Introduction
In cancer biology, the transition of a cell from a
normal state to a neoplastic one is a process through which the
cell must overcome the anti-oncogenic checkpoints. Based on these
checkpoints, a list of six hallmarks of cancer has been created.
These hallmarks include the possibility of unlimited cell
proliferation, prolonged angiogenesis, resistance to cell death,
the possibility of invasion and metastasis, evading growth
inhibitors and self-sufficiency in growth signals (1). In recent years, two other hallmarks
of cancer have been added to the aforementioned list, which include
the deregulation of metabolism, a process that plays a key role in
cellular stress responses, and the avoidance of the immune system
(2).
Overcoming environmental pressures, such as hypoxia,
nutrient depletion and DNA-damaging factors is one of the key
abilities of cancer cells. Cellular stress is an environmental
factor that affects the growth and development of cancer, and
includes oxidative stress induced by reactive oxygen species (ROS),
metabolic stress due to increased metabolic procedures and
genotoxic stress, including DNA damage. In general, cellular stress
activates the process of cell death. However, cancer cells are able
to resist cellular stress by altering their gene expression and
metabolic pathways, and avoiding growth inhibition signals
(3). Key factors in these altered
mechanisms are non-coding RNAs (ncRNAs).
As it is known, the coding regions of the human
genome constitute only 1–2% of the whole genome, while the
remaining ~98% consists of non-coding regions (4). For a number of years, these
non-coding parts of the genome were considered noise and were
termed ‘junk DNA’. However, it has been shown that the majority of
these non-coding regions are transcribed into RNA molecules, ncRNAs
(5). According to the literature,
these molecules are involved in various cell functions and are
involved in numerous diseases, including cancer (6). ncRNAs are divided into two broad
categories, the small ones, which consist of <200 nucleotides,
and the long ones, which consist of >200 nucleotides. The first
category includes microRNAs (miRNAs/miRs), short interference RNAs
(siRNAs), Piwi-interacting (piRNAs) and small nucleolar RNAs
(snoRNAs), all of which participate in either the positive or
negative regulation of gene expression through the epigenetic and
post-transcriptional regulation of target mRNAs (7,8).
The second category includes long ncRNAs (lncRNAs) that promote and
inhibit gene expression through a variety of mechanisms (9). From the aforementioned categories,
miRNAs and lncRNAs have been identified mainly as critical
regulators of the cellular stress response, and are thus involved
in the maintenance of human cancer (10). From this point of view, the
present review summarizes the current evidence on the
cancer-specific role and functions of these two types of ncRNAs in
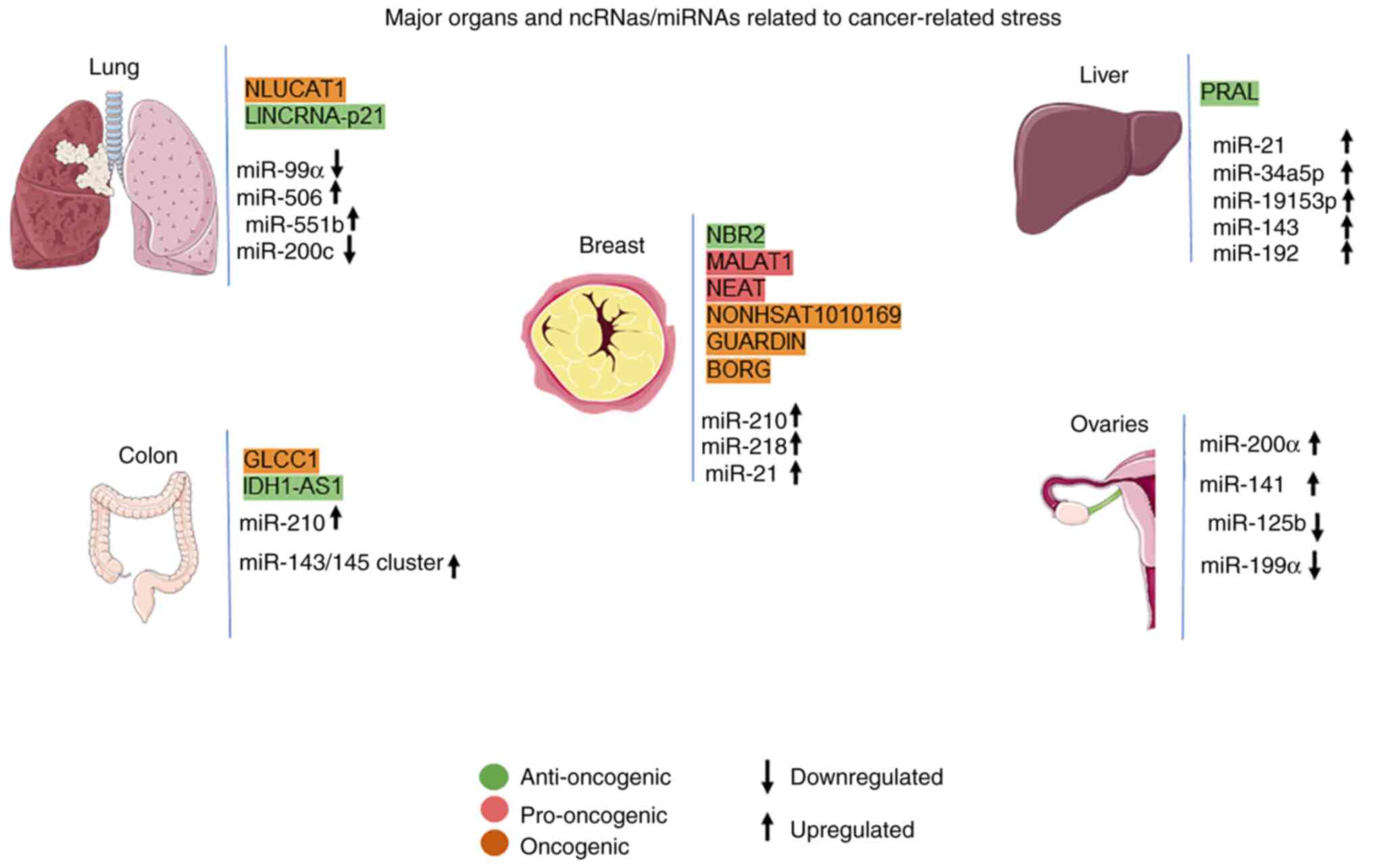
cellular stress (Fig. 1).
