Introduction
Poor decidualization of endometrial stromal cells is
an important cause for the decreased implantation rate of
endometriosis (1). Implantation
depends on endometrial receptivity and embryo quality (2,3), and
the synchronization of both is needed for the guarantee of a
successful pregnancy. This scrupulous biological process allows the
embryo to complete the orientation, adhesion and penetration into
the basal layer of epidermal cells and intrusion in the endometrial
matrix (4). Decidualization refers
to the differentiation of endometrial stromal cells into decidual
cells with secretory function during the mid-secretory period of
the normal menstrual cycle, which provides the basic conditions for
embryo implantation and placental formation (5,6). It
only occurs at the window of implantation, which is short and
decisive. This usually occurs at days 20–24 of the normal menstrual
cycle (7), 7–10 days after
ovulation, which is equivalent to the mid-secretory phase when the
uterus accepts embryo implantation. Barragan et al (8) have reported that patients with
endometriosis had significant amount of damage to the
decidualization of eutopic endometrial stroma cells following in
vitro induction compared with that of the control group.
Decidual impairment is an important factor in
endometriosis-associated sterility. Infertility resulting in
endometriosis is due to morphological changes in the endometrium,
hormonal disorders (9), immunity
rejection (10) and abnormal state
of gene expression (9,11,12).
Long non-coding RNAs (lncRNAs) are ncRNAs >200
nucleotides in length with extensive biological functions, such as
regulation of growth and development. Similar to microRNAs
(miRNAs/miRs) (13,14), lncRNAs play a role at the
post-transcriptional level, and achieve biological action by
regulating the transcriptional products of their target mRNA
(15). Several studies have
demonstrated that lncRNAs and miRNAs are closely associated with
endometriosis-associated infertility (13,16–18). A
previous study revealed that decreased expression of H19 in
endometrial stromal cells in patients with endometriosis increases
the biological activity of miRNA let-7, while inhibiting the
expression of insulin-like growth factor-I (IGF-I) at the
post-transcriptional level, thus reducing the proliferation of
endometrial stromal cells. This indicated that H19-let-7-IGF-I
signaling is likely to be a key element to the declined endometrial
receptivity of endometriosis (13).
The exact mechanism responsible for infertility in
endometriosis is still unknown, but its severity has endangered
women's physical and mental health, and has influenced their
quality of life, and even created a socio-economic burden. There
are 4 million women suffering from different degrees of infertility
in China (19), and 7 million
worldwide (20), among which the
major reason for infertility is endometriosis (20). A previous study has indicated that
25–50% of infertile patients have endometriosis, and 30–50% of
patients with endometriosis have clinical symptoms of infertility
(21). In order to improve women's
overall fertility status, it is of great significance to explore
the infertility of endometriosis. At present, there are few studies
on the problems associated with the regulation of deficient
decidualization with endometriosis. In particular, to the best of
our knowledge, studies on the post-transcriptional regulation of
decidualization of endometrial stromal cells have not been reported
to date.
Based on our previous results (22), long intergenic non-protein coding
RNA (LINC)01960-201 was selected as the target. According to
bioinformatical analysis, it was demonstrated that hsa-miR-760 and
hsa-miR-608 have a common target gene, ADAMTS7, with LINC01960-201.
It was also confirmed by target point analysis that there is a
target site between hsa-miR-608 and ADAMTS7. In the present study,
clinical samples (endometrial tissues) were collected from five
female patients with infertility due to endometriosis. Endometrial
stroma cells were isolated for primary cell culture, and in
vitro decidualization was induced. The present study aimed to
explore the biological function of LINC01960-201, and the
regulatory association between ADAMTS7, hsa-miR-760 and hsa-miR-608
in the decidualization of endometrial stromal cells with
endometriosis in order to identify novel ways to solve problems
caused by endometriosis, such as pelvic pain and infertility.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committees of Peking Union Medical College Hospital and the Chinese
Academy of Medical Sciences (Beijing, China; approval no. S-k332).
Written informed consent was obtained from the five patients and
all the specimens were acquired with the knowledge of the
patients.
Bioinformatics analysis
The potential binding sites between LINC01960-201
and hsa-miR-760, LINC01960-201 and hsa-miR-608, ADAMTS7 and
hsa-miR-760, ADAMTS7 and hsa-miR-608 were analyzed using miRanda
(Memorial Sloan-Kettering; http://www.microrna.org/microrna/home.do), PITA
(SegalLab; http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html)
and RNAhybrid (Behmsmeier; http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
Vector construction and plasmid
extraction
DNAMAN software 8.0 (Lynnon Biosoft) analyses of the
3′-untranslated region (UTR) sequences and vector (pmiR-RB-REPORT™
dual luciferase reporter vector) of the target genes LINC01960-201
and ADAMTS7 were performed. Primer design was based on the base
pairing principle (Table I), and
293T cell genomic DNA was used as the template. The 3′-UTR
fragments of LINC01960-201 and ADAMTS7 were amplified using PCR.
The PCR products were subjected to 1.5% agarose gel electrophoresis
and then purified. Two restriction enzymes, XhoI and
NotI, were used for digesting the target gene segment and
vector, respectively. The target gene fragment was cloned into the
vector using PCR. The junction products were transformed into DH5α
receptive cells (Tiangen Biotech Co., Ltd.), amplicillin-containing
antibiotic Luria-Bertani (LB) broth (Tinagen Biotech Co., Ltd.) was
added (antibiotic concentration, 100 µg/ml), and five colonies were
selected for PCR. The products were examined using 1.5% agarose
electrophoresis, and the positive clones were further identified by
sanger sequencing (23). Next, the
colonies were expanded and cultured to extract the plasmid. The
sequencing results of the recombinant plasmid were evaluated using
the SnapGene® Viewer 2.8.3 (from GSL Biotech; available
at snapgene.com) through Basic Local Alignment Search Tool
comparison (Genbank; NM_014272.4) (Figs. S1 and S2). The properly sequenced bacterial
liquid was transferred to 10 ml LB liquid medium containing the
corresponding antibiotics and cultured overnight at 37°C. Plasmid
extraction was carried out with the TIANprep Mini Plasmid kit
(Tiangen Biotech Co., Ltd.) according to the manufacturer's
instructions.
 | Table I.Primer sequence used to construct a
luciferase vector containing the target gene 3′ untranslated
region. |
Table I.
Primer sequence used to construct a
luciferase vector containing the target gene 3′ untranslated
region.
| Primer name | Sequence
(5′-3′) | Length (bp) |
|---|
|
h-LINC01960-201-F-(1) (XhoI) |
CGG(CTCGAG)CTCTGGTCTCCTGACTCTGG |
1 |
|
h-LINC01960-201-R-(1884) (NotI) |
AAT(GCGGCCGC)TTATATTTCACCCTTAAATACTTT | 1,884 |
|
h-LINC01960-201-MUT2 (278) |
GCCTTGTC(GGGTGGGC)CTTGGCTCGCGTTCTGAA |
278 |
|
h-LINC01960-201-MUT2 (304) |
GAGCCAAG(GCCCACCC)GACAAGGCTCCTCCGGGG |
304 |
|
h-LINC01960-201-MUT1 (754) |
CTCCGCAG(GGTCCCGG)CAGGAGGCCGCTCTTCCG |
754 |
|
h-LINC01960-201-MUT1 (780) |
GCCTCCTG(CCGGGACC)CTGCGGAGGCGACAGGCT |
780 |
|
h-ADAMTS7-3UTR-F-(1) (XhoI) | CGG (CTCGAG)
CTGCGCCAGGATGCACAGA |
1 |
|
h-ADAMTS7-3UTR-R-(218) (NotI) |
AAT(GCGGCCGC)TGCTTTGGAATGGTAGATGCT |
218 |
|
h-ADAMTS7-3UTR-MUT1-F (27) |
ACAGACCT(GTCACGG)CACCACGGGCTGTGGCGG |
27 |
|
h-ADAMTS7-3UTR-MUT1-R (52) |
CCGTGGTG(CCGTGAC)AGGTCTGTCGGTCGGTCT |
52 |
|
h-ADAMTS7-3UTR-MUT2-F (59) |
GGAGCTCC(GCGGGG)CTGCGCCCTAATGGTGCT |
59 |
|
h-ADAMTS7-3UTR-MUT2-R (81) |
GGGCGCAG(CCCCGC)GGAGCTCCGCCACAGCCC |
81 |
Dual luciferase reporting
experiment
According to the manufacturer's instructions, miRNA
mimics or non-targeting control (24,25)
(hsa-miR-608 mimic sequence:
5′-AGGGGUGGUGUUGGGACAGCUCCGUUCCCCACCACAACCCUGUCGAGGCA-3′; Guangzhou
RiboBio Co., Ltd.), target gene 3′-UTR dual reporter vector or
mutant vector plasmid (Guangzhou RiboBio Co., Ltd.) and
Lipofectamine™ 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) were mixed and transfected into A549 cells, cells were plated
in 96-well plates at a density of 1.5×104 cells/ml at
37°C. All reactions were performed in triplicate. At 48 h
post-transfection, Dual-Glo® Stop & Glo buffer
(Promega Corporation) was added to start the Dual-Glo Luciferase
Assay System (Promega Corporation) and a luminometer (Veritas
9100-002; Promega Corporation) was used to measure the fluorescence
value. The reported fluorescence of the vector was Renilla
luciferase (hRluc), and the corrected fluorescence was luciferase
(hluc). The 3′-UTR region of the gene was cloned downstream of the
hRluc gene. Since miRNAs act on the target gene through the 3′-UTR
region, miRNAs were co-transformed with the constructed reporter
vector, and the interaction between miRNA and target gene was
verified by the decrease in the relative fluorescence value of the
reporter gene.
Patients and samples
From June to September 2017, the endometrial tissues
of female patients mainly affected by ‘infertility due to
endometriosis’ who underwent hysteroscopy and laparoscopy at Peking
Union Medical College Hospital (Beijing, China) were collected.
Their mean age was 30±2.9 years. All patients underwent
laparoscopic excision of ovarian endometriomas and hysteroscopy
examination at mid-secretion time (19–24 days of the normal
menstrual cycle). According to the revised American Fertility
Society (American Society of Reproductive Medicine Staging, as
amended in 1997), all the patients were clinical stage III–IV
(26). The exclusion criteria
included other endocrine diseases such as diseases of the adrenal
and pituitary glands, uterine fibroids and genital tuberculosis.
None of the patients had been treated with hormonal drugs for ~3
months. The patients were diagnosed with endometriosis by
laparoscopy and verified by clinical pathology. All the tissues
were collected under sterile conditions, washed with physiological
saline and placed into aseptic freezers without enzymes at −20°C.
The primary cells were extracted for culture on the day of dry ice
transportation.
Primary cell culture and in vitro
induction of decidualization
Endometrial tissue was placed in a cell culture dish
with its 2–3 times volume (ml) of DMEM/F12 phenol red-free medium
(HyClone; Cytiva), cut into pieces of 0.5-1.0 mm with ophthalmic
scissors until visually mushy and then placed in a 15-ml centrifuge
tube with the appropriate quantity of medium. Next, 0.1%
collagenase I (Gibco; Thermo Fisher Scientific, Inc.) was added and
digested at 37°C for 1 h. After filtering through a 150-µm 100-mesh
cell strainer, the filtered endometrial stromal cells were
immediately centrifuged at 1,000 × g for 5 min at room temperature.
