0.0033 |
| Sult3a2 | 1.7457 | 0.0031 |
|
| B, Downregulated
genes |
|
| Gene
symbol | Log10
FC | P-value |
|
| Hsd3b5 | −2.6038 | 0.0001 |
| Hsd3b4 | −2.0468 | 0.0031 |
| Elovl3 | −1.8459 | 0.0009 |
| Slco1a1 | −1.4250 | 0.0159 |
| Ugt2b38 | −1.2663 | 0.0043 |
 | Table III.Polymerase chain reaction variation
of top five downregulated and upregulated DEGs. |
Table III.
Polymerase chain reaction variation
of top five downregulated and upregulated DEGs.
| A, Upregulated
DEGs |
|---|
|
|---|
| Gene symbol |
2−ΔΔCq | Log10
FC | P-value |
|---|
| Fmo3 | 17.354 | 1.239 | 0.0026 |
| Cyp2b9 | 410.518 | 2.613 | 0.0003 |
| Sult3a2 | 19.962 | 1.300 | 0.0014 |
| Sult1e1 | 15.666 | 1.195 | 0.0088 |
| 9530077C05Rik | 4.396 | 0.643 | 0.0192 |
|
| B, Downregulated
DEGs |
|
| Gene
symbol |
2−ΔΔCq | Log10
FC | P-value |
|
| Hsd3b5 | 0.003 | −2.492 | 0.0001 |
| Hsd3b4 | 0.026 | −1.585 | 0.0015 |
| Elovl3 | 0.072 | −1.145 | 0.0079 |
| Slco1a1 | 0.166 | −0.780 | 0.0312 |
| Ugt2b38 | 0.117 | −0.931 | 0.0217 |
GO and KEGG pathway analysis
By performing GO enrichment analysis of DEGs, genes
can be classified based on different functions to achieve the
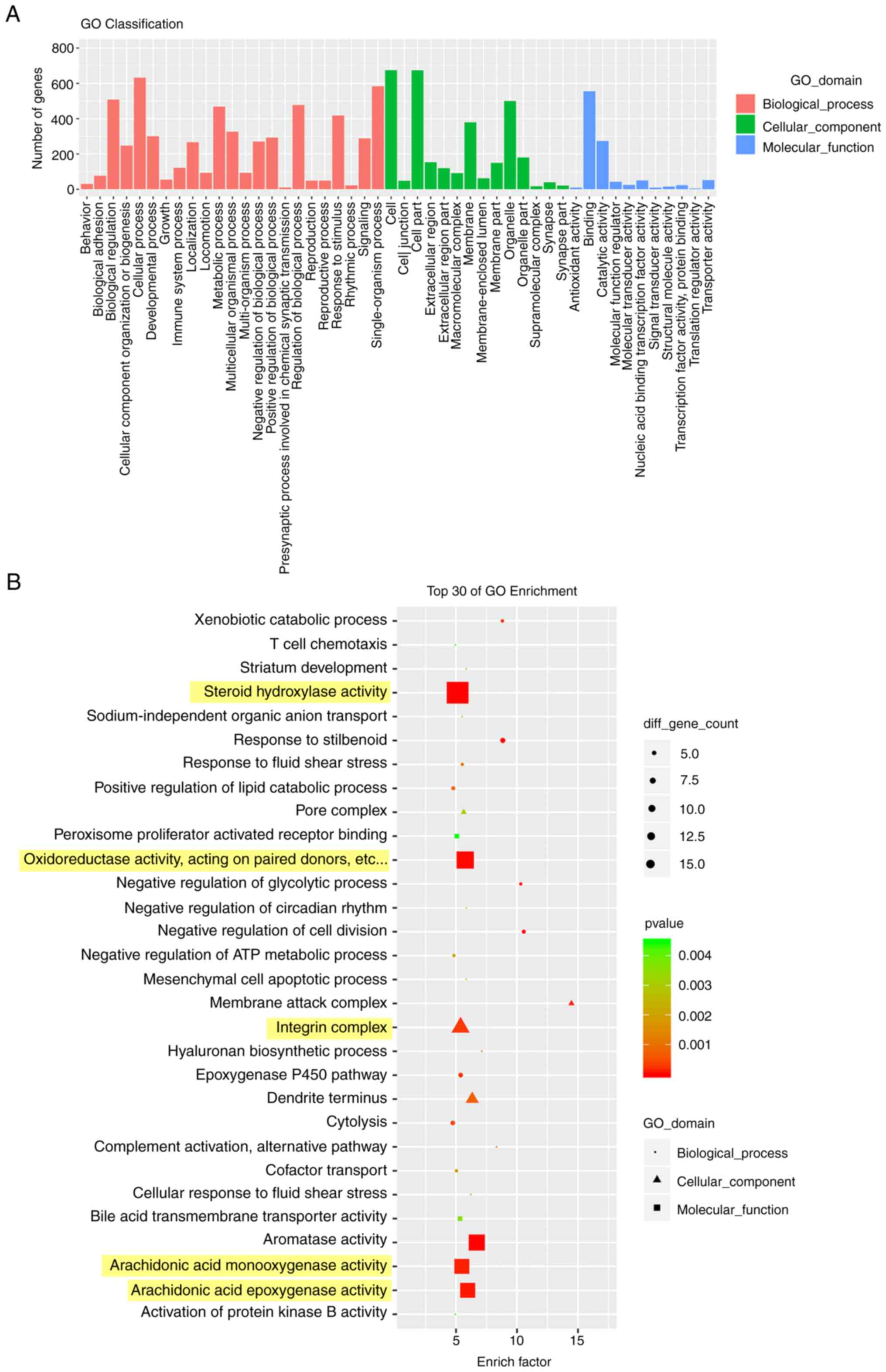
purpose of annotation and classification of the genes (29). As shown in Fig. 5, DEGs between the control group
and 10 mg/kg OXA group were mostly enriched in biological processes
(Fig. 5A). In addition, the GO
analysis of the top 30 enrichment factors showed that DEGs were
mainly enriched in ‘steroid hydroxylase activity’, ‘oxidoreductase
activity, acting on paired donors’, ‘integrin complex’,
‘arachidonic acid monooxygenase activity’ and ‘arachidonic acid
epoxygenase activity’ (Fig.
