Introduction
Hepatitis B virus (HBV) infection is a serious
condition that endangers patients worldwide. Approximately 250
million people have chronic hepatitis B (1) and are at a high risk of developing
liver cirrhosis and hepatocellular carcinoma (2). Therefore, there is an urgent need for
additional studies determining the mechanism of and novel
treatments for HBV infection. Although current therapeutic regimens
for hepatitis B are continuously being optimized, the primary
treatment measures are still antiviral treatments with varying
efficacy and prognostic outcomes. The limited efficacy is primarily
associated with the ability of HBV to evade innate immunity through
various mechanisms (3–6), including destroying the corresponding
recognition receptors (7) and
depleting the inflammatory factors important for the activation of
adaptive immunity (8). Studies on
HBV are becoming increasingly diverse, ranging from studies at the
molecular and protein level, to studying the impact of the
occurrence and development of viral hepatitis (9–11).
However, to the best of our knowledge, only one study (12) has investigated the effects of
transcription factors on the invasive activity of HBV in
hepatocytes.
ATOH8, a transcription factor of the basic
helix-loop-helix (bHLH) superfamily of proteins, is involved in the
occurrence and development of various types of cancer, exhibiting
cancer-specific effects. For example, upregulated expression of
ATOH8 in the development of breast cancer is associated with a
poorer prognosis (13). In
contrast, in liver cancer, its overexpression plays a positive role
in suppressing tumor development (14), consistent with our previous
studies, which showed that overexpression of ATOH8 inhibited the
migration and metastasis of liver cancer (15). However, to the best of our
knowledge, there are no studies assessing the immunoregulatory
effects of ATOH8 and its role in liver cancer development.
Unlike apoptosis, pyroptosis is a form of
inflammatory cell death. It involves a chain action of
inflammasomes, Caspase-1, GSDMD, and other molecules that induce
programmed inflammatory cell death, playing an important role in
various infectious and immune diseases (16–18).
Similar to the hepatitis C virus (HCV), which can induce hepatocyte
pyroptosis (19); HBV can also
induce several inflammatory responses; however, there are few
studies on the relationship between HBV and pyroptosis.
Therefore, this study focused on the effects of
ATOH8 on immune regulation in response to HBV infection. Our
previous study revealed that ATOH8 enables chemotactic monocytes to
secrete inflammatory factors (15). To further explore the role of ATOH8
in immune regulation, HepG2.2.15 liver cancer cells stably
expressing HBV were sued to investigate whether ATOH8 interfered
with the activity of HBV through pyroptosis and further explore the
invasive mechanism of HBV.
Materials and methods
The experimental methods used in the present study
primarily included collecting clinical samples (DNA and RNA),
protein extraction, cDNA reverse transcription, qPCR, western
blotting, and ELISA. In addition, a recombinant lentiviral vector
was used to infect HepG2.2.15 and Huh7 cells to overexpress ATOH8.
Relative RT-qPCR was used to detect the mRNA expression levels in
various cell lines. Western blotting was used to detect the
relative expression levels of the target proteins in the cell lines
used in the present study, and ELISA was used to detect the
expression of various proteins or cytokines in the cell culture
supernatant. The detection of HBV expression in HepG2.2.15 cells
was divided into the detection of HBV DNA in HepG2.2.15 cells
(absolute RT-qPCR detection) and human hepatitis B surface antigen
(HBsAg) in the culture supernatant of HepG2.2.15 cells (ELISA
detection). Signed informed consent was obtained from all patients
for all clinical samples used in the present study.
Clinical samples
Fresh tissue specimens of human liver cancer and
adjacent non-tumor tissues were collected from patients with
hepatocellular carcinoma (HCC) (9 males and 3 females, aged 35–60
years old; median age, 54.5 years) who underwent hepatectomy at the
Shanghai Public Health Clinical Center, Fudan University, between
March 2018 and September 2020. Blood samples were collected from
patients with HBV (1 female and 3 males, aged 40–65 years old)
between April 2021 and June 2022, who underwent antiviral treatment
at the same hospital. Normal blood samples (2 females and 2 males,
aged 30–40 years old) were collected from volunteers during the
same period of time at the same hospital. The present study was
approved by the Ethics Committee (approval no. 2016-S026-08) of the
Shanghai Public Health Clinical Center; all patients provided
written informed consent.
Cell culture
The HepG2.2.15 and Huh7 liver cancer cell lines used
in the present study were all purchased from ATCC. HepG2.2.15 cells
were cultured in complete MEM (BI) supplemented with 10% FBS (BI),
1% penicillin-streptomycin solution (BI) and 5 µg/ml puromycin
[Yeason Biotechnology (Shanghai) Co., Ltd.] in a humidified
incubator at 37°C supplied with 5% CO2. Huh7 cells were
cultured in complete DMEM supplemented with 10% FBS, 1% PSA and 2
µg/ml puromycin. The identity of HepG2.2.15 cells used in the
present study was verified by STR profiling.
Construction of ATOH8 overexpressing
cells
Huh7 cells overexpressing ATOH8 that were used in
the present study were constructed in a previous study (15), and the same protocol as that
described in the previous study was used to construct
HepG2.2.15-ATOH8 overexpressing cells, using the same
pHBLV-CMVIE-ZsGreen-Puro vector. After 48 h of infection, the
fluorescence status of the cells was observed under a fluorescence
microscope (×10 magnification), and the cells were screened for
successful infection using puromycin [Yeason Biotechnology
(Shanghai) Co., Ltd.]. Validation of the successful construction of
ATOH8-overexpressing cells was performed using qPCR; verification
results of successful construction of Huh7-ATOH8 cells were shown
in our previous study (15).
