Introduction
Osteonecrosis of the femoral head (ONFH) frequently
occurs in young adults and can lead to a high disability rate. The
primary mechanism responsible for ONFH development is osteocyte
death resulting from damage to microvascular circulation (1). In addition, cell hypoxia due to
vascular disruption can give rise to mitochondrial permeability,
which may result in apoptosis in smooth muscle, vascular
endothelial cells and osteoblasts (2). Furthermore, a number of studies have
shown that hypoxia can negatively affect osteoblast proliferation,
differentiation and expression of osteoblast-specific genes and
proteins (3,4). Therefore, effective prevention of
osteoblast apoptosis caused by hypoxia has become a feasible mean
for necrosis treatment.
Proteins belonging to the fibroblast growth factor
(FGF) family are defined as humoral factors with β-trefoil
structures. Humans have at least 22 FGF proteins grouped into
several subfamilies (5). FGF1-5
sub-protein molecules are produced in a paracrine form and
endocrine-type FGF19, FGF21 and FGF23 regulate activity in
vivo in a target tissue Klotho protein-dependent manner
(6). FGF23 is secreted by
osteocytes and osteoblasts and is a phosphophilic hormone that
binds to the co-receptor formed by FGFR-αKlotho on the cell
membranes of specific tissues and regulates the metabolism of
systemic vitamin D and phosphorus (7,8).
Phosphorus is one of the major minerals in the body and is involved
in bone formation, cell signal transduction, energy metabolism and
acid-base balance (9). Studies
have reported the regulatory effects of FGF23 on various metabolic
and apoptotic processes (10–12).
However, the effect of FGF23 on osteoblast apoptosis induced by
external injury, especially hypoxia, has not yet been explored.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs 18–22 nucleotides in length that repress the expression of
target genes post-transcriptionally by complementary pairing with
incomplete bases in the bases 3′ untranslated regions, resulting in
the translational arrest of target gene mRNAs (13,14).
miRNAs are widely involved in developing and progressing various
human diseases, including ONFH (15). miR-17-5p, as a member of the miR-17
family, is involved in regulating the progression of
osteoarthritis, osteoporosis, ONFH, myocardial ischemia and various
tumors (16–20). In addition, studies have shown that
miR-17-5p can regulate osteoblast cell differentiation and cell
proliferation in non-traumatic ONFH (18,21).
The present study aimed to investigate the role of
FGF23 in osteoblasts in vitro and the potential mechanisms
involved. TargetScan online database predicted miR-17-5p as a
target of FGF23. Therefore, it was hypothesized that FGF23 might
affect osteoblast apoptosis by targeting miR-17-5p to regulate the
autophagy-signaling pathway.
Materials and methods
Cell culture
The MC3T3-E1 osteoblast cell line was obtained from
Procell Life Science & Technology Co., Ltd. and the primary
osteoblasts were obtained from C57BL/6 mice (22). A total of 12 C57BL/6 mice were used
in this study and all animals were housed in the SPF-grade Animal
Experiment Center of the Institute of Sports Medicine, Shandong
First Medical University, with a room temperature of 22°C, relative
humidity of 55% and 12-h light/dark cycle, standard mice food
feeding, free water intake. Chloral hydrate (3%) was used to
anesthetize the mice by intraperitoneal injection. Neonatal C57BL/6
mice were sacrificed by cervical dislocation method and disinfected
with 70% ethanol and calvaria was predigested in 0.25% trypsin
(MilliporeSigma) at 37°C for 20 min. After the bone fragments were
cut into small pieces of ~1 mm2 and digested in 0.1%
collagenase type II (MilliporeSigma) at 37°C for 50 min to collect
the digested product. Digestion of residual bone fragments was
repeated once with fresh collagenase. The digested product
collected twice was centrifuged at 90 × g at room temperature for 5
min, the supernatant was discarded and the pellet was resuspended
and inoculated with Dulbecco's Modified Eagle's Medium (DMEM;
Gibco; cat. no. C11995500BT; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (FBS; cat. no. 04-001-1ACS;
Biological Industries) and 1% penicillin-streptomycin
(MilliporeSigma). The Biomedical Research Ethics Committee of
Shandong First Medical University approved this study (Shandong
Academy of Medical Sciences; ethics approval no. W202210070243).
All cells were cultured in DMEM at 37°C in a 5% carbon dioxide
incubator. The medium was changed two to three times per week.
Transfection
Primary osteoblasts and MC3T3-E1 cells were plated
at a density of 1×105 cells/ml in 6-well plates at 37°C
in a 5% CO2 constant temperature incubator. When the
cells were 80% confluent, the plasmids were transfected according
to the instructions of the Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The short interfering
(si)RNA-FGF23 plasmid (5′-GCTATCACCTACAGATCCATA-3′), empty vector
plasmid (5′-UUCUCCGAACGUGUCACGU-3′), FGF23 overexpression plasmid
(5′-UCCAUGCAGAGGUUAUCUUC-3′) and miR-17-5p inhibitor
(5′-CAAACACCUUACACACCAGGUAG-3′) were synthesized by Hanbio
Biotechnology Co., Ltd. To prepare the transfection mixture, 4 µg
of plasmid was added to 500 µl of serum-free medium and 5 µl of
Lipofectamine 2000 (cat. no. 11668019; Thermo Fisher) was added to
500 µl of serum-free medium and mixed well and then allowed to
stand at room temperature for 20 min. The cells were washed with
phosphate-buffered saline (PBS) three times and then 1 ml of
Lipofectamine® 2000 transfection mixture and 1.5 ml serum-free
medium were added and the cells were incubated for 6 h at room
temperature. The cells were then switched to the regular medium and
continued to be incubated for 48 h at 37°C in a 5% carbon dioxide
incubator. The transfection efficiency of FGF23 was verified by
western blotting. Transfection with an empty vector served as the
control for plasmid transfection (23).
Cell model and grouping
The incubators were gassed with nitrogen to maintain
an anoxic atmosphere of 5% CO2 and 95% N2
(24). The experimental subjects
were divided into five groups as follows: Normal control group
(NC), hypoxia control group (con), empty vector group (EV), FGF23
overexpression group (FGF23) and FGF23 silencing group
(si-FGF23).
MTT assay
Primary osteoblasts and MC3T3-E1 cells were
collected from each group, digested with trypsin, plated at
1×104 cells/well in 96-well plates and cultured for 48
h. Three replicate wells were used for each group. Then, 10 µl of 5
mg/ml MTT solution (cat. no. Top0191T; Beijing Biotuoda) were
added. After incubation at 37°C for 4 h, 150 µl dimethyl sulfoxide
was added and incubated for 10 min. The absorbance value
(OD490 nm) at a wavelength of 490 nm was measured using
a microplate reader. Three replicate blank samples were used for
testing, three times for each group (25).
Detection of caspase-3 activity
Follow the instructions for using the caspase-3
protein activation assay kit (Nanjing KeyGen Biotech Co., Ltd.), 50
µl of each group of sample proteins were taken, 50 µl 2X reaction
buffer and 5 µl caspase-3 substrate were added and then incubated
at 37°C for 4 h in the dark. The absorbance at 405 nm was measured
using a microplate reader (2).
Flow cytometry
The influence of hypoxia on apoptosis in the
osteoblasts was tested by flow cytometry (FCM; Beckman CytoFLEX
FCM; Beckman Coulter) using a phospholipid-binding protein V
(Annexin V-FITC)/propidium iodide (PI) kit (US Everbright Inc.).
After the experimental hypoxia treatment ended, the cells were
washed three times with phosphate buffer, collected by
centrifugation at 90 × g at 4°C for 5 min and resuspended at
5×104 cells/ml in 500 µl binding buffer. The cells were
then transferred to Eppendorf tubes and incubated with 5 µl Annexin
V-FITC and 5 µl PI for 30 min at room temperature in the dark.
Results were analyzed by FlowJo v10.6.2 software. The apoptosis
rate is the sum of early apoptosis rate and late apoptosis rate
(26).
Fluorescein diacetate (FDA) and
ethidium bromide (EB) staining
Transfected cells were seeded in 6-well plates at
1×105 cells/well and washed twice with PBS after 48 h
incubation. Then, 100 µl of FDA and EB (MilliporeSigma) were added
to each well and incubated at 37°C in the dark for 10 min before
imaging with a fluorescence microscope (27).
Autophagy staining assay
Cellular autophagy was detected with the Cellular
Autophagy Staining Assay Kit (cat. no. C3018S; Beyotime Institute
of Biotechnology). Monodansylcadaverine (MDC) is an eosinophilic
fluorescent probe that can specifically label autophagosomes by ion
capture and specific binding to membrane lipids. After the cells
were treated accordingly, 1 ml of MDC staining solution was added
to each well and incubated for 30 min at 37°C in a cell incubator
protected from light. After washing, 1 ml of Assay Buffer was added
and green fluorescence was observed under a fluorescence
microscope.