5B).
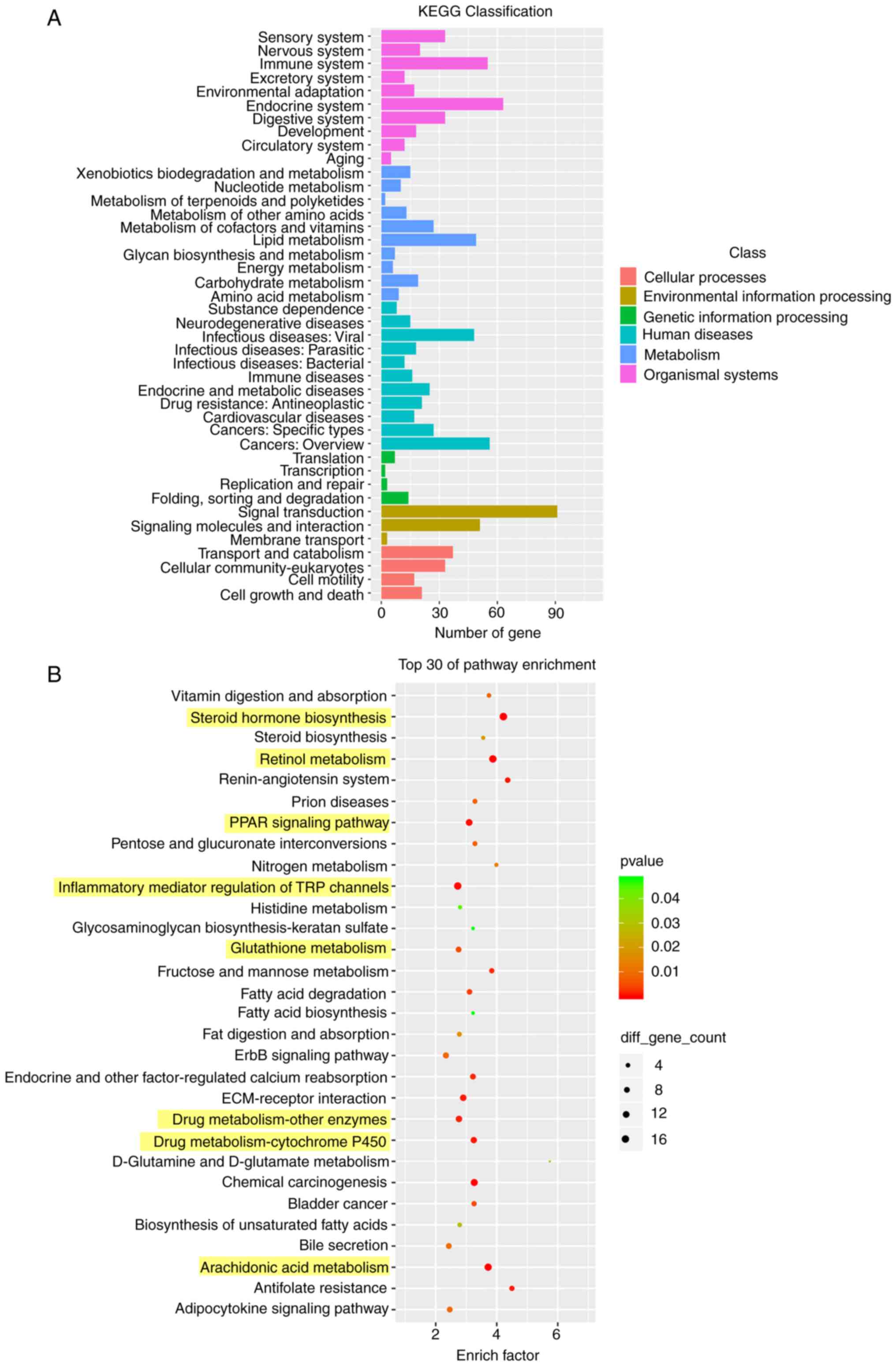
KEGG enrichment analysis of the DEGs showed the
enrichment levels of the these genes in different pathways
(29), which may identify the
biological regulatory pathways that were significantly different
under the experimental conditions. The results provided directions
for subsequent research on identifying the underlying mechanisms
involved. According to the KEGG enrichment results, most of the
enriched pathways were related to human diseases (Fig. 6A). Compared with the control
group, the enriched pathways of DEGs in the 10 mg/kg OXA group were
mainly involved in ‘steroid hormone biosynthesis’, ‘retinol
metabolism’, ‘PPAR signaling pathway’, ‘inflammatory mediator
regulation of TRP channels’, ‘glutathione metabolism’, ‘drug
metabolism-other enzymes’, ‘drug metabolism-cytochrome P450’ and
‘arachidonic acid metabolism’ (Fig.