Western blotting
A total of 1×106−107 cells
were collected and lysed using RIPA lysis buffer on ice for 15–30
min and centrifuged at 4°C, at 12,000-14,000 × g for 10 min. The
supernatant was collected, 1× Loading buffer was added, and the
samples were denatured by heating in a metal bath at 100°C for 10
min and stored at −20°C for later use. Equal quantities of protein
were loaded on a 12% SDS-gel (YESEN, China), resolved using
SDS-PAGE, transferred to methanol-activated PVDF membranes, blocked
with 5% skimmed milk for 15–30 min, and incubated with different
primary antibodies at 4°C overnight. The following day, the
membranes were washed and incubated with horseradish
peroxidase-conjugated secondary antibody for 45–60 min at 25°C.
Signals were visualized using an Immobilon Western Kit
(MilliporeSigma). The antibodies used in this study were:
Anti-GSDMD (1:2,000; cat. no. AF4012; Affinity Biosciences);
Anti-Cleaved-Caspase-1 (1:2,000; cat. no. AF4005, Affinity
Biosciences); anti-Caspase-1 (1:1,000; cat. no. ab138483; Abcam);
anti-β-actin [1:2,000; cat. no. 30102ES60; Yeason Biotechnology
(Shanghai) Co., Ltd.], and anti-rabbit mAb (1:4,000, cat. no.
5571S; Cell Signaling Technology, Inc.)
RT-qPCR
Total RNA was extracted using Direct-ZOL RNA
Miniprep Kits, according to the manufacturer's instructions (ZYMO
Research Corp.). cDNA was synthesized by reverse transcription of 1
µg RNA and qPCR was performed using a NovoStart SYNR®
qPCR SuperMix Plus kit, both according to the manufacturer's
protocol (Novoprotein). mRNA expression analysis was performed
using Bio-Rad CFX Maestro version 4.0 (supported by the CFX-96 qPCR
instrument; Bio-Rad Laboratories, Inc.). GAPDH was used as the
housekeeping gene, and expression was calculated using the
2−∆∆Cq method. The sequences of the primers used in the
present study were: GAPDH forward, ACGGATTTGGTCGTATTGGG and
reverse, ATCTCGCTCCTGGAAGATGG; ATOH8 forward, CAGGTGCCGTGCTACTCATA
and reverse, CAGGTGCCGTGCTACTCATA; GSDMD forward,
GTGTGTCAACCTGTCTATCAAGG and reverse, CATGGCATCGTAGAAGTGGAAG;
Caspase-1 forward, CCTTAATATGCAAGACTCTCAAGGA and reverse,
TAAGCTGGGTTGTCCTGCACT; IL-18 forward, TCTTCATTGACCAAGGAAATCGG and
reverse, TCCGGGGTGCATTATCTCTAC; IL-1β forward,
ATGATGGCTTATTACAGTGGCAA and reverse, GTCGGAGATTCGTAGCTGGA; TNF-α
forward, TCAGGATCATCTTCTCGAACC and reverse, GAGTCCTTCTCACATTGTCTC;
INF-α forward, AGAATCACTCTCTATCTGAAAGAGAAG and reverse,
TCATGATTTCTGCTCTGACAACCT.
ELISA
The expression of HBsAg in the cell culture
supernatant of HepG2.2.15 cells was detected using a human HBsAg
ELISA kit (cat. no. JM-05282H1, JINGMEI). Absorbance (OD values)
was measured at 450 nm according to the manufacturer's
instructions.
HBV DNA extraction and RT-qPCR viral
load analysis
The intracellular HBV DNA of HepG2.2.15 cells was
extracted according to the manufacturer's instructions of the Viral
DNA/RNA Kit (YESEN). The extracted DNA was directly used for qPCR
(Hepatitis B Virus Nucleic Acid Assay Kit, Sansure Biotech) or
stored at −20°C (but it is recommended to proceed to the next step
of qPCR immediately). qPCR was performed as above for evaluation of
intracellular or extracellular HBV DNA (HBV DNA in cell culture
supernatant) using the HepG2.2.15 cells. The relative HBV
expression values in HepG2.2.15 cells are presented using a Log
scale, as shown in Table SI.
Statistical analysis
Data were compared using a paired Student's t-tests.
Densitometry analysis of the western blot bands was performed using
ImageJ (version 2.3.0; National Institutes of Health). Normally
distributed are presented as the mean ± SD, whereas those of skewed
data are presented as the median (interquartile range). P<0.05
was considered to indicate a statistically significant difference.
Data were analyzed using GraphPad Prism version 9 (GraphPad
Software, Inc.).
Results
Pyroptosis is increased in liver
cancer tissues and PBMCs of patients with HBV
As previously reported, the expression of ATOH8 was
decreased in liver cancer (15).
To detect the relationship between ATOH8 and pyroptosis, liver
cancer tissues from HCC patients were assessed and it was found
that pyroptosis was more common in cancerous tissues than in
paracancerous tissues (Fig. 1A).
To detect the relationship between HBV and pyroptosis, PBMCs were
isolated from the blood samples of patients with HBV to detect the
expression of pyroptotic molecules. The expression of pyroptotic
molecules in the PBMCs of patients with HBV was higher than that in
normal PBMCs (Fig. 1B and C).
