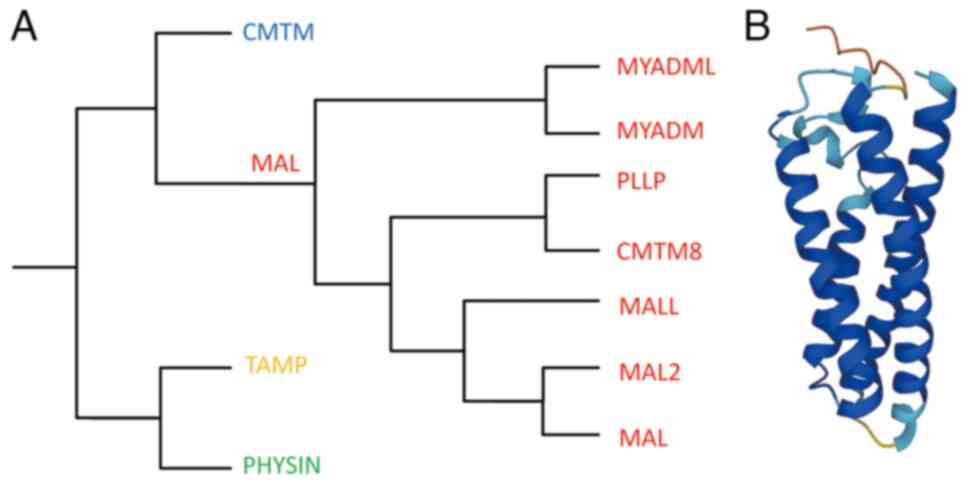
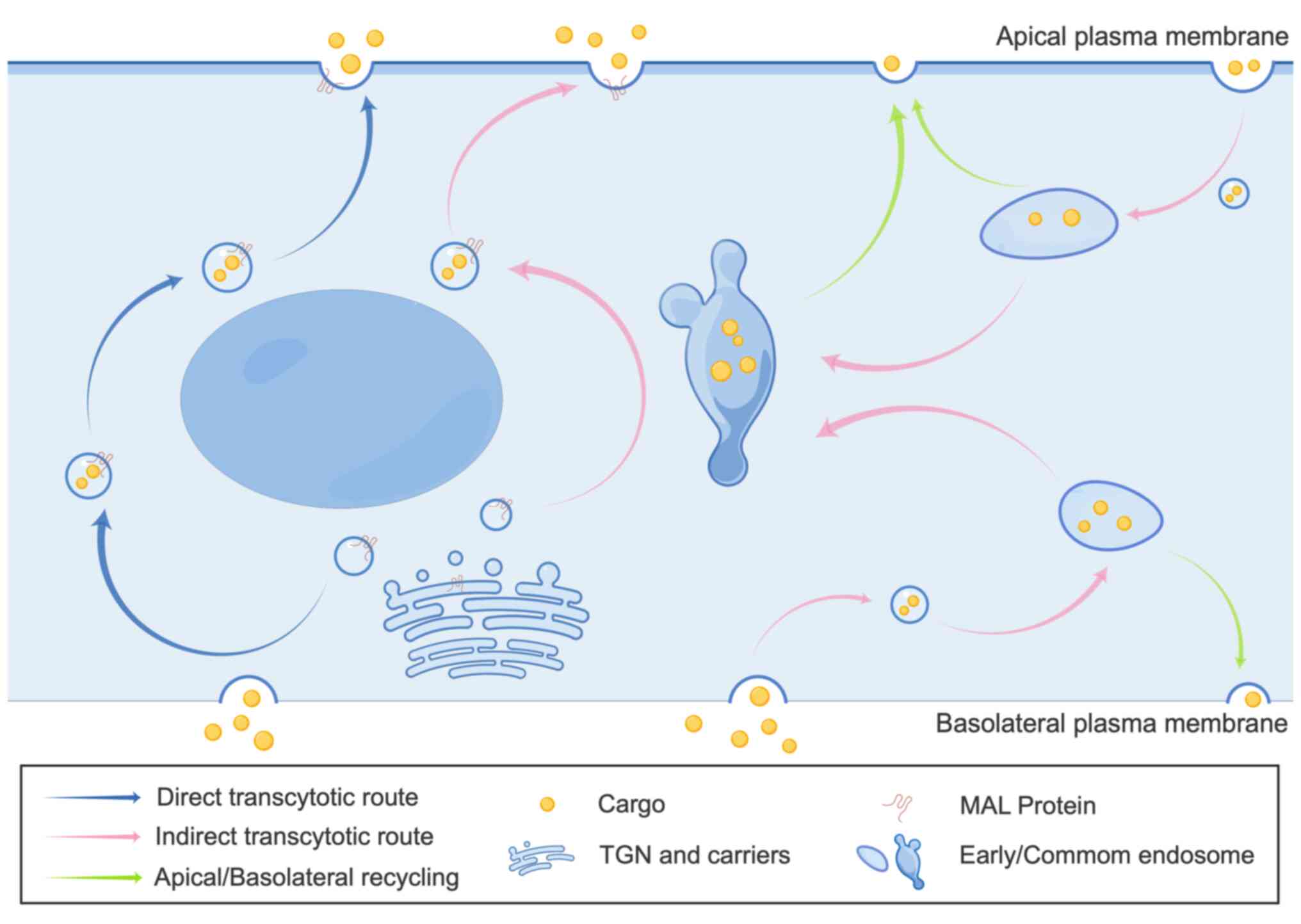
|
1
|
Rubio-Ramos A, Labat-de-Hoz L, Correas I
and Alonso MA: The MAL protein, an integral component of
specialized membranes, in normal cells and cancer. Cells.
10:10652021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sánchez-Pulido L, Martín-Belmonte F,
Valencia A and Alonso MA: MARVEL: A conserved domain involved in
membrane apposition events. Trends Biochem Sci. 27:599–601. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alonso MA and Weissman SM: cDNA cloning
and sequence of MAL, a hydrophobic protein associated with human
T-cell differentiation. Proc Natl Acad Sci USA. 84:1997–2001. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheong KH, Zacchetti D, Schneeberger EE
and Simons K: VIP17/MAL, a lipid raft-associated protein, is
involved in apical transport in MDCK cells. Proc Natl Acad Sci USA.
96:6241–6248. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martín-Belmonte F, Puertollano R, Millán J
and Alonso MA: The MAL proteolipid is necessary for the overall
apical delivery of membrane proteins in the polarized epithelial
Madin-Darby canine kidney and fischer rat thyroid cell lines. Mol
Biol Cell. 11:2033–2045. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martín-Belmonte F, Arvan P and Alonso MA:
MAL mediates apical transport of secretory proteins in polarized
epithelial Madin-Darby canine kidney cells. J Biol Chem.
276:49337–49342. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Puertollano R, Martín-Belmonte F, Millán
J, de Marco MC, Albar JP, Kremer L and Alonso MA: The MAL
proteolipid is necessary for normal apical transport and accurate
sorting of the influenza virus hemagglutinin in Madin-Darby canine
kidney cells. J Cell Biol. 145:141–151. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martín-Belmonte F, Kremer L, Albar JP,
Marazuela M and Alonso MA: Expression of the MAL gene in the
thyroid: The MAL proteolipid, a component of glycolipid-enriched
membranes, is apically distributed in thyroid follicles.