Role of lncRNAs in cancer-related
stress
lncRNAs can be classified according to their
location in the genome, their biogenesis, their structure, their
protein-binding pattern (k-mers) and their mechanisms of action.
lncRNAs function as epigenetic regulators, promoting or inhibiting
the transcription, splicing, translation and the modulation of
protein function (11). In
addition, lncRNAs can function as oncogenes or tumor suppressors
through a mechanism wherein a gene encoding a lncRNA has the
ability to either directly promote or inhibit oncogenesis,
respectively. According to the literature, lncRNAs are involved in
the regulation of cancer cell stress, which includes oxidative,
metabolic and genotoxic stress, as they participate in a variety of
cancer-related signaling pathways (12) (Table
I).
 | Table I.lncRNAs in cancer-related stress. |
Table I.
lncRNAs in cancer-related stress.
| lncRNA | Cancer type | Stress | Function | (Refs.) |
|---|
| MACC1-AS1 | Gastric | Metabolic | Pro-oncogenic | (16) |
| GLCC1 | Colorectal | Metabolic | Pro-oncogenic | (17) |
| SAMMSON | Melanoma | Metabolic | Pro-oncogenic | (18) |
| FILNC1 | Renal | Metabolic | Anti-oncogenic | (19) |
| IDH1-AS1 | Colon/Cervical | Metabolic | Anti-oncogenic | (20) |
| NBR2 | Breast | Metabolic | Anti-oncogenic | (21) |
| HAND2-AS1 | Osteosarcoma | Metabolic | Anti-oncogenic | (22) |
| H19 |
Cholangiocarcinoma/pituitary, breast |
Metabolic/oxidative |
Pro/anti-oncogenic | (23,24) |
| HULC |
Cholangiocarcinoma | Oxidative | Pro-oncogenic | (23) |
| NLUCAT1 | Lung
adenocarcinoma | Oxidative | Pro-oncogenic | (25) |
| MIAT | Neuroblastoma and
glioblastoma | Oxidative | Oncogenic | (26) |
| MALAT1 | Breast | Oxidative | Oncogenic | (27) |
| NEAT | Breast | Oxidative | Oncogenic | (27) |
| NONHSAT1010169 | Breast | Genotoxic | Pro-oncogenic | (31) |
| GUARDIN | Breast | Genotoxic | Pro-oncogenic | (32) |
| NEAT1 | Multiple
myeloma | Genotoxic | Pro-oncogenic | (33) |
| BORG | Breast | Genotoxic | Pro-oncogenic | (34) |
| PRAL | Hepatocellular
carcinoma | Genotoxic | Anti-oncogenic | (36) |
| LOC572558 | Bladder | Genotoxic | Anti-oncogenic | (36) |
| LincRNA-p21 |
Lung/sarcoma/lymphoma | Genotoxic | Anti-oncogenic | (37) |
| PANDA | Bone | Genotoxic | Anti-oncogenic | (38) |
lncRNAs in cancer-related metabolic
stress
A well-recognized characteristic of cancer is
metabolic reprogramming. This hallmark of cancer cells was
recognized by Otto Warburg in the 1920s, who observed that cancer
cells exhibit higher rates of glucose uptake and lactic acid
secretion, even in the presence of oxygen, compared to normal
cells. These characteristics indicate that cancer cells use aerobic
glycolysis for energy production, which is termed as the ‘Warburg
effect’ (13). The result of this
abnormal metabolic pathway is, on the one hand, the production of
high energy levels which are required for the rapid proliferation
of cancer cells, while on the other hand, an increase in stress.
Cancer cells overexpress key proteins of energy production and
metabolite transport pathways in order to address this increased
stress, such as 5′AMP-activated protein kinase (AMPK), PKM2 and
MYC, while downregulating metabolic suppressors, such as p53
(14). lncRNAs are key factors in
the treatment of metabolic stress and in the metabolic
reprogramming of cancer cells (15).
As previously mentioned, lncRNAs can function as
oncogenes and may aid in the treatment of metabolic stress of
cancer cells. One such lncRNA is MACC1-AS1, which has been detected
in gastric cancer cells and induces the stabilization of MACC1 RNA
and increases the post-transcriptional expression of MACC1
(16). Subsequently, MACC1,
through the AMPL/Lin28 pathway, contributes to metabolic plasticity
by maintaining the expression of the metabolic enzymes, GLUT1, HK2
and LDH during glucose deprivation (16). In this context, lncRNA GLCC1
appears to have a similar function to MACC1-AS1 in colorectal
cancer cells where it is expressed during glucose starvation. More
extensively, this lncRNA stabilizes the oncogenic transcription
factor c-MYC through its direct binding to HSP90, promoting cell
survival at high glycolysis and lactation rates (17). In addition, lncRNA SAMMSON is also
related to metabolic stress resistance in cancer since it promotes
mitochondrial stability in melanoma (18). The aforementioned process is
carried out by isolating a key regulator of mitochondrial
homeostasis and metabolism, p32, to the cytoplasm, promoting cell
proliferation (18).