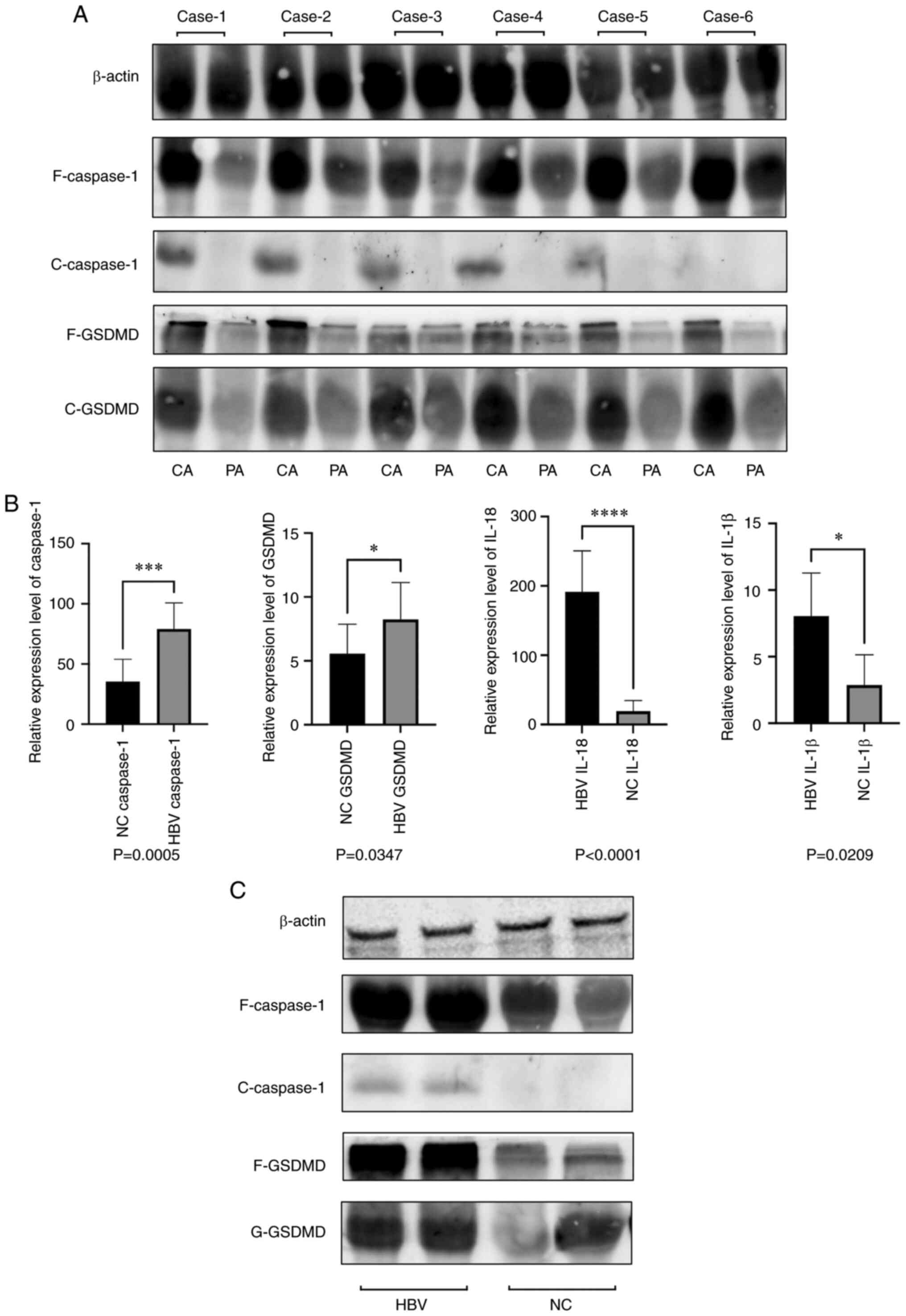 | Figure 1.Expression of pyroptosis in liver
cancer tissues and PBMC of HBV patients. (A) Relative protein
expression levels of full-length Caspase-1, cleaved-Caspase-1,
cleaved-GSDMD, and full-length GSDMD in liver cancer tissues. (B)
Relative mRNA expression levels of Caspase-1, GSDMD, IL-18, and
IL-1β in the PBMCs of HBV patients. (C) Relative protein expression
levels of full-length Caspase-1, cleaved-Caspase-1, cleaved-GSDMD,
and full-length GSDMD in the PBMCs of HBV patients. *P<0.05,
***P<0.001 and ****P<0.0001. PBMC, peripheral blood
mononuclear cells; HBV, hepatitis B virus; F, full-length; C,
cleaved; CA, cancerous; PA, paracancerous; NC, negative
control/normal. |
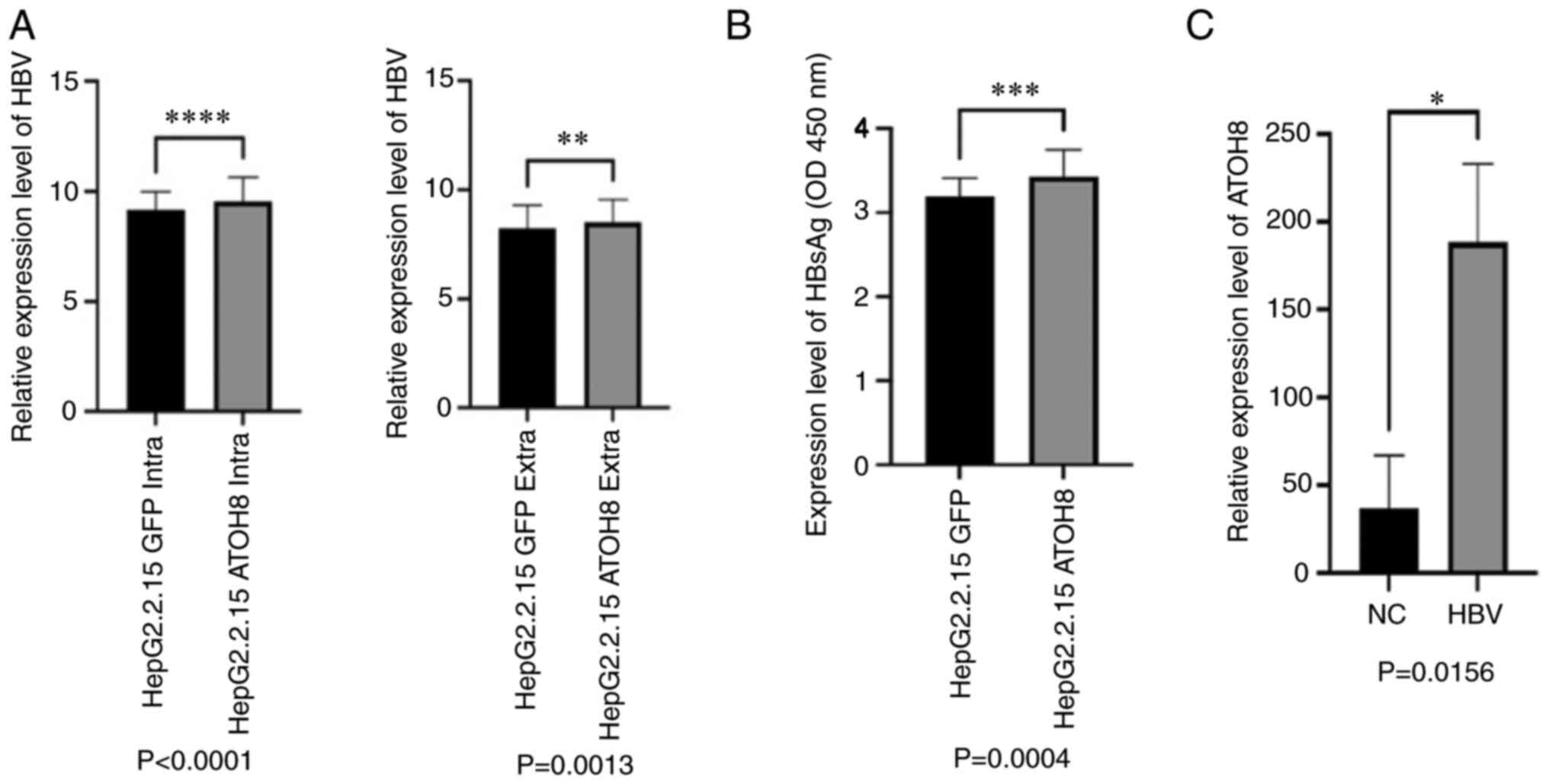
ATOH8 increases HBV DNA expression
levels in HepG2.2.15 cells
Based on our previous screening results that showed
that the expression levels of ATOH8 in the HepG2.2.15 cells were
very low (15), whether ATOH8
inhibited the expression of HBV, and in the process inhibited tumor
proliferation and migration was assessed. The results of the
establishment of the HepG2.2.15-ATOH8 cells are shown in Fig. S1. HBV DNA was extracted from
HepG2.2.15 cells and qPCR was performed. Compared with the control
group, HepG2.2.15 cells in the ATOH8 exhibited increased HBV DNA
levels. Similar results were obtained regarding both intracellular
and extracellular HBV DNA levels (Fig.
2A and B). For further verification, the cell culture
supernatant of HepG2.2.15 cells was collected and the relative