Endocrinology. 139:2077–2084. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Puertollano R and Alonso MA: MAL, an
integral element of the apical sorting machinery, is an itinerant
protein that cycles between the trans-Golgi network and the plasma
membrane. Mol Biol Cell. 10:3435–3447. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mostov KE, Verges M and Altschuler Y:
Membrane traffic in polarized epithelial cells. Curr Opin Cell
Biol. 12:483–490. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Simons K and Wandinger-Ness A: Polarized
sorting in epithelia. Cell. 62:207–210. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matter K and Mellman I: Mechanisms of cell
polarity: Sorting and transport in epithelial cells. Curr Opin Cell
Biol. 6:545–554. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Raleigh DR, Marchiando AM, Zhang Y, Shen
L, Sasaki H, Wang Y, Long M and Turner JR: Tight
junction-associated MARVEL proteins marveld3, tricellulin, and
occludin have distinct but overlapping functions. Mol Biol Cell.
21:1200–1213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duan HJ, Li XY, Liu C and Deng XL:
Chemokine-like factor-like MARVEL transmembrane domain-containing
family in autoimmune diseases. Chin Med J (Engl). 133:951–958.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Magyar JP, Ebensperger C, Schaeren-Wiemers
N and Suter U: Myelin and lymphocyte protein (MAL/MVP17/VIP17) and
plasmolipin are members of an extended gene family. Gene.
189:269–275. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Janz R, Südhof TC, Hammer RE, Unni V,
Siegelbaum SA and Bolshakov VY: Essential roles in synaptic
plasticity for synaptogyrin I and synaptophysin I. Neuron.
24:687–700. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adams DJ, Arthur CP and Stowell MH:
Architecture of the synaptophysin/synaptobrevin complex: Structural
evidence for an entropic clustering function at the synapse. Sci
Rep. 5:136592015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han W, Ding P, Xu M, Wang L, Rui M, Shi S,
Liu Y, Zheng Y, Chen Y, Yang T and Ma D: Identification of eight
genes encoding chemokine-like factor superfamily members 1–8
(CKLFSF1-8) by in silico cloning and experimental validation.
Genomics. 81:609–617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Wu QQ, Zhou YB, Zhang KH, Pang BX,
Li L, Sun N, Wang HS, Zhang S, Li WJ, et al: Cancer research
advance in CKLF-like MARVEL transmembrane domain containing member
family (review). Asian Pac J Cancer Prev. 17:2741–2744.
2016.PubMed/NCBI
|
|
20
|
Pérez P, Puertollano R and Alonso MA:
Structural and biochemical similarities reveal a family of proteins
related to the MAL proteolipid, a component of detergent-insoluble
membrane microdomains. Biochem Biophys Res Commun. 232:618–621.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Marco MC, Puertollano R,
Martínez-Menárguez JA and Alonso MA: Dynamics of MAL2 during
glycosylphosphatidylinositol-anchored protein transcytotic
transport to the apical surface of hepatoma HepG2 cells. Traffic.
7:61–73. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marazuela M, Acevedo A, García-López MA,
Adrados M, de Marco MC and Alonso MA: Expression of MAL2, an
integral protein component of the machinery for
basolateral-to-apical transcytosis, in human epithelia. J Histochem
Cytochem. 52:243–252. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Muppa P, Dao L, McCann B, Mcphail E, Shi M
and Rech K: Co-expression of MAL and PD-L2 by Immunohistochemistry
is Specific for Primary Mediastinal Large B-Cell Lymphoma. Lab
Invest. 99 (Suppl 1):972019.
|
|
24
|
Marazuela M, Acevedo A, Adrados M,
García-López MA and Alonso MA: Expression of MAL, an integral
protein component of the machinery for raft-mediated pical
transport, in human epithelia. J Histochem Cytochem. 51:665–674.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang B, Xiao J, Cheng X and Liu T: MAL2
interacts with IQGAP1 to promote pancreatic cancer progression by
increasing ERK1/2 phosphorylation. Biochem Biophys Res Commun.
554:63–70. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng C, Wang J, Zhang J, Hou S, Zheng Y
and Wang Q: Myelin and lymphocyte protein 2 regulates cell
proliferation and metastasis through the Notch pathway in prostate
adenocarcinoma. Transl Androl Urol. 10:2067–2077. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu J, Shimizu E, Zhang X, Partridge NC
and Qin L: EGFR signaling suppresses osteoblast differentiation and
inhibits expression of master osteoblastic transcription factors
Runx2 and Osterix. J Cell Biochem. 112:1749–1760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bello-Morales R, Pérez-Hernández M, Rejas
MT, Matesanz F, Alcina A and López-Guerrero JA: Interaction of PLP
with GFP-MAL2 in the human oligodendroglial cell line HOG. PLoS
One. 6:e193882011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lian Z, Yan X, Diao Y, Cui D and Liu H: T
cell differentiation protein 2 facilitates cell proliferation by
enhancing mTOR-mediated ribosome biogenesis in non-small cell lung
cancer. Discov Oncol. 13:262022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Byrne JA, Maleki S, Hardy JR, Gloss BS,
Murali R, Scurry JP, Fanayan S, Emmanuel C, Hacker NF, Sutherland
RL, et al: MAL2 and tumor protein D52 (TPD52) are frequently
overexpressed in ovarian carcinoma, but differentially associated
with histological subtype and patient outcome. BMC Cancer.
10:4972010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Li H, Yao D, Zou Q, Yu W and Zhou
L: The novel circ_0084904/miR-802/MAL2 axis promotes the
development of cervical cancer. Reprod Biol. 22:1006002022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mimori K, Shiraishi T, Mashino K, Sonoda
H, Yamashita K, Yoshinaga K, Masuda T, Utsunomiya T, Alonso MA,
Inoue H and Mori M: MAL gene expression in esophageal cancer
suppresses motility, invasion and tumorigenicity and enhances
apoptosis through the Fas pathway. Oncogene. 22:3463–3471. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zorzan E, Elgendy R, Guerra G, Da Ros S,
Gelain ME, Bonsembiante F, Garaffo G, Vitale N, Piva R, Marconato
L, et al: Hypermethylation-mediated silencing of CIDEA, MAL and
PCDH17 tumour suppressor genes in canine DLBCL: From multi-omics
analyses to mechanistic studies. Int J Mol Sci. 23:40212022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao
Y, Fan J, Cao M and Zhou Z: FTO modifies the m6A level of MALAT and
promotes bladder cancer progression. Clin Transl Med. 11:e3102021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Edwards JR, Yarychkivska O, Boulard M and
Bestor TH: DNA methylation and DNA methyltransferases. Epigenetics
Chromatin. 10:232017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang JS, Guo M, Montgomery EA, Thompson
RE, Cosby H, Hicks L, Wang S, Herman JG and Canto MI: DNA promoter
hypermethylation of p16 and APC predicts neoplastic progression in
Barrett's esophagus. Am J Gastroenterol. 104:2153–2160. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Labat-de-Hoz L, Rubio-Ramos A, Correas I
and Alonso MA: The MAL family of proteins: Normal function,
expression in cancer, and potential use as cancer biomarkers.