Next, 2 ml 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.)
was added, and the cells were digested for 10 min. The reaction was
terminated by adding 2 ml dextran charcoal stripped-fetal bovine
serum (DCC-FBS; HyClone; Cytiva). Upon centrifugation at 1,000 × g
for 5 min at room temperature, the supernatant was discarded and
the pellet was added to a 25-mm culture bottle. Next, an
appropriate quantity of DMEM/F12 phenol red-free medium (HyClone;
Cytiva) with 10% DCC-FBS and 1% penicillin-streptomycin was added,
and the cells were cultured at 37°C in a 5% CO2
incubator (Thermo Fisher Scientific, Inc.). After 24 h, the
adherent cells were observed under an optical microscope
(magnification, ×40), while the non-adherent cells and the blood
cells were discarded. The cells were replaced with DMEM/F12 phenol
red-free medium (HyClone; Cytiva), which was then replaced every
2–3 days. Microscopic observation (Nikon Corporation) revealed that
the cells were overgrown in culture vials, and were subcultured by
using 0.25% trypsin at a 1:2 ratio for 3–4 generations. Phenol
red-free DMEM/F12 with 2% DCC-FBS, 0.5 mmol/l 8-Bromoadenosine
3′,5′-cyclic monophosphate (8-Br-cAMP; Sigma-Aldrich; Merck KGaA)
and 1 µmol/l medroxyprogesterone acetate (MPA; Sigma-Aldrich; Merck
KGaA) were added to induce decidualization in vitro, and for
concomitant knockdown and transfection.
LINC01960-201-knockdown and
hsa-miR-608 mimic transfection
siRNA and miRNA mimic sequences are presented in
Table II. Endometrial stroma cells
were digested with trypsin, and DMEM/F12 phenol red-free medium
(HyClone; Cytiva) was added. Cells at a density of 2×105
cells were cultured in a 24-well plate. In total, three replicates
of each sample and three negative controls (NCs) were prepared.
Briefly, Ribo™ lncRNA Smart Silencer (Guangzhou RiboBio Co., Ltd.)
or the NC siN0000001-1-5 siR NC #1 (Guangzhou RiboBio Co., Ltd.)
was added to a 24-well plate at a concentration of 100 nM, and
riboFECT™ CP reagent (Guangzhou RiboBio Co., Ltd.) was added. After
being lightly agitated until thoroughly mixed, the samples were
incubated at room temperature for 15 min. Next, an appropriate
quantity of medium was added to the mixture and shaken gently until
fully mixed. Subsequently, 1 µmol/l MPA and 0.5 mmol/l 8-Br-cAMP
were added to induce the decidualization of endometrial cells in
vitro, and the well-mixed 24-well plate was placed into a
CO2 cell incubator at 37°C for 48 h. The differential
expression levels of hsa-miR-608 between the
LINC01960-201-knockdown groups and the negative controls, as
measured using reverse transcription-quantitative PCR (RT-qPCR),
were used to evaluate the effect of lncRNA-knockdown on the
expression of hsa-miR-608 in the positive control group, in order
to explore the mutual regulation between LINC01960-201 and
hsa-miR-608.
 | Table II.Sequences of LINC01960-201 silencer
and hsa-miR-608 mimic. |
Table II.
Sequences of LINC01960-201 silencer
and hsa-miR-608 mimic.
| Gene | Target sequence
(5′-3′) |
|---|
| Ribo™
h-LINC01960-201 |
ATTCCTCCCAACAGCTGGAT |
| Smart Silencer |
GCTTAAGAGGGTCAACGTGT |
|
|
TCCAGACCTTAGTCACTCTG |
|
|
TGCACAACCTCAGGAAACA |
|
|
AGACCTTAGTCACTCTGCT |
|
|
CACGTAACAACCAAATGCA |
| hsa-miR-608
mimic |
AGGGGUGGUGUUGGGAC |
|
| AGCUCC |
|
|
GUUCCCCACCACAACCCU |
|
| GUCGAGGCA |
Next, hsa-miR-608 mimic (Guangzhou RiboBio Co.,
Ltd.) or the NC miR1N0000001-1-5 micrON mimic NC #22 (Guangzhou
RiboBio Co., Ltd.) was added to a 24-well plate at a concentration
of 50 nM, followed by addition of riboFECT™ CP reagent (Guangzhou
RiboBio Co., Ltd.). After being lightly shaken until thoroughly
mixed, the mixture was incubated at room temperature for 15 min.
Next, an appropriate quantity of medium (464.5 µl) was added to the
mixture and shaken gently until fully mixed. Subsequently, 1 µmol/l
MPA and 0.5 mmol/l 8-Br-cAMP were added to induce the
decidualization of endometrial cells in vitro and the
well-mixed 24-well plate was placed into a CO2 cell
incubator at 37°C for 48 h. Western blotting was used to detect the
expression of ADAMTS7 after hsa-miR-608 mimics transfection.
RT-qPCR
According to the manufacturer's protocol, TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract
the total RNA from the decidual cells. The primers used in the
present study are presented in Table
III. For miRNA quantification, Bulge-loop™ miRNA qRT-PCR Primer
Sets (one RT primer and a pair of qPCR primers for each set)
specific for hsa-miR-608 were designed by Guangzhou RiboBio Co.,
Ltd. (cat. no. MQPS0001954-1-100). The FastQuant RT kit (Tiangen
Biotech Co., Ltd.) was used for the RT of total RNA. Total RNA (1
µg) and 20 µl reaction system (10X Fast-RT Buffer, 2 µl; FQ-RT
Primer Mix, 2 µl; RT Enzyme Mix, 1 µl; RNase-free water, 5 µl; and
buffer, 10 µl) were subjected to 42°C for 15 min and 95°C for 3
min, followed by cooling on ice and RT. The same quantity of RNA
was used for all samples. The qPCR reaction system (10 µl)
consisted of 5 µl 2X Power SYBR® Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), 0.5 µl
complementary DNA sample, 0.25 µl forward primer (10 µM), 0.25 µl
reverse primer (10 µM) and 4 µl nuclease-free water. A MicroAmp
Optical 96-well reaction plate (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used. Denaturation was carried out at 95°C
for 10 min in a QuantStudio™ 7 Flex Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Next, 40 cycles of
95°C for 15 sec and 60°C for 1 min, and a final extension at 60°C
for 10 min were performed, before subjecting the samples to 95°C
for melting curve analysis (temperature ramp of 2%). All reactions
were performed in triplicate. Electrophoresis (2.0% agarose gel)
was used to detect the amplification specificity of the products.
U6 was selected as the internal reference gene. Expression was
measured using the 2−ΔΔCq method (27).
 | Table III.Primers for reverse
transcription-quantitative PCR. |
Table III.
Primers for reverse
transcription-quantitative PCR.
| Primer name | Sequence
(5′-3′) |
|---|
| lncDETECT™ | F:
CTATTGCACAACCTCAGGAAACAG |
|
h-LINC01960_qPCR_1085 bp | R:
TGGGAAAGGAAAACACACTTCA |
| GAPDH | F:
GAACGGGAAGCTCACTGG |
|
| R:
GCCTGCTTCACCACCTTCT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
Protein extraction and western
blotting
The decidualized endometrial cells were washed two
to three times with PBS. Next, 250 µl RIPA lysis buffer (containing
freshly added protease inhibitors) was added to the plate for 3–5
min. The culture plate was shaken repeatedly to allow the reagent
to be in full contact with the cells. Next, cells and reagents were
scraped off with a cell scraper and collected into a 1.5-ml
centrifuge tube and cooled on ice for 30 min. Repeated blowing with
a pipette ensured that the cells were completely lysed. After
centrifugation at 13,000 × g for 10 min at 4°C, the supernatant was
kept and a bicinchoninic acid kit (Wuhan Servicebio Technology,
Co., Ltd.) was used to determine the protein concentration. The
protein solution [mixed at a 4:1 ratio with 5X protein sample
buffer (Wuhan Servicebio Technology, Co., Ltd.)] was boiled for 15
min and stored at −20°C until use. Proteins (30 µg per lane) were
separated using 10% SDS-PAGE and then transferred to polyvinylidene
difluoride membranes, which were blocked with 5% skimmed milk for 1
h at room temperature. Next, the membranes were incubated with
primary antibodies against ADAMTS7 (1:1,000; cat. no. 250456;
Abbiotec, Inc.) at 4°C overnight, followed by washing with TBS
containing 0.05% Tween-20 (TBST) three times at room temperature.
Next, the secondary antibody HRP-labeled goat anti-rabbit (1:3,000;
cat. no. GB23303; Wuhan Servicebio Technology, Co., Ltd.) was added
and incubated at room temperature for 30 min. The membranes were
then washed in TBST, and detection by chemiluminescence (ECLA and
ECLB; cat. no. G2014; Wuhan Servicebio Technology, Co., Ltd.) was
carried out. Adobe Photoshop CS5 (Adobe Systems, Inc.) was used for
image analysis. β-actin (1:3,000; cat. no. GB12001; Wuhan
Servicebio Technology, Co., Ltd.) was selected as an internal
reference. Grey value analysis (alphaEaseFC4.0; ProteinSimple) was
conducted.
Statistical analysis
SPSS 19.0 (IBM Corp.) was used for data processing
and statistical analysis. All RT-qPCR reactions were performed in
triplicate. All data are expressed as the mean ± standard deviation
and were analyzed using ANOVA with post hoc Tukey's test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Bioinformatics analysis
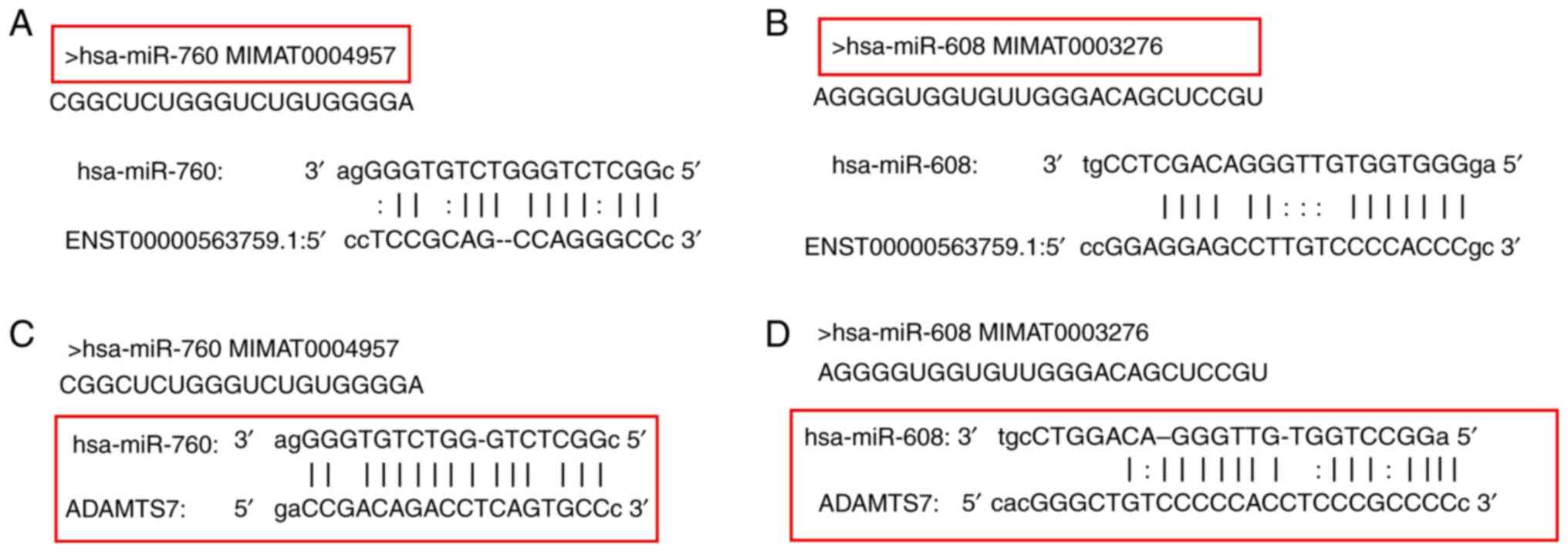
The potential target sites between LINC01960-201 and
hsa-miR-760, LINC01960-201 and hsa-miR-608, ADAMTS7 and hsa-miR-760
and ADAMTS7 and hsa-miR-608 were predicted using bioinformatical
analysis of the data using miRanda, PITA and RNAhybrid. Binding
sites were identified in the 3′-UTR of LINC01960-201 and ADAMTS7.