expression levels of HBsAg were measured using ELISA and the
results were consistent with those of qPCR (Fig. 2B). Similarly, the expression of
ATOH8 in the PBMCs of patients with HBV was assessed; the
expression of ATOH8 in patients with HBV was higher than that of
normal samples, further confirming the positive association between
ATOH8 and HBV (Fig. 2C).
ATOH8 inhibits the expression of
pyroptosis-related molecules in HepG2.2.15 and Huh7 cells
GO analysis performed of next-generation sequencing
results in Huh7 cells, performed in our previous study, showed that
the activity of the NF-κB pathway regulated by AKT was inhibited in
ATOH8-overexpressing Huh7 cells (15). Therefore, it was speculated that
ATOH8 may assist in HBV immune escape by inhibiting the activity of
inflammatory pathways. Since NF-κB is a typical trigger pathway of
pyroptosis, qPCR and western blotting were used to detect the
expression of pyroptotic molecules in the HepG2.2.15 cells.
Consistent results were observed between the qPCR and western
blotting analyses, showing that the ATOH8 overexpression group
exhibited a lower degree of pyroptosis (Fig. 3A-C). Similarly, to further verify
whether ATOH8 affected hepatocyte pyroptosis, the expression of
pyroptosis markers in the Huh7 cell line was detected (the hepatoma
cell line with the most apparent decrease in ATOH8 expression).
Compared with the ATOH8-overexpression group, the expression levels
of GSDMD and Caspase-1 in the Huh7-GFP group were significantly
higher (Fig. 3D-F).