Cancers (Basel). 15:28012023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lara-Lemus R: On the role of myelin and
lymphocyte protein (MAL) in cancer: A puzzle with two faces. J
Cancer. 10:2312–2318. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Overmeer RM, Louwers JA, Meijer CJ, van
Kemenade FJ, Hesselink AT, Daalmeijer NF, Wilting SM, Heideman DA,
Verheijen RH, Zaal A, et al: Combined CADM1 and MAL promoter
methylation analysis to detect (pre-)malignant cervical lesions in
high-risk HPV-positive women. Int J Cancer. 129:2218–2225. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Holubekova V, Mersakova S, Grendar M,
Snahnicanova Z, Kudela E, Kalman M, Lasabova Z, Danko J and Zubor
P: The role of CADM1 and MAL promoter methylation in inflammation
and cervical intraepithelial neoplasia. Genet Test Mol Biomarkers.
24:256–263. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
de Marco MC, Martín-Belmonte F, Kremer L,
Albar JP, Correas I, Vaerman JP, Marazuela M, Byrne JA and Alonso
MA: MAL2, a novel raft protein of the MAL family, is an essential
component of the machinery for transcytosis in hepatoma HepG2
cells. J Cell Biol. 159:37–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang Y, Wang L, Wan C, Sun Y, Van der
Jeught K, Zhou Z, Dong T, So KM, Yu T, Li Y, et al: MAL2 drives
immune evasion in breast cancer by suppressing tumor antigen
presentation. J Clin Invest. 131:e1408372021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hamacher M, Pippirs U, Köhler A, Müller HW
and Bosse F: Plasmolipin: Genomic structure, chromosomal
localization, protein expression pattern, and putative association
with Bardet-Biedl syndrome. Mamm Genome. 12:933–937. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Aranda JF, Reglero-Real N, Kremer L,
Marcos-Ramiro B, Ruiz-Sáenz A, Calvo M, Enrich C, Correas I, Millán
J and Alonso MA: MYADM regulates Rac1 targeting to ordered
membranes required for cell spreading and migration. Mol Biol Cell.
22:1252–1262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Alonso MA, Barton DE and Francke U:
Assignment of the T-cell differentiation gene MAL to human
chromosome 2, region cen-q13. Immunogenetics. 27:91–95. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Marazuela M and Alonso MA: Expression of
MAL and MAL2, two elements of the protein machinery for
raft-mediated transport, in normal and neoplastic human tissue.
Histol Histopathol. 19:925–933. 2004.PubMed/NCBI
|
|
50
|
Michel V and Bakovic M: Lipid rafts in
health and disease. Biol Cell. 99:129–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Patra SK and Bettuzzi S: Epigenetic
DNA-methylation regulation of genes coding for lipid
raft-associated components: A role for raft proteins in cell
transformation and cancer progression (review). Oncol Rep.
17:1279–1290. 2007.PubMed/NCBI
|
|
52
|
Leitner J, Mahasongkram K, Schatzlmaier P,
Pfisterer K, Leksa V, Pata S, Kasinrerk W, Stockinger H and
Steinberger P: Differentiation and activation of human CD4 T cells
is associated with a gradual loss of myelin and lymphocyte protein.
Eur J Immunol. 51:848–863. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Antón OM, Andrés-Delgado L, Reglero-Real
N, Batista A and Alonso MA: MAL protein controls protein sorting at
the supramolecular activation cluster of human T lymphocytes. J
Immunol. 186:6345–6356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ventimiglia LN, Fernández-Martín L,
Martínez-Alonso E, Antón OM, Guerra M, Martínez-Menárguez JA,
Andrés G and Alonso MA: Cutting edge: Regulation of exosome
secretion by the integral MAL protein in T cells. J Immunol.
195:810–814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim T, Fiedler K, Madison DL, Krueger WH
and Pfeiffer SE: Cloning and characterization of MVP17: A
developmentally regulated myelin protein in oligodendrocytes. J
Neurosci Res. 42:413–422. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schaeren-Wiemers N, Schaefer C, Valenzuela
DM, Yancopoulos GD and Schwab ME: Identification of new
oligodendrocyte- and myelin-specific genes by a differential
screening approach. J Neurochem. 65:10–22. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zacchetti D, Peränen J, Murata M, Fiedler
K and Simons K: VIP17/MAL, a proteolipid in apical transport
vesicles. FEBS Lett. 377:465–469. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Millán J, Puertollano R, Fan L and Alonso
MA: Caveolin and MAL, two protein components of internal
detergent-insoluble membranes, are in distinct lipid
microenvironments in MDCK cells. Biochem Biophys Res Commun.
233:707–712. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Frank M, Schaeren-Wiemers N, Schneider R
and Schwab ME: Developmental expression pattern of the myelin
proteolipid MAL indicates different functions of MAL for immature
Schwann cells and in a late step of CNS myelinogenesis. J
Neurochem. 73:587–597. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Frank M: MAL, a proteolipid in
glycosphingolipid enriched domains: Functional implications in
myelin and beyond. Prog Neurobiol. 60:531–544. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Copie-Bergman C, Plonquet A, Alonso MA,
Boulland ML, Marquet J, Divine M, Möller P, Leroy K and Gaulard P:
MAL expression in lymphoid cells: further evidence for MAL as a
distinct molecular marker of primary mediastinal large B-cell
lymphomas. Mod Pathol. 15:1172–1180. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Overmeer RM, Henken FE, Bierkens M,
Wilting SM, Timmerman I, Meijer CJ, Snijders PJ and Steenbergen RD:
Repression of MAL tumour suppressor activity by promoter
methylation during cervical carcinogenesis. J Pathol. 219:327–336.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cao W, Zhang ZY, Xu Q, Sun Q, Yan M, Zhang
J, Zhang P, Han ZG and Chen WT: Epigenetic silencing of MAL, a
putative tumor suppressor gene, can contribute to human epithelium
cell carcinoma. Mol Cancer. 9:2962010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wilting SM, de Wilde J, Meijer CJ, Berkhof
J, Yi Y, van Wieringen WN, Braakhuis BJ, Meijer GA, Ylstra B,
Snijders PJ and Steenbergen RD: Integrated genomic and
transcriptional profiling identifies chromosomal loci with altered
gene expression in cervical cancer. Genes Chromosomes Cancer.
47:890–905. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Maruya S, Kim HW, Weber RS, Lee JJ, Kies
M, Luna MA, Batsakis JG and El-Naggar AK: Gene expression screening
of salivary gland neoplasms: Molecular markers of potential
histogenetic and clinical significance. J Mol Diagn. 6:180–190.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Suzuki M, Shiraishi K, Eguchi A, Ikeda K,
Mori T, Yoshimoto K, Ohba Y, Yamada T, Ito T, Baba Y and Baba H:
Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small
cell lung cancer. Oncol Rep. 29:1308–1314. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ostrow KL, Park HL, Hoque MO, Kim MS, Liu
J, Argani P, Westra W, Van Criekinge W and Sidransky D:
Pharmacologic unmasking of epigenetically silenced genes in breast
cancer. Clin Cancer Res. 15:1184–1191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li D, Zhang J, Wu L, Yang X, Chen Z and
Yuan J: Myelin and lymphocyte protein (MAL): A novel biomarker for
uterine corpus endometrial carcinoma. Cancer Manag Res.