The predicted binding sites are presented in Fig. 1A-D. The potential target sites were
analyzed by bioinformatics.
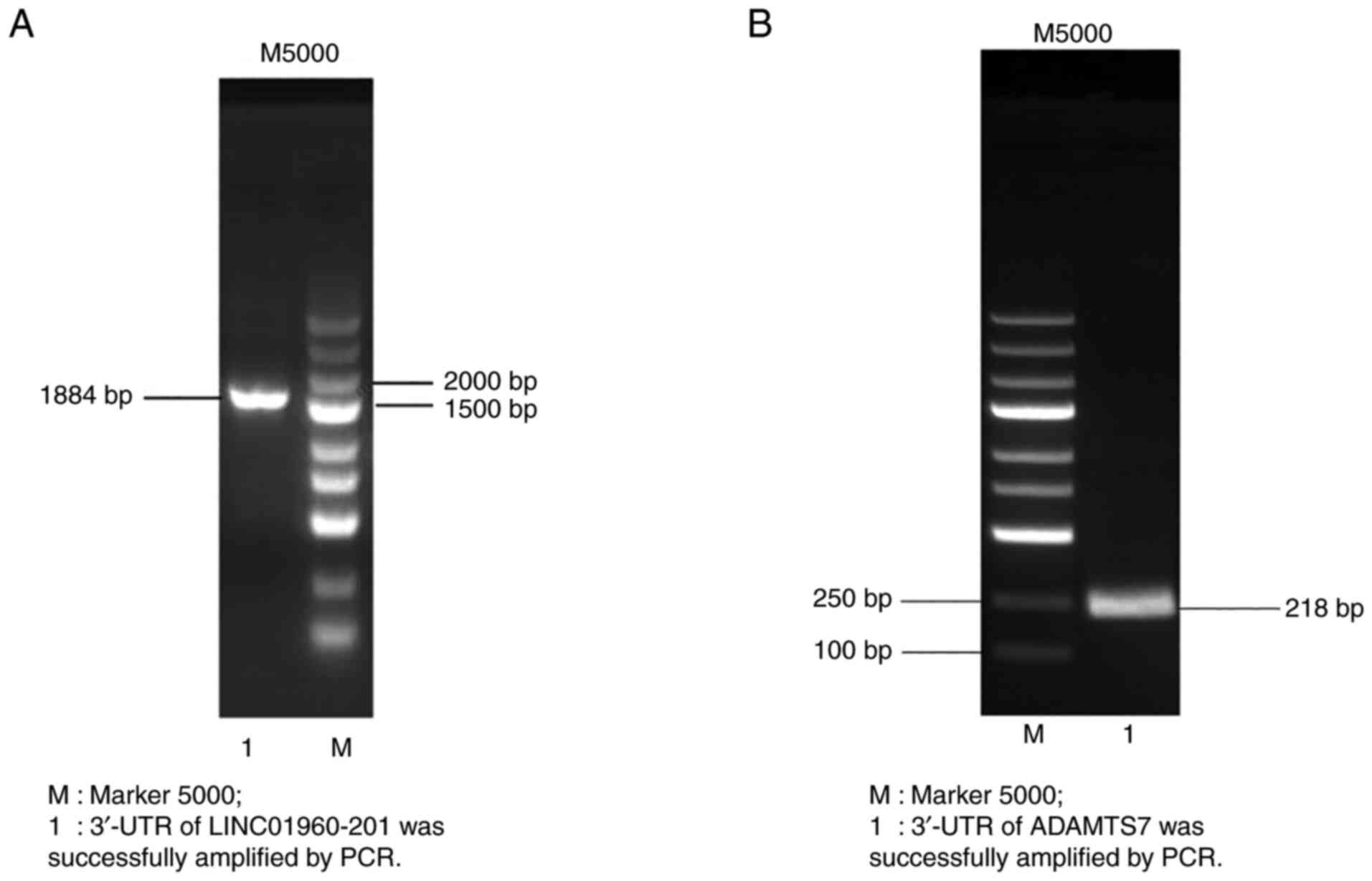
PCR amplification of the target
gene
The primer length of LINC01960-201 was 1,884 bp, and
the PCR product of lncRNA sequencing was consistent with the size
of the LINC01960-201 band. A DNA band was generated at ~1.884 bp
(Fig. 2A) that was approximately
the expected size, indicating that the 3′-UTR of the LINC01960-201
gene was successfully amplified.
The target fragment of ADAMTS7 was amplified, and
the amplified product was subjected to agarose gel electrophoresis.
A single band was observed at ~218 bp (Fig. 2B), and the size was uniform with the
target gene ADAMTS7. Overall, PCR amplification of the target gene
was successful.
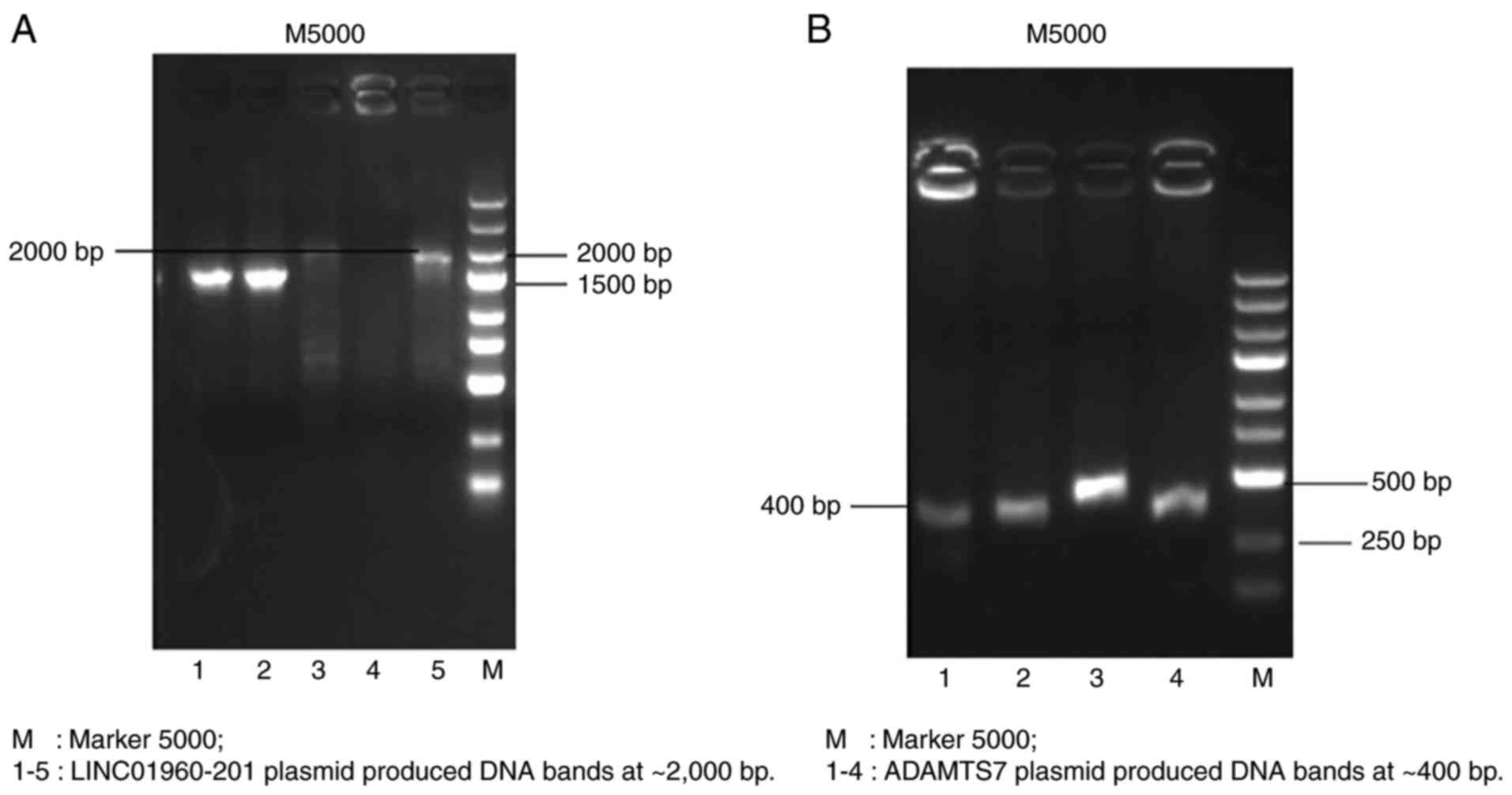
PCR and enzymatic digestion-mediated
identification of the recombinant plasmid
As one of the primers was located in the target gene
and the other one was located in the vector, it was verified that
the vector plasmid was successfully linked to the target gene. The
emolic electrophoretic bands illustrated that a splicing reaction
in the constructed LINC01960-201 plasmid produced DNA bands at
~2,000 bp (Fig. 3A) and 400 bp
(Fig. 3B) at ADAMTS7, which are the
sizes of the expected fragments. Overall, the recombinant vector
plasmid was successfully linked to the target gene.