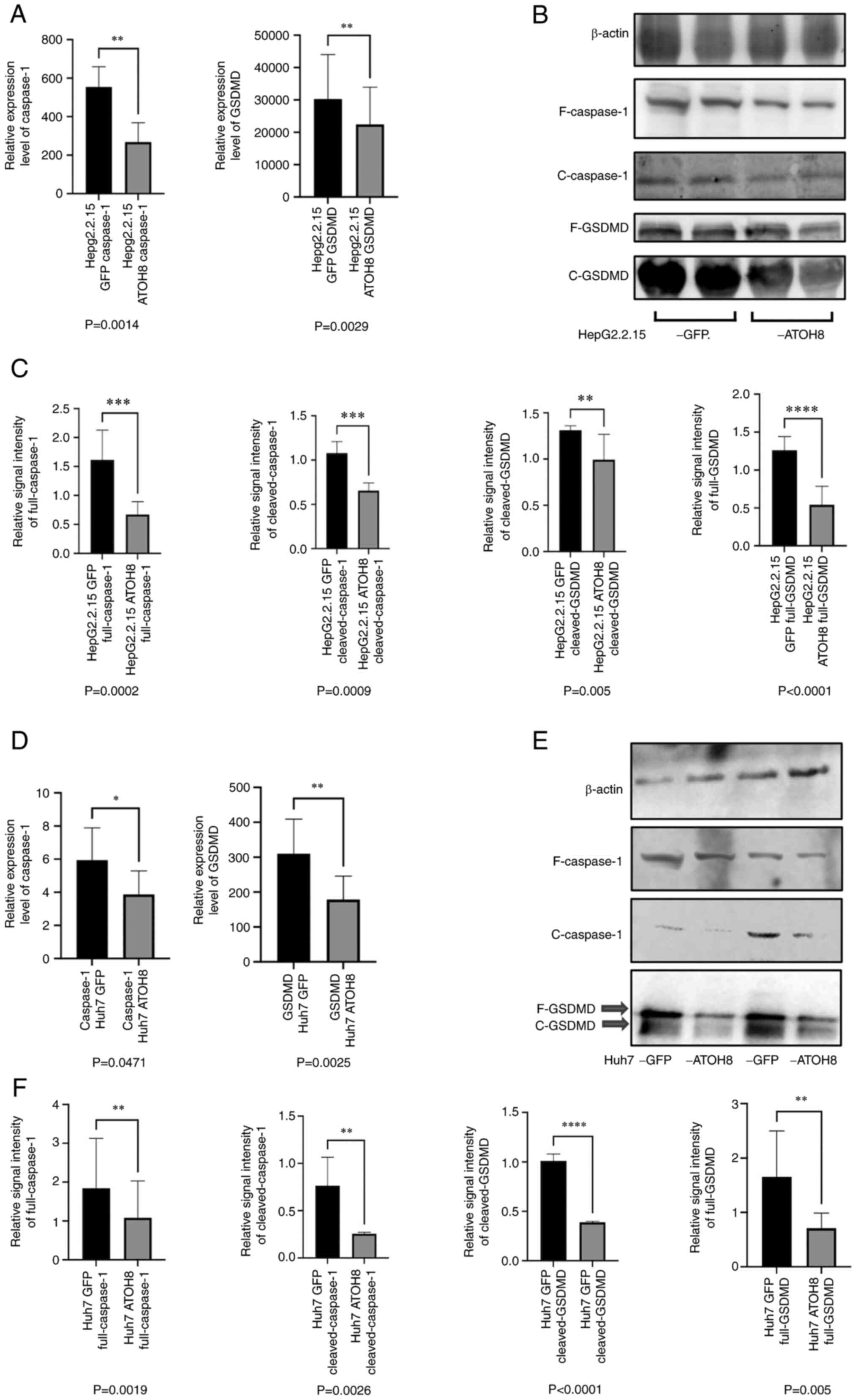 | Figure 3.Effects of ATOH8 on the levels of
pyroptosis-associated proteins in HepG2.2.15 and Huh7 cells. (A)
Relative mRNA expression levels of GSDMD and Caspase-1 in
HepG2.2.15 overexpressing ATOH8. (B and C) Relative protein
expression levels of full-length Caspase-1, cleaved-Caspase-1,
cleaved-GSDMD, and full-length GSDMD in HepG2.2.15 cells
overexpressing ATOH8. (D) Relative mRNA expression levels of GSDMD
and Caspase-1 in Huh7-GFP and Huh7-ATOH8 cells. (E and F) Relative
protein expression levels of full-length Caspase-1,
cleaved-Caspase-1, cleaved-GSDMD, and full-length GSDMD in Huh7-GFP
and Huh7-ATOH8 cells. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. F, full-length; C, cleaved. |
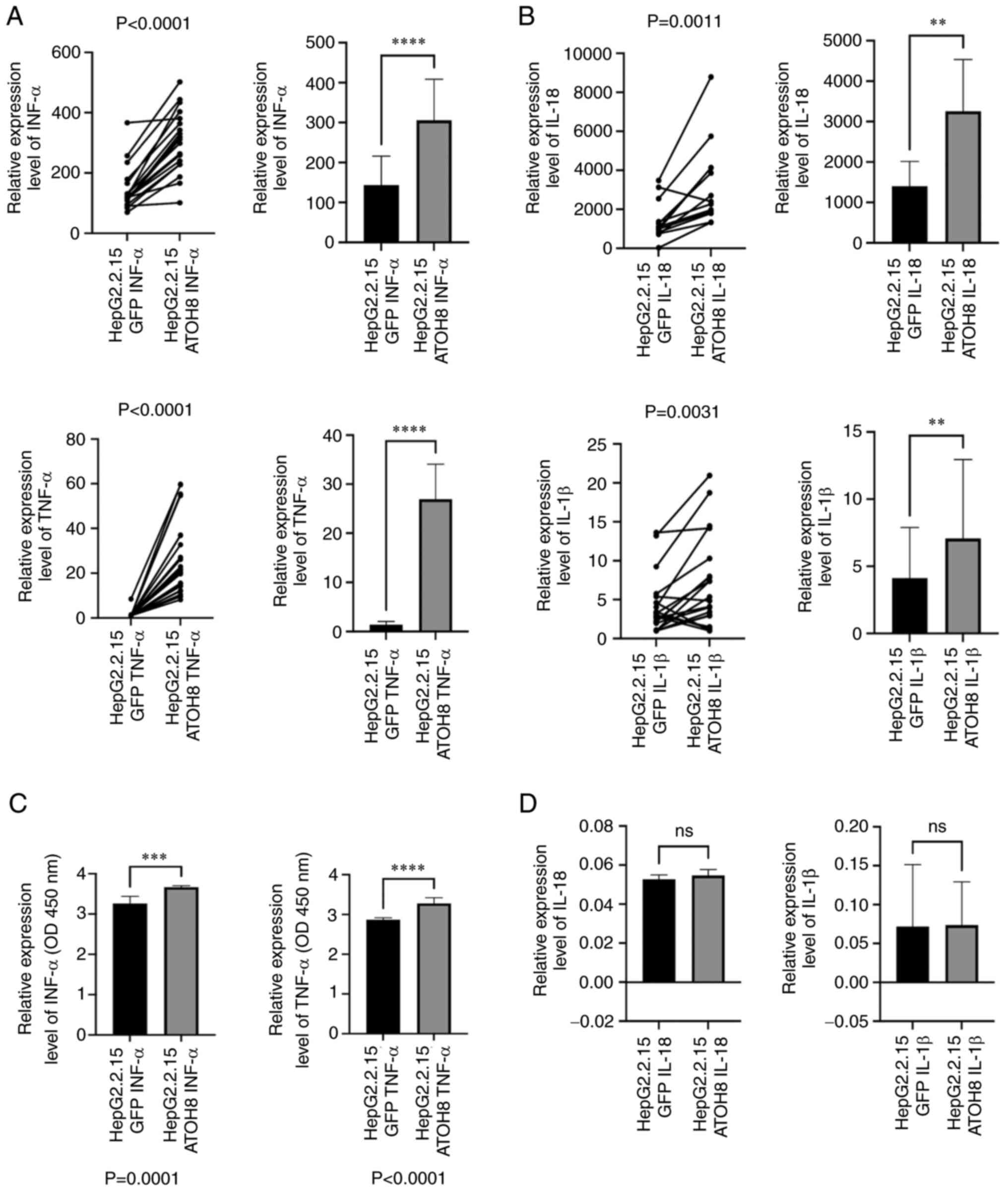
ATOH8 promotes the secretion of
antiviral factors in HepG2.2.15 cells
According to our previous study, ATOH8 affects the
chemotaxis of monocytes and induces them to secrete inflammatory
factors (15). Therefore, whether
ATOH8 affected the secretion of antiviral inflammatory factors in
HepG2.2.15 cells was assessed. The expression levels of INF-α and
TNF-α in HepG2.2.15 using qPCR; the results were consistent with
our previous study, showing INF-α and TNF-α secretion from
HepG2.2.15 cells was significantly higher in the ATOH8
overexpression group than in the control group (Fig. 4A and C). Furthermore, the
expression of anti-HBV inflammatory factors, such as IL18 and
IL-1β, which are primarily released during pyroptosis, were
assessed; and the results were consistent with the above. The
expression levels of IL-18 and IL-1β in the control group were
lower, but the differences were not statistically significant
(Fig. 4B and D). This result
contradicts the result regarding the inhibition of pyroptosis in
the ATOH8 overexpression group.
Discussion
ATOH8 is a transcription factor in the basic
helix-loop-helix (bHLH) superfamily that affects physiological
growth and development. In the present study, the relationship
between pyroptosis and HBV was assessed. It was shown that HBV
escaped immune surveillance by inhibiting hepatocyte pyroptosis.
Previous studies on ATOH8 have primarily focused on tumors,
assessing the effect of upregulation or downregulation on tumor
development. Moreover, the upregulation of ATOH8 has opposing
effects in different types of cancer (13,14).
However, to the best of our knowledge, only one study (20) has investigated the role of ATOH8 on