6B).
Oxidative stress-related indicators in
OXA-induced HSOS in mice
The microarray results suggested that oxidative
stress may serve an important role in OXA-induced HSOS. Therefore,
the levels of common oxidative stress markers were determined and
changes in ROS were observed by DHE staining. MDA is a lipid
peroxidation product in vivo, which can cause cytotoxicity
(30). The MDA levels in the mice
livers were significantly increased following the administration of
10 mg/kg OXA (P<0.05; Fig.
7A). SOD is an antioxidant metal enzyme that can catalyze the
superoxide anion radical and provide cellular defenses against ROS
(22). CAT is also an antioxidant
enzyme that protects cells from the toxicity of
H2O2 (22).
GSH has antioxidant and integrative detoxification effects
(22). OXA markedly reduced the
levels of SOD, CAT and GSH in the mice livers (P<0.05; Fig. 7B-D). Furthermore, DHE probe
technology was used to analyze changes in ROS levels. The results
showed that the ROS levels in the livers of the mice in the OXA
group were increased in a dose-dependent manner (Fig. 7E). Taken together, these data
confirmed that oxidative stress may have an important role in the
liver damage of mice with OXA-induced HSOS.
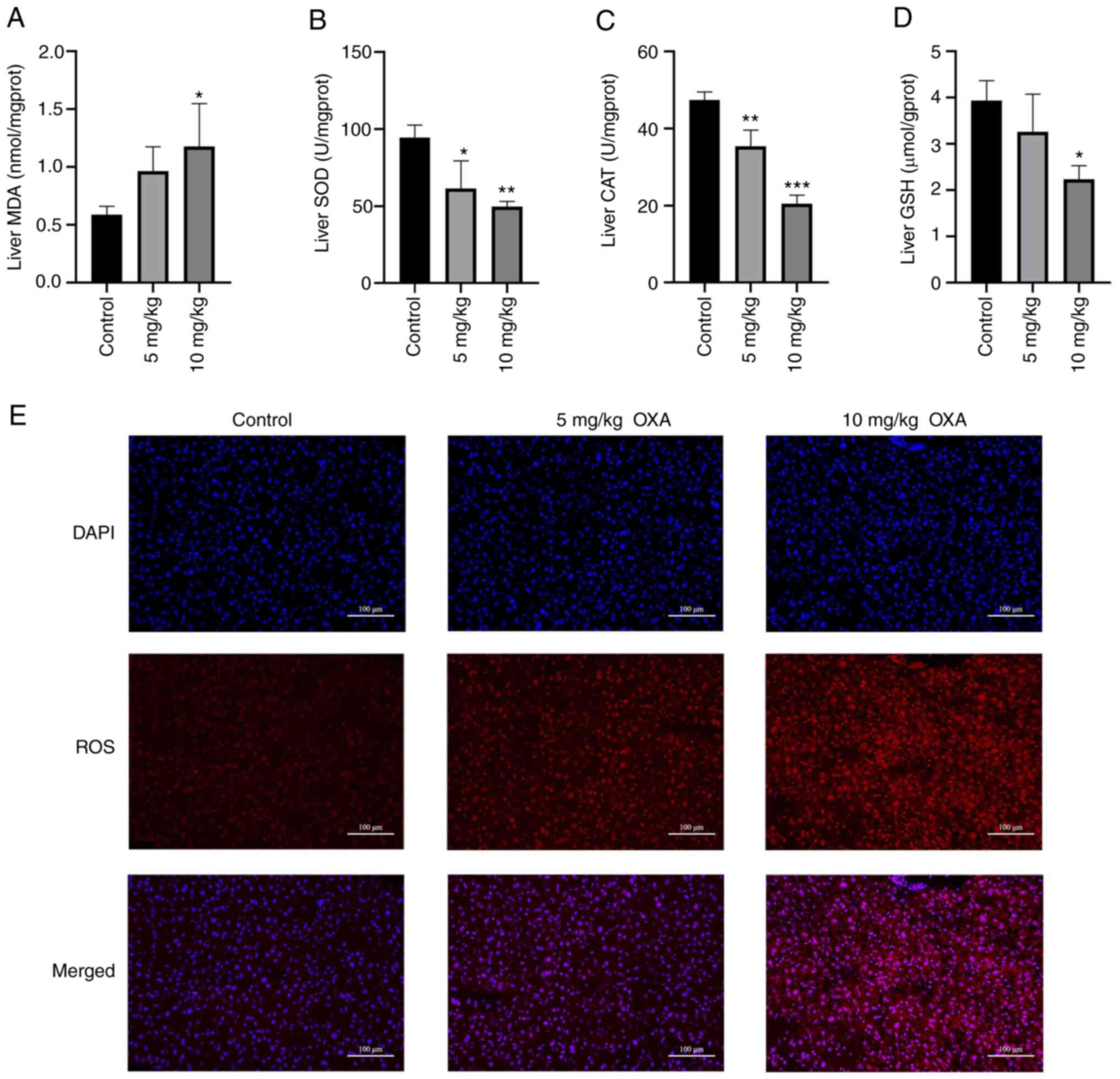 | Figure 7.OXA-induced oxidative stress in the
liver. (A) MDA levels, and activity of (B) SOD, (C) CAT and (D) GSH
in each group of mice were measured. (E) Detection of ROS levels
(DHE probe technology; magnification, ×400; scale bar=50 µm). Data
are shown as the mean ± standard deviation for each group and
analyzed with a one-way ANOVA followed by Dunnett's post hoc test.