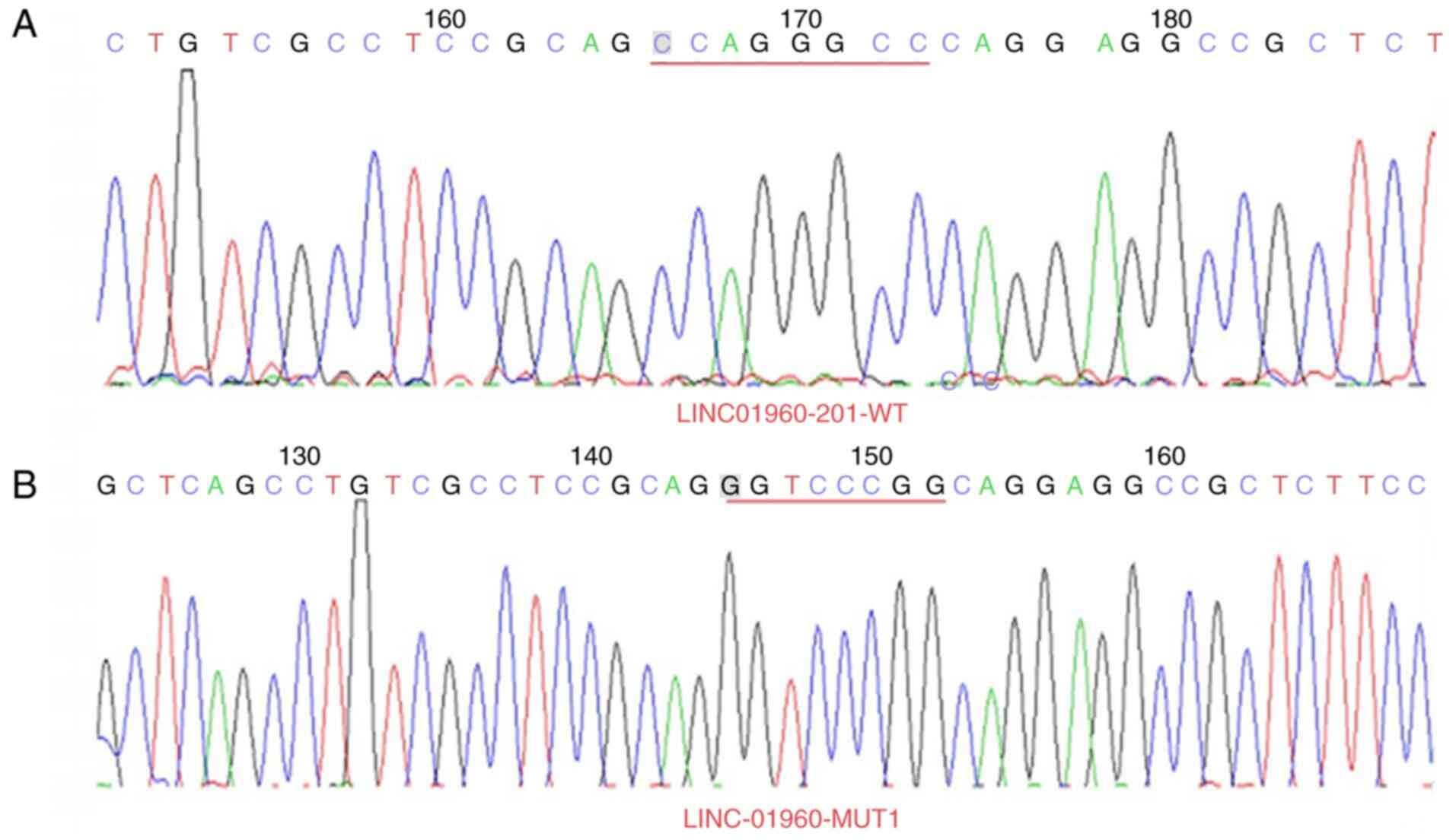
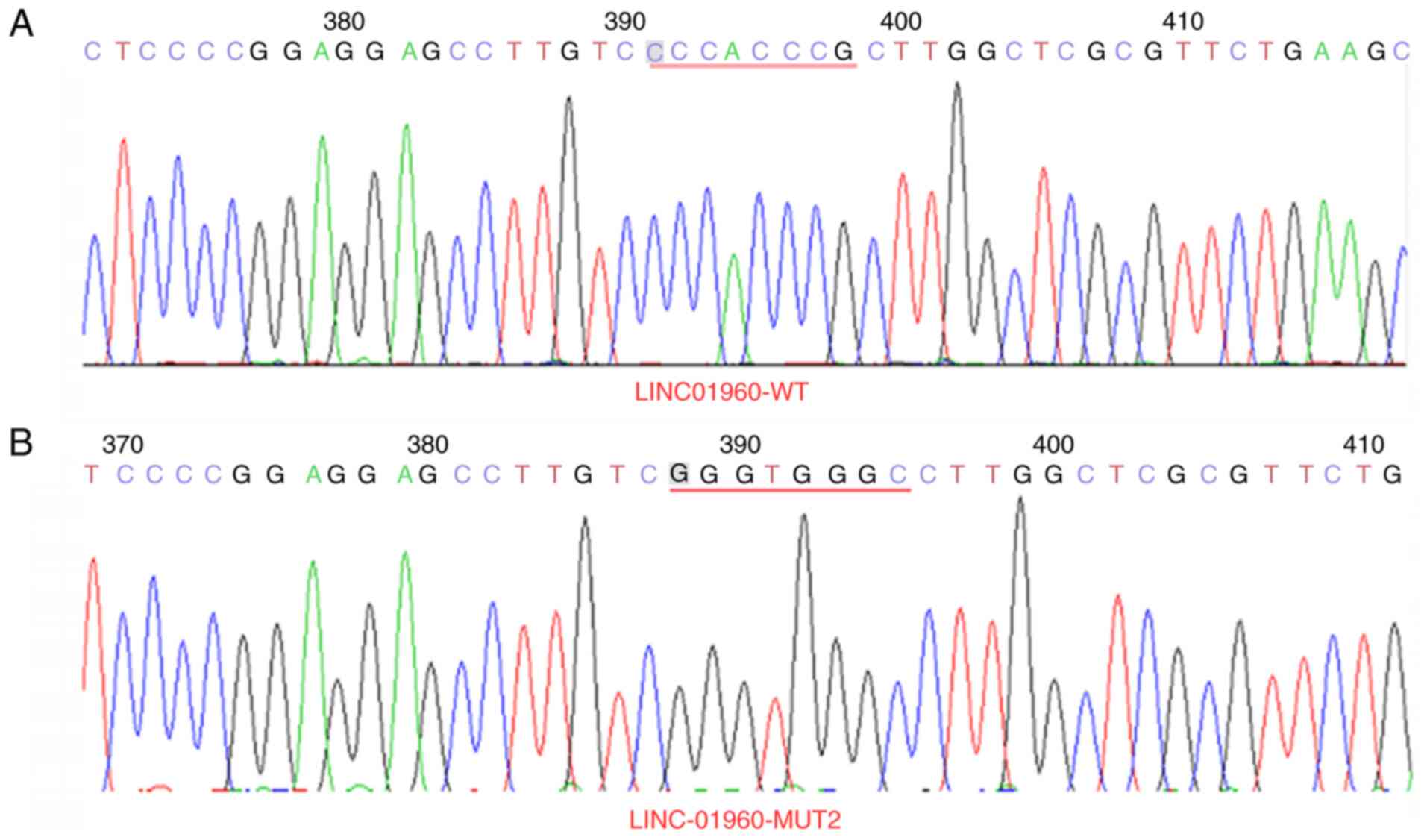
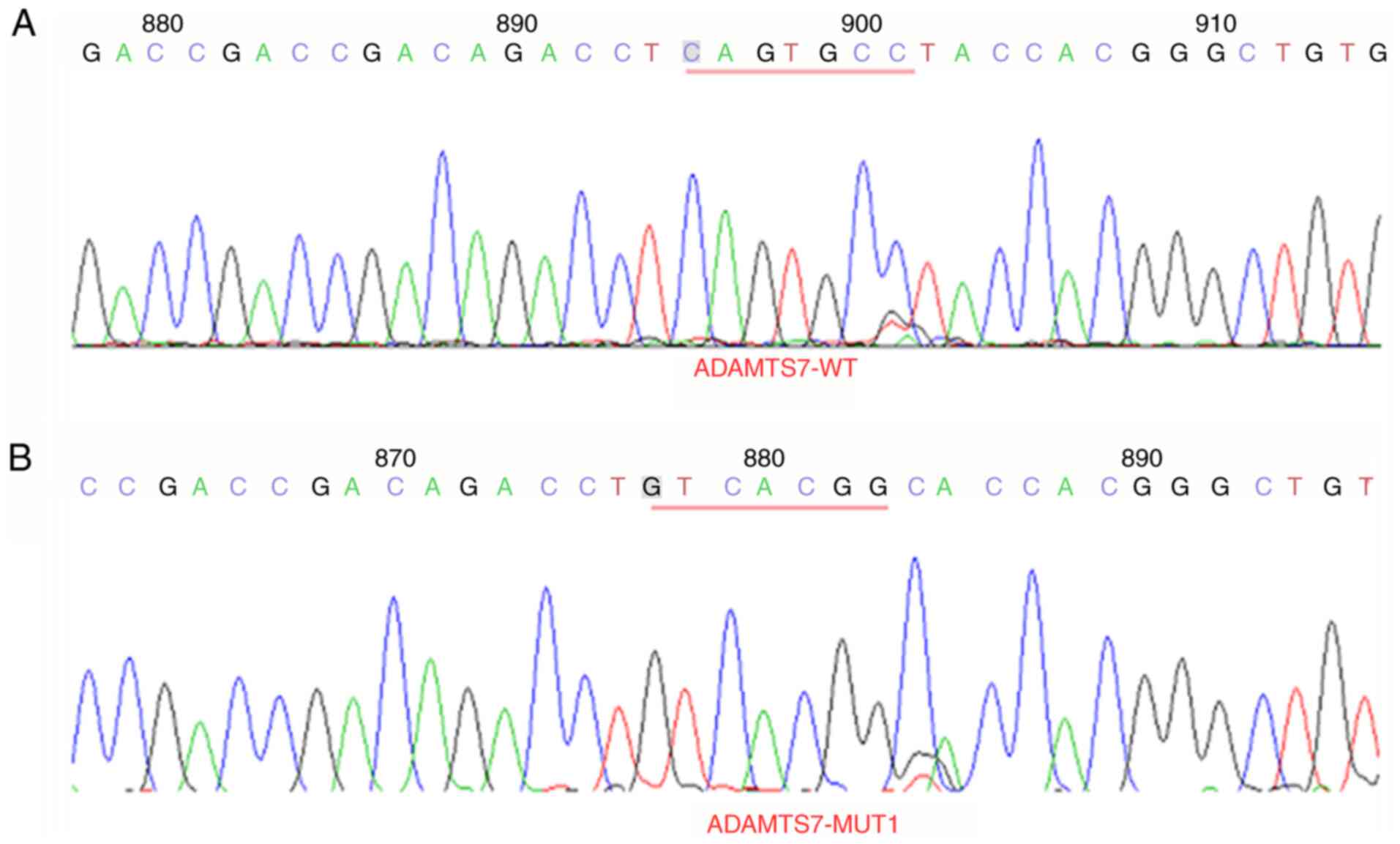
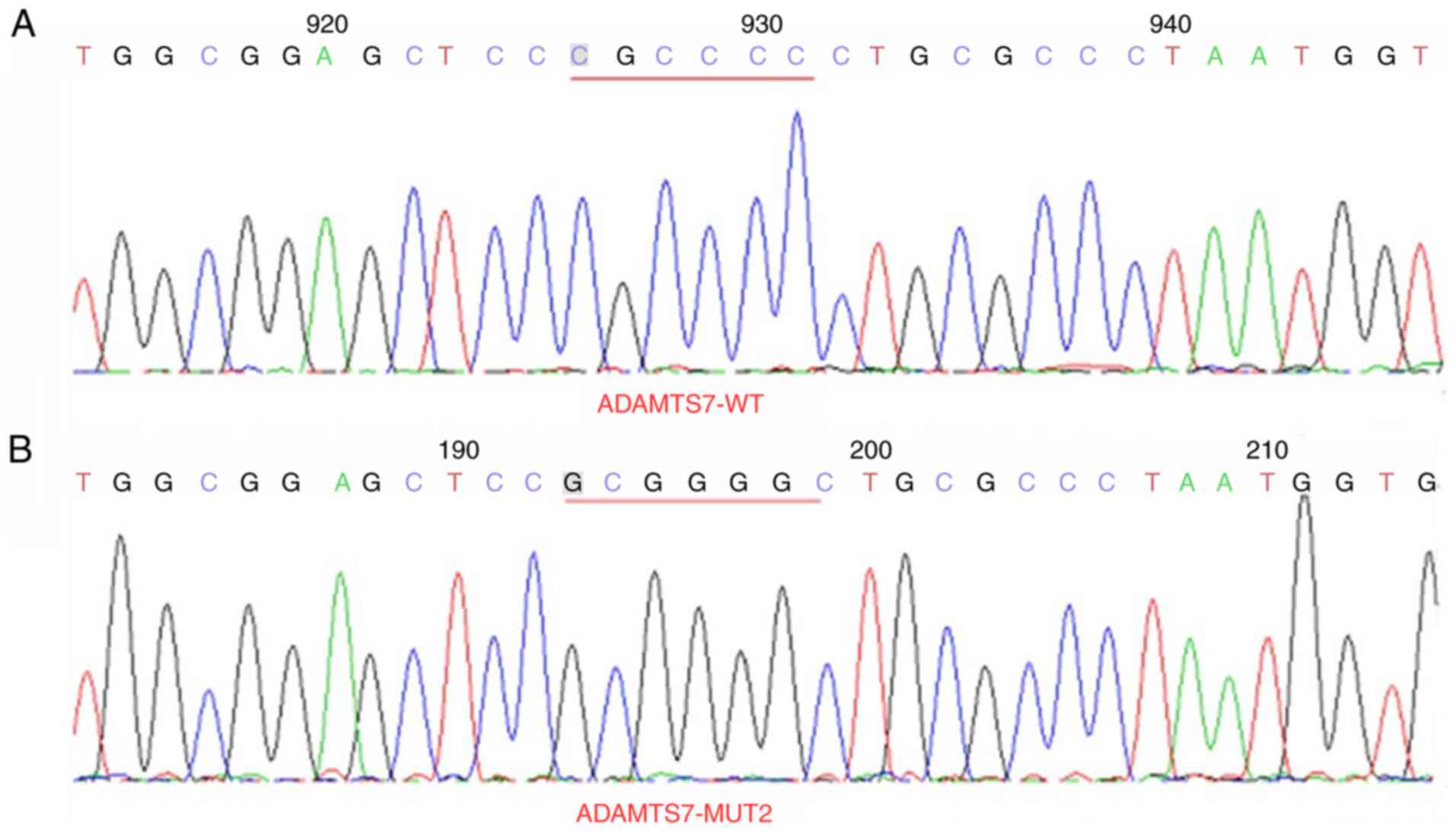
Plasmid sequencing results
The 3′-UTR of LINC01960-201 sites 1 (CCAGGGCC muted
to GGTCCCGG) and sites 2 (CCCACCCG muted to GGGTGGGC) (Figs. 4 and 5) and the 3′-UTR of ADAMTS7 sites 1
(CAGTGCC muted to GTCACGG) and sites 2 (CGCCCC muted to GCGGGG)
(Figs. 6 and 7) were successfully mutated. Overall, the
mutation of 3′-UTR of LINC01960-201 and the 3′-UTR of ADAMTS7 was
successful. Following sequencing of the recombinant plasmid
LINC01960-201, the following SNP was detected: C/G at 539 bp on
NCBI (SNP number: rs11693010; http://www.ncbi.nlm.nih.gov/snp/). In addition, the A
was mutated to C at 422 bp (the non-SNP site) (Fig. S1). Following sequencing of the
recombinant plasmid ADAMTS7, the following SNP was detectedt: C/T
at 42 bp on NCBI (SNP number: rs1045130) (Fig. S2).