inflammation.
In our previous study in which Huh7 liver carcinoma
cells were used, it was shown that ATOH8 played a role in the tumor
immune microenvironment. For example, ATOH8 can positively affect
monocyte chemotaxis and induce the secretion of inflammatory
factors (15), Therefore, to
further explore the role of ATOH8 on immune regulation, HepG2.2.15
liver carcinoma cells, which endogenously express low levels of
ATOH8 and stably express HBV, were used. Compared with other types
of liver cancer, HBV-induced liver cancer can influence the liver
microenvironment, which may be more representative of immune
regulation studies (21).
It was initially hypothesized that if ATOH8 played
an active role in immune regulation, there would be a decrease in
the levels of HBV released from HepG2.2.15-ATOH8 cells. However,
the negative results of HBV DNA in HepG2.2.15 cells and HBsAg
expression in the cell culture supernatant showed higher levels of
HBV in the ATOH8-overexpressing group, suggesting that ATOH8 may
contribute to the immune escape of HBV. GO analysis was performed
on the results of next-generation sequencing (NGS) in Huh7 cells in
our previous study (15) and it
was found that ATOH8 played an active role in regulating Huh7
inflammation, manifested as decreased in AKT expression and altered
regulation of the NF-κB pathway activity in the Huh7-ATOH8 group.
Thus, considering the important role of NF-κB during pyroptosis, it
was speculated that ATOH8 assisted HBV immune escape by inhibiting
HepG2.2.15 cell pyroptosis. Currently, there is only one study on
the relationship between HBV and pyroptosis, to the best of our
knowledge; only Yu et al (22) has shown that the immune tolerance
of HBV was related to its action on the pyroptotic pathway,
consistent with the findings of the present study. Therefore, to
further verify our hypothesis, the expression of pyroptosis-related
molecules in the HepG2.2.15 and Huh7 cells were determined; the
results were consistent with the results of GO analysis in our
previous study, showing decreased expression of pyroptosis-related
molecules in the ATOH8-overexpressing cell group. Furthermore,
these results were confirmed using clinical samples. Compared with
the paracancerous tissues, liver cancer tissues with low ATOH8
expression showed increased expression of molecules related to
pyroptosis, showing that ATOH8 could affect the expression of
pyroptotic molecules.