13:7311–7323. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
His ED, Sup SJ, Alemany C, Tso E, Skacel
M, Elson P, Alonso MA and Pohlman B: MAL is expressed in a subset
of Hodgkin lymphoma and identifies a population of patients with
poor prognosis. Am J Clin Pathol. 125:776–782. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kohno T, Moriuchi R, Katamine S, Yamada Y,
Tomonaga M and Matsuyama T: Identification of genes associated with
the progression of adult T cell leukemia (ATL). Jpn J Cancer Res.
91:1103–1110. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chan JK: Mediastinal large B-cell
lymphoma: New evidence in support of its distinctive identity. Adv
Anat Pathol. 7:201–209. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Copie-Bergman C, Gaulard P,
Maouche-Chrétien L, Brière J, Haioun C, Alonso MA, Roméo PH and
Leroy K: The MAL gene is expressed in primary mediastinal large
B-cell lymphoma. Blood. 94:3567–3575. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lind GE, Ahlquist T, Kolberg M, Berg M,
Eknaes M, Alonso MA, Kallioniemi A, Meling GI, Skotheim RI, Rognum
TO, et al: Hypermethylated MAL gene-a silent marker of early colon
tumorigenesis. J Transl Med. 6:132008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jin Z, Wang L, Zhang Y, Cheng Y, Gao Y,
Feng X, Dong M, Cao Z, Chen S, Yu H, et al: MAL hypermethylation is
a tissue-specific event that correlates with MAL mRNA expression in
esophageal carcinoma. Sci Rep. 3:28382013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Horne HN, Lee PS, Murphy SK, Alonso MA,
Olson JA Jr and Marks JR: Inactivation of the MAL gene in breast
cancer is a common event that predicts benefit from adjuvant
chemotherapy. Mol Cancer Res. 7:199–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Buffart TE, Overmeer RM, Steenbergen RD,
Tijssen M, van Grieken NC, Snijders PJ, Grabsch HI, van de Velde
CJ, Carvalho B and Meijer GA: MAL promoter hypermethylation as a
novel prognostic marker in gastric cancer. Br J Cancer.
99:1802–1807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Geng Z, Li J, Li S, Wang Y, Zhang L, Hu Q,
Wang X, Zuo L, Song X, Zhang X, et al: MAL protein suppresses the
metastasis and invasion of GC cells by interfering with the
phosphorylation of STAT3. J Transl Med. 20:502022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Deng F and Han Bae Y: Lipid raft-mediated
and upregulated coordination pathways assist transport of
glycocholic acid-modified nanoparticle in a human breast cancer
cell line of SK-BR-3. Int J Pharm. 617:1215892022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bierkens M, Hesselink AT, Meijer CJ,
Heideman DA, Wisman GB, van der Zee AG, Snijders PJ and Steenbergen
RD: CADM1 and MAL promoter methylation levels in hrHPV-positive
cervical scrapes increase proportional to degree and duration of
underlying cervical disease. Int J Cancer. 133:1293–1299. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Salta S, Lobo J, Magalhães B, Henrique R
and Jerónimo C: DNA methylation as a triage marker for colposcopy
referral in HPV-based cervical cancer screening: A systematic
review and meta-analysis. Clin Epigenetics. 15:1252023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
De Strooper LM, Hesselink AT, Berkhof J,
Meijer CJ, Snijders PJ, Steenbergen RD and Heideman DA: Combined
CADM1/MAL methylation and cytology testing for colposcopy triage of
high-risk HPV-positive women. Cancer Epidemiol Biomarkers Prev.
23:1933–1937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Leffers M, Herbst J, Kropidlowski J,
Prieske K, Bohnen AL, Peine S, Jaeger A, Oliveira-Ferrer L, Goy Y,
Schmalfeldt B, et al: Combined liquid biopsy methylation analysis
of CADM1 and MAL in cervical cancer patients. Cancers (Basel).
14:39542022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Phillips S, Cassells K, Garland SM,
Machalek DA, Roberts JM, Templeton DJ, Jin F, Poynten IM, Hillman
RJ, Grulich AE, et al: Gene methylation of CADM1 and MAL identified
as a biomarker of high grade anal intraepithelial neoplasia. Sci
Rep. 12:35652022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Berchuck A, Iversen ES, Lancaster JM,
Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M,
et al: Patterns of gene expression that characterize long-term
survival in advanced stage serous ovarian cancers. Clin Cancer Res.
11:3686–3696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zanotti L, Romani C, Tassone L, Todeschini
P, Tassi RA, Bandiera E, Damia G, Ricci F, Ardighieri L, Calza S,
et al: MAL gene overexpression as a marker of high-grade serous
ovarian carcinoma stem-like cells that predicts chemoresistance and
poor prognosis. BMC Cancer. 17:3662017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lee PS, Teaberry VS, Bland AE, Huang Z,
Whitaker RS, Baba T, Fujii S, Secord AA, Berchuck A and Murphy SK:
Elevated MAL expression is accompanied by promoter hypomethylation
and platinum resistance in epithelial ovarian cancer. Int J Cancer.
126:1378–1389. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Iwasaki T, Matsushita M, Nonaka D, Nagata
K, Kato M, Kuwamoto S, Murakami I and Hayashi K: Lower expression
of CADM1 and higher expression of MAL in Merkel cell carcinomas are
associated with Merkel cell polyomavirus infection and better
prognosis. Hum Pathol. 48:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Su C, Huang R, Yu Z, Zheng J, Liu F, Liang
H and Mo Z: Myelin and lymphocyte protein serves as a prognostic
biomarker and is closely associated with the tumor microenvironment
in the nephroblastoma. Cancer Med. 11:1427–1438. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Pal SK, Noguchi S, Yamamoto G, Yamada A,
Isobe T, Hayashi S, Tanaka J, Tanaka Y, Kamijo R, Yamane GY and
Tachikawa T: Expression of myelin and lymphocyte protein (MAL) in
oral carcinogenesis. Med Mol Morphol. 45:222–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Beder LB, Gunduz M, Hotomi M, Fujihara K,
Shimada J, Tamura S, Gunduz E, Fukushima K, Yaykasli K, Grenman R,
et al: T-lymphocyte maturation-associated protein gene as a
candidate metastasis suppressor for head and neck squamous cell
carcinomas. Cancer Sci. 100:873–880. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Luo Y, Zhou LQ, Yang F, Chen JC, Chen JJ
and Wang YJ: Construction and analysis of a conjunctive diagnostic
model of HNSCC with random forest and artificial neural network.
Sci Rep. 13:67362023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yue Y, Song M, Qiao Y, Li P, Yuan Y, Lian
J, Wang S and Zhang Y: Gene function analysis and underlying
mechanism of esophagus cancer based on microarray gene expression
profiling. Oncotarget. 8:105222–105237. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang X, Li G, Luo Q and Gan C:
Identification of crucial genes associated with esophageal squamous
cell carcinoma by gene expression profile analysis. Oncol Lett.