Reverse transcription-quantitative
(RT-q) PCR
RT-qPCR was used to assess FGF23 expression. First,
total RNA was isolated from primary osteoblasts (1×106)
and MC3T3-E1 cells (1×106) using an RNA extraction kit
(cat. no. BSC52M1; BioFlux) and complementary DNA was synthesized
using RT kit (cat. no. BSB07M2B, BioFlux) according to the
manufacturer's protocols. The Hieff® qPCR SYBR Green Master Mix
(cat. no. 11184ES03; Shanghai Yeasen Biotechnology Co., Ltd.) was
used for qPCR. PCR primer sequences are shown in Table I. The prepared cDNA, GAPDH and U6
were used as a template and reference for RT-qPCR reactions.
Amplification conditions were as follows: 95°C for 5 min, 95°C for
10 sec, 60°C for 30 sec, 72°C for 30 sec, for a total of 35 cycles.
Finally, the relative FGF23 expression was analyzed using the
2−ΔΔCq method [ΔCq=Cq (target gene)-Cq (reference gene)]
(28). Two replicate blank samples
were used for each group (29).
 | Table I.Sequences of primers for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers for reverse
transcription-quantitative PCR.
| Gene | Sequence,
5′à3′ |
|---|
| FGF23 | FOR:
ATGCTAGGGACCTGCCTTAGA |
|
| REV:
GGAGCCAAGCAATGGGGAA |
| Bax | FOR:
AGACAGGGGCCTTTTTGCTAC |
|
| REV:
AATTCGCCGGAGACACTCG |
| Bcl-2 | FOR:
TGACTTCTCTCGTCGCTACCGT |
|
| REV:
CCTGAAGAGTTCCTCCACCACC |
| caspase-3 | FOR:
CTCGCTCTGGTACGGATGTG |
|
| REV:
TCCCATAAATGACCCCTTCATCA |
| caspase-9 | FOR:
GGCTGTTAAACCCCTAGACCA |
|
| REV:
TGACGGGTCCAGCTTCACTA |
| Beclin-1 | FOR:
GAGATTGGACCAGGAGGAAGCT |
|
| REV:
GTGCCAAACTGTCCGCTGTG |
| Light chain 3 | FOR:
GGCTACGGCTACTATCGCAC |
|
| REV:
GGAGAAGGTTTTGCGGTTGAAA |
| GAPDH | FOR:
AATGGATTTGGACGCATTGGT |
|
| REV:
TTTGCACTGGTACGTGTTGAT |
| U6 | FOR:
ATGGGTCGAAGTCGTAGCC |
|
| REV:
TTCTCGGCGTCTTCTTTCTCG |
| microRNA 17-5p | FOR:
ACACTCCAGCTGGGCAAAGTGCTTACAGTG |
|
| REV:
CTCAACTGGTGTCGTGGAGTCGG |
Western blotting
Total protein of each group was lysed with RIPA
lysis solution (cat. no. R0010; Beijing Solarbio Science &
Technology Co., Ltd.). The protein concentration was determined by
bicinchoninic acid (BCA; cat. no. PC0020; Beijing Solarbio Science
& Technology Co., Ltd.) method. RIPA lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd.) was added to extract
the total protein fully and ~30 µg per lane of protein samples were
resolved by 10% sodium dodecyl sulfate-polyacrylamide gel, after
which the proteins were transferred to polyvinylidene
(MilliporeSigma) membranes for 1 h at 100 V and blocked for 1 h in
5% skimmed milk powder at room temperature. Primary antibodies
against, anti-hypoxia-inducible factor-1α (HIF-1α; 1:1,000; cat.
no. 14179; Cell Signaling Technology, Inc.), anti-Bax (1:1,000;
cat. no. GB11007; Wuhan Servicebio Technology Co., Ltd.),
anti-Bcl-2 (1:1,000; cat. no. sc-492; Santa Cruz Biotechnology,
Inc.), anti-caspases-3 (1:1,000; cat. no. sc-7148; Santa Cruz
Biotechnology, Inc.) and −9 (1:1,000; cat. no. 9504; Cell Signaling
Technology, Inc.), anti-LC3B (1:1,000; cat. no. 3868; Cell
Signaling Technology, Inc.), anti-LC3A (1:1,000; cat. no. 4599;
Cell Signaling Technology, Inc.) anti-Beclin-1 (1:1,000; cat. no.
3738; Cell Signaling Technology, Inc.) and anti-β-actin (1:1,000;
cat. no. GB15001; Wuhan Servicebio Technology Co., Ltd.) were added
followed by overnight incubation at 4°C. After washing with TBST
(0.1% Tween 20; cat. no. ST673, Beyotime Institute of
Biotechnology), the secondary antibodies, goat anti-mouse IgG
(1:2,000; cat. no. GB23301; Wuhan Servicebio Technology Co., Ltd.)
and goat anti-rabbit IgG (1:2,000; cat. no. GB23303; Wuhan
Servicebio Technology Co., Ltd.), were incubated for 1 h at room
temperature. Protein bands were visualized using an ECL kit (cat.
no. P10200; New Cell & Molecular Biotech Co., Ltd.). The
relative expression levels of the proteins were analyzed by ImageJ
v1.8.0 software using β-actin as a reference (30).
Dual-luciferase reporter assay
Complementary binding sites between miR-17-5p and
FGF23 were predicted using the TargetScan database (http://www.tragetscan.org/) and StarBase database
(http://www.starbase.sysu.edu.cn/).
Nucleotide sequences containing miR-17-5p binding sites in
wild-type (WT) or mutant (MUT) FGF23 3′-UTR were cloned into the
pGL3 luciferase reporter plasmid (Invitrogen; Thermo Fisher
Scientific, Inc.). MC3T3-E1 cells in the logarithmic growth phase
were seeded in 24-well plates (2×105 cells/well) and the
miR-17-5p mimic and miR-NC were co-transfected with WT or MUT
reporter plasmids into MC3T3-E1 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and cultured for 48 h.
The fluorescence activity was detected 48 h post-transfection using
a dual-luciferase kit (cat.no.E1910; Promega). The firefly and
Renilla Luciferase activity were detected by the dual
luciferase reporter gene detection system and the luciferase
activity of the reporter genes was calculated and the luciferase
value=Firefly Luciferase activity/Renilla Luciferase
activity (31).
Statistical analysis
Each experiment was repeated three times. GraphPad
Prism v6.02 statistical software (Dotmatics) was used to analyze
the data. Normally distributed measurement data are expressed as
mean ± standard deviation. The unpaired Student's t test was used
for two samples and on way analysis of variance (ANOVA) was used
for multiple samples. Tukey's post hoc tests were performed using
the Bonferroni method for two samples comparisons between groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
The present study verified the hypothesis that FGF23
targets miR-17-5p to regulate hypoxia-induced osteoblast apoptosis
by regulating the autophagy-signaling pathway. FGF23 expression was
upregulated in osteoblasts following hypoxia treatment. FGF23
overexpression can promote apoptosis and autophagy in osteoblasts.
miR-17-5p is a potential biological target of FGF23. It was found
that miR-17-5p reversed the biological effects of FGF23. The
present study provided a theoretical basis for the clinical
treatment and early intervention of ONFH.