The studies by Xie et al (23) and Yu et al (22) have results that contradict each
other; the present study assessed the ATOH8 transcription factor
and may explain the opposing phenomenon observed in the two
studies. Xie et al (23)
showed that HBV activates the NLRP3 inflammasome under hypoxic
conditions, whereas Yu et al (22) showed that HBV achieved escape from
immune surveillance by inhibiting the NF-κB pathway. The difference
in findings may be explained by the differential expression of
ATOH8 in the development of liver inflammation. During the
development from normal liver cells, which rarely undergo
pyroptosis, to chronic hepatitis, and then to liver cancer,
expression of ATOH8 increases as the cancer develops. Following
ATOH8 inhibition, HBV immune escape was reduced. Therefore, the
difference between the results of the present study and those of
previous studies may be due to the study of different stages of
hepatitis B, and the varying roles of ATOH8 in each stage.
Regarding the lower inflammasome expression levels in HBV-induced
liver cancer than that in the adjacent tissues (23,24),
this may be explained by the fact that following
hepatocarcinogenesis, pyroptosis in liver cells is inhibited to
maintain the survival state of cancer cells. In contrast, the
adjacent tissues maintain a high degree of inflammatory response
under HBV stimulation. This explanation does not contradict the
results of the present study, in that the incidence of pyroptosis
in liver cancer tissues was higher than that in adjacent tissues.
Although pyroptosis is inhibited in cancer cells compared to normal
tissues, cancer antigens in cancer tissues still stimulate cancer
cells to induce inflammatory responses; such inflammation may be
more evident without the inhibitory effect of ATOH8.
In the present study, in addition to assessing the
effect of ATOH8 on HBV immune escape, the expression levels of
certain antiviral factors, such as TNF-α, INF-α, IL-18, and IL-1β
in HepG2.2.15 cells were also determined. Notably, the expression
of these antiviral factors was higher in the ATOH8 overexpressing
cells compared with the control group. In addition, the expression
of certain inflammatory factors (IL-18 and IL-1β) was higher in the
ATOH8 overexpressing cells, and these were likely primarily
produced during pyroptosis. However, only differences in the mRNA
levels of IL-18 and IL-1β were detected, and these are associated
with pyroptosis primarily due to their activation by Caspase-1
(25,26). There was no significant difference
in IL-18 and IL-1β with regard to the extracellular expression
levels detected using ELISA between the two groups as they required
cleavage of Caspase-1, and this process was blocked by ATOH8.
Therefore, the expression of these antiviral factors may be a
compensatory increase in response to HBV invasion, indirectly
aggravating the inflammatory state during HBV invasion.
To further understand the primary antiviral
mechanism of normal hepatocytes during HBV invasion, the expression
of antiviral inflammatory factors and pyroptosis-related molecules
in PBMCs of patients with HBV was detected. The results showed that
PBMCs from patients with HBV exhibited higher expression levels of
pyroptotic molecules and increased secretion of antiviral
inflammatory factors. However, the expression of ATOH8 in PBMCs of
patients with HBV was higher than that in normal samples,
indicating that ATOH8 did not inhibit pyroptosis in PBMCs, only in
hepatocytes. Therefore, considering the positive role of pyrolytic
inflammatory molecules in anti-HBV activity, it is hypothesized
that when HBV invades, ATOH8 inhibits hepatocyte pyroptosis, the
PBMCs of patients exert antiviral effects, and the antiviral
effects of hepatocytes are limited.
The present study has some limitations. For example,
the findings of the present study were not validated in animal
experiments. Additionally, the specific mechanism by which ATOH8
suppressed pyroptosis requires further elucidation.
In conclusion, ATOH8 interfered with the host's
innate immune system by inhibiting hepatocyte pyroptosis and
assisting HBV immune escape. The results of the present study may
provide novel avenues for research on the mechanisms of hepatitis
caused by HBV and for the development of specific immune inducers
for the treatment of hepatitis B. For example, creating an
inflammatory environment conducive to the expression of ATOH8 to
further interfere with the progression of hepatitis B and assist in
treating patients with hepatitis B with antiviral drugs.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by funding from the Shanghai Public
Health Clinical Center Fund Project (grant no. KY-GW-2021-31) and
the National Science and Technology Major Project (grant no.
2017ZX10203202-003-007).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and LC designed the study. JC helped designed the
study. XL, ZF, and JY performed the experiments. XL and ZF analyzed
the data. XL and JC wrote the manuscript. XL and JC confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol used in the present study
conformed to the guidelines described in the Declaration of
Helsinki, and was approved by the Human Ethics Committee of the
Shanghai Public Health Clinical Center (approval no.
2016-S026-08).
Patient consent for publication
All patients included in the present study provided
signed informed consent and agreed to the publication of their
data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nicolini LA, Orsi A, Tatarelli P, Viscoli
C, Icardi G and Sticchi L: A global view to HBV chronic infection:
Evolving strategies for diagnosis, treatment and prevention in
immunocompetent individuals. Int J Environ Res Public Health.
16:33072019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pollicino T and Caminiti G:
HBV–Integration studies in the clinic: Role in the natural history
of infection. Viruses. 13:3682021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou L, He R, Fang P, Li M, Yu H, Wang Q,
Yu Y, Wang F, Zhang Y, Chen A, et al: Hepatitis B virus rigs the
cellular metabolome to avoid innate immune recognition. Nat Commun.