*P<0.05, **P<0.01 and ***P<0.001 vs. control group. OXA,
oxaliplatin; MDA, malondialdehyde; CAT, catalase; GSH, reduced
glutathione; ROS, reactive oxygen species; DHE,
dihydroethidium. |
Discussion
The main side effects of OXA are neurotoxicity, bone
marrow suppression and gastrointestinal reaction. Recently, hepatic
sinusoidal injury has garnered attention as a hepatotoxic reaction
that occurs in patients with CRC liver metastases after receiving
OXA-based neoadjuvant chemotherapy, with a worldwide incidence rate
of 48–79% (10,31,32). The typical pathological features
of OXA-induced HSOS include hepatic sinusoidal dilatation, platelet
aggregation in the hepatic sinusoids and some typical clinical
features, such as blue liver, splenomegaly and thrombocytopenia
(33). A number of clinical
reports have proven the universality of OXA-induced HSOS, which
usually only focused on clinical features and diagnosis (10,13,18,19). Reports on the mechanism of its
pathogenesis are limited. There is currently no recognized
OXA-induced hepatotoxicity model.
In previous studies on OXA-induced hepatotoxicity,
the doses of OXA ranged between 5 and 30 mg/kg (26,34–36). In the present study, a gradient of
OXA doses for modeling was screened and it was revealed that the
mortality rate of mice in the 15 and 20 mg/kg OXA groups reached
78–100%, which indicated a strong toxic reaction from high-dose
OXA. Furthermore, the present study showed that a suitable HSOS
model with an acceptable survival rate could be established after 6
weeks of 10 mg/kg OXA administration. The OXA dose the present
study recommended for HSOS modeling was much lower than the dose
presented in other studies (34,35). It was hypothesized that this
difference may be due to the dosing frequency and modeling time.
According to the National Comprehensive Cancer Network guidelines,
the optimal duration of OXA plus 5-fluorouracil and leucovorin or
oral fluoropyrimidine (capecitabine) in patients with CRC is 3 or 6
months, with a cycle of 2 weeks (37). Instead of the continuous daily
administration used in other studies (23,38), the present study decided to
administer OXA by intraperitoneal injection once a week for 6
consecutive weeks to simulate the clinical protocol (22). Which also took into consideration
the growth cycle of the mice.
Nam et al (12) demonstrated that patients with CRC
who received OXA chemotherapy were more likely to have abnormal
liver functions, and increased serum levels of AST, ALT and total
bilirubin. The results of the present study showed that after the
long-term administration of 10 mg/kg OXA, the body weight of the
mice decreased significantly, whereas the liver weight-to-body
weight ratio, and the serum levels of ALT and AST were increased.
Overman et al (17)
revealed that liver sinusoidal injury caused by OXA-based
chemotherapy may cause a dose-dependent increase in spleen volume.
El Chediak et al (28)
reported that an increase in spleen volume was an independent
predictor of HSOS in 79 patients treated with OXA. The current
study revealed that the spleens were enlarged in mice in the 10
mg/kg OXA group compared with in the control group, which was
consistent with the clinical characteristics of patients with
cancer and OXA-induced HSOS.
HSOS is a clinical syndrome in which hepatic
sinusoidal endothelial cells are shed by drugs or toxins, resulting
in the obstruction of the hepatic outflow tract and downstream
hepatic small veins, thus leading to intrahepatic pre-sinusoidal or
post-sinusoidal portal hypertension in patients who undergo
chemotherapy (39). Rubbia-Brandt
et al (13) showed that
the pathology of liver tissue resected from 274 patients with CRC
liver metastases treated with OXA-based chemotherapy exhibited
hepatic sinusoidal dilation and obstruction, nodular changes in the
liver, hepatic lobular central fibrosis, hepatocyte steatosis and
hemorrhage. To further confirm liver changes in mice with
OXA-induced HSOS, in addition to basic histological staining
analysis, the present study performed SEM to observe the changes in
hepatic sinusoids in mice, which is an important indicator of
whether OXA-induced HSOS has been produced. The results showed
hepatic sinusoidal dilatation and increased hepatocyte space
following treatment with OXA and the severity of the liver injury
was proportional to the dose of OXA received. SEM analysis showed
that OXA treatment resulted in the enlargement of hepatic
sinusoidal endothelial spaces and expansion of the fenestra. The
results confirmed that an OXA-induced HSOS model was successfully
established with typical pathological characteristics. In addition,
liver fibrosis was also found in mice in the 10 mg/kg group, which
may aggravate the course of liver injury and prolong its recovery
time (16). Based on these
results, a typical HSOS model could be established after 6 weeks of
OXA administration at a dose of 10 mg/kg.