15:8983–8990. 2018.PubMed/NCBI
|
|
94
|
Visser E, Franken IA, Brosens LA, Ruurda
JP and van Hillegersberg R: Prognostic gene expression profiling in
esophageal cancer: A systematic review. Oncotarget. 8:5566–5577.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Choi B, Han TS, Min J, Hur K, Lee SM, Lee
HJ, Kim YJ and Yang HK: MAL and TMEM220 are novel DNA methylation
markers in human gastric cancer. Biomarkers. 22:35–44. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kurashige J, Sawada G, Takahashi Y, Eguchi
H, Sudo T, Ikegami T, Yoshizumi T, Soejima Y, Ikeda T, Kawanaka H,
et al: Suppression of MAL gene expression in gastric cancer
correlates with metastasis and mortality. Fukuoka Igaku Zasshi.
104:344–349. 2013.PubMed/NCBI
|
|
97
|
Ahlquist T, Lind GE, Costa VL, Meling GI,
Vatn M, Hoff GS, Rognum TO, Skotheim RI, Thiis-Evensen E and Lothe
RA: Gene methylation profiles of normal mucosa, and benign and
malignant colorectal tumors identify early onset markers. Mol
Cancer. 7:942008. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Patai ÁV, Valcz G, Hollósi P, Kalmár A,
Péterfia B, Patai Á, Wichmann B, Spisák S, Barták BK, Leiszter K,
et al: Comprehensive DNA methylation analysis reveals a common
ten-gene methylation signature in colorectal adenomas and
carcinomas. PLoS One. 10:e01338362015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sambuudash O, Kim HS and Cho MY: Lack of
aberrant methylation in an adjacent area of left-sided colorectal
cancer. Yonsei Med J. 58:749–755. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu X, Bi H, Ge A, Xia T, Fu J, Liu Y, Sun
H, Li D and Zhao Y: DNA hypermethylation of MAL gene may act as an
independent predictor of favorable prognosis in patients with
colorectal cancer. Transl Cancer Res. 8:1985–1996. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ma R, Xu YE, Wang M and Peng W:
Suppression of MAL gene expression is associated with colorectal
cancer metastasis. Oncol Lett. 10:957–961. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Huang KT, Mikeska T, Li J, Takano EA,
Millar EK, Graham PH, Boyle SE, Campbell IG, Speed TP, Dobrovic A
and Fox SB: Assessment of DNA methylation profiling and copy number
variation as indications of clonal relationship in ipsilateral and
contralateral breast cancers to distinguish recurrent breast cancer
from a second primary tumour. BMC Cancer. 15:6692015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Vasiljević N, Scibior-Bentkowska D,
Brentnall AR, Cuzick J and Lorincz AT: Credentialing of DNA
methylation assays for human genes as diagnostic biomarkers of
cervical intraepithelial neoplasia in high-risk HPV positive women.
Gynecol Oncol. 132:709–714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ki EY, Lee KH, Hur SY, Rhee JE, Kee MK,
Kang C and Park JS: Methylation of cervical neoplastic cells
infected with human papillomavirus 16. Int J Gynecol Cancer.
26:176–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Deng J, Wang Y, Zhang S and Chen L: A
novel long noncoding RNA located on the antisense strand of MAL
promotes the invasion and metastasis of oral squamous cell
carcinoma. Arch Oral Biol. 155:1057902023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Meršaková S, Holubeková V, Grendár M,
Višňovský J, Ňachajová M, Kalman M, Kúdela E, Žúbor P, Bielik T,
Lasabová Z and Danko J: Methylation of CADM1 and MAL together with
HPV status in cytological cervical specimens serves an important
role in the progression of cervical intraepithelial neoplasia.
Oncol Lett. 16:7166–7174. 2018.PubMed/NCBI
|
|
107
|
Ondič O, Němcová J, Alaghehbandan R, Černá
K, Gomolčáková B, Kinkorová-Luňáčková I, Chytra J, Šidlová H, Májek
O and Bouda J: The detection of DNA methylation of tumour
suppressor genes in cervical high-grade squamous intraepithelial
lesion: A prospective cytological-histological correlation study of
70 cases. Cytopathology. 30:426–431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Berchuck A, Iversen ES, Luo J, Clarke JP,
Horne H, Levine DA, Boyd J, Alonso MA, Secord AA, Bernardini MQ, et
al: Microarray analysis of early stage serous ovarian cancers shows
profiles predictive of favorable outcome. Clin Cancer Res.
15:2448–2455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kulbe H, Otto R, Darb-Esfahani S, Lammert
H, Abobaker S, Welsch G, Chekerov R, Schäfer R, Dragun D, Hummel M,
et al: Discovery and validation of novel biomarkers for detection
of epithelial ovarian cancer. Cells. 8:7132019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Vasiljević N, Ahmad AS, Thorat MA, Fisher
G, Berney DM, Møller H, Foster CS, Cuzick J and Lorincz AT: DNA
methylation gene-based models indicating independent poor outcome
in prostate cancer. BMC Cancer. 14:6552014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ahmad AS, Vasiljević N, Carter P, Berney
DM, Møller H, Foster CS, Cuzick J and Lorincz AT: A novel DNA
methylation score accurately predicts death from prostate cancer in
men with low to intermediate clinical risk factors. Oncotarget.
7:71833–71840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Choi P and Chen C: Genetic expression
profiles and biologic pathway alterations in head and neck squamous
cell carcinoma. Cancer. 104:1113–1128. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Riva G, Biolatti M, Pecorari G, Dell'Oste
V and Landolfo S: PYHIN proteins and HPV: Role in the pathogenesis
of head and neck squamous cell carcinoma. Microorganisms. 8:142019.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Misawa K, Imai A, Matsui H, Kanai A,
Misawa Y, Mochizuki D, Mima M, Yamada S, Kurokawa T, Nakagawa T and
Mineta H: Identification of novel methylation markers in
HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue
verification and validation testing in ctDNA. Oncogene.
39:4741–4755. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Jiang Y, Chen Y, Gao L, Ye Q and Alonso
MA: Expression pattern of MAL in normal epithelial cells, benign
tumor, and squamous cell carcinoma of larynx. Lin Chuang Er Bi Yan
Hou Tou Jing Wai Ke Za Zhi. 23:451–453. 2009.(In Chinese).
PubMed/NCBI
|
|
116
|
Wilson SH, Bailey AM, Nourse CR, Mattei MG
and Byrne JA: Identification of MAL2, a novel member of the mal
proteolipid family, though interactions with TPD52-like proteins in
the yeast two-hybrid system. Genomics. 76:81–88. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
In JG, Striz AC, Bernad A and Tuma PL:
Serine/threonine kinase 16 and MAL2 regulate constitutive secretion
of soluble cargo in hepatic cells. Biochem J. 463:201–213. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Marazuela M, Martín-Belmonte F,
García-López MA, Aranda JF, de Marco MC and Alonso MA: Expression
and distribution of MAL2, an essential element of the machinery for
basolateral-to-apical transcytosis, in human thyroid epithelial
cells. Endocrinology. 145:1011–1016. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Llorente A, de Marco MC and Alonso MA:
Caveolin-1 and MAL are located on prostasomes secreted by the
prostate cancer PC-3 cell line. J Cell Sci. 117:5343–5351. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Fanayan S, Shehata M, Agterof AP, McGuckin
MA, Alonso MA and Byrne JA: Mucin 1 (MUC1) is a novel partner for
MAL2 in breast carcinoma cells. BMC Cell Biol. 10:72009. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
In JG and Tuma PL: MAL2 selectively
regulates polymeric IgA receptor delivery from the Golgi to the
plasma membrane in WIF-B cells. Traffic. 11:1056–1066. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
López-Coral A, Del Vecchio GJ, Chahine JJ,
Kallakury BV and Tuma PL: MAL2-induced actin-based protrusion
formation is anti-oncogenic in hepatocellular carcinoma. Cancers
(Basel). 12:4222020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Farazi PA and DePinho RA: The genetic and
environmental basis of hepatocellular carcinoma. Discov Med.