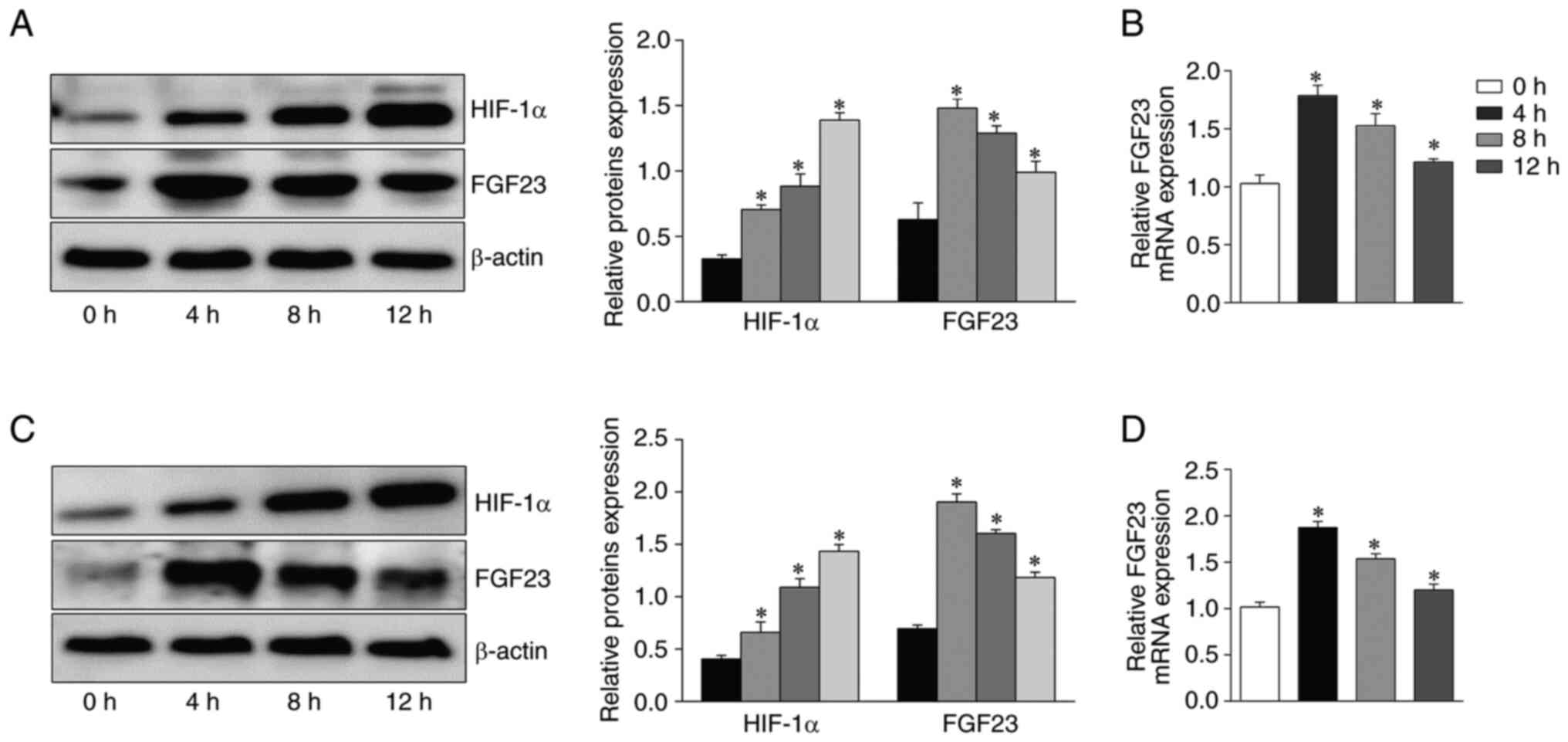
Effect of the alteration of hypoxia
time on FGF23 expression level in osteoblasts
The levels of FGF23 in the cells after apoptosis
induction under different hypoxic conditions were evaluated by
western blotting and RT-qPCR. Due to alterations in hypoxia time,
the expression levels of FGF23 protein and mRNA in osteoblasts
showed significant fluctuations. Western blotting showed that FGF23
protein expression was the highest at 4 h and then gradually
decreased with the extension of hypoxia time in in MC3T3-E1 cells
(P<0.05; Fig. 1A). The RT-qPCR
results were consistent with the western blotting results (Fig. 1B). Additionally, HIF-1α expression
increased under prolonged hypoxia for up to 12 h (Fig. 1A). FGF23 protein and mRNA
expression in primary osteoblasts was highest at 4 h and then
gradually decreased (Fig. 1C and
D). These results indicated that the expression of FGF23 is
influenced by hypoxic environments and associated with time.
Therefore, hypoxia for 4 h was selected for the following
experiments. MC3T3-E1 cells, a mouse osteogenic precursor cell
line, are commonly used to study ONFH (32,33).
Effects of hypoxia and FGF23 on
autophagy-associated gene and protein expression in primary
osteoblasts and MC3T3-E1 cells
Increased autophagosomes were found in primary
osteoblasts and MC3T3-E1 cells following hypoxia, and FGF23
knockdown partially reversed this result (Fig. 2A). Western blotting demonstrated
that the protein levels of FGF23 were altered by transfection
(Fig. 2B and D). Significant
increases were observed in the levels of microtubule-associated
protein light chain-phosphatidylethanolamine conjugate (LC3-II),
Beclin-1 and LC3-II/LC3-I ratio, under hypoxic conditions
(P<0.05). Furthermore, compared with the EV group, Beclin-1,
LC3-II and the LC3-II/LC3-I ratio were increased in the FGF23
group; however, they were significantly decreased in the si-FGF23
group (Fig. 2B and D). In
addition, autophagy-related gene expression also showed
corresponding alterations (Fig. 2C and
E). Taken together, these results indicate that FGF23 can
promote hypoxia-induced autophagy levels in osteoblasts.
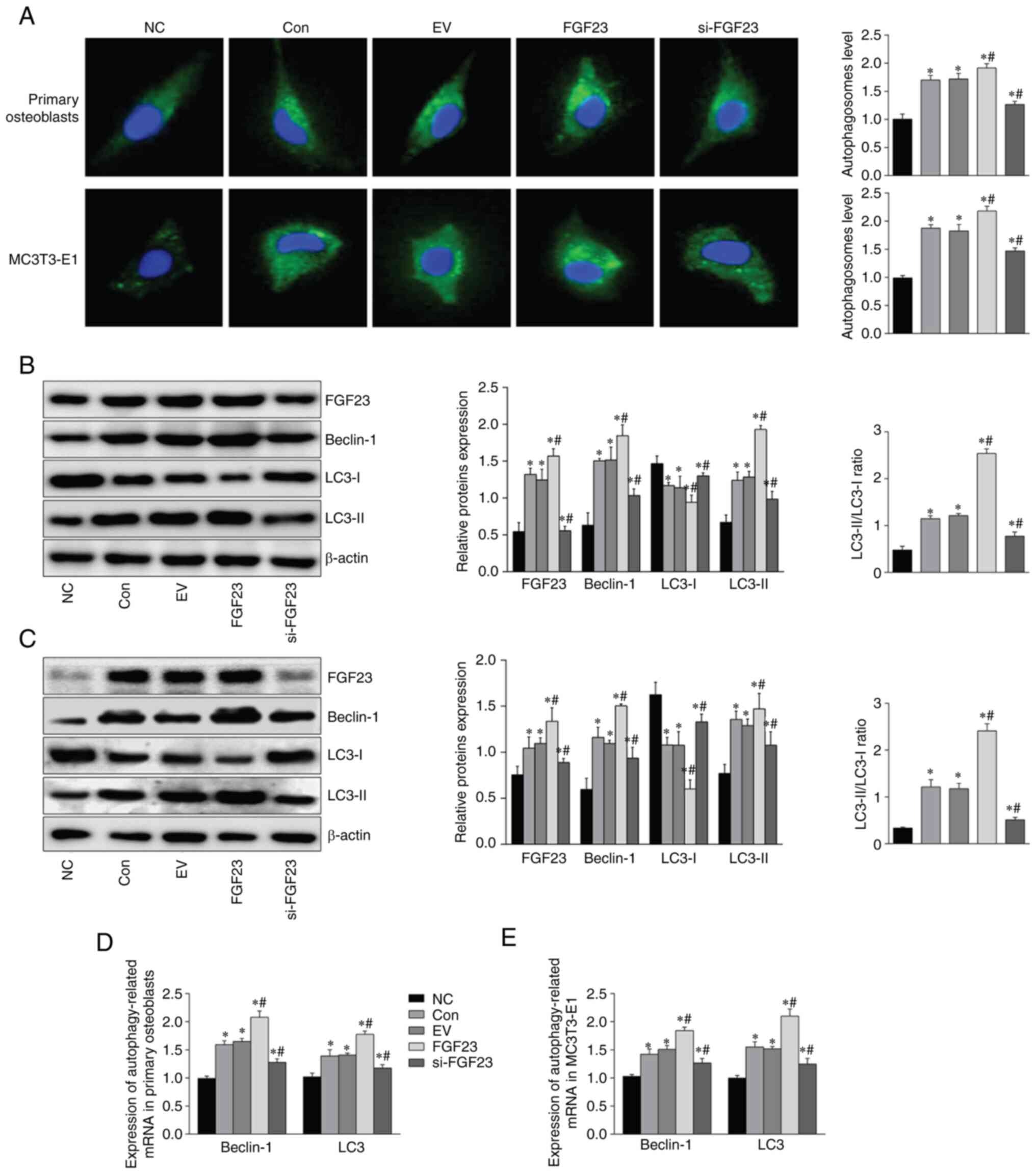 | Figure 2.FGF23 promotes hypoxia-induced
autophagy in osteoblasts. (A) Detection of intracellular
autophagosomes by Monodansylcadaveriner staining (magnification,
×400). The expression of FGF23, Beclin-1, LC3-I and LC3-II in (B)
primary osteoblasts and (C) MC3T3-E1 cells was detected via western
blotting. The mRNA level of Beclin-1 and LC3 of (D) primary
osteoblasts and (E) MC3T3-E1 cells. B-actin and GAPDH expression
were used to determine the target protein's expression or gene.