Results of relative luciferase
activity detection
In the present study, There was no significant
difference in the double luciferase activity of
human-LINC01960-201-WT and hsa-miR-760 co-transfected with the NC
group. There was no significant difference in the double luciferase
activity of LINC01960-201-MUT1 and hsa-miR-760 co-transfected with
the NC group (Fig. 8A).
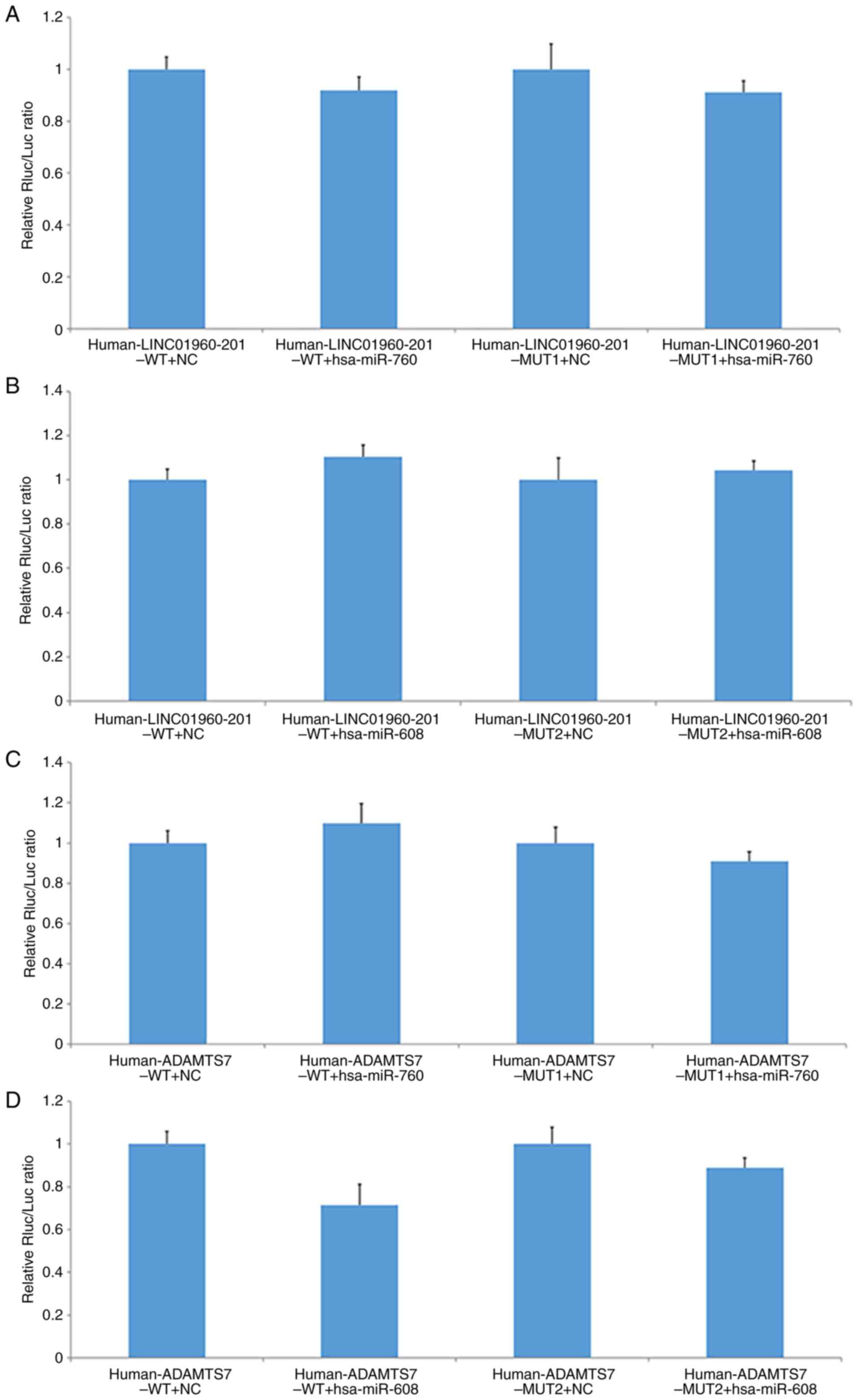 | Figure 8.Relative fluorescence activity in
each group of cells. Measurements of the relative relationship
between (A) LINC01960-201 and hsa-miR-760, (B) LINC01960-201 and
hsa-miR-608, (C) ADAMTS7 and hsa-miR-760 and (D) ADAMTS7 and
hsa-miR-608. miR, microRNA; ADAMTS7, a disintegrin and
metalloproteinase with thrombospondin motifs 7; MUT, mutant; WT,
wild-type; NC, negative control; Rluc, Renilla luciferase;
Luc, luciferase; LINC, long intergenic non-protein coding RNA. |
The double luciferase activity of
human-LINC01960-201-WT co-transfected with hsa-miR-608 was not
significantly different from that of the NC group. There was no
significant difference in the double luciferase activity of the
LINC01960-201-MUT2 and hsa-miR-608 co-transfected with the NC
group. These results indicated that hsa-miR-760 and hsa-miR-608 had
no significant interaction with this segment of the 3′-UTR side of
human-LINC01960-201 (Fig. 8B).
The hsa-miR-760 mimic had no significant effect on
the reported fluorescence expression of human-ADAMTS7-WT or
human-ADAMTS7-MUT1. There was no significant difference in the
double luciferase activity of human-ADAMTS7-WT co-transfected with
hsa-miR-760 compared with the NC group, and there was no
significant difference in the double luciferase activity of
human-ADAMTS7-MUT1 co-transfected with hsa-miR-760 compared with
the NC group. The results indicated that hsa-miR-760 mimic had no
significant interaction with the 3′-UTR of the human-ADAMTS7 gene
(Fig. 8C).
The double luciferase activity of human-ADAMTS7-WT
co-transfected with hsa-miR-608 was significantly lower than that
of the NC group, while the double luciferase activity of
human-ADAMTS7-MUT2 co-transfected with hsa-miR-608 was not
significantly different from that of the NC group. Compared with
that of the NC, hsa-miR-608 mimic had a more pronounced downward
effect on human-ADAMTS7-WT report fluoridation. After mutating its
predicted target point, the reported fluorescence in the mutant
vector human-ADAMTS7-MUT2 was restored. These results suggested
that hsa-miR-608 mimic may regulate the expression of the gene
through this site on the 3′-UTR of human-ADAMTS7 (Fig. 8D). Overall, there was evidence of
significant regulatory sites between the 3′-UTR of ADAMTS7 and
hsa-miR-608.
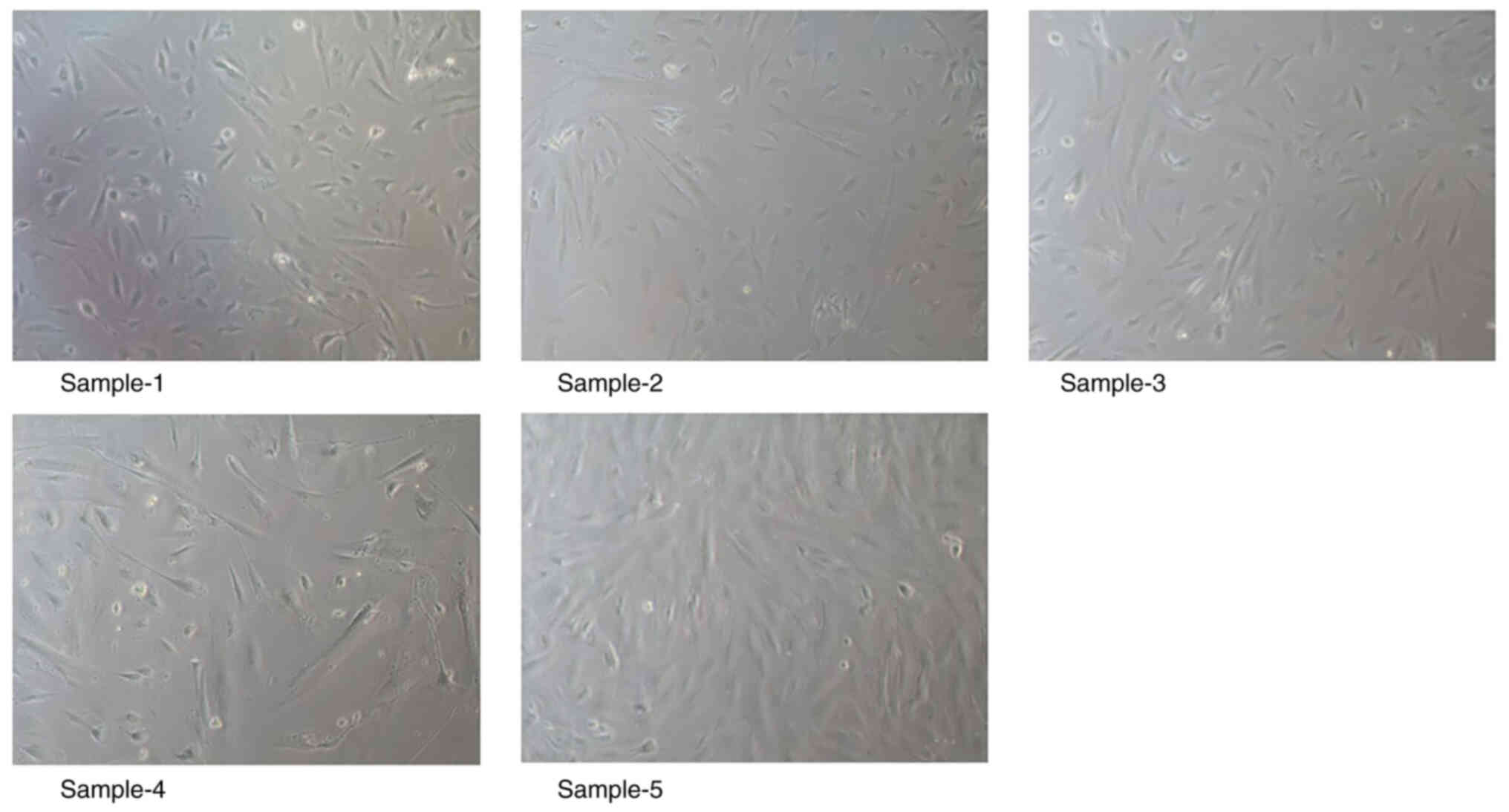
Primary culture of endometrial stromal
cells in endometriosis
A total of five samples of eutopic endometrial
tissues in women with endometriosis were collected, and the success
rate of endometrial stromal cell isolation and culture was 100%.