6:182–186. 2006.PubMed/NCBI
|
|
124
|
Bhandari A, Shen Y, Sindan N, Xia E,
Gautam B, Lv S and Zhang X: MAL2 promotes proliferation, migration,
and invasion through regulating epithelial-mesenchymal transition
in breast cancer cell lines. Biochem Biophys Res Commun.
504:434–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
An L, Gong H, Yu X, Zhang W, Liu X, Yang
X, Shu L, Liu J and Yang L: Downregulation of MAL2 inhibits breast
cancer progression through regulating β-catenin/c-Myc axis. Cancer
Cell Int. 23:1442023. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhong Y, Zhuang Z, Mo P, Shang Q, Lin M,
Gong J, Huang J, Mo H and Huang M: Overexpression of MAL2
correlates with immune infiltration and poor prognosis in breast
cancer. Evid Based Complement Alternat Med. 2021:55578732021.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Yuan J, Jiang X, Lan H, Zhang X, Ding T,
Yang F, Zeng D, Yong J, Niu B and Xiao S: Multi-omics analysis of
the therapeutic value of MAL2 based on data mining in human
cancers. Front Cell Dev Biol. 9:7366492022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Jeong J, Shin JH, Li W, Hong JY, Lim J,
Hwang JY, Chung JJ, Yan Q, Liu Y, Choi J and Wysolmerski J: MAL2
mediates the formation of stable HER2 signaling complexes within
lipid raft-rich membrane protrusions in breast cancer cells. Cell
Rep. 37:1101602021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Dersh D and Yewdell JW: Immune
MAL2-practice: Breast cancer immunoevasion via MHC class I
degradation. J Clin Invest. 131:e1443442021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhu X, Bu J, Zhu T and Jiang Y: Targeting
KK-LC-1 inhibits malignant biological behaviors of triple-negative
breast cancer. J Transl Med. 21:1842023. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Eguchi D, Ohuchida K, Kozono S, Ikenaga N,
Shindo K, Cui L, Fujiwara K, Akagawa S, Ohtsuka T, Takahata S, et
al: MAL2 expression predicts distant metastasis and short survival
in pancreatic cancer. Surgery. 154:573–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Xiong F, Wu GH, Wang B and Chen YJ:
Plastin-3 is a diagnostic and prognostic marker for pancreatic
adenocarcinoma and distinguishes from diffuse large B-cell
lymphoma. Cancer Cell Int. 21:4112021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen Y, Zheng B, Robbins DH, Lewin DN,
Mikhitarian K, Graham A, Rumpp L, Glenn T, Gillanders WE, Cole DJ,
et al: Accurate discrimination of pancreatic ductal adenocarcinoma
and chronic pancreatitis using multimarker expression data and
samples obtained by minimally invasive fine needle aspiration. Int
J Cancer. 120:1511–1517. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Gao X, Chen Z, Li A, Zhang X and Cai X:
MiR-129 regulates growth and invasion by targeting MAL2 in
papillary thyroid carcinoma. Biomed Pharmacother. 105:1072–1078.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li J, Li Y, Liu H, Liu Y and Cui B: The
four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are
highly expressed in colorectal cancer and correlated with poor
prognosis. PLoS One. 12:e01785152017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Wang K, Yang Y, Zheng S and Hu W:
Association mining identifies MAL2 as a novel tumor suppressor in
colorectal cancer. Onco Targets Ther. 15:761–769. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Shao B, Fu X, Li X, Li Y and Gan N:
RP11-284F21.9 promotes oral squamous cell carcinoma development via
the miR-383-5p/MAL2 axis. J Oral Pathol Med. 49:21–29. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Dasgupta S, Tripathi PK, Qin H,
Bhattacharya-Chatterjee M, Valentino J and Chatterjee SK:
Identification of molecular targets for immunotherapy of patients
with head and neck squamous cell carcinoma. Oral Oncol. 42:306–316.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Weis VG, Petersen CP, Mills JC, Tuma PL,
Whitehead RH and Goldenring JR: Establishment of novel in vitro
mouse chief cell and SPEM cultures identifies MAL2 as a marker of
metaplasia in the stomach. Am J Physiol Gastrointest Liver Physiol.
307:G777–G792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Smolarz B, Zadrożna-Nowak A and Romanowicz
H: The role of lncRNA in the development of tumors, including
breast cancer. Int J Mol Sci. 22:84272021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
He J, Yan H, Wei S and Chen G: LncRNA
ST8SIA6-AS1 promotes cholangiocarcinoma progression by suppressing
the miR-145-5p/MAL2 axis. Onco Targets Ther. 14:3209–3223. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhang C, Xu L, Li X, Chen Y, Shi T and
Wang Q: LINC00460 facilitates cell proliferation and inhibits
ferroptosis in breast cancer through the miR-320a/MAL2 axis.
Technol Cancer Res Treat. 22:153303382311643592023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Le Guelte A and Macara IG: Plasmolipin-a
new player in endocytosis and epithelial development. EMBO J.
34:1147–1148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Rodríguez-Fraticelli AE, Bagwell J,
Bosch-Fortea M, Boncompain G, Reglero-Real N, García-León MJ,
Andrés G, Toribio ML, Alonso MA, Millán J, et al: Developmental
regulation of apical endocytosis controls epithelial patterning in
vertebrate tubular organs. Nat Cell Biol. 17:241–250. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
You J, Corley SM, Wen L, Hodge C,
Höllhumer R, Madigan MC, Wilkins MR and Sutton G: RNA-Seq analysis
and comparison of corneal epithelium in keratoconus and myopia
patients. Sci Rep. 8:3892018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Yaffe Y, Hugger I, Yassaf IN,
Shepshelovitch J, Sklan EH, Elkabetz Y, Yeheskel A, Pasmanik-Chor
M, Benzing C, Macmillan A, et al: The myelin proteolipid
plasmolipin forms oligomers and induces liquid-ordered membranes in
the Golgi complex. J Cell Sci. 128:2293–2302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Fischer I, Cochary EF, Konola JT and
Romano-Clark G: Expression of plasmolipin in oligodendrocytes. J
Neurosci Res. 28:81–89. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Fischer I, Durrie R and Sapirstein VS:
Plasmolipin: The other myelin proteolipid. A review of studies on
its structure, expression, and function. Neurochem Res. 19:959–966.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Shea TB, Fischer I and Sapirstein V:
Expression of a plasma membrane proteolipid during differentiation
of neuronal and glial cells in primary culture. J Neurochem.
47:697–706. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Hasse B, Bosse F and Müller HW: Proteins
of peripheral myelin are associated with
glycosphingolipid/cholesterol-enriched membranes. J Neurosci Res.