*P<0.05 vs. NC group. #P<0.05 vs. EV group. Among
them, the FGF23 group had the lowest cell survival rate. FGF23,
fibroblast growth factor; LC3, light chain 3; NC, normal control;
con, control; EV, empty vector group; si, short interfering. |
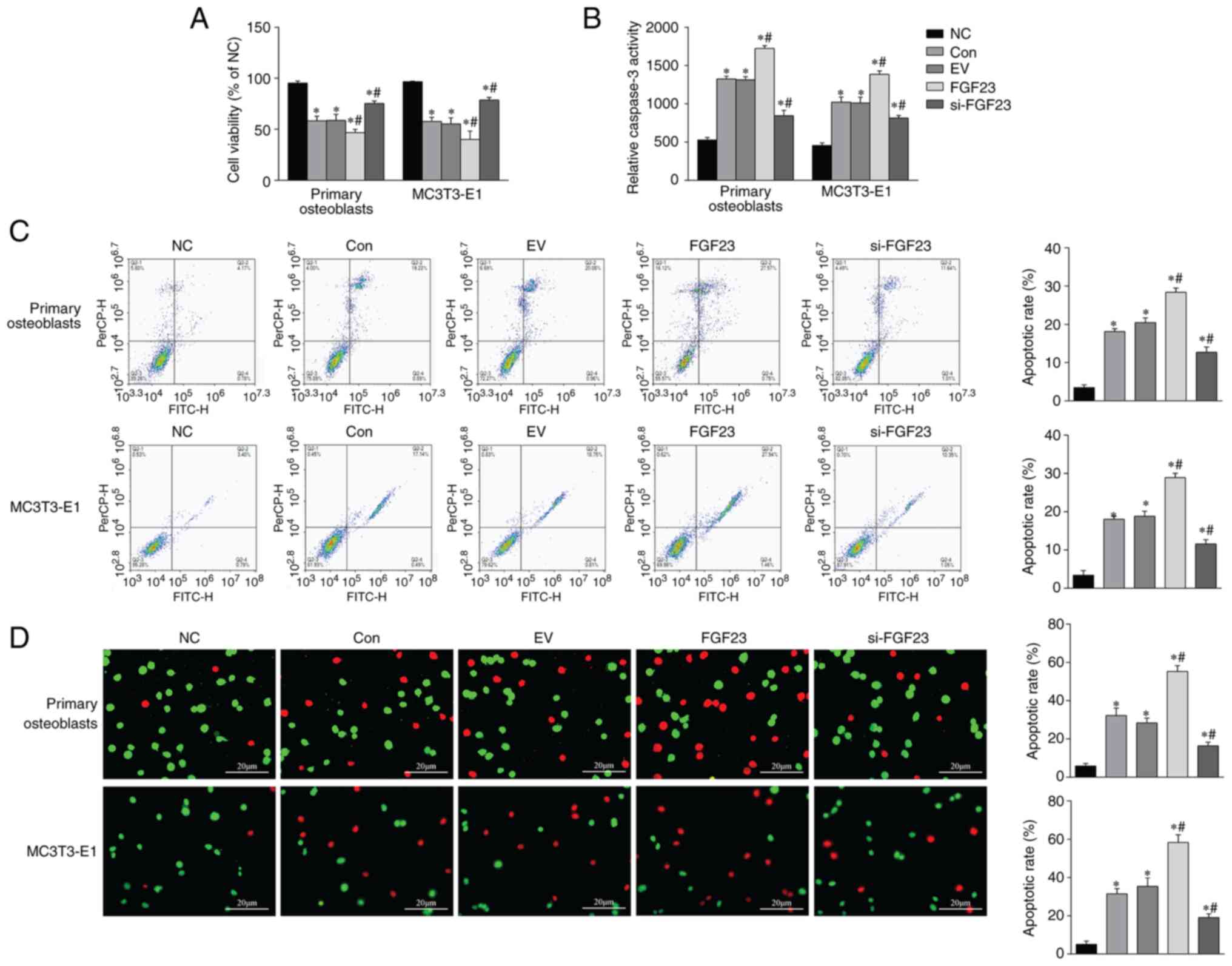
Increased FGF23 exacerbates hypoxic
apoptosis of osteoblasts
MTT assay was used to measure the influence of
different factors on cell viability. The viability was reduced in
the con, FGF23 overexpression and si-FGF23 groups compared with the
NC group. The FGF23 overexpression group had the lowest cell
survival rate (P<0.05; Fig.
3A). FGF23 could significantly increase activity of caspase-3
compared with that in the EV group (P<0.05; Fig. 3B). FCM and FDA/EB staining showed
increased apoptosis of osteoblasts in the EV group. However, the
FGF23 group showed significantly increased apoptosis compared with
the EV group, whereas apoptosis decreased slightly in the si-FGF23
group (Fig. 3C-D). These results
indicated that FGF23 might activate the apoptosis pathway and
promote cell death during hypoxic cellular stress.
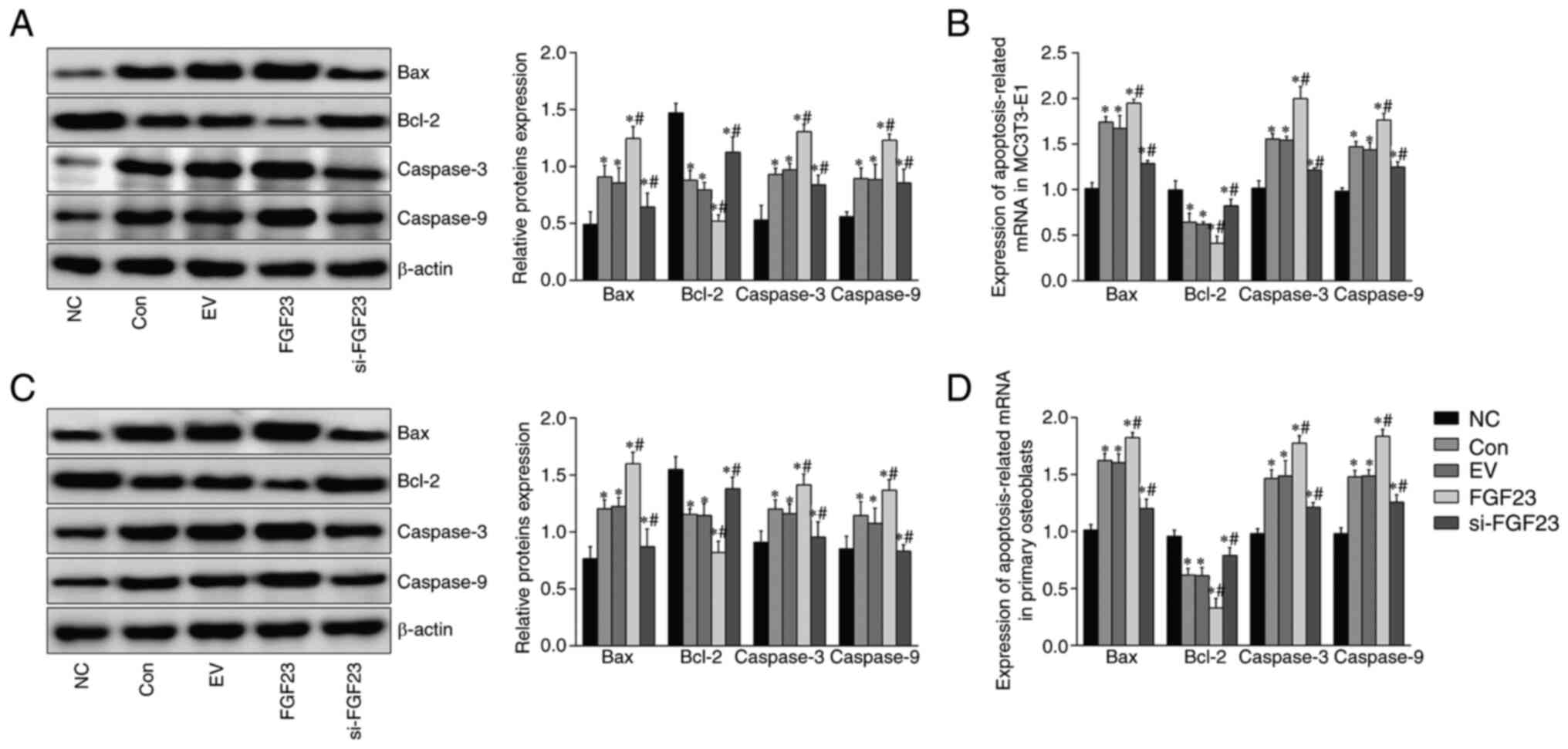
Expression of apoptosis-associated
genes and proteins in hypoxic primary osteoblasts and MC3T3-E1
cells
Western blotting results showed that hypoxia led to
a significant increase in levels of pro-apoptotic proteins Bax,
caspases-3 and −9 and a decrease in levels of anti-apoptotic
protein Bcl-2 in primary osteoblasts and MC3T3-E1 cells. Western
blotting results showed significantly increased levels of the
pro-apoptotic proteins Bax and caspases-3 and −9 and decreased
levels of anti-apoptotic Bcl-2 (Fig.