In a previous study, a preliminary analysis was
performed based on the comparison of gene expression profiles of 11
human livers with OXA-induced HSOS and 12 histologically normal
livers (25). The main genes in
the following pathways were upregulated in patients with HSOS:
Acute-phase reaction, coagulation system disorder, liver fibrosis
and oxidative stress (25).
However, in this previous study, only a simple microarray analysis
of a few genes was made and the study lacked an analysis of the
mechanism of the pathways involved. In addition, to the best of our
knowledge, no mRNA microarray study on an animal model of
OXA-induced HSOS has been performed. Therefore, the present study
used mRNA microarray technology to explore changes in the gene
expression profiles of livers with OXA-induced HSOS, hoping to
provide possible directions for subsequent studies of the disease
mechanism. The GO database classifies gene function into three
broad categories: Cellular components, molecular functions and
biological processes. The present study proved that DEGs between
the control group and the 10 mg/kg OXA group were expressed in all
three categories but were mainly concentrated in the biological
processes category. Among the top 30 enriched GO terms, the DEGs
were mainly enriched in ‘steroid hydroxylase activity’,
‘oxidoreductase activity, acting on paired donors’, ‘integrin
complex’, ‘arachidonic acid monooxygenase activity’ and
‘arachidonic acid epoxygenase activity’. The integrin complex
connects the extracellular matrix and the cytoskeleton, controls
intracellular signal events related to proliferation,
differentiation, migration and other cellular processes (40), and may be involved in the
regulation of fibrosis (41). OXA
has been reported to accelerate the activation of hepatic stellate
cells and collagen synthesis by upregulating the expression of type
I collagen and transforming growth factor, thereby aggravating the
deposition of extracellular matrix components and promoting the
development of hepatic fibrosis (16,35). At present, no studies, to the best
of our knowledge, have described whether the integrin complex could
participate in the regulation of fibrosis in OXA-induced HSOS. OXA
can promote platelet aggregation and adhesion through the release
of matrix metalloproteinases and a variety of growth factors, thus
leading to the aggravation of hepatic sinusoidal obstruction
(16). The arachidonic acid
oxidation pathway can be used as a target for the regulation of
blood coagulation and platelet activation (42) and also participates in
inflammatory reaction (43),
which may affect the state of platelet activation in OXA-induced
HSOS. Steroid hydroxylase, mainly cytochrome P-450, plays a role in
maintaining homeostasis in the synthesis and metabolism of
cholesterol and bile acids (44,45). However, no studies related to bile
acid and lipid balance in OXA-induced HSOS are available, to the
best of our knowledge.
KEGG enrichment analysis of DEGs identified the
significantly enriched pathways, which is helpful for identifying
biological regulatory pathways with significant differences under
experimental conditions. The present study revealed that the DEGs
between the control group and the 10 mg/kg OXA group were mostly
enriched in pathways related to human diseases. Among the top 30
enriched KEGG pathways, the regulation of ‘steroid hormone
biosynthesis’, ‘retinol metabolism’, ‘drug metabolism cytochrome
P450’ and ‘arachidonic acid metabolism’ were consistent with the
main functional categories enriched in GO function analysis, which
further confirmed their regulatory role in steroid anabolism,
changes in coagulation function and ligand activation. Liver
inflammation has been considered a possible driving factor for
liver damage caused by OXA (16).
A previous study demonstrated that in OXA-induced animal models,
the inflammatory factors interleukin 16 and C-C motif chemokine
ligand 2 were upregulated (46).
The microarray results of the present study demonstrated that the
inflammatory mediator regulation of TRP channels may serve a role
in OXA-induced HSOS, which needs to be further explored.
Notably, ‘oxidoreductase activity’ in GO analysis
and the ‘glutathione metabolism’ pathway in KEGG analysis indicated
that oxidative stress may be involved in OXA-induced HSOS. The
activity of oxidoreductase affects the regulation of redox
homeostasis in the body. A previous study (35) preliminarily found that after the
injection of OXA, the levels of antioxidant enzymes SOD and
glutathione peroxidase were decreased in the liver tissue of mice
with nonalcoholic fatty liver disease. Glutathione metabolism
affects the changes in ROS levels in a variety of liver diseases
and can aggravate the state of oxidative stress (47). Moreover, it has been reported that
oxidative stress caused by GSH depletion may be one of the reasons
for OXA-induced hepatic sinusoid injury (16). Tabassum et al (48) evaluated the GSH content in liver
mitochondria after OXA treatment and demonstrated that a decrease
in GSH content may be related to the binding of OXA to free or
protein-bound sulfhydryl groups. The current study also revealed
that after the long-term administration of OXA, the MDA liver
oxidation index in mice increased, whereas the levels of
antioxidants SOD, CAT and GSH were significantly decreased. In
addition, ROS levels in the liver of mice in the OXA group were
increased in a dose-dependent manner, indicating that OXA could
lead to an imbalance in the redox system.