On the other hand, as previously mentioned, lncRNAs
may also have tumor suppressor functions. One such lncRNA is
FILNC1, the expression of which is significantly reduced in renal
cancer cells. The function of FILNC1 involves binding to a c-MYC
mRNA binding protein, AUF1, leading to the inhibition of c-MYC
protein and metabolic plasticity (19). Another lncRNA with a similar
activity is IDH1-AS1. The expression of this lncRNA induces a
decrease in the proliferation of cancer cells of the colon and
cervix, while it is inhibited via transcription by c-MYC, thus
promoting the ‘Warburg effect’ (20). Apart from the regulation of the
expression of c-MYC protein and its functions by these two lncRNAs,
the function of AMPK is regulated by lncRNA NBR2 (21). This lncRNA is reported in breast
cancer cells and is induced under energy stress by the LKB1-AMPK
pathway. It normally functions as a sensor of cellular energy, thus
maintaining the control of metabolic pathways in the cell. However,
the reduced expression of this lncRNA leads to increased cell
proliferation, decreased apoptosis and the maintenance of cell
function under conditions of high energy stress (21). Another tumor suppressor that
promotes apoptosis due to metabolic stress is lncRNA HAND2-AS1
(22). It has been found in
osteosarcoma, and its normal expression leads to the inhibition of
glucose uptake and lactate production, as well as the expression of
metabolic enzymes through the isolation of an inhibitory enzyme of
the HIF1α metabolic gene, FBP1. Nevertheless, the inhibition
of this lncRNA has been shown to result in a reduction of apoptosis
induced in cases of metabolic stress, thus promoting the survival
of cancer cells (22). Finally,
in addition to the two categories mentioned above, there are also
lncRNAs that have a dual function, sometimes acting as oncogenes
and sometimes as tumor suppressors. One such lncRNA is H19, which,
while under conditions of oxidative stress, promotes the growth of
cholangiocarcinoma cells (23),
whereas in the case of pituitary tumors, it acts as a suppressor by
inhibiting the ability of cells to respond to metabolic stress
(24).
lncRNAs in cancer-related oxidative
stress
Apart from metabolic reprogramming, another
characteristic of cancer cells is their hypoxic microenvironment,
where high levels of reactive oxygen species (ROS), which are
by-products of aerobic metabolism in the cell and are produced by
mitochondria, are required for increased proliferation and
metabolism. According to previous studies, lncRNAs play a crucial
role in the ability of cancer cells to respond to oxidative stress.
Two of these lncRNAs are H19 and HULC, which are upregulated in
cholangiocarcinoma cells under conditions of oxidative stress. They
regulate the expression of cytokine IL-6, which, through in
vitro assays, has been shown to promote metastasis and cell
invasion by sponging and regulating let-7a, let-7b and
miR-372/miR-373 (23). Another
lncRNA with a similar function is NLUCAT1, which has been detected
in lung adenocarcinoma. This lncRNA upregulates the expression of
the oxidative homeostasis genes, ALDH3A1, GPX2, GLRX and
PDK4, increasing the resistance of cancer cells to
ROS-induced apoptosis (25).
Myocardial infarction-associated transcript (MIAT)
is a subnuclear lncRNA that interferes with alternative splicing
and is associated with an increased risk of various heart
conditions and nervous system tumors. In the study by Bountali
et al (26), this lncRNA
was found to be involved in the regulation of oxidative stress and
its downstream effects in neuroblastoma and glioblastoma cell
lines. In this regard, various other lncRNAs have been found to be
implicated in oxidative stress and consequent hypoxia, including
nuclear enrichment abundant transcript 1 (NEAT1), lincRNA-p21,
urothelial cancer associated 1 (UCA1) and metastasis-associated
lung adenocarcinoma transcript 1 (MALAT1) (27).
lncRNAs in cancer-related genotoxic
stress
Genotoxic stress results from damage to DNA
structure and genome instability due to the deregulation of key
regulatory pathways in cancer cells (28). Cancer cells are capable of
resisting cell death induced by genotoxic stress through a variety
of mechanisms, including the inhibition of tumor suppression genes,
the upregulation of cell growth factors and the omission of cell
cycle control points. Notably, the basis of several chemotherapies
is the induction of genotoxic stress to kill cancer cells through
cell death. However, resistance to genotoxic stress also results in
resistance to drugs and therapies. According to previous studies,
the modified expression of lncRNAs also contributes to this process
(29,30).
One such mode of function of lncRNAs in the response
to genotoxic stress is through the isolation of several miRNAs. One
of these lncRNAs is NONHSAT1010169 which has been studied in breast
cancer cells (31). The
overexpression of this ncRNA leads to resistance to treatment with
the anthracycline, epirubicin, via the isolation of miR-129, which
inhibits the expression of the oncogenic protein Twist1, and
promotes the migration and invasion of breast cancer cells
(31). From this point of view,
lncRNA GUARDIN is another lncRNA that has been studied in breast
cancer with a similar function. This lncRNA responds to p53, and
its mechanism of action involves the isolation of miR-23a where it
leads to the stabilization of TRF2 and functions as an RNA scaffold
for the oncoprotein BRCA1, thus protecting cancer cells from
apoptosis induced by genotoxic stress (32). In addition to those two lncRNAs,
NEAT1 is a lncRNA that promotes genotoxic stress resistance in
multiple myeloma cells through a different pathway. This lncRNA
regulates DNA repair proteins, and its reduction leads to reduced
DNA repair and sensitization of cells to therapies (33).
In several different cases, the direct induction of
lncRNAs by genotoxic stress has also been recorded. lncRNA BORG is
an example of this category, which is found in breast cancer cells
(34); the exposure of these
cells to doxorubicin induces the expression of this lncRNA. This
process is driven by NF-κB. leading to chemical resistance. The
aforementioned mechanism of expression of lncRNA BORG underlines
the rapid and immediate synthesis of lncRNAs, rendering them the
ideal study tools in cases of cellular stress (34).
By contrast, the inhibition of cancer cell
resistance to genotoxic stress can be induced equally by lncRNAs.
The major mechanism promoting apoptosis and the DNA-damage response
pathway is the tumor suppression gene TP53. The p53 protein,
resulting from the expression of this gene, acts as a regulator of
genes involved in repairing DNA damage and apoptosis (35). One p53-associated lncRNA is PRAL
(36). This lncRNA has been
identified in hepatocellular carcinoma, where it induces p53
apoptosis in both in vitro and in vivo assays. lncRNA
LOC572558 belongs to the same category. The overexpression of this
ncRNA in bladder cancer cells enhances p53 phosphorylation, thus
enhancing p53 signaling and inhibiting cancer cell proliferation
(36). LincRNA-p21 is another
activator of p53 in DNA damage. Its mechanism of action involves
the uptake of hnRNP-K to increase the p53-dependent transcription
of p21, which is a control protein of the p53 pathway (37). Finally, lncRNA PANDA is another
ncRNA that stabilizes the p53 protein and protects it from
proteasome degradation, although its mechanism of action is still
unknown (38).