4A and C). The RT-qPCR data showed enhanced expression of the
pro-apoptotic (Bax and caspases-3 and −9) mRNA. By contrast, the
apoptosis suppressor Bcl-2 mRNA was diminished in osteoblasts
compared with the NC group in response to FGF23. The differences
were statistically significant (Fig.
4B and D). Apoptosis levels were further elevated following
FGF23 overexpression, whereas, FGF23 knockdown attenuated the
apoptosis level in osteoblasts compared with the con group. The
above results indicated that knockdown of FGF23 can ameliorate
hypoxia-induced apoptosis in osteoblasts.
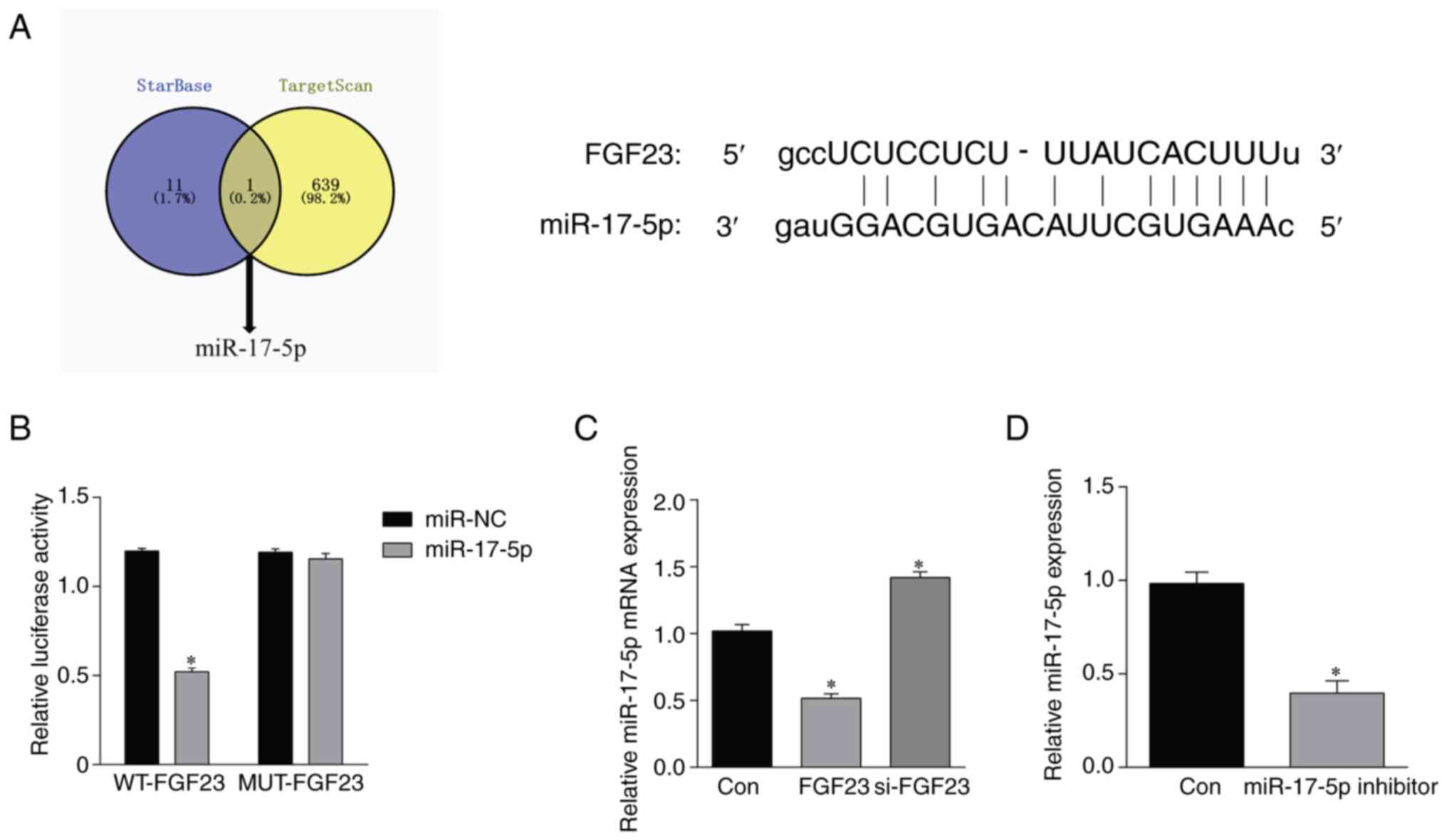
FGF23 targets miR-17-5p
To further investigate the molecular mechanisms by
which FGF23 regulates osteoblast apoptosis, a potential binding
site between FGF23 and miR-17-5p was identified by the online
database TargetScan and StarBase (Fig.
5A). Subsequent overexpression or inhibition of miR-17-5p was
performed for dual luciferase reporter gene assay. As shown in
Fig. 5B, luciferase analysis
revealed that the FGF23 wild-type luciferase activity in the
miR-17-5p group was lower than that in the miR-NC group
(P<0.05). By contrast, there was no statistically significant
difference in the FGF23 MUT luciferase activity between the
miR-17-5p and miR-NC groups (P>0.05). In addition, RT-qPCR
results showed that the expression level of miR-17-5p was
downregulated after overexpression of FGF23; it was significantly
upregulated after the silencing of FGF23 (Fig. 5C). Detection of cell transfection
efficiency revealed that transfection with miR-17-5p inhibitor
downregulated miR-17-5p expression (Fig. 5D).