69:227–232. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Fredriksson K, Van Itallie CM, Aponte A,
Gucek M, Tietgens AJ and Anderson JM: Proteomic analysis of
proteins surrounding occludin and claudin-4 reveals their proximity
to signaling and trafficking networks. PLoS One. 10:e01170742015.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Sapirstein VS, Nolan C, Stern R, Ciocci M
and Masur SK: Identification of the plasma membrane proteolipid
protein as a constituent of brain coated vesicles and synaptic
plasma membrane. J Neurochem. 51:925–933. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Sapirstein VS, Nolan CE, Stern R,
Gray-Board G and Beard ME: Identification of plasmolipin as a major
constituent of white matter clathrin-coated vesicles. J Neurochem.
58:1372–1378. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Azzaz F, Mazzarino M, Chahinian H, Yahi N,
Scala CD and Fantini J: Structure of the myelin sheath proteolipid
plasmolipin (PLLP) in a ganglioside-containing lipid raft. Front
Biosci (Landmark Ed). 28:1572023. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Gillen C, Gleichmann M, Greiner-Petter R,
Zoidl G, Kupfer S, Bosse F, Auer J and Müller HW: Full-lenth
cloning, expression and cellular localization of rat plasmolipin
mRNA, a proteolipid of PNS and CNS. Eur J Neurosci. 8:405–414.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Fischer I and Sapirstein VS: Molecular
cloning of plasmolipin. Characterization of a novel proteolipid
restricted to brain and kidney. J Biol Chem. 269:24912–24919. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Bosse F, Hasse B, Pippirs U,
Greiner-Petter R and Müller HW: Proteolipid plasmolipin:
Localization in polarized cells, regulated expression and lipid
raft association in CNS and PNS myelin. J Neurochem. 86:508–518.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Dannaeus K, Bessonova M and Jönsson JI:
Characterization of the mouse myeloid-associated differentiation
marker (MYADM) gene: Promoter analysis and protein localization.
Mol Biol Rep. 32:149–157. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Takematsu E, Spencer A, Auster J, Chen PC,
Graham A, Martin P and Baker AB: Genome wide analysis of gene
expression changes in skin from patients with type 2 diabetes. PLoS
One. 15:e02252672020. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zheng P, Sun S, Wang J, Cheng ZJ, Lei KC,
Xue M, Zhang T, Huang H, Zhang XD and Sun B: Integrative omics
analysis identifies biomarkers of idiopathic pulmonary fibrosis.
Cell Mol Life Sci. 79:662022. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Lautner-Rieske A, Thiebe R and Zachau HG:
Searching for non-V kappa transcripts from the human immunoglobulin
kappa locus. Gene. 159:199–202. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
de Marco MC, Kremer L, Albar JP,
Martinez-Menarguez JA, Ballesta J, Garcia-Lopez MA, Marazuela M,
Puertollano R and Alonso MA: BENE, a novel raft-associated protein
of the MAL proteolipid family, interacts with caveolin-1 in human
endothelial-like ECV304 cells. J Biol Chem. 276:23009–23017. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Rubio-Ramos A, Bernabé-Rubio M,
Labat-de-Hoz L, Casares-Arias J, Kremer L, Correas I and Alonso MA:
MALL, a membrane-tetra-spanning proteolipid overexpressed in
cancer, is present in membraneless nuclear biomolecular
condensates. Cell Mol Life Sci. 79:2362022. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Kim K, Park U, Wang J, Lee J, Park S, Kim
S, Choi D, Kim C and Park J: Gene profiling of colonic serrated
adenomas by using oligonucleotide microarray. Int J Colorectal Dis.
23:569–580. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Kim HA, Kim KH and Lee RA: Expression of
caveolin-1 is correlated with Akt-1 in colorectal cancer tissues.
Exp Mol Pathol. 80:165–170. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wang X, Fan J, Yu F, Cui F, Sun X, Zhong
L, Yan D, Zhou C, Deng G, Wang B, et al: Decreased MALL expression
negatively impacts colorectal cancer patient survival. Oncotarget.
7:22911–22927. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Li DX, Yu QX, Zeng CX, Ye LX, Guo YQ, Liu
JF, Zheng HH, Feng D and Wei W: A novel endothelial-related
prognostic index by integrating single-cell and bulk RNA sequencing
data for patients with kidney renal clear cell carcinoma. Front
Genet. 14:10964912023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Aranda JF, Reglero-Real N, Marcos-Ramiro
B, Ruiz-Sáenz A, Fernández-Martín L, Bernabé-Rubio M, Kremer L,
Ridley AJ, Correas I, Alonso MA and Millán J: MYADM controls
endothelial barrier function through ERM-dependent regulation of
ICAM-1 expression. Mol Biol Cell. 24:483–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Wang Q, Li N, Wang X, Shen J, Hong X, Yu
H, Zhang Y, Wan T, Zhang L, Wang J and Cao X: Membrane protein
hMYADM preferentially expressed in myeloid cells is up-regulated
during differentiation of stem cells and myeloid leukemia cells.
Life Sci. 80:420–429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Cui W, Yu L, He H, Chu Y, Gao J, Wan B,
Tang L and Zhao S: Cloning of human myeloid-associated
differentiation marker (MYADM) gene whose expression was
up-regulated in NB4 cells induced by all-trans retinoic acid. Mol
Biol Rep. 28:123–138. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Pettersson M, Dannaeus K, Nilsson K and
Jönsson JI: Isolation of MYADM, a novel hematopoietic-associated
marker gene expressed in multipotent progenitor cells and
up-regulated during myeloid differentiation. J Leukoc Biol.
67:423–431. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Dy ABC, Langlais PR, Barker NK, Addison
KJ, Tanyaratsrisakul S, Boitano S, Christenson SA, Kraft M, Meyers
D, Bleecker ER, et al: Myeloid-associated differentiation marker is
a novel SP-A-associated transmembrane protein whose expression on
airway epithelial cells correlates with asthma severity. Sci Rep.
11:233922021. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Sun L, Lin P, Chen Y, Yu H, Ren S, Wang J,
Zhao L and Du G: miR-182-3p/Myadm contribute to pulmonary artery
hypertension vascular remodeling via a KLF4/p21-dependent
mechanism. Theranostics. 10:5581–5599. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
de Wit NJ, Rijntjes J, Diepstra JH, van
Kuppevelt TH, Weidle UH, Ruiter DJ and van Muijen GN: Analysis of
differential gene expression in human melanocytic tumour lesions by
custom made oligonucleotide arrays. Br J Cancer. 92:2249–2261.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Megger DA, Naboulsi W, Meyer HE and Sitek
B: Proteome analyses of hepatocellular carcinoma. J Clin Transl
Hepatol. 2:23–30. 2014.PubMed/NCBI
|
|
177
|
Zhou M, Chen Y, Gu X and Wang C: A
comprehensive bioinformatic analysis for identification of
myeloid-associated differentiation marker as a potential negative
prognostic biomarker in non-small-cell lung cancer. Pathol Oncol
Res. 28:16105042022. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Wang X, Dou X, Ren X, Rong Z, Sun L, Deng
Y, Chen P and Li Z: A ductal-cell-related risk model integrating
single-cell and bulk sequencing data predicts the prognosis of
patients with pancreatic adenocarcinoma. Front Genet.