Adherent proliferation of the cells was observed a common optical
microscope, which revealed a triangular shuttle shape of the cells
with the cell nucleus circular centered. There was no significant
difference in cell morphology between each group. The results of
cell culture are presented in Fig.
9. Overall, primary culture of endometrial stromal cells in
endometriosis was successful.
Changes in gene expression of after
LINC01960-201-knockdown
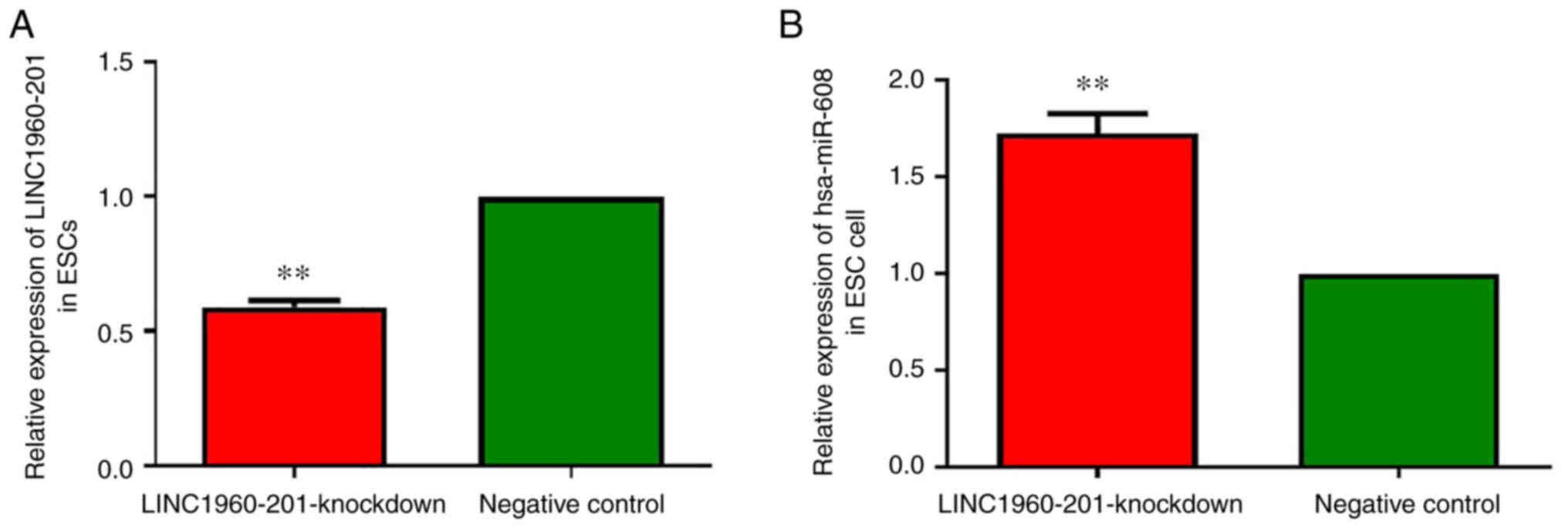
The expression of LINC01960-201 was decreased during
decidualization induced by 1 µmol/l MPA and 0.5 mmol/l 8-Br-cAMP in
endometrial stromal cells in vitro. The expression of
LINC01960-201 in the experimental group was significantly
downregulated after LINC01960-201-knockdown (P<0.01),
demonstrating that LINC01960-201 was successfully knocked down
(Fig. 10A).
The results of RT-qPCR demonstrated that the
expression of hsa-miR-608 in the experimental group was
significantly higher compared with that in the NC group (P<0.01;
Fig. 10B). After devaluation of
LINC01960-201 dependence in the endometrial stromal cells,
hsa-miR-608 expression was moderately rebounded (Fig. 10B), which demonstrated that there
was a mutual regulatory association between LINC01960-201 and
hsa-miR-608 in the process of induction of decidualization of
endometrial stroma cells in vitro. Overall, the expression
of hsa-miR-608 was upregulated after LINC01960-201-knockdown.
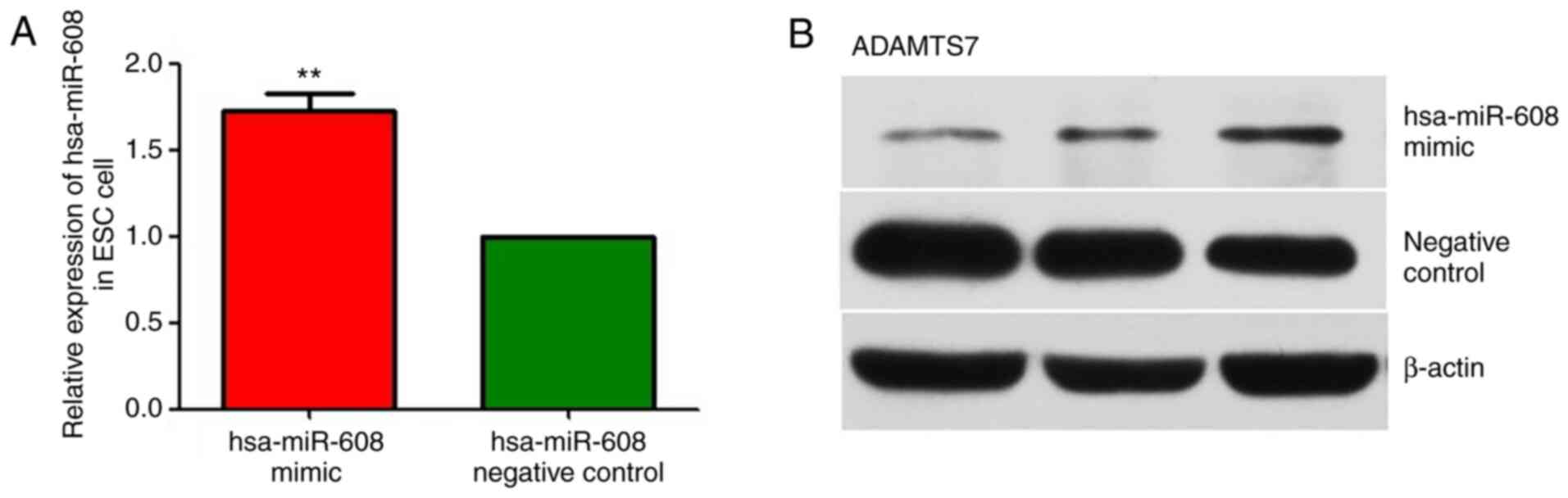
Effects of hsa-miR-608 mimic
transfection during in vitro decidualization
As presented in Fig.
11A, compared with that of the NC group, the expression level
of hsa-miR-608 in the experimental group was significantly
upregulated (P<0.01), indicating that hsa-miR-608 mimic
transfection was successful. Subsequently, the relative protein
expression changes of ADAMTS7 were detected by western blotting
using β-actin as an internal reference. As presented in Fig. 11B, the strip color detected in the
hsa-miR-608 group was markedly lighter compared with that of the NC
group (Fig. 11B). With the
upregulation of miR-608 expression, the expression of ADAMTS7 in
the hsa-miR-608 mimic group was reduced, suggesting that miR-608
has a reverse regulatory effect on ADAMTS7. Overall, the expression
of ADAMTS7 was downregulated after hsa-miR-608 mimic transfection
during in vitro decidualization.
Discussion
The present study used bioinformatics software
(miRanda, PITA and RNAhybrid) to demonstrate that hsa-miR-760 and
hsa-miR-608 share a common target gene, ADAMTS7, with
LINC01960-201. Upon dual luciferase vector construction, plasmid
extraction and relative fluorescence value detection, it was
revealed that there might be regulatory targets between hsa-miR-608
and the 3′-UTR of ADAMTS7, while there was almost no possibility of
regulatory sites between hsa-miR-608 and LINC01960-201, hsa-miR-760
and LINC01960-201 or hsa-miR-760 and the 3′-UTR of ADAMTS7.
In the present study, five samples of endometrial
tissues were collected from women suffering from infertility due to
endometriosis who underwent laparoscopic surgery. Once the
endometrial stromal cells had been extracted for primary cell
culture, in vitro decidualization was performed with
LINC01960-201-knockdown and hsa-miR-608 mimic transfection. RT-qPCR
confirmed that LINC01960-201-knockdown was successful. It was
revealed that the expression of hsa-miR-608 in the
LINC01960-201-knockdown group was increased compared with that of
the NC group. After transfection with hsa-miR-608 mimic, RT-qPCR
demonstrated that hsa-miR-608 mimic was successfully transfected.
Subsequently, the results of western blotting revealed that the
band of the hsa-miR-608 mimic transfection group was significantly
lighter compared with that of the NC group. With the increase in
hsa-miR-608 expression, the expression of ADAMTS7 was decreased,
suggesting that hsa-miR-608 has a reverse regulatory effect on
ADAMTS7.