Inhibition of miR-17-5p reverses the
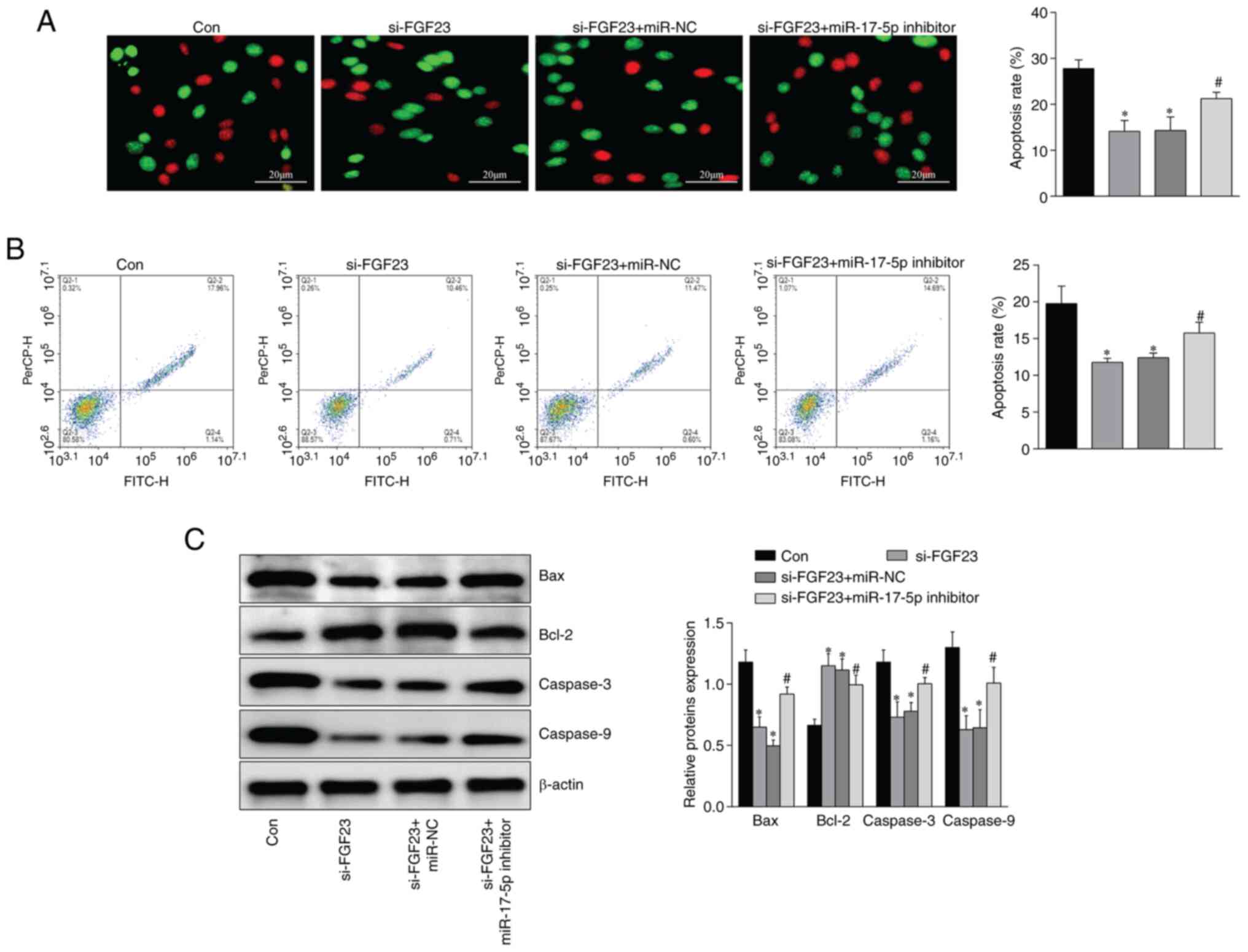
effect of silencing FGF23 on apoptosis of MC3T3-E1 cells
Compared with the con group, the apoptosis rate
(Fig. 6A and B), Bax, caspase-3
and caspase-9 protein (Fig. 6C)
expression were significantly decreased, and Bcl-2 protein
expression was increased, in MC3T3-E1 cells in the si-FGF23 group.
Conversely, compared with the si-FGF23 group, Bax, caspase-3 and
caspase-9 protein expression and apoptosis rate were significantly
increased. Bcl-2 expression was downregulated in MC3T3-E1 cells in
the si-FGF23 + miR-17-5p inhibitor group (Fig. 6).
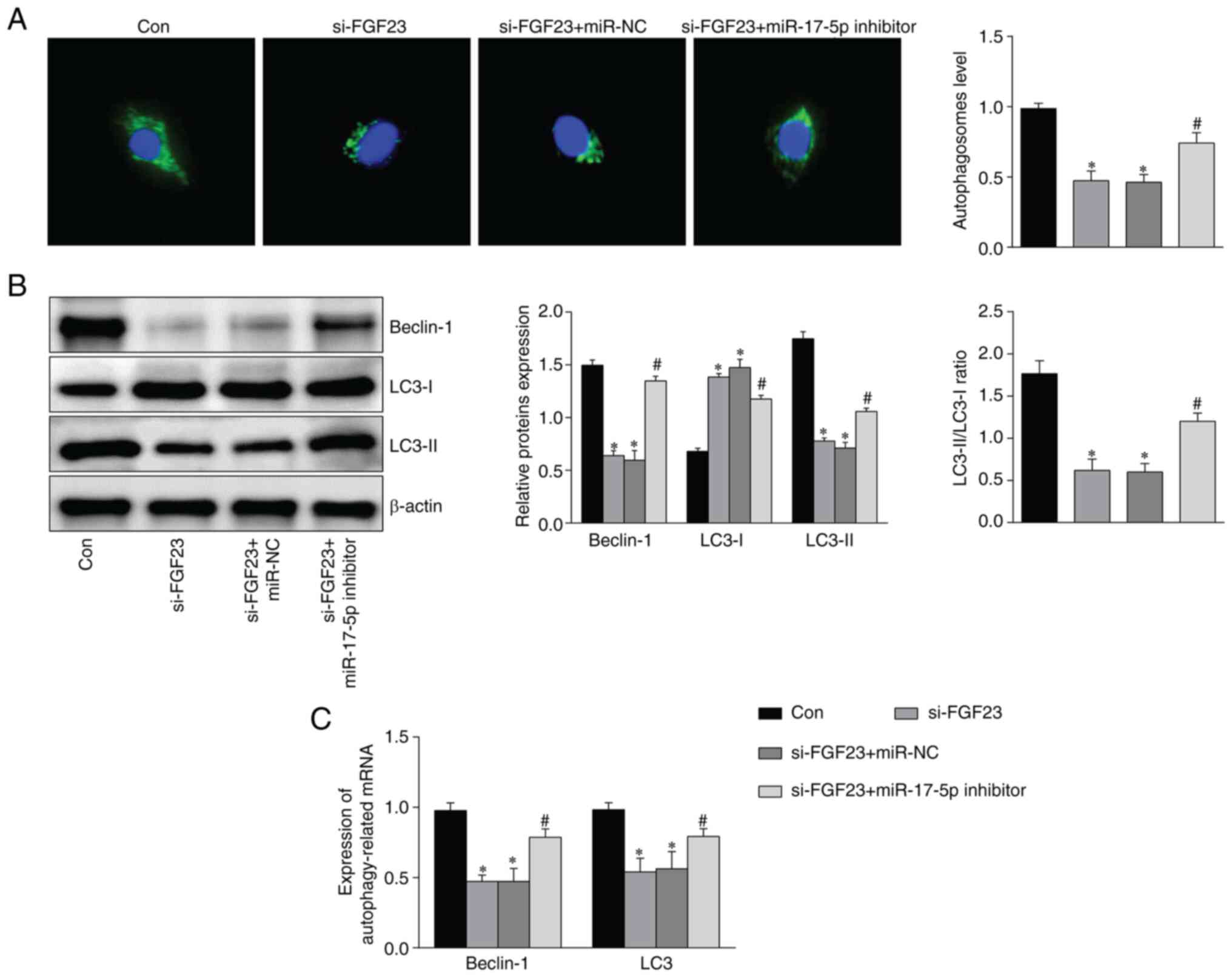
FGF23 activates the autophagy
signaling pathway via targeted regulation of miR-17-5p
FGF23 knockdown reduced the number of
autophagosomes, which was partly reversed by the addition of
miR-17-5p inhibitor (Fig. 7A). The
expression of the autophagy-associated proteins Beclin-1 and LC3
was detected by western blotting and RT-qPCR. The results showed
that Beclin-1 and LC3-II/LC3-I expression decreased after FGF23
silencing, whereas this result was partially reversed after
miR-17-5p inhibition (Fig. 7B and
C).
Discussion
The present study found that hypoxia led to
apoptosis and increased FGF23 expression in osteoblasts, whereas
FGF23 overexpression further aggravated osteoblast apoptosis. To
the best of the authors' knowledge, the present study is the first
to demonstrate that FGF23 regulates hypoxia-induced apoptosis of
osteoblasts by regulating the activation of the autophagy-signaling
pathway.
FGF is the most widely distributed growth factor
in vivo. FGF family members are involved in numerous
cellular functions, including metabolic and transcriptional
regulation (34). FGF23 modulates
cellular energy metabolism and apoptosis. The present study
observed that hypoxia increased LC3-II and Beclin-1 in MC3T3-E1
osteoblasts, whereas FGF23 treatment further increased LC3-II and
Beclin-1 expression. The microtubule-associated protein light chain
3 (LC3) is an autophagy-related gene expressed in the initial
stages of autophagosome formation, promoting autophagy; LC3-I to
LC3-II conversion is necessary for completing the process (35). Beclin-1 is a specific
autophagy-related gene and its expression level directly reflects
autophagic activity (36). The
present study indicated that the autophagic activity of MC3T3-E1
osteoblasts was induced in a hypoxic environment and was enhanced
in response to FGF23. It has been demonstrated that osteoblast
apoptosis and autophagy occur in the early stage of avascular ONFH
and that the protein and mRNA expression of LC3B and caspase-3 are
significantly upregulated. By contrast, inhibition of autophagy can
protect osteoblasts from apoptosis (37,38).
Zheng et al (39) reported
that the hypoxic microenvironment activates autophagy in
periodontal ligament tissue, inhibits osteogenic differentiation
and promotes apoptosis. Therefore, apoptosis has an intricate
relationship with autophagy (40).
Normally, autophagy occurs at low levels; however, when cells are
stimulated by hypoxia, hormones, or other factors, the process is
activated, resulting in the breakdown of the damaged material into
small molecules that can participate in energy metabolism (41,42).