12:7636362021. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Papasotiriou I, Apostolou P, Ntanovasilis
DA, Parsonidis P, Osmonov D and Jünemann KP: Study and detection of
potential markers for predicting metastasis into lymph nodes in
prostate cancer. Biomark Med. 14:1317–1327. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Echevarria MI, Awasthi S, Cheng CH,
Berglund AE, Rounbehler RJ, Gerke TA, Takhar M, Davicioni E, Klein
EA, Freedland SJ, et al: African American specific gene panel
predictive of poor prostate cancer outcome. J Urol. 202:247–255.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Zhang B, Ren Z, Zheng H, Lin M, Chen G,
Luo OJ and Zhu G: CRISPR activation screening in a mouse model for
drivers of hepatocellular carcinoma growth and metastasis.
iScience. 26:1060992023. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Li D, Jin C, Yin C, Zhang Y, Pang B, Tian
L, Han W, Ma D and Wang Y: An alternative splice form of CMTM8
induces apoptosis. Int J Biochem Cell Biol. 39:2107–2119. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Jin C, Ding P, Wang Y and Ma D: Regulation
of EGF receptor signaling by the MARVEL domain-containing protein
CKLFSF8. FEBS Lett. 579:6375–6382. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Jin C, Wang Y, Han W, Zhang Y, He Q, Li D,
Yin C, Tian L, Liu D, Song Q and Ma D: CMTM8 induces
caspase-dependent and -independent apoptosis through a
mitochondria-mediated pathway. J Cell Physiol. 211:112–120. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Yan M, Zhu X, Qiao H, Zhang H, Xie W and
Cai J: Downregulated CMTM8 correlates with poor prognosis in
gastric cancer patients. DNA Cell Biol. 40:791–797. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wu K, Li X, Gu H, Yang Q, Liu Y and Wang
L: Research advances in CKLF-like MARVEL transmembrane
domain-containing family in non-small cell lung cancer. Int J Biol
Sci. 15:2576–2583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Hu H, Chen JW, Xu KX, Wang D, Wang Y, Wang
GW, Zhang SY and Wang XF: Expressions of CMTM8 and E-cadherin in
primary and metastatic clear cell renal cell carcinoma. Beijing Da
Xue Xue Bao Yi Xue Ban. 45:537–541. 2013.(In Chinese). PubMed/NCBI
|
|
188
|
Zhang W, Mendoza MC, Pei X, Ilter D,
Mahoney SJ, Zhang Y, Ma D, Blenis J and Wang Y: Down-regulation of
CMTM8 induces epithelial-to-mesenchymal transition-like changes via
c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol
Chem. 287:11850–11858. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Zeng X, Ma X, Guo H, Wei L, Zhang Y, Sun
C, Han N, Sun S and Zhang N: MicroRNA-582-5p promotes
triple-negative breast cancer invasion and metastasis by
antagonizing CMTM8. Bioengineered. 12:10126–10135. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Both J, Krijgsman O, Bras J, Schaap GR,
Baas F, Ylstra B and Hulsebos TJ: Focal chromosomal copy number
aberrations identify CMTM8 and GPR177 as new candidate driver genes
in osteosarcoma. PLoS One. 9:e1158352014. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Zhang S, Pei X, Hu H, Zhang W, Mo X, Song
Q, Zhang Y, Xu K, Wang Y and Na Y: Functional characterization of
the tumor suppressor CMTM8 and its association with prognosis in
bladder cancer. Tumour Biol. 37:6217–6225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Gao D, Hu H, Wang Y, Yu W, Zhou J, Wang X,
Wang W, Zhou C and Xu K: CMTM8 inhibits the carcinogenesis and
progression of bladder cancer. Oncol Rep. 34:2853–2863. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Kang N, Xie X, Zhou X, Wang Y, Chen S, Qi
R, Liu T and Jiang H: Identification and validation of
EMT-immune-related prognostic biomarkers CDKN2A, CMTM8 and ILK in
colon cancer. BMC Gastroenterol. 22:1902022. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhang M, Wang J, Yue H and Zhang L:
Identification of prognostic biomarkers in the CMTM family genes of
human ovarian cancer through bioinformatics analysis and
experimental verification. Front Genet. 13:9183192022. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen
L, Liu L, Chen Z and Meng Z: CMTM8 as an LPA1-associated partner
mediates lysophosphatidic acid-induced pancreatic cancer
metastasis. Ann Transl Med. 9:422021. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Liu Y, Chew MH, Tham CK, Tang CL, Ong SY
and Zhao Y: Methylation of serum SST gene is an independent
prognostic marker in colorectal cancer. Am J Cancer Res.
6:2098–2108. 2016.PubMed/NCBI
|
|
197
|
Guerrero-Preston R, Hadar T, Ostrow KL,
Soudry E, Echenique M, Ili-Gangas C, Pérez G, Perez J,
Brebi-Mieville P, Deschamps J, et al: Differential promoter
methylation of kinesin family member 1a in plasma is associated
with breast cancer and DNA repair capacity. Oncol Rep. 32:505–512.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Hentschel AE, Nieuwenhuijzen JA,
Bosschieter J, Splunter APV, Lissenberg-Witte BI, Voorn JPV,
Segerink LI, Moorselaar RJAV and Steenbergen RDM: Comparative
analysis of urine fractions for optimal bladder cancer detection
using DNA methylation markers. Cancers (Basel). 12:8592020.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Lind GE, Danielsen SA, Ahlquist T, Merok
MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff
G, et al: Identification of an epigenetic biomarker panel with high
sensitivity and specificity for colorectal cancer and adenomas. Mol
Cancer. 10:852011. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Obermayr E, Sanchez-Cabo F, Tea MK, Singer
CF, Krainer M, Fischer MB, Sehouli J, Reinthaller A, Horvat R,
Heinze G, et al: Assessment of a six gene panel for the molecular
detection of circulating tumor cells in the blood of female cancer
patients. BMC Cancer. 10:6662010. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Obermayr E, Maritschnegg E, Agreiter C,
Pecha N, Speiser P, Helmy-Bader S, Danzinger S, Krainer M, Singer C
and Zeillinger R: Efficient leukocyte depletion by a novel
microfluidic platform enables the molecular detection and
characterization of circulating tumor cells. Oncotarget. 9:812–823.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Ding W, Li Z, Wang C, Dai J, Ruan G and Tu
C: Anthracycline versus nonanthracycline adjuvant therapy for early
breast cancer: A systematic review and meta-analysis. Medicine
(Baltimore). 97:e129082018. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Willson ML, Burke L, Ferguson T, Ghersi D,
Nowak AK and Wilcken N: Taxanes for adjuvant treatment of early
breast cancer. Cochrane Database Syst Rev.
9:CD0044212019.PubMed/NCBI
|
|
204
|
Arumugam T, Ramachandran V, Fournier KF,
Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey
DJ and Choi W: Epithelial to mesenchymal transition contributes to
drug resistance in pancreatic cancer. Cancer Res. 69:5820–5828.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Bracker TU, Sommer A, Fichtner I, Faus H,
Haendler B and Hess-Stumpp H: Efficacy of MS-275, a selective
inhibitor of class I histone deacetylases, in human colon cancer
models. Int J Oncol. 35:909–920. 2009.PubMed/NCBI
|