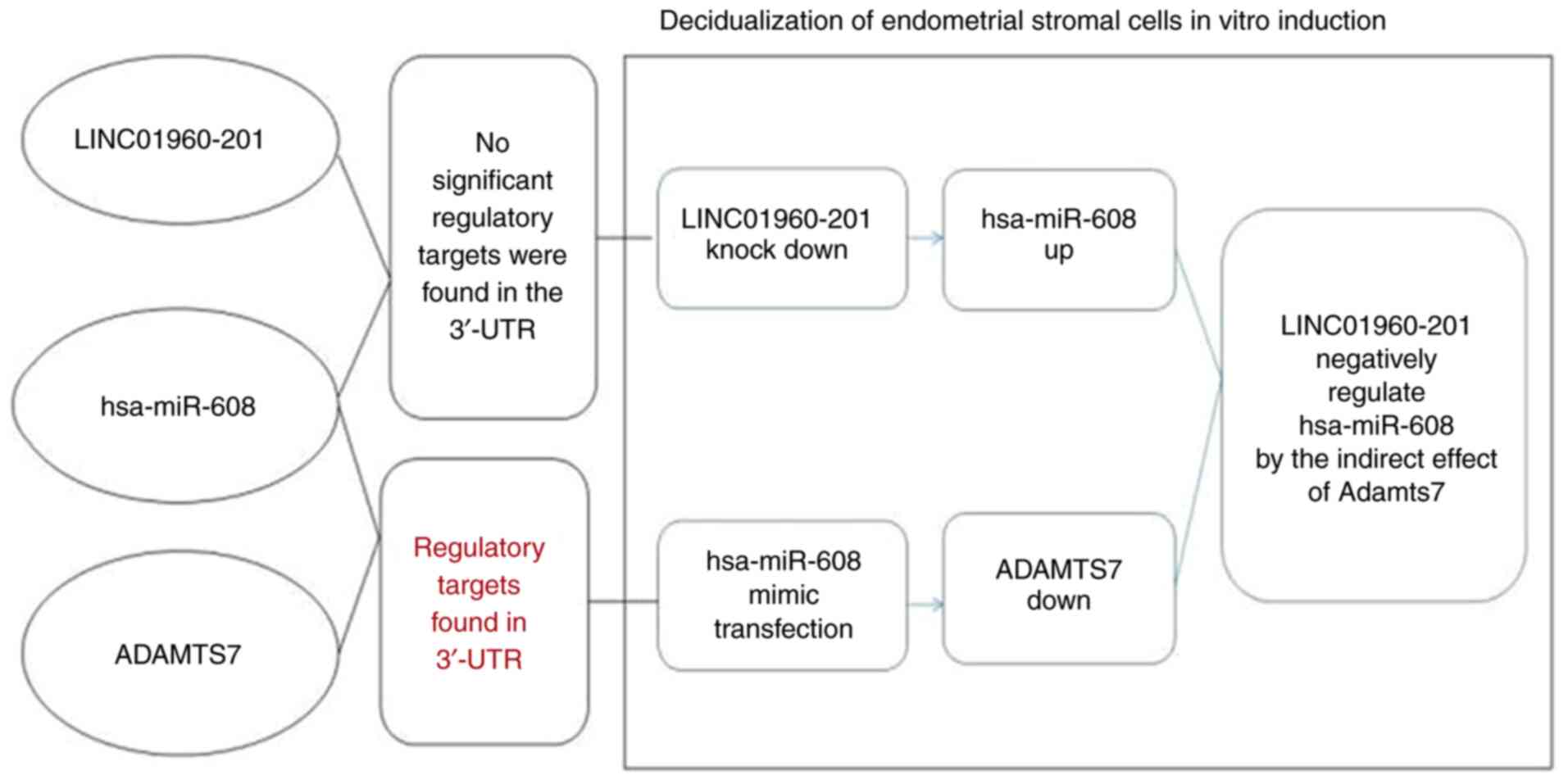
The present study also demonstrated that
downregulation of LINC01960-201 increased the expression of
hsa-miR-608 in vitro after induction of decidualization of
endometrial stromal cells in women with endometriosis during the
period of implantation; while no significant regulatory points were
revealed between them in the target verification experiment.
hsa-miR-608 expression increased while ADAMTS7 expression
decreased, and the target verification experiment revealed that
there was a target between the two. The present study indicated
that LINC01960-201 played a notable regulatory role in
decidualization of the endometrial stromal cells in vitro
induction in endometriosis, and the expression of LINC01960-201
could not be separated from the regulation of hsa-miR-608 and its
ADAMTS7. Therefore, it was hypothesized that the
LINC01960-201/hsa-miR-608/ADAMTS7 regulatory pathway plays an
important role upon the decidualization of endometrial stromal
cells in endometriosis (Fig.
12).
The decidualization does not depend on pregnancy,
but can exist independently in humans, and changes in the
morphology and activity of endometrial stromal cells via the
biological behavior of estrogen and progesterone secreted by the
ovaries (1). Decidualization is the
process of morphological and biochemical differentiation of
endometrial stromal cells into decidual cells (28). Decidualization of endometrial
stromal cells is a pivotal element in embryo implantation and
pregnancy maintenance (29). It has
been demonstrated at the transcriptional and protein level that it
is a complex gene regulatory network (30,31);
however, the regulatory process at the post-transcriptional level
is unclear thus far.
Both miRNAs and lncRNAs are non-coding RNAs that
play a regulatory role in growth and development, and their common
characteristics are to perform their biological functions by the
action of mRNA. However, they also have mutual regulatory effects
in each other (5,8,32–35).
miRNAs can be negatively regulated by lncRNAs in a mechanism
similar to that of mRNAs (36).
lncRNAs can also be positively regulated by other epigenetic
pathways. There are three main mechanisms by which lncRNAs regulate
miRNAs (37,38). First, lncRNAs can competitively bind
the 3′-UTR of their target gene mRNA to inhibit the negative
regulatory mechanism of miRNAs (of note, this signaling pathway is
similar to the one described in the present study). The study by
Pang et al (39) also
indicated that lncRNA (by the antisense RNA of BACE1) could be
transcribed from the β-secretase-encoded gene locus. This antisense
RNA can be complementary to the mRNA of the BACE1 gene and
competitively prevent the degradation of miRNA on the BACE1 gene.
Secondly, certain lncRNAs can form the precursors of miRNAs through
intracellular shear action, thereby processing and producing
specific miRNAs, and regulating the expression and function of
target genes. Third, certain lncRNAs can act as endogenous miRNA
sponges, which in turn can inhibit miRNA expression. Such
biological functions indirectly affect the malignant invasion
behavior of tumor cells (40). The
association between lncRNA ZEB1-AS1 and miR-101/ZEB1 is involved in
the proliferation and migration of colon cancer cells (41). The expression of H19 lncRNA and that
of integrin β3 in the endometrium of patients with recurrent
miscarriages was reduced (42).
lncRNA LINC00261 prevents cell proliferation and migration in
endometrial stromal cells in patients with endometriosis (17), while lncRNA HOXD cluster antisense
RNA 1 promotes the proliferation, migration and invasion of ovarian
cancer cells by binding to the miR-608/frizzled family receptor-4
(43).
lncRNAs and miRNAs play an important role in the
development of diseases in the female reproductive system. A
previous study has stated that exosomal lncRNA CHL1-AS1 derived
from peritoneal macrophage can promote the proliferation, invasion
and migration of ectopic endometrial stromal cells and inhibit
their apoptosis by downregulating miR-610 and upregulating MDM2,
which may be a potential target for the treatment of endometriosis
(44). Hou et al have shown
that lncRNA TMPO-AS1 is located in the nucleus and cytoplasm of
granulosa-like tumor cells in follicular fluid of women with
polycystic ovary syndrome (PCOS), which is likely to inhibit the
maturation of miR-335-5p, thereby resulting in PCOS (45). A study of inter-modulation among
lncRNA, miRNA and mRNA in estrogen receptor (ER)-positive breast
cancer tissues revealed that LINC0092, hsa-miR-449a and
hsa-miR-452-5p regulates the co-expression of mRNAs (secreted
frizzled-related protein 1 and repulsive guidance molecule A) at
the open reading frame (C2orf71) of chromosome 2. This orf contains
a 14-node ER-related subtype of the lncRNA-miRNA-mRNA network. It
was speculated that LINC0092 and C2orf71 could be used as
prognostic indicators for breast cancer (46). Zhou et al reported that
lncRNA urothelial cancer-associated 1 elevates the expression of
HOXA9 by competitively binding with miR-184 in cardiac muscle cells
of patients with hypertrophic heart disease, which results in the
development and progression of the disease (47). Wambecke et al have
demonstrated that lncRNA ‘UCA1’ regulates ovarian cancer response
to chemotherapy by binding directly to miR-27a-5p and controlling
UBE2N level (48). It was also
reported that lncRNA brain cytoplasmic RNA 1 promotes the
proliferation, metastasis and invasion of cervical cancer cells
(SiHa, HeLa and CaSki) by preventing the expression of miR-138,
which is regulated by metalloproteinase-2 (49).
ADAMTS7 is a member of the ADAMTS family, which
comprises the family of disintegrin and metalloproteinase
associated with thrombospondin motifs. It was reported in aprevious
study that this gene plays a main biological role in coronary heart
disease (50). Multiple studies
have confirmed the regulatory association between lncRNAs, miRNAs
and the ADAMTS family (51,52). Dou et al have reported that
lncRNA HOX transcript antisense RNA promotes the expression of
ADAMTS5 in human osteoarthritis chondrocytes by improving the
stability of ADAMTS5 mRNA (53). It
was hypothesized that the upregulation of ADAMTS7 affects vascular
calcification by inhibiting the expression of miR-29a/b, and
predicted that this conduction pathway can be possibly used to
reduce the incidence and mortality of cardiovascular diseases
(54). Hanin et al reported
that miR-608 is controlled by acetylcholinesterase (AChE) during
the occurrence and development of hypertension. Moreover, the
expression level of AChE is suppressed due to the interaction
between the single nucleotide polymorphism of AChE allele
rs17228616 in the 3′-UTR region and miR-608 (55). At the cellular level, attenuating
the restraining effects of miR-608 on its negatively regulated cell
division control protein 42 homolog and interleukin-6, thus
affecting the anxiety of the individual and increasing the risk of
hypertension (55).
In our previous studies, abnormal changes were
observed in the miRNA, lncRNA and mRNA expression profiles of rats
with endometriosis during the implantation window period (22,56).
Alignment of the abnormally expressed rat lncRNA sequence with
human lncRNA revealed that the similarity between rat lncRNA
gi|672045999 |ref|XR_591544.1| and human lncRNA p10107
(LINC01960-201) was estimated to be 83.29% (22). Therefore, LINC01960-201 was selected
as the target gene in the present study. The difference between the
current study results and the expected ones was that there were no
regulatory sites among between hsa-miR-760, LINC01960-201 and
ADAMTS7. A limitation of the present study is that the regulatory
association between them in the process of decidualization of
endometrial stroma cells induced in vitro was not further
verified. Moreover, the expression of hsa-miR-760 should be further
verified. In addition, the expression of ADAMTS7 should be measured
after LINC01960-201-knockdown in order to clearly demonstrate the
association between the LINC01960-201 and ADAMTS7. Our future
studies intend to explore the regulatory role of LINC01960-201 in
the decidualization of endometrial stromal cells in the
implantation window of healthy women.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Shan Deng
(Department of Obstetrics and Gynecology, Peking Union Medical
College Hospital, Peking Union Medical College and Chinese Academy
of Medical Sciences, Beijing, China) for their help with sample
collection.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JL designed the study. HC designed the study,
performed all experiments and analysis, and wrote the paper. HC and
JL confirm the authenticity of all the raw data. Both authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Peking Union Medical College Hospital and the Chinese
Academy of Medical Sciences (Beijing, China; approval no. S-k332).
Written informed consent was obtained from the five patients and
all the specimens were acquired with the knowledge of the
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Minici F, Tiberi F, Tropea A, Orlando M,
Gangale MF, Romani F, Campo S, Bompiani A, Lanzone A and Apa R:
Endometriosis and human infertility: A new investigation into the
role of eutopic endometrium. Hum Reprod. 23:530–537. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gellersen B, Brosens IA and Brosens JJ:
Decidualization of the human endometrium: Mechanisms, functions,
and clinical perspectives. Semin Reprod Med. 25:445–453. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He D, Zeng H, Chen J, Xiao L, Zhao Y and
Liu N: H19 regulates trophoblastic spheroid adhesion by
competitively binding to let-7. Reproduction. 157:423–430. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Staun-Ram E and Shalev E: Human
trophoblast function during the implantation process. Reprod Biol
Endocrinol. 3:562005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gellersen B and Brosens JJ: Cyclic
decidualization of the human endometrium in reproductive health and
failure. Endocr Rev. 35:851–905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lessey BA, Castelbaum AJ, Buck CA, Lei Y,
Yowell CW and Sun J: Further characterization of endometrial
integrins during the menstrual cycle and in pregnancy. Fertil
Steril. 62:497–506. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barragan F, Irwin JC, Balayan S, Erikson
DW, Chen JC, Houshdaran S, Piltonen TT, Spitzer TLB, George A,
Rabban JT, et al: Human endometrial fibroblasts derived from
mesenchymal progenitors inherit progesterone resistance and acquire
an inflammatory phenotype in the endometrial niche in
endometriosis. Biol Reprod. 94:1182016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takano M, Lu Z, Goto T, Fusi L, Higham J,
Francis J, Withey A, Hardt J, Cloke B, Stavropoulou AV, et al:
Transcriptional cross talk between the forkhead transcription
factor forkhead box O1A and the progesterone receptor coordinates
cell cycle regulation and differentiation in human endometrial
stromal cells. Mol Endocrinol. 21:2334–2349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lessey BA and Kim JJ: Endometrial
receptivity in the eutopic endometrium of women with endometriosis:
It is affected, and let me show you why. Fertil Steril. 108:19–27.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Atkins HM, Lombardini ED, Caudell DL, Appt
SE, Dubois A and Cline JM: Decidualization of endometriosis in
macaques. Vet Pathol. 53:1252–1258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Petracco R, Dias ACO, Taylor H, Petracco
A, Badalotti M, Da Rosa Michelon J, Marinowic DR, Hentschke M, De
Azevedo PN, Zanirati G and Machado DC: Evaluation of miR-135a/b
expression in endometriosis lesions. Biomed Rep. 11:181–187.