From the information presented above, the key role
of lncRNAs in cancer cell resistance to genotoxic stress responses
is highlighted, as well as their critical roles as biomarkers and
drug targets in cancer treatment (39).
Role of microRNAs in cancer-related
stress
The role of miRNAs has been elucidated and studied
mainly in oxidative stress conditions in cancer cells. Studies have
demonstrated that the expression of miRNAs can be affected under
the influence of stressful conditions, such as hypoxia, through
mechanisms involving a change in the function or expression of
enzymes involved in the biogenesis of miRNAs (40,41). An example is the inhibition of
DROSHA and DICER1 expression in cancer cells under hypoxic
conditions, leading to the incomplete biogenesis of miRNAs
(42).
As previously mentioned, as the microenvironment of
cancer cells is hypoxic, increased ROS production is observed,
inhibiting antioxidant activity in an uncontrolled state. ROS is
generally considered a carcinogenic factor that promotes
carcinogenesis (43). There is a
significant increase in ROS levels due to the accumulation of
oxidative stress, thus activating the oncogenic signaling pathway,
mutagenesis and genomic instability in cancer cells, promoting
cancer progression (44). In this
section, the miRNAs associated with oxidative stress in different
types of cancer are summarized (Table II).
 | Table II.Condition and functions of oxidative
stress-related microRNAs in different types of cancer. |
Table II.
Condition and functions of oxidative
stress-related microRNAs in different types of cancer.
| miRNA | Cancer type | Condition in
cancer | Function | (Refs.) |
|---|
| miR-210 | Breast | Upregulated | Oncogenic | (47) |
| miR-28 | Breast | Upregulated | Oncogenic | (48) |
| miR-21 | Breast | Upregulated | Oncogenic | (49) |
| miR-143/miR-145
cluster | Colon | Downregulated | Oncogenic | (51) |
| miR-210 | Colon | Upregulated | Oncogenic | (54) |
| miR-34a-5p | Hepatocellular | Upregulated | Oncogenic | (55,56) |
| miR-1915-3p | Hepatocellular | Upregulated | Oncogenic | (55) |
| miR-143 | Hepatocellular | Upregulated | Oncogenic | (58) |
| miR-21 | Hepatocellular | Upregulated | Oncogenic | (60) |
| miR-92 | Hepatocellular | Upregulated | Oncogenic | (61) |
| miR-99a | Lung | Downregulated | Oncogenic | (62) |
| miR-506 | Lung | Upregulated | Oncogenic | (64) |
| miR-551b | Lung | Upregulated | Oncogenic | (65) |
| miR-200c | Lung | Downregulated | Oncogenic | (66) |
| miR-200a | Ovarian | Upregulated | Oncogenic | (69) |
| miR-141 | Ovarian | Upregulated | Oncogenic | (69) |
| miR-125b | Ovarian | Downregulated | Anti-oncogenic | (72) |
| miR-199a | Ovarian | Downregulated | Anti-oncogenic | (72) |
| miR-193a-5p | Prostate | Upregulated | Oncogenic | (73) |
| miR-21 | Prostate | Upregulated | Oncogenic | (74) |
| miR-137 | Prostate | Downregulated | Anti-oncogenic | (76) |
| miR-96 | Prostate | Downregulated | Oncogenic | (79) |
| miR-494 | Pancreatic | Downregulated | Anti-oncogenic | (80) |
| miR-155 | Pancreatic | Upregulated | Oncogenic | (82) |
| miR-29c | Pancreatic | Downregulated | Anti-oncogenic | (83) |
Breast cancer
Several miRNAs associated with oxidative stress have
been identified in breast cancer. One of these is miR-500a-5p, the
expression of which is induced by H2O2
treatment, and it leads to the targeting of oxidative stress
response genes (45). In
addition, the induction of oxidative stress, DNA damage and
apoptosis have been observed with the simultaneous induction of
miR-139-5p and radiation both in vitro and in vivo
(46). miR-210 is another ncRNA
that can increase ROS production and mitochondrial metabolism
levels by targeting cytochrome c oxidase assembly protein
(COX10) and transferrin receptor 1, thereby increasing
carcinogenesis (47).
Another mechanism through which miRNAs affect
intracellular ROS levels is through the targeting of antioxidant
defense factors. One such factor is Nrf2, which causes an increase
in the transcription of catalase and dismutase peroxide. This
factor has been shown to inhibit miR-28, a miRNA that increases
cell proliferation. The result of this inhibition is increased
levels of intracellular ROS and increased oncogenesis. In addition,
the increased expression of oncomiRs has been observed due to
increased ROS levels in cancer cells (48). In addition, one such miRNA is
miR-21. Its expression is affected by the factor NF-κB, which in
conjunction with STAT3, induces miR-21 expression, thereby
inhibiting the expression of PTEN and PDCD4. The aforementioned
pathway results in the escape of cancer cells from apoptosis and
increased metastasis in breast cancer (49).
Colorectal cancer
As with breast cancer, miRNAs associated with
oxidative stress have been found in cases of colon cancer. One of
these is miR-1915-3p, which targets genes that affect oxidative
stress and is involved in chemotherapy-induced DNA damage, which is
achieved by targeting Bcl-2 (50). The miR-143/145 complex comprises
two more miRNAs that are downregulated in solid tumors. However,
the overexpression of these ncRNAs induces apoptosis and reduces
the proliferation of cancer cells, while rendering the cells
sensitive to chemotherapy (51).
In addition, due to the increased regulation of miR-143, there is
an increased activation of ROS production, which indicates
oxidative stress, leading to the sensitization of cancer cells to
oxaliplatin (52).
In another study, the activation of ROS production
and the induction of aging by four miRNAs, miR-186, miR-216b,
miR-37-3p and miR-760, which target protein kinase 2, were
identified, leading to the inhibition of oncogenesis (53). Another miRNA is miR-210, which
increases ROS production and inhibits the iron-sulfur cluster
scaffold homolog and COX10 genes, which are part of the electron
transport chain (54).
Hepatocellular carcinoma
In the case of hepatocellular carcinoma, an increase
in four miRNAs under conditions of oxidative stress has been
identified, specifically miR-34a-5p, miR-1915-3p, miR-638 and
miR-150-3p. The expression of the first two is dependent on p53,
while the expression of the latter two is independent (55). The aforementioned miRNAs
negatively control the oxidative stress pathway where more
specifically, miR-34a-5p promotes ROS production by inhibiting
mitochondrial antioxidant enzymes (56), and miR-638, which is located in
the Dnm2 gene, induces oxidative stress (57).