Notably, in the present study, GO and KEGG analyses
suggested that other pathways may be involved in OXA-induced HSOS,
such as mesenchymal cell apoptotic process and bile secretion.
Therefore, the data provided novel directions for the study of the
pathogenetic mechanisms of OXA-induced HSOS.
There were some limitations to the present study.
First, the aim of the present study was to establish an animal
model of OXA-induced HSOS, so in vivo experiments were
preferentially conducted. Corresponding in vitro experiments
using hepatic sinusoidal endothelial cells to explore the
pathogenetic mechanisms of OXA-induced HSOS will be conducted in
the future. Second, the symptoms caused by OXA may be associated
with its systemic effects, such as pro-inflammatory effects, and
effects on the nervous system and circulatory system, which may
also aggravate liver injuries and is worth exploring in the future.
Third, more advanced detection equipment and indicators, such as
ultrasound, angiography and the assessment of portal hypertension,
will be employed in the confirmation of HSOS.
In conclusion, the present study established a mouse
model of OXA-induced HSOS. Through microarray analysis, it verified
the role of the oxidative stress pathway in this disease and
identified some potential mechanistic pathways, which could lay the
foundation for identifying the mechanisms of OXA-induced HSOS.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Clinical Toxicology
Foundation of the Chinese Society of Toxicology (grant no.
CST2020CT107), the Research Project of the Drug Clinical Evaluation
Professional Committee of China Pharmaceutical Association (grant
no. CPA-Z06-CZ-2021-004) and the Chen Xiao-ping Foundation for the
Development of Science and Technology of Hubei Province (grant no.
CXPJJH121003-2122).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211859.
All other data sets in the present study are available from the
corresponding author on reasonable request.
Authors' contributions
CZhu, DL, XR and CZha designed the study. CZhu, QG
and XW performed the experiments. CZhu, XC, DL and PG contributed
to the data acquisition and analysis. CZhu wrote the manuscript.
CZhu, PG and CZha contributed to the critical revision of the
manuscript. CZhu and XC confirm the authenticity of all the raw
data. All authors made significant contributions to this study, and
all authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal
Experiment Ethics Committee of Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China; approval no.
2646).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ALT
|
alanine aminotransferase
|
|
AST
|
aspartate aminotransferase
|
|
CAT
|
catalase
|
|
CRC
|
colorectal cancer
|
|
DEG
|
differentially expressed genes
|
|
DHE
|
dihydroethidium
|
|
GO
|
Gene Ontology
|
|
GSH
|
reduced glutathione
|
|
H&E
|
hematoxylin and eosin
|
|
HSOS
|
hepatic sinusoidal obstruction
syndrome
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
LSEC
|
liver sinusoidal endothelial cell
|
|
MDA
|
malondialdehyde
|
|
OXA
|
oxaliplatin
|
|
ROS
|
reactive oxygen species
|
|
SEM
|
scanning electron microscopy
|
|
SOD
|
superoxide dismutase
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soulié P, Raymond E, Brienza S and
Cvitkovic E: Oxaliplatin: The first DACH platinum in clinical
practice. Bull Cancer. 84:665–673. 1997.(In French). PubMed/NCBI
|
|
3
|
Adam R, Wicherts DA, de Haas RJ, Ciacio O,
Lévi F, Paule B, Ducreux M, Azoulay D, Bismuth H and Castaing D:
Patients with initially unresectable colorectal liver metastases:
Is there a possibility of cure? J Clin Oncol. 27:1829–1835. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duwe G, Knitter S, Pesthy S, Beierle AS,
Bahra M, Schmelzle M, Schmuck RB, Lohneis P, Raschzok N, Öllinger
R, et al: Hepatotoxicity following systemic therapy for colorectal
liver metastases and the impact of chemotherapy-associated liver
injury on outcomes after curative liver resection. Eur J Surg
Oncol. 43:1668–1681. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang XB, Hou SH, Li YP, Wang LC, Zhang X
and Yang J: Irinotecan or oxaliplatin combined with 5-fluorouracil
and leucovorin as first-line therapy for advanced colorectal
cancer: A meta-analysis. Chin Med J (Engl). 123:3314–3318.
2010.PubMed/NCBI
|
|
6
|
Puente A, Fortea JI, Del Pozo C, Huelin P,
Cagigal ML, Serrano M, Cabezas J, Arias LM, Iruzubieta P, Cuadrado
A, et al: Porto-sinusoidal vascular disease associated to
oxaliplatin: An entity to think about it. Cells. 8:15062019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
André T, Boni C, Navarro M, Tabernero J,
Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F
and de Gramont A: Improved overall survival with oxaliplatin,
fluorouracil, and leucovorin as adjuvant treatment in stage II or
III colon cancer in the MOSAIC trial. J Clin Oncol. 27:3109–3116.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Erdem GU, Dogan M, Demirci NS and Zengin
N: Oxaliplatin-induced acute thrombocytopenia. J Cancer Res Ther.