12:982021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wieland S, Thimme R, Purcell RH and
Chisari FV: Genomic analysis of the host response to hepatitis B
virus infection. Proc Natl Acad Sci USA. 101:6669–6674. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lebossé F, Testoni B, Fresquet J,
Facchetti F, Galmozzi E, Fournier M, Hervieu V, Berthillon P, Berby
F, Bordes I, et al: Intrahepatic innate immune response pathways
are downregulated in untreated chronic hepatitis B. J Hepatol.
66:897–909. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luangsay S, Gruffaz M, Isorce N, Testoni
B, Michelet M, Faure-Dupuy S, Maadadi S, Ait-Goughoulte M, Parent
R, Rivoire M, et al: Early inhibition of hepatocyte innate
responses by hepatitis B virus. J Hepatol. 63:1314–1322. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Visvanathan K, Skinner NA, Thompson AJV,
Riordan SM, Sozzi V, Edwards R, Rodgers S, Kurtovic J, Chang J,
Lewin S, et al: Regulation of toll-like receptor-2 expression in
chronic hepatitis B by the precore protein. Hepatology. 45:102–110.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Sun R, Xu L, Yin W, Chen Y, Zheng X,
Lian Z, Wei H and Tian Z: Kupffer cells support hepatitis B
virus-mediated CD8+ T cell exhaustion via hepatitis B core
antigen-TLR2 interactions in mice. J Immunol. 195:3100–3109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teng Y, Xu Z, Zhao K, Zhong Y, Wang J,
Zhao L, Zheng Z, Hou W, Zhu C, Chen X, et al: Novel function of
SART1 in HNF4α transcriptional regulation contributes to its
antiviral role during HBV infection. J Hepatol. 75:1072–1082. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Chen X, Cao Y and Yang Z: Roles
of APOBEC3 in hepatitis B virus (HBV) infection and
hepatocarcinogenesis. Bioengineered. 12:2074–2086. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lak R, Yaghobi R and Garshasbi M:
Importance of miR-125a-5p and miR-122-5p expression in patients
with HBV infection. Cell Mol Biol (Noisy-le-grand). 66:1–8. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oropeza CE, Tarnow G, Sridhar A, Taha TY,
Shalaby RE and McLachlan A: The regulation of HBV transcription and
replication. Adv Exp Med Biol. 1179:39–69. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu M, Huang S, Dong X, Chen Y, Li M, Shi
W, Wang G, Huang C, Wang Q, Liu Y, et al: A novel isoform of ATOH8
promotes the metastasis of breast cancer by regulating RhoC. J Mol
Cell Biol. 13:59–71. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song Y, Pan G, Chen L, Ma S, Zeng T, Chan
THM, Li L, Lian Q, Chow R, Cai X, et al: Loss of ATOH8 increases
stem cell features of hepatocellular carcinoma cells.
Gastroenterology. 149:1068–1081.e5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Yang J, Wang Y, Wu N, Li X, Li J,
Huang Y and Cheng J: ATOH8 overexpression inhibits the tumor
progression and monocyte chemotaxis in hepatocellular carcinoma.
Int J Clin Exp Pathol. 13:2534–2543. 2020.PubMed/NCBI
|
|
16
|
Yu P, Zhang X, Liu N, Tang L, Peng C and
Chen X: Pyroptosis: Mechanisms and diseases. Signal Transduct
Target Ther. 6:1282021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song L, Pei L, Yao S, Wu Y and Shang Y:
NLRP3 inflammasome in neurological diseases, from functions to
therapies. Front Cell Neurosci. 11:632017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pezuk JA: Pyroptosis in combinatorial
treatment to improve cancer patients' outcome, is that what we
want? EBioMedicine. 41:17–18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Zeyu W, Liu B, Jang S, Zhang Z
and Jiang Y: Pyroptosis in liver disease. Rev Esp Enferm Dig.
113:280–285. 2021.PubMed/NCBI
|
|
20
|
Ługowska A, Hetmańczyk-Sawicka K,
Iwanicka-Nowicka R, Fogtman A, Cieśla J, Purzycka-Olewiecka JK,
Sitarska D, Płoski R, Filocamo M, Lualdi S, et al: Gene expression
profile in patients with Gaucher disease indicates activation of
inflammatory processes. Sci Rep. 9:60602019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Y, Han Q, Zhao H and Zhang J: The
mechanisms of HBV-induced hepatocellular carcinoma. J Hepatocell
Carcinoma. 8:435–450. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu X, Lan P, Hou X, Han Q, Lu N, Li T,
Jiao C, Zhang J, Zhang C and Tian Z: HBV inhibits LPS-induced NLRP3
inflammasome activation and IL-1β production via suppressing the
NF-κB pathway and ROS production. J Hepatol. 66:693–702. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie WH, Ding J, Xie XX, Yang XH, Wu XF,
Chen ZX, Guo QL, Gao WY, Wang XZ and Li D: Hepatitis B virus X
protein promotes liver cell pyroptosis under oxidative stress
through NLRP3 inflammasome activation. Inflamm Res. 69:683–696.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X,
Zhao W, Huai W, Guo P and Han L: Deregulation of the NLRP3
inflammasome in hepatic parenchymal cells during liver cancer
progression. Lab Invest. 94:52–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zarković G: Population growth and planning
in Yugoslavia. Nar Zdrav. 28:41–48. 1972.(In Serbian). PubMed/NCBI
|
|
26
|
Whitsett TL, Levin DC and Manion CV:
Comparison of the beta 1 and beta 2 adrenoceptor blocking
properties of acebutolol and propranolol. Chest. 82:668–673. 1982.
View Article : Google Scholar : PubMed/NCBI
|