As oxidative stress and ROS production frequently
occur in inflammatory conditions by activating NF-κB, the
regulation of the expression of a number of miRNAs is affected by
this signaling pathway. miR-143 is one such miRNA in hepatocellular
carcinoma, the expression of which is increased by NF-κB, and its
mechanism of function involves targeting the fibronectin type III
domain-containing 3B (FNDC3B), thus promoting metastasis (58). miR-224 is another miRNA the
expression of which is NF-κB-dependent. In this case, this factor
also causes an increase in the expression of this miRNA, which is
equally associated with metastasis and cell invasion (59). Another miRNA that promotes
metastasis, proliferation and invasion in liver cancer is miR-21.
Its expression is increased by ROS and its upregulation leads to a
significant reduction in PTEN expression (60). Finally, the increased expression
of miR-92 is responsible for the development of chronic liver
disease into hepatocellular carcinoma (61). ROS and oxidative stress, in
combination with the induction of telomerase activity, lead to DNA
damage. It has also been found that miR-92 inhibits apoptosis in
hepatocellular carcinoma by targeting the Bad and Bax
genes (61).
Lung cancer
In the case of lung cancer, the decreased expression
of miR-99a is associated with a poor prognosis, as it is associated
with metastasis and increased cell proliferation. By contrast, the
increased expression of this miRNA leads to the targeting of NADPH
oxidase 4 (NOX4), which causes an increase in ROS levels, leading
to metastasis and cell proliferation (62). Thus, the increased expression of
miR-99a leads to a significant reduction in ROS levels. Otherwise,
the inhibition of this miRNA and ROS production lead to the
activation of the PI3K/Akt pathway, and the regulation of MMP2 and
MMP9, which are the basic proteases of cell migration in this type
of cancer (63). In contrast to
miR-99a, miR-506 expression is increased in lung cancer. This miRNA
inhibits NF-κB p65 expression, thus increasing ROS production
(64).
In general, miRNAs, which inhibit enzymes involved
in oxidative stress and ROS production, are responsible for
carcinogenesis. One such miRNA is miR-551b, which is upregulated in
apoptosis-resistant cancer cells (65). This increase causes an
accumulation of ROS, which leads to the activation of the mucin-1
oncogene, resulting in increased survival and the resistance of
lung cancer cells to drugs (65).
miR-200c is another ncRNA whose expression can affect the level of
intracellular ROS. More specifically, it functions as a regulator
of the oxidative stress response genes by downregulating their
expression, thus increasing the levels of ROS and p21 (66). Finally, the increased expression
of miR-200c has been shown to predispose cancer cells to
radiotherapy, thus setting an example of the potential of using
miRNAs as an effective strategy in the treatment of lung cancer
(66).
Ovarian cancer
In this type of cancer, one of the miRNAs studied is
miR-29b. This miRNA functions as a tumor suppressor, as it is
associated with apoptosis and the inhibition of cancer cell
viability. Its mode of action involves targeting the SIRT1
gene, which is involved in oxidative stress, cell survival and
differentiation. This increased ROS production inhibits miR-29b
expression (67,68). miR-200a and miR-141 are two other
miRNAs that target the P38α gene, which is a
stress-activated kinase, thus modulating oxidative stress responses
(69). The result of the
expression of miR-200a and miR-141 is the inhibition of this
kinase, resulting in the increase of the tumor in animal models,
but also in the increase of the response to chemotherapy (70). Another miRNA whose expression
increases in response to increased ROS production is miR-182. The
expression of this miRNA occurs due to the increase in β-catenin
levels induced by ROS. The upregulation of miR-182 in this type of
cancer leads to increased p53-mediated expression of p21, although
miR-182 may function as an oncomiR and increase oncogenicity in the
case of a mutation in p53 (71).
Finally, miR-125b and miR-199a are two miRNAs that are inhibited by
ROS in ovarian cancer. Their increased expression leads to the
inhibition of tumor-induced angiogenesis (72).
Prostate cancer
In cell lines of this type of cancer, such as LNCap,
PC3 and DU145, the increased expression of miR-193a-5p has been
observed. miR-193a-5p induces cancer progression by inhibiting the
BACH2 gene and increasing heme oxygenase-1 expression, which
leads to the resistance of cancer cells to apoptosis. By contrast,
the inhibition of this miRNA leads to increased sensitivity to
chemotherapy (73). miR-21 is
another miRNA in prostate cancer, the expression of which is
regulated by the NADPH oxidase enzyme, which is the main source of
ROS. Increased ROS production leads to an increase in miR-21
expression via the Akt pathway (74). In contrast to the aforementioned
miRNAs, miR-137 expression is decreased in prostate cancer
(75). The expression of this
ncRNA leads to the inhibition of oncogenesis by targeting NOX4,
thereby inhibiting glycolysis and cancer cell proliferation
(76).
In addition, according to a previous study, an
increase in the expression of miR-708-5p has been observed by the
effects of metformin (77),
inducing the apoptosis of cancer cells, thus being a possible
therapeutic mechanism in the treatment of prostate cancer (78). Finally, the dual function of
miR-96 has been observed in hypoxic conditions in prostate cancer
cells. More specifically, its increased expression inhibits mTOR
protein expression, inducing autophagy; however, the overexpression
of this miRNA leads to the inhibition of ATG4 and consequently, to
the inhibition of autophagy (79). In general, autophagy is a
condition that has been found to reduce the apoptosis of cancer
cells, contributing to their survival under conditions of hypoxic
stress (80).
Pancreatic cancer
c-MYC and SIRT1 have been identified as regulators
of oxidative stress in pancreatic cancer and are targets of
miR-494. The increase in the expression of this miRNA inhibits cell
proliferation and promotes apoptosis (81). On the other hand, miR-155 has been
found to significantly increase ROS levels in cancer cells by
downregulating the basic enzymes in the defense mechanism of
oxidative stress, namely catalase and superoxide dismutase 2
(82). Finally, the decreased
expression of miR-29c in cancer cells leads to an enhanced invasive
capacity. This miRNA targets MMP9, thus leading to the suppression
of migration and invasion (83).