12:509–514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao J, van Mierlo K, Gomez-Ramirez J, Kim
H, Pilgrim CHC, Pessaux P, Rensen SS, van der Stok EP, Schaap FG,
Soubrane O, et al: Systematic review of the influence of
chemotherapy-associated liver injury on outcome after partial
hepatectomy for colorectal liver metastases. Br J Surg.
104:990–1002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rubbia-Brandt L, Audard V, Sartoretti P,
Roth AD, Brezault C, Le Charpentier M, Dousset B, Morel P, Soubrane
O, Chaussade S, et al: Severe hepatic sinusoidal obstruction
associated with oxaliplatin-based chemotherapy in patients with
metastatic colorectal cancer. Ann Oncol. 15:460–466. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nassereddine S, Alsubait S and Tabbara I:
Sinusoidal obstruction syndrome (veno-occlusive disease) following
hematopoietic stem cell transplant: Insights and therapeutic
advances. Anticancer Res. 38:2597–2605. 2018.PubMed/NCBI
|
|
12
|
Nam SJ, Cho JY, Lee HS, Choe G, Jang JJ,
Yoon YS, Han HS and Kim H: Chemotherapy-associated hepatopathy in
korean colorectal cancer liver metastasis patients:
Oxaliplatin-based chemotherapy and sinusoidal injury. Korean J
Pathol. 46:22–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rubbia-Brandt L, Lauwers GY, Wang H, Majno
PE, Tanabe K, Zhu AX, Brezault C, Soubrane O, Abdalla EK, Vauthey
JN, et al: Sinusoidal obstruction syndrome and nodular regenerative
hyperplasia are frequent oxaliplatin-associated liver lesions and
partially prevented by bevacizumab in patients with hepatic
colorectal metastasis. Histopathology. 56:430–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan CQ and Crawford JM: Sinusoidal
obstruction syndrome (hepatic veno-occlusive disease). J Clin Exp
Hepatol. 4:332–346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Russolillo N, Langella S, Perotti S, Lo
Tesoriere R, Forchino F and Ferrero A: Preoperative assessment of
chemotherapeutic associated liver injury based on indocyanine green
retention test. Int J Surg. 31:80–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu C, Ren X, Liu D and Zhang C:
Oxaliplatin-induced hepatic sinusoidal obstruction syndrome.
Toxicology. 460:1528822021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Overman MJ, Maru DM, Charnsangavej C,
Loyer EM, Wang H, Pathak P, Eng C, Hoff PM, Vauthey JN, Wolff RA
and Kopetz S: Oxaliplatin-mediated increase in spleen size as a
biomarker for the development of hepatic sinusoidal injury. J Clin
Oncol. 28:2549–2555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cayet S, Pasco J, Dujardin F, Besson M,
Orain I, De Muret A, Miquelestorena-Standley E, Thiery J, Genet T
and Le Bayon AG: Diagnostic performance of contrast-enhanced
CT-scan in sinusoidal obstruction syndrome induced by chemotherapy
of colorectal liver metastases: Radio-pathological correlation. Eur
J Radiol. 94:180–190. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han NY, Park BJ, Kim MJ, Sung DJ and Cho
SB: Hepatic parenchymal heterogeneity on contrast-enhanced CT scans
following oxaliplatin-based chemotherapy: Natural history and
association with clinical evidence of sinusoidal obstruction
syndrome. Radiology. 276:766–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Couto M and Cates C: Laboratory guidelines
for animal care. Methods Mol Biol. 1920:407–430. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Y, Wu S, Xiang B, Li L and Lin Y:
Curcumin attenuates oxaliplatin-induced liver injury and oxidative
stress by activating the Nrf2 pathway. Drug Des Devel Ther.
14:73–85. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Zhang ZS, Zhang XH, Yang SN, Liu D,
Diao CR, Wang H and Zheng FP: Cyanidin inhibits EMT induced by
oxaliplatin via targeting the PDK1-PI3K/Akt signaling pathway. Food
Funct. 10:592–601. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyagi A, Kawashiri T, Shimizu S,
Shigematsu N, Kobayashi D and Shimazoe T: Dimethyl fumarate
attenuates oxaliplatin-induced peripheral neuropathy without
affecting the anti-tumor activity of oxaliplatin in rodents. Biol
Pharm Bull. 42:638–644. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rubbia-Brandt L, Tauzin S, Brezault C,
Delucinge-Vivier C, Descombes P, Dousset B, Majno PE, Mentha G and
Terris B: Gene expression profiling provides insights into pathways
of oxaliplatin-related sinusoidal obstruction syndrome in humans.