However, excessive autophagy can potentially affect metabolism and
cause cell death (43).
To further validate the function of FGF23 in the
apoptosis pathway, hypoxia-induced expression of
apoptosis-associated proteins in osteoblasts was assessed.
Caspase-3 activity assay indicated that FGF23 was involved in
hypoxia-induced apoptosis. Caspase family proteases are core
proteases that mediate apoptosis and can induce apoptosis in a
cascading manner, directly leading to cell disassembly. Caspase-9
initiates the apoptotic program, whereas caspase-3 executes it
(44–46). In the present study, western
blotting results showed that MC3T3-E1 cells treated with FGF23
overexpression showed increased expression levels of the
pro-apoptotic proteins caspases-3 and −9 and Bax and decreased
expression levels of the anti-apoptotic protein Bcl-2. This result
was reversed following intervention with FGF23 silencing. Bcl-2
stabilizes mitochondria, blocks the release of calcium ions from
the endoplasmic reticulum, regulates the transduction of
mitochondrial signaling molecules, interrupts DNA apoptosis and
protects cells from apoptotic damage (47). When cells receive signaling stimuli
such as death, Bax translocates from the cytoplasm into the
mitochondria and interacts with the anti-apoptotic factor Bcl-2 on
the mitochondrial membrane; thus, the anti-apoptotic factor is
inactivated, the release of pro-apoptotic factors is increased, the
mitochondrial structure and function are destroyed and, finally,
the caspase pathway is activated to initiate apoptosis (48). Therefore, Bcl-2 and Bax are
essential for cell survival and death (49). In the present study, MTT results
revealed that FGF23 led to a decrease in MC3T3-E1 cell viability.
Concurrently, FGF23 promoted hypoxia-induced osteoblast apoptosis
directly by FC and FDA/EB staining. In addition, in osteoarthritis,
FGF23 can regulate 1,25-dihydroxyvitamin D3 and extracellular
inorganic phosphate, which significantly upregulates caspase-9
expression and increases apoptosis in normal chondrocytes (50).
Using the TargetScan database, the binding of
miR-17-5p and FGF23 was predicted. A luciferase reporter assay
confirmed that miR-17-5p is a target of FGF23. miR-17-5p
overexpression decreased the luciferase activity of WT-FGF23 but
had no effect on MUT-FGF23. Furthermore, it has been shown that
miR-17-5p plays an important role in cell proliferation,
differentiation and apoptosis (51,52).
miR-17-5p regulates osteoblast differentiation and cell
proliferation by inhibiting SMAD7 in non-traumatic ONFH (18). In addition, miR-17-5p inhibits
chondrocyte apoptosis by targeting enhancer of zeste homolog 2,
inhibiting osteoarthritis progression (16). Therefore, the present study
investigated whether miR-17-5p inhibitor is a direct target after
discovering that it can regulate osteoblast apoptosis. This
conjecture was confirmed by FDA/EB staining, FCM and western
blotting, suggesting that the miR-17-5p inhibitor partly reversed
the effect of FGF23 silencing on osteoblast apoptosis. According to
previous studies, miR-17-5p may be involved in the autophagy
signaling pathway (53,54). Therefore, it was hypothesized that
FGF23 might regulate the autophagy signaling pathway by targeting
miR-17-5p. The results showed that FGF23 silencing significantly
decreased the expression of Beclin-1 protein and the LC3-II/LC3-I
ratio, whereas miR-17-5p had the opposite effect on the expression
of these proteins after inhibition. The results showed that FGF23
could affect osteoblast apoptosis by targeting miR-17-5p via the
autophagy signaling pathway.
The present study demonstrated that FGF23 regulates
hypoxia-induced osteoblast apoptosis by targeting miR-17-5p through
the autophagy-signaling pathway. The findings provided foundational
support for the clinical prevention and treatment of orthopedic
diseases, such as ONFH, caused by various injuries. However, only
in vitro tests were performed in the present study, which
were not able to simulate the in vivo state.
Acknowledgements
Not applicable.
Funding
The present study was supported by Shandong Provincial Natural
Science Foundation of China (grant no. ZR2019MH120) and the
Projects of Medical and Health Technology Development Program in
Shandong Province (grant no. 2019WS397).
Availability of data and materials
The datasets generated and analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
QY, HY and LZ designed the study and performed the
experiments, LF and QW collected the data, SG and YW analyzed the
data and QY and HY prepared the manuscript. QY and LZ confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Shandong First Medical University (Shandong
Academy of Medical Sciences; grant no. W202210070243).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cohen-Rosenblum A and Cui Q: Osteonecrosis
of the femoral head. Orthop Clin North Am. 50:139–149. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou L, Wang SI, Moon YJ, Kim KM, Lee KB,
Park BH, Jang KY and Kim JR: Overexpression of SIRT1 prevents
hypoxia-induced apoptosis in osteoblast cells. Mol Med Rep.
16:2969–2975. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang G, Wang J, Sun D, Xin J, Wang L,
Huang D, Wu W and Xian CJ: Short-term hypoxia accelerates bone loss
in ovariectomized rats by suppressing osteoblastogenesis but
enhancing osteoclastogenesis. Med Sci Monit. 22:2962–2971. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma HP, Ma XN, Ge BF, Zhen P, Zhou J, Gao
YH, Xian CJ and Chen KM: Icariin attenuates hypoxia-induced
oxidative stress and apoptosis in osteoblasts and preserves their
osteogenic differentiation potential in vitro. Cell Prolif.
47:527–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Itoh N and Ornitz DM: Evolution of the Fgf
and Fgfr gene families. Trends Genet. 20:563–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gohil A and Imel EA: FGF23 and associated
disorders of phosphate wasting. Pediatr Endocrinol Rev. 17:17–34.
2019.PubMed/NCBI
|
|
7
|
Chen G, Liu Y, Goetz R, Fu L, Jayaraman S,
Hu MC, Moe OW, Liang G, Li X and Mohammadi M: α-Klotho is a
non-enzymatic molecular scaffold for FGF23 hormone signalling.
Nature. 553:461–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andrukhova O, Zeitz U, Goetz R, Mohammadi
M, Lanske B and Erben RG: FGF23 acts directly on renal proximal
tubules to induce phosphaturia through activation of the
ERK1/2-SGK1 signaling pathway. Bone. 51:621–628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prié D, Forand A, Francoz C, Elie C, Cohen
I, Courbebaisse M, Eladari D, Lebrec D, Durand F and Friedlander G:
Plasma fibroblast growth factor 23 concentration is increased and
predicts mortality in patients on the liver-transplant waiting
list. PLoS One. 8:e661822013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fukumoto S: FGF23 and bone and mineral
metabolism. Handb Exp Pharmacol. 262:281–308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Millar SA, Anderson SI and O'Sullivan SE:
Osteokines and the vasculature: A review of the in vitro effects of
osteocalcin, fibroblast growth factor-23 and lipocalin-2. PeerJ.
7:e71392019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Domazetovic V, Falsetti I, Ciuffi S,
Iantomasi T, Marcucci G, Vincenzini MT and Brandi ML: Effect of
oxidative stress-induced apoptosis on active FGF23 levels in MLO-Y4
cells: The protective role of 17-β-estradiol. Int J Mol Sci.
23:21032022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ramzan F, Vickers MH and Mithen RF:
Epigenetics, microRNA and metabolic syndrome: A comprehensive
review. Int J Mol Sci. 22:50472021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen J, Huang Y, Li H, Zhang X, Cheng P,
Deng D, Peng Z, Luo J, Zhao W, Lai Y and Liu Z: Over-expression of
miR-196b-5p is significantly associated with the progression of
myelodysplastic syndrome. Int J Hematol. 105:777–783. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu GZ, Chen C, Kong N, Tian R, Li YY, Li
Z, Wang KZ and Yang P: Identification of potential miRNA biomarkers
for traumatic osteonecrosis of femoral head. J Cell Physiol.
235:8129–8140. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Yuan F, Song Y and Guan X: miR-17-5p
and miR-19b-3p prevent osteoarthritis progression by targeting
EZH2. Exp Ther Med. 20:1653–1663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su Y, Meng X, Wang W, Gu G and Chen Y:
LncRNA HOTAIR regulates fracture healing in osteoporotic rats
through inhibition on MiR-17-5p. Minerva Med. 112:525–527. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu
X, Feng Y and Dai Z: MiR-17-5p modulates osteoblastic
differentiation and cell proliferation by targeting SMAD7 in
non-traumatic osteonecrosis. Exp Mol Med. 46:e1072014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou W, Song L, Zhao Y, Liu Q and Zhang S:
Inhibition of beclin-1-mediated autophagy by MicroRNA-17-5p
enhanced the radiosensitivity of glioma cells. Oncol Res. 25:43–53.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen B, Yang Y, Wu J, Song J and Lu J:
microRNA-17-5p downregulation inhibits autophagy and myocardial
remodelling after myocardial infarction by targeting STAT3.