Conclusion
In conclusion, cancer cells have the ability to
resist the mechanisms of cellular stress, resulting in tumor
progression and resistance to treatment. These mechanisms involve a
variety of ncRNAs that sometimes function beneficially, while in
other instances, they inhibit tumor progression, with the two main
categories being lncRNAs and miRNAs. Further research is therefore
required to clarify the roles of these molecules in cancer-stress
responses in order to provide important additional information
about their functions. This may aid in the utilization of these
non-coding molecules in therapeutic strategies against various
types of cancer.
Acknowledgements
Not applicable.
Funding
The authors would like to acknowledge funding from the following
organizations: i) AdjustEBOVGP-Dx (RIA2018EF-2081): Biochemical
Adjustments of native EBOV Glycoprotein in Patient Sample to Unmask
target Epitopes for Rapid Diagnostic Testing. A European and
Developing Countries Clinical Trials Partnership (EDCTP2) under the
Horizon 2020 ‘Research and Innovation Actions’ DESCA; and ii)
‘MilkSafe: A novel pipeline to enrich formula milk using omics
technologies’, a research co-financed by the European Regional
Development Fund of the European Union and Greek national funds
through the Operational Program Competitiveness, Entrepreneurship
and Innovation, under the call RESEARCH-CREATE-INNOVATE (project
code: T2EDK-02222).
Availability of data and materials
Not applicable.
Authors' contributions
KP, EP, LP, ID, TM, KD, DAS, FB, GPC, GNG, EE and DV
contributed to the conceptualization, design, writing, drafting,
revising, editing and reviewing of the manuscript. All authors have
read and approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
References
|
1
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Connerty P, Lock RB and de Bock CE: Long
Non-coding RNAs: Major regulators of cell stress in cancer. Front
Oncol. 10:285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium, . Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Svoboda P: Renaissance of mammalian
endogenous RNAi. FEBS Lett. 588:2550–2556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roufayel R and Kadry S: MicroRNAs: Crucial
regulators of stress. Microrna. 9:93–100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dahariya S, Paddibhatla I, Kumar S,
Raghuwanshi S, Pallepati A and Gutti RK: Long non-coding RNA:
Classification, biogenesis and functions in blood cells. Mol
Immunol. 112:82–92. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu PF, Zheng X, Fan X and Lin AF: Role of
cytoplasmic lncRNAs in regulating cancer signaling pathways. J
Zhejiang Univ Sci B. 20:1–8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warburg O: The metabolism of carcinoma
cells. J Cancer Res. 9:148–163. 1925. View Article : Google Scholar
|
|
14
|
Papakonstantinou E, Vlachakis D, Thireou
T, Vlachoyiannopoulos PG and Eliopoulos E: A Holistic Evolutionary
and 3D pharmacophore modelling study provides insights into the
metabolism, function, and substrate selectivity of the human
monocarboxylate transporter 4 (hMCT4). Int J Mol Sci. 22:29182021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia D, Lu M, Jung KH, Park JH, Yu L,
Onuchic JN, Kaipparettu BA and Levine H: Elucidating cancer
metabolic plasticity by coupling gene regulation with metabolic
pathways. Proc Natl Acad Sci USA. 116:3909–3918. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: LncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Leucci E, Vendramin R, Spinazzi M,
Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli
C, Vanderheyden K, et al: Melanoma addiction to the long non-coding
RNA SAMMSON. Nature. 531:518–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y,
Baddour J, Nagrath D, Wood CG, Gu J, Wu X, et al: Energy
stress-induced lncRNA FILNC1 represses c-Myc-mediated energy
metabolism and inhibits renal tumor development. Nat Commun.
8:7832017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiang S, Gu H, Jin L, Thorne RF, Zhang XD
and Wu M: LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α
via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci USA.
115:E1465–E1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Xiao ZD, Han L, Zhang J, Lee SW,
Wang W, Lee H, Zhuang L, Chen J, Lin HK, et al: LncRNA NBR2 engages
a metabolic checkpoint by regulating AMPK under energy stress. Nat
Cell Biol. 18:431–442. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang Y, Zhu X, Xu Y, Tang Q, Huang Z, Zhao
Z, Lu J, Song G, Xu H, Deng C and Wang J: Energy stress-induced
lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and
inhibits osteosarcoma progression. Am J Cancer Res. 8:526–537.
2018.PubMed/NCBI
|
|
23
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu ZR, Yan L, Liu YT, Cao L, Guo YH, Zhang
Y, Yao H, Cai L, Shang HB, Rui WW, et al: Inhibition of mTORC1 by
lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary
tumours. Nat Commun. 9:46242018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moreno Leon L, Gautier M, Allan R, Ilié M,
Nottet N, Pons N, Paquet A, Lebrigand K, Truchi M, Fassy J, et al:
The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA
contributes to an aggressive phenotype in lung adenocarcinoma
through regulation of oxidative stress. Oncogene. 38:7146–7165.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bountali A, Tonge DP and
Mourtada-Maarabouni M: RNA sequencing reveals a key role for the
long non-coding RNA MIAT in regulating neuroblastoma and
glioblastoma cell fate. Int J Biol Macromol. 130:878–891. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Choudhry H, Schödel J, Oikonomopoulos S,
Camps C, Grampp S, Harris AL, Ratcliffe PJ, Ragoussis J and Mole
DR: Extensive regulation of the non-coding transcriptome by
hypoxia: Role of HIF in releasing paused RNApol2. EMBO Rep.
15:70–76. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Campisi J and d'Adda di Fagagna F:
Cellular senescence: When bad things happen to good cells. Nat Rev
Mol Cell Biol. 8:729–740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji X, Lu Y, Tian H, Meng X, Wei M and Cho
WC: Chemoresistance mechanisms of breast cancer and their
countermeasures. Biomed Pharmacother. 114:1088002019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Velegzhaninov IO, Ievlev VA, Pylina YI,
Shadrin DM and Vakhrusheva OM: Programming of cell resistance to
genotoxic and oxidative stress. Biomedicines. 6:52018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y,
Xia T and Wang S: Long non-coding RNA NONHSAT101069 promotes
epirubicin resistance, migration, and invasion of breast cancer
cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene.