Mol Cancer Ther. 10:687–696. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou X, Wang Y, Peng C, Wang B, Niu Z, Li Z
and Niu J: Magnesium isoglycyrrhizinate has hepatoprotective
effects in an oxaliplatin-induced model of liver injury. Int J Mol
Med. 42:2020–2030. 2018.PubMed/NCBI
|
|
27
|
Kim MJ, Han SW, Lee DW, Cha Y, Lee KH and
Kim TY, Oh DY, Kim SH, Im SA, Bang YJ and Kim TY: Splenomegaly and
its associations with genetic polymorphisms and treatment outcome
in colorectal cancer patients treated with adjuvant FOLFOX. Cancer
Res Treat. 48:990–997. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
El Chediak A, Haydar AA, Hakim A, Massih
SA, Hilal L, Mukherji D, Temraz S and Shamseddine A: Increase in
spleen volume as a predictor of oxaliplatin toxicity. Ther Clin
Risk Manag. 14:653–657. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang YF, Huang Y, Ni YH and Xu ZM:
Systematic elucidation of the mechanism of geraniol via network
pharmacology. Drug Des Devel Ther. 13:1069–1075. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsikas D: Assessment of lipid peroxidation
by measuring malondialdehyde (MDA) and relatives in biological
samples: Analytical and biological challenges. Anal Biochem.
524:13–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nordlinger B, Sorbye H, Glimelius B,
Poston GJ, Schlag PM, Rougier P, Bechstein WO, Primrose JN, Walpole
ET, Finch-Jones M, et al: Perioperative FOLFOX4 chemotherapy and
surgery versus surgery alone for resectable liver metastases from
colorectal cancer (EORTC 40983): Long-term results of a randomised,
controlled, phase 3 trial. Lancet Oncol. 14:1208–1215. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vreuls CP, Van Den Broek MA, Winstanley A,
Koek GH, Wisse E, Dejong CH, Olde Damink SW, Bosman FT and Driessen
A: Hepatic sinusoidal obstruction syndrome (SOS) reduces the effect
of oxaliplatin in colorectal liver metastases. Histopathology.
61:314–318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tajima H, Ohta T, Miyashita T, Nakanuma S,
Matoba M, Miyata T, Sakai S, Okamoto K, Makino I, Kinoshita J, et
al: Oxaliplatin-based chemotherapy induces extravasated platelet
aggregation in the liver. Mol Clin Oncol. 3:555–558. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Robinson SM, Mann J, Vasilaki A, Mathers
J, Burt AD, Oakley F, White SA and Mann DA: Pathogenesis of FOLFOX
induced sinusoidal obstruction syndrome in a murine chemotherapy
model. J Hepatol. 59:318–326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu Y, Lin Y, Huang X, Wu S, Wei J and Yang
C: Oxaliplatin aggravates hepatic oxidative stress, inflammation
and fibrosis in a non-alcoholic fatty liver disease mouse model.
Int J Mol Med. 43:2398–2408. 2019.PubMed/NCBI
|
|
36
|
Yang L, Ding Y, Rao S, Chen C and Zeng M:
T1 mapping on Gd-EOB-DTPA-enhanced MRI for the
prediction of oxaliplatin-induced liver injury in a mouse model. J
Magn Reson Imaging. 53:896–902. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oneda E and Zaniboni A: Adjuvant treatment
of colon cancer with microsatellite instability-the state of the
art. Crit Rev Oncol Hematol. 169:1035372022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia T, Zhang J, Han L, Jin Z, Wang J, Li
X, Man S, Liu C and Gao W: Protective effect of magnolol on
oxaliplatin-induced intestinal injury in mice. Phytother Res.
33:1161–1172. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Valla DC and Cazals-Hatem D: Sinusoidal
obstruction syndrome. Clin Res Hepatol Gastroenterol. 40:378–385.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McCarty JH: αvβ8 integrin adhesion and
signaling pathways in development, physiology and disease. J Cell
Sci. 133:jcs2394342020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Munger JS, Huang X, Kawakatsu H, Griffiths
MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et
al: The integrin alpha v beta 6 binds and activates latent TGF beta
1: A mechanism for regulating pulmonary inflammation and fibrosis.
Cell. 96:319–328. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Trostchansky A, Moore-Carrasco R and
Fuentes E: Oxidative pathways of arachidonic acid as targets for
regulation of platelet activation. Prostaglandins Other Lipid
Mediat. 145:1063822019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Capdevila JH, Falck JR and Harris RC:
Cytochrome P450 and arachidonic acid bioactivation. Molecular and
functional properties of the arachidonate monooxygenase. J Lipid
Res. 41:163–181. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sheets JJ, Mason JI, Wise CA and Estabrook
RW: Inhibition of rat liver microsomal cytochrome P-450 steroid
hydroxylase reactions by imidazole antimycotic agents. Biochem
Pharmacol. 35:487–491. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pikuleva IA: Cholesterol-metabolizing
cytochromes P450. Drug Metab Dispos. 34:513–520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jarzabek MA, Proctor WR, Vogt J, Desai R,
Dicker P, Cain G, Raja R, Brodbeck J, Stevens D, van der Stok EP,
et al: Interrogation of transcriptomic changes associated with
drug-induced hepatic sinusoidal dilatation in colorectal cancer.
PLoS One. 13:e1980992018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu SC, Mato JM, Espinosa-Diez C and Lamas
S: MicroRNA-mediated regulation of glutathione and methionine
metabolism and its relevance for liver disease. Free Radic Biol
Med. 100:66–72. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tabassum H, Waseem M, Parvez S and Qureshi
MI: Oxaliplatin-induced oxidative stress provokes toxicity in
isolated rat liver mitochondria. Arch Med Res. 46:597–603. 2015.
View Article : Google Scholar : PubMed/NCBI
|