2019.PubMed/NCBI
|
|
13
|
Ghazal S, McKinnon B, Zhou J, Mueller M,
Men Y, Yang L, Mueller M, Flannery C, Huang Y and Taylor HS: H19
lncRNA alters stromal cell growth via IGF signaling in the
endometrium of women with endometriosis. EMBO Mol Med. 7:996–1003.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu S, Qiu J, Tang X, Cui H, Zhang Q and
Yang Q: lncRNA-H19 regulates cell proliferation and invasion of
ectopic endometrium by targeting ITGB3 via modulating miR-124-3p.
Exp Cell Res. 381:215–222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li K, Wu Y, Yang H, Hong P, Fang X and Hu
Y: H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation
process in human adipose tissue-derived mesenchymal stem cells. J
Cell Physiol. 234:20925–20934. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Z, Chen Y, Zhao Y, Xu C, Zhang A,
Zhang Q, Wang D, He J, Hua W and Duan P: miR-200c suppresses
endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell
Res Ther. 8:2512017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sha L, Huang L, Luo X, Bao J, Gao L, Pan
Q, Guo M, Zheng F and Wang H: Long non-coding RNA LINC00261
inhibits cell growth and migration in endometriosis. J Obstet
Gynaecol Res. 43:1563–1569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang H, Shi G, Li M, Fan H, Ma H and Sheng
L: Correlation of IL-1 and HB-EGF with endometrial receptivity. Exp
Ther Med. 16:5130–5136. 2018.PubMed/NCBI
|
|
19
|
Cai QF, Wan F, Dong XY, Liao XH, Zheng J,
Wang R, Wang L, Ji LC and Zhang HW: Fertility clinicians and
infertile patients in China have different preferences in fertility
care. Hum Reprod. 29:712–719. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grechukhina O, Petracco R, Popkhadze S,
Massasa E, Paranjape T, Chan E, Flores I, Weidhaas JB and Taylor
HS: A polymorphism in a let-7 microRNA binding site of KRAS in
women with endometriosis. EMBO Mol Med. 4:206–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meuleman C, Vandenabeele B, Fieuws S,
Spiessens C, Timmerman D and D'Hooghe T: High prevalence of
endometriosis in infertile women with normal ovulation and
normospermic partners. Fertil Steril. 92:68–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai H, Zhu X, Li Z, Zhu Y and Lang J:
lncRNA/mRNA profiling of endometriosis rat uterine tissues during
the implantation window. Int J Mol Med. 44:2145–2160.
2019.PubMed/NCBI
|
|
23
|
Crossley BM, Bai J, Glaser A, Maes R,
Porter E, Killian ML, Clement T and Toohey-Kurth K: Guidelines for
sanger sequencing and molecular assay monitoring. J Vet Diagn
Invest. 32:767–775. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen L, Hu P, Zhang Y, Ji Z, Shan X, Ni L,
Ning N, Wang J, Tian H, Shui G, et al: Serine metabolism
antagonizes antiviral innate immunity by preventing
ATP6V0d2-mediated YAP lysosomal degradation. Cell Metab.
33:971–987.e6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lv H, Lv G, Chen C, Zong Q, Jiang G, Ye D,
Cui X, He Y, Xiang W, Han Q, et al: NAD (+) metabolism maintains
inducible PD-L1 expression to drive tumor immune evasion. Cell
Metab. 33:110–127.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roux P, Perrin J, Mancini J, Agostini A,
Boubli L and Courbiere B: Factors associated with a poor prognosis
for the IVF-ICSI live birth rate in women with rAFS stage III and
IV endometriosis. J Assist Reprod Genet. 34:921–928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szwarc MM, Hai L, Gibbons WE, Peavey MC,
White LD, Mo Q, Lonard DM, Kommagani R, Lanz RB, DeMayo FJ and
Lydon JP: Human endometrial stromal cell decidualization requires
transcriptional reprogramming by PLZF. Biol Reprod. 98:15–27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rytkonen KT, Erkenbrack EM, Poutanen M,
Elo LL, Pavlicev M and Wagner GP: Decidualization of human
endometrial stromal fibroblasts is a multiphasic process involving
distinct transcriptional programs. Reprod Sci. 26:323–336. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vasquez YM, Mazur EC, Li X, Kommagani R,
Jiang L, Chen R, Lanz RB, Kovanci E, Gibbons WE and DeMayo FJ:
FOXO1 is required for binding of PR on IRF4, novel transcriptional
regulator of endometrial stromal decidualization. Mol Endocrinol.
29:421–433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feng C, Shen JM, Lv PP, Jin M, Wang LQ,
Rao JP and Feng L: Construction of implantation failure related
lncRNA-mRNA network and identification of lncRNA biomarkers for
predicting endometrial receptivity. Int J Biol Sci. 14:1361–1377.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cazalla D, Yario T and Steitz JA:
Down-regulation of a host microRNA by a herpesvirus
saimiriNoncoding RNA. Science. 328:1563–1566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Braza-Boils A, Mari-Alexandre J, Gilabert
J, Sánchez-Izquierdo D, España F, Estellés A and Gilabert-Estellés
J: MicroRNA expression profile in endometriosis: Its relation to
angiogenesis and fibrinolytic factors. Hum Reprod. 29:978–988.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu JH, Chang WH, Fu HW, Yuan T and Chen P:
The mRNA, miRNA and lncRNA networks in hepatocellular carcinoma: An
integrative transcriptomic analysis from gene expression omnibus.
Mol Med Rep. 17:6472–6482. 2018.PubMed/NCBI
|
|
37
|
Song X, Cheng L, Zhou T, Guo X, Zhang X,
Chen YPP, Han P and Sha J: Predicting miRNA-mediated gene silencing
mode based on miRNA-target duplex features. Comput Biol Med.
42:1–7. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li F, Orban R and Baker B: SoMART: A web
server for plant miRNA, tasiRNA and target gene analysis. Plant J.
70:891–901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pang M, Xing C, Adams N, Rodriguez-Uribe
L, Hughs SE, Hanson SF and Zhang J: Comparative expression of miRNA
genes and miRNA-based AFLP marker analysis in cultivated tetraploid
cottons. J Plant Physiol. 168:824–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brodersen P, Sakvarelidze-Achard L,
Schaller H, Khafif M, Schott G, Bendahmane A and Voinnet O:
Isoprenoid biosynthesis is required for miRNA function and affects
membrane association of ARGONAUTE 1 in Arabidopsis. Proc
Natl Acad Sci USA. 109:1778–1783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiong WC, Han N, Wu N, Zhao KL, Han C,
Wang HX, Ping GF, Zheng PF, Feng H, Qin L and He P: Interplay
between long noncoding RNA ZEB1-AS1 and miR-101/ZEB1 axis regulates
proliferation and migration of colorectal cancer cells. Am J Transl
Rese. 10:605–617. 2018.PubMed/NCBI
|
|
42
|
Zeng H, Fan X and Liu N: Expression of H19
imprinted gene in patients with repeated implantation failure
during the window of implantation. Arch Gynecol Obstet.
296:835–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Y, Zhang W, Wang Y and Wang S:
HOXD-AS1 promotes cell proliferation, migration and invasion
through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res.
8:170–182. 2018.PubMed/NCBI
|
|
44
|
Liu T, Liu M, Zheng C, Zhang D, Li M and
Zhang L: Exosomal lncRNA CHL1-AS1 derived from peritoneal
macrophages promotes the progression of endometriosis via the
miR-610/MDM2 Axis. Int J Nanomedicine. 16:5451–5464. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hou F, Li J, Peng J, Teng Z, Feng J and
Xia W: lncRNA TMPO-AS1 suppresses the maturation of miR-335-5p to
participate in polycystic ovary syndrome. J Ovarian Res. 14:992021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xiao B, Zhang W, Chen L, Hang J, Wang L,
Zhang R, Liao Y, Chen J, Ma Q, Sun Z and Li L: Analysis of the
miRNA-mRNA-lncRNA network in human estrogen receptor-positive and
estrogen receptor-negative breast cancer based on TCGA data. Gene.
658:28–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou G, Li C, Feng J, Zhang J and Fang Y:
lncRNA UCA1 is a novel regulator in cardiomyocyte hypertrophy
through targeting the miR-184/HOXA9 axis. Cardiorenal Med.
8:130–139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wambecke A, Ahmad M, Morice PM, Lambert B,
Weiswald LB, Vernon M, Vigneron N, Abeilard E, Brotin E, Figeac M,
et al: The lncRNA ‘UCA1’ modulates the response to chemotherapy of
ovarian cancer through direct binding to miR-27a-5p and control of
UBE2N levels. Mol Oncol. 15:3659–3678. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Peng J, Hou F, Feng J, Xu SX and Meng XY:
Long non-coding RNA BCYRN1 promotes the proliferation and
metastasis of cervical cancer via targeting microRNA-138 in
vitro and in vivo. Oncol Lett. 15:5809–5818.
2018.PubMed/NCBI
|
|
50
|
Mizoguchi T, MacDonald BT, Bhandary B,
Popp NR, Laprise D, Arduini A, Lai D, Zhu QM, Xing Y, Kaushik VK,
et al: Coronary disease association with ADAMTS7 is due to protease
activity. Circ Res. 129:458–470. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao N, Peng S, Wu H, Liu W, Cai D and
Huang D: Long noncoding RNA PVT1 promotes chondrocyte extracellular
matrix degradation by acting as a sponge for miR-140 in
IL-1β-stimulated chondrocytes. J Orthop Surg Res. 17:2182022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang X, Li D, Jia C, Cai H, Lv Z and Wu
B: METTL14 promotes tumorigenesis by regulating lncRNA
OIP5-AS1/miR-98/ADAMTS8 signaling in papillary thyroid cancer. Cell
Death Dis. 12:6172021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dou P, Hu R, Zhu W, Tang Q, Li D, Li H and
Wang W: Long non-coding RNA HOTAIR promotes expression of ADAMTS-5
in human osteoarthritic articular chondrocytes. Pharmazie.
72:113–117. 2017.PubMed/NCBI
|
|
54
|
Du Y, Gao C, Liu Z, Wang L, Liu B, He F,
Zhang T, Wang Y, Wang X, Xu M, et al: Upregulation of a disintegrin
and metalloproteinase with thrombospondin motifs-7 by miR-29
repression mediates vascular smooth muscle calcification.
Arterioscler Thromb Vasc Biol. 32:2580–2588. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hanin G, Shenhar-Tsarfaty S, Yayon N, Yau
YH, Bennett ER, Sklan EH, Rao DC, Rankinen T, Bouchard C,
Geifman-Shochat S, et al: Competing targets of microRNA-608 affect
anxiety and hypertension. Hum Mol Genet. 23:4569–4580. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cai H, Zhu XX, Li ZF and Lang JH: MicroRNA
dysregulation and steroid hormone receptor expression in uterine
tissues of rats with endometriosis during the implantation window.
Chin Med J (Engl). 131:2193–2204. 2018. View Article : Google Scholar : PubMed/NCBI
|