38:7216–7233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu WL, Jin L, Xu A, Wang YF, Thorne RF,
Zhang XD and Wu M: GUARDIN is a p53-responsive long non-coding RNA
that is essential for genomic stability. Nat Cell Biol. 20:492–502.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Taiana E, Favasuli V, Ronchetti D,
Todoerti K, Pelizzoni F, Manzoni M, Barbieri M, Fabris S,
Silvestris I, Gallo Cantafio ME, et al: Long non-coding RNA NEAT1
targeting impairs the DNA repair machinery and triggers anti-tumor
activity in multiple myeloma. Leukemia. 34:234–244. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gooding AJ, Zhang B, Gunawardane L, Beard
A, Valadkhan S and Schiemann WP: The lncRNA BORG facilitates the
survival and chemoresistance of triple-negative breast cancers.
Oncogene. 38:2020–2041. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hafner A, Bulyk ML, Jambhekar A and Lahav
G: The multiple mechanisms that regulate p53 activity and cell
fate. Nat Rev Mol Cell Biol. 20:199–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F,
Yuan JH, Bi FR, Lin KY, Yin JH, Cao GW, et al: Systemic genome
screening identifies the outcome associated focal loss of long
noncoding RNA PRAL in hepatocellular carcinoma. Hepatology.
63:850–863. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kotake Y, Kitagawa K, Ohhata T, Sakai S,
Uchida C, Niida H, Naemura M and Kitagawa M: Long Non-coding RNA,
PANDA, contributes to the stabilization of p53 tumor suppressor
protein. Anticancer Res. 36:1605–1611. 2016.PubMed/NCBI
|
|
39
|
Jiang MC, Ni JJ, Cui WY, Wang BY and Zhuo
W: Emerging roles of lncRNA in cancer and therapeutic
opportunities. Am J Cancer Res. 9:1354–1366. 2019.PubMed/NCBI
|
|
40
|
Wang Z, Liu Y, Han N, Chen X, Yu W, Zhang
W and Zou F: Profiles of oxidative stress-related microRNA and mRNA
expression in auditory cells. Brain Res. 1346:14–25. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen J, Xia W, Khotskaya YB, Huo L,
Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al: EGFR
modulates microRNA maturation in response to hypoxia through
phosphorylation of AGO2. Nature. 497:383–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rupaimoole R, Wu SY, Pradeep S, Ivan C,
Pecot CV, Gharpure KM, Nagaraja AS, Armaiz-Pena GN, McGuire M, Zand
B, et al: Hypoxia-mediated downregulation of miRNA biogenesis
promotes tumour progression. Nat Commun. 5:52022014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saha SK, Lee SB, Won J, Choi HY, Kim K,
Yang GM, Dayem AA and Cho SG: Correlation between oxidative stress,
nutrition, and cancer initiation. Int J Mol Sci. 18:15442017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Degli Esposti D, Aushev VN, Lee E, Cros
MP, Zhu J, Herceg Z, Chen J and Hernandez-Vargas H: miR-500a-5p
regulates oxidative stress response genes in breast cancer and
predicts cancer survival. Sci Rep. 7:159662017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pajic M, Froio D, Daly S, Doculara L,
Millar E, Graham PH, Drury A, Steinmann A, de Bock CE,
Boulghourjian A, et al: miR-139-5p modulates radiotherapy
resistance in breast cancer by repressing multiple gene networks of
DNA repair and ROS defense. Cancer Res. 78:501–515. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Devlin C, Greco S, Martelli F and Ivan M:
miR-210: More than a silent player in hypoxia. IUBMB Life.
63:94–100. 2011.PubMed/NCBI
|
|
48
|
Yang M, Yao Y, Eades G, Zhang Y and Zhou
Q: MiR-28 regulates Nrf2 expression through a Keap1-independent
mechanism. Breast Cancer Res Treat. 129:983–991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Niu J, Shi Y, Tan G, Yang CH, Fan M,
Pfeffer LM and Wu ZH: DNA damage induces NF-κB-dependent
microRNA-21 up-regulation and promotes breast cancer cell invasion.
J Biol Chem. 287:21783–21795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakazawa K, Dashzeveg N and Yoshida K:
Tumor suppressor p53 induces miR-1915 processing to inhibit Bcl-2
in the apoptotic response to DNA damage. FEBS J. 281:2937–2944.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Das AV and Pillai RM: Implications of miR
cluster 143/145 as universal anti-oncomiRs and their dysregulation
during tumorigenesis. Cancer Cell Int. 15:922015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gomes SE, Pereira DM, Roma-Rodrigues C,
Fernandes AR, Borralho PM and Rodrigues CMP: Convergence of miR-143
overexpression, oxidative stress and cell death in HCT116 human
colon cancer cells. PLoS One. 13:e0191607. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting α subunit of protein kinase CKII in human
colorectal cancer cells. Biochem Biophys Res Commun. 429:173–179.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen Z, Li Y, Zhang H, Huang P and Luthra
R: Hypoxia-regulated microRNA-210 modulates mitochondrial function
and decreases ISCU and COX10 expression. Oncogene. 29:4362–4368.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wan Y, Cui R, Gu J, Zhang X, Xiang X, Liu
C, Qu K and Lin T: Identification of four oxidative
stress-responsive MicroRNAs, miR-34a-5p, miR-1915-3p, miR-638, and
miR-150-3p, in Hepatocellular carcinoma. Oxid Med Cell Longev.
2017:51891382017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bai XY, Ma Y, Ding R, Fu B, Shi S and Chen
XM: miR-335 and miR-34a Promote renal senescence by suppressing
mitochondrial antioxidative enzymes. J Am Soc Nephrol.