Autoimmunity. 55:43–51. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang T, Wu Q, Zhou L, Mu S and Fu Q:
miR-106b-5p and miR-17-5p suppress osteogenic differentiation by
targeting Smad5 and inhibit bone formation. Exp Cell Res.
347:74–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hou Z, Wang Z, Tao Y, Bai J, Yu B, Shen J,
Sun H, Xiao L, Xu Y, Zhou J, et al: KLF2 regulates osteoblast
differentiation by targeting of Runx2. Lab Invest. 99:271–280.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang X, Chen C, Chen Y, Xu J and Liu L:
Omentin-1 alleviate interleukin-1β(IL-1β)-induced nucleus pulposus
cells senescence. Bioengineered. 13:13849–13859. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, Bian H, Dou QL, Huang XW, Tao WY,
Liu WH, Li N and Zhang WW: Ginkgetin alleviates inflammation,
oxidative stress, and apoptosis induced by hypoxia/reoxygenation in
h9c2 cells via caspase-3 dependent pathway. Biomed Res Int.
2020:19284102020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang W, Zhu M, Xu Z, Li W, Dong X, Chen Y,
Lin B and Li M: Ropivacaine promotes apoptosis of hepatocellular
carcinoma cells through damaging mitochondria and activating
caspase-3 activity. Biol Res. 52:362019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiong H, Li Y and Liu M: DEPDC1B is
involved in the proliferation, metastasis, cell cycle arrest and
apoptosis of colon cancer cells by regulating NUP37. Mol Med Rep.
27:1262023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qi J, Cao F, Han Y, Xie D, Song H, Chen B
and Zhou L: Reliability of cartilage digestion and FDA-EB
fluorescence staining for the detection of chondrocyte viability in
osteochondral grafts. Cell Tissue Bank. 19:399–404. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres- sion data using real-time quantitative PCR
and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yi WR, Tu MJ, Yu AX, Lin J and Yu AM:
Bioengineered miR-34a modulates mitochondrial inner membrane
protein 17 like 2 (MPV17L2) expression toward the control of cancer
cell mitochondrial functions. Bioengineered. 13:12489–12503. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pang X, Wang SS, Zhang M, Jiang J, Fan HY,
Wu JS, Wang HF, Liang XH and Tang YL: OSCC cell-secreted exosomal
CMTM6 induced M2-like macrophages polarization via ERK1/2 signaling
pathway. Cancer Immunol Immunother. 70:1015–1029. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song J, Liu Y, Wang T, Li B and Zhang S:
MiR-17-5p promotes cellular proliferation and invasiveness by
targeting RUNX3 in gastric cancer. Biomed Pharmacother.
128:1102462020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu P, Shen YM, Hua T, Pan T, Chen G, Dai T
and Shi KQ: Overexpression of FGF2 delays the progression of
osteonecrosis of the femoral head activating the PI3K/Akt signaling
pathway. J Orthop Surg Res. 16:6132021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bai Y, Liu Y, Jin S, Su K, Zhang H and Ma
S: Expression of microRNA-27a in a rat model of osteonecrosis of
the femoral head and its association with TGF-β/Smad7 signalling in
osteoblasts. Int J Mol Med. 43:850–860. 2019.PubMed/NCBI
|
|
34
|
Savchenko E, Teku GN, Boza-Serrano A, Russ
K, Berns M, Deierborg T, Lamas NJ, Wichterle H, Rothstein J,
Henderson CE, et al: FGF family members differentially regulate
maturation and proliferation of stem cell-derived astrocytes. Sci
Rep. 9:96102019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mancias JD and Kimmelman AC: Mechanisms of
selective autophagy in normal physiology and cancer. J Mol Biol.
428:1659–1680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Musumeci G, Castrogiovanni P, Trovato FM,
Weinberg AM, Al-Wasiyah MK, Alqahtani MH and Mobasheri A:
Biomarkers of chondrocyte apoptosis and autophagy in
osteoarthritis. Int J Mol Sci. 16:20560–20575. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng L, Wang W, Ni J, Mao X, Song D, Liu
T, Wei J and Zhou H: Role of autophagy in tumor necrosis
factor-α-induced apoptosis of osteoblast cells. J Investig Med.
65:1014–1020. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng LW, Wang WC, Mao XZ, Luo YH, Tong ZY
and Li D: TNF-α regulates the early development of avascular
necrosis of the femoral head by mediating osteoblast autophagy and
apoptosis via the p38 MAPK/NF-κB signaling pathway. Cell Biol Int.
44:1881–1889. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zheng J, Zhu X, He Y, Hou S, Liu T, Zhi K,
Hou T and Gao L: CircCDK8 regulates osteogenic differentiation and
apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia.
Ann N Y Acad Sci. 1485:56–70. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang T, Li Y, Park KA, Byun HS, Won M,
Jeon J, Lee Y, Seok JH, Choi SW, Lee SH, et al: Cucurbitacin
induces autophagy through mitochondrial ROS production which
counteracts to limit caspase-dependent apoptosis. Autophagy.
8:559–576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ho TT, Warr MR, Adelman ER, Lansinger OM,
Flach J, Verovskaya EV, Figueroa ME and Passegué E: Autophagy
maintains the metabolism and function of young and old stem cells.
Nature. 543:205–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Manai F, Azzalin A, Gabriele F, Martinelli
C, Morandi M, Biggiogera M, Bozzola M and Comincini S: The in vitro
effects of enzymatic digested gliadin on the functionality of the
autophagy process. Int J Mol Sci. 19:6352018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Z, Xie Q, Zhou H, Zhang M, Shen J and
Ju D: Amino acid degrading enzymes and autophagy in cancer therapy.
Front Pharmacol. 11:5825872021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi Y: Apoptosome: The cellular engine for
the activation of caspase-9. Structure. 10:285–288. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fan TJ, Han LH, Cong RS and Liang J:
Caspase family proteases and apoptosis. Acta Biochim Biophys Sin
(Shanghai). 37:719–727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Araya LE, Soni IV, Hardy JA and Julien O:
Deorphanizing caspase-3 and caspase-9 substrates in and out of
apoptosis with deep substrate profiling. ACS Chem Biol.
16:2280–2296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Perini GF, Ribeiro GN, Pinto Neto JV,
Campos LT and Hamerschlak N: BCL-2 as therapeutic target for
hematological malignancies. J Hematol Oncol. 11:652018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ly JD, Grubb DR and Lawen A: The
mitochondrial membrane potential (deltapsi(m)) in apoptosis; an
update. Apoptosis. 8:115–128. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shu Y, Yang Y, Zhao Y, Ma L, Fu P, Wei T
and Zhang L: Melittin inducing the apoptosis of renal tubule
epithelial cells through upregulation of Bax/Bcl-2 expression and
activation of TNF-α signaling pathway. Biomed Res Int.
2019:94503682019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Orfanidou T, Malizos KN, Varitimidis S and
Tsezou A: 1,25-Dihydroxyvitamin D(3) and extracellular inorganic
phosphate activate mitogen-activated protein kinase pathway through
fibroblast growth factor 23 contributing to hypertrophy and
mineralization in osteoarthritic chondrocytes. Exp Biol Med
(Maywood). 237:241–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shi YP, Liu GL, Li S and Liu XL: miR-17-5p
knockdown inhibits proliferation, autophagy and promotes apoptosis
in thyroid cancer via targeting PTEN. Neoplasma. 67:249–258. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li H, Li T, Wang S, Wei J, Fan J, Li J,
Han Q, Liao L, Shao C and Zhao RC: miR-17-5p and miR-106a are
involved in the balance between osteogenic and adipogenic
differentiation of adipose-derived mesenchymal stem cells. Stem
Cell Res. 10:313–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hao MX, Wang X and Jiao KL: MicroRNA-17-5p
mediates hypoxia-induced autophagy and inhibits apoptosis by
targeting signal transducer and activator of transcription 3 in
vascular smooth muscle cells. Exp Ther Med. 13:935–941. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yuan Y, Li X and Li M: Overexpression of
miR-17-5p protects against high glucose-induced endothelial cell
injury by targeting E2F1-mediated suppression of autophagy and
promotion of apoptosis. Int J Mol Med. 42:1559–1568.
2018.PubMed/NCBI
|