22:1252–1261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li D, Wang Q, Liu C, Duan H, Zeng X, Zhang
B, Li X, Zhao J, Tang S, Li Z, et al: Aberrant expression of
miR-638 contributes to Benzo(a)pyrene-induced human cell
transformation. Toxicol Sci. 125:382–391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Scisciani C, Vossio S, Guerrieri F,
Schinzari V, De Iaco R, D'Onorio de Meo P, Cervello M, Montalto G,
Pollicino T, Raimondo G, et al: Transcriptional regulation of
miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J
Hepatol. 56:855–861. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cardin R, Piciocchi M, Sinigaglia A,
Lavezzo E, Bortolami M, Kotsafti A, Cillo U, Zanus G, Mescoli C,
Rugge M and Farinati F: Oxidative DNA damage correlates with cell
immortalization and mir-92 expression in hepatocellular carcinoma.
BMC Cancer. 12:1772012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun M, Hong S, Li W, Wang P, You J, Zhang
X, Tang F, Wang P and Zhang C: miR-99a regulates ROS-mediated
invasion and migration of lung adenocarcinoma cells by targeting
NOX4. Oncol Rep. 35:2755–2766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee KR, Lee JS, Song JE, Ha SJ and Hong
EK: Inonotus obliquus-derived polysaccharide inhibits the migration
and invasion of human non-small cell lung carcinoma cells via
suppression of MMP-2 and MMP-9. Int J Oncol. 45:2533–2540. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yin M, Ren X, Zhang X, Luo Y, Wang G,
Huang K, Feng S, Bao X, Huang K, He X, et al: Selective killing of
lung cancer cells by miRNA-506 molecule through inhibiting NF-κB
p65 to evoke reactive oxygen species generation and p53 activation.
Oncogene. 34:691–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xu X, Wells A, Padilla MT, Kato K, Kim KC
and Lin Y: A signaling pathway consisting of miR-551b, catalase and
MUC1 contributes to acquired apoptosis resistance and
chemoresistance. Carcinogenesis. 35:2457–2466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cortez MA, Valdecanas D, Zhang X, Zhan Y,
Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al:
Therapeutic delivery of miR-200c enhances radiosensitivity in lung
cancer. Mol Ther. 22:1494–1503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dai F, Zhang Y and Chen Y: Involvement of
miR-29b signaling in the sensitivity to chemotherapy in patients
with ovarian carcinoma. Hum Pathol. 45:1285–1293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hou M, Zuo X, Li C, Zhang Y and Teng Y:
Mir-29b regulates oxidative stress by targeting SIRT1 in ovarian
cancer cells. Cell Physiol Biochem. 43:1767–1776. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kozak J, Wdowiak P, Maciejewski R and
Torres A: Interactions between microRNA-200 family and Sestrin
proteins in endometrial cancer cell lines and their significance to
anoikis. Mol Cell Biochem. 459:21–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu Y, Qiang W, Xu X, Dong R, Karst AM,
Liu Z, Kong B, Drapkin RI and Wei JJ: Role of miR-182 in response
to oxidative stress in the cell fate of human fallopian tube
epithelial cells. Oncotarget. 6:38983–38998. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
He J, Jing Y, Li W, Qian X, Xu Q, Li FS,
Liu LZ, Jiang BH and Jiang Y: Roles and mechanism of miR-199a and
miR-125b in tumor angiogenesis. PLoS One. 8:e566472013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang Z, Chen JS, Wen JK, Gao HT, Zheng B,
Qu CB, Liu KL, Zhang ML, Gu JF, Li JD, et al: Silencing of
miR-193a-5p increases the chemosensitivity of prostate cancer cells
to docetaxel. J Exp Clin Cancer Res. 36:1782017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jajoo S, Mukherjea D, Kaur T, Sheehan KE,
Sheth S, Borse V, Rybak LP and Ramkumar V: Essential role of NADPH
oxidase-dependent reactive oxygen species generation in regulating
microRNA-21 expression and function in prostate cancer. Antioxid
Redox Signal. 19:1863–1876. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nilsson EM, Laursen KB, Whitchurch J,
McWilliam A, Ødum N, Persson JL, Heery DM, Gudas LJ and Mongan NP:
MiR137 is an androgen regulated repressor of an extended network of
transcriptional coregulators. Oncotarget. 6:35710–35725. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lu JP, Monardo L, Bryskin I, Hou ZF,
Trachtenberg J, Wilson BC and Pinthus JH: Androgens induce
oxidative stress and radiation resistance in prostate cancer cells
though NADPH oxidase. Prostate Cancer Prostatic Dis. 13:39–46.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Queiroz EA, Puukila S, Eichler R, Sampaio
SC, Forsyth HL, Lees SJ, Barbosa AM, Dekker RF, Fortes ZB and
Khaper N: Metformin induces apoptosis and cell cycle arrest
mediated by oxidative stress, AMPK and FOXO3a in MCF-7 breast
cancer cells. PLoS One. 9:e982072014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang J, Wei J, Wu Y, Wang Z, Guo Y, Lee P
and Li X: Metformin induces ER stress-dependent apoptosis through
miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis.
4:e1582015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ma Y, Yang HZ, Dong BJ, Zou HB, Zhou Y,
Kong XM and Huang YR: Biphasic regulation of autophagy by miR-96 in
prostate cancer cells under hypoxia. Oncotarget. 5:9169–9182. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mazure NM and Pouysségur J:
Hypoxia-induced autophagy: Cell death or cell survival? Curr Opin
Cell Biol. 22:177–180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu Y, Li X, Zhu S, Zhang JG, Yang M, Qin
Q, Deng SC, Wang B, Tian K, Liu L, et al: Ectopic expression of
miR-494 inhibited the proliferation, invasion and chemoresistance
of pancreatic cancer by regulating SIRT1 and c-Myc. Gene Ther.
22:729–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang P, Zhu CF, Ma MZ, Chen G, Song M,
Zeng ZL, Lu WH, Yang J, Wen S, Chiao PJ, et al: Micro-RNA-155 is
induced by K-Ras oncogenic signal and promotes ROS stress in
pancreatic cancer. Oncotarget. 6:21148–21158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Binker-Cosen MJ, Richards D, Oliver B,
Gaisano HY, Binker MG and Cosen-Binker LI: Palmitic acid increases
invasiveness of pancreatic cancer cells AsPC-1 through
TLR4/ROS/NF-κB/MMP-9 signaling pathway. Biochem Biophys Res Commun.
484:152–158. 2017. View Article : Google Scholar : PubMed/NCBI
|