|
1
|
Fritz AJ, Sehgal N, Piss A, Xu J and
Berezney R: Chromosome territories and the global regulation of the
genome. Genes Chromosomes Cancer. 58:407–426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rajapakse I, Perlman MD, Scalzo D,
Kooperberg C, Groudine M and Kosak ST: The emergence of
lineage-specific chromosomal topologies from coordinate gene
regulation. Proc Natl Acad Sci USA. 106:6679–6684. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cremer T, Cremer M, Dietzel S, Müller S,
Solovei I and Fakan S: Chromosome territories-a functional nuclear
landscape. Curr Opin Cell Biol. 18:307–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chuang CH, Carpenter AE, Fuchsova B,
Johnson T, de Lanerolle P and Belmont AS: Long-range directional
movement of an interphase chromosome site. Curr Biol. 18:825–831.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu M and Cook PR: Similar active genes
cluster in specialized transcription factories. J Cell Biol.
181:615–623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sutherland H and Bickmore WA:
Transcription factories: Gene expression in unions? Nat Rev Genet.
10:457–466. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pombo A, Hollinshead M and Cook PR:
Bridging the resolution gap: Imaging the same transcription
factories in cryosections by light and electron microscopy. J
Histochem Cytochem. 47:471–480. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Faro-Trindade I and Cook PR: A conserved
organization of transcription during embryonic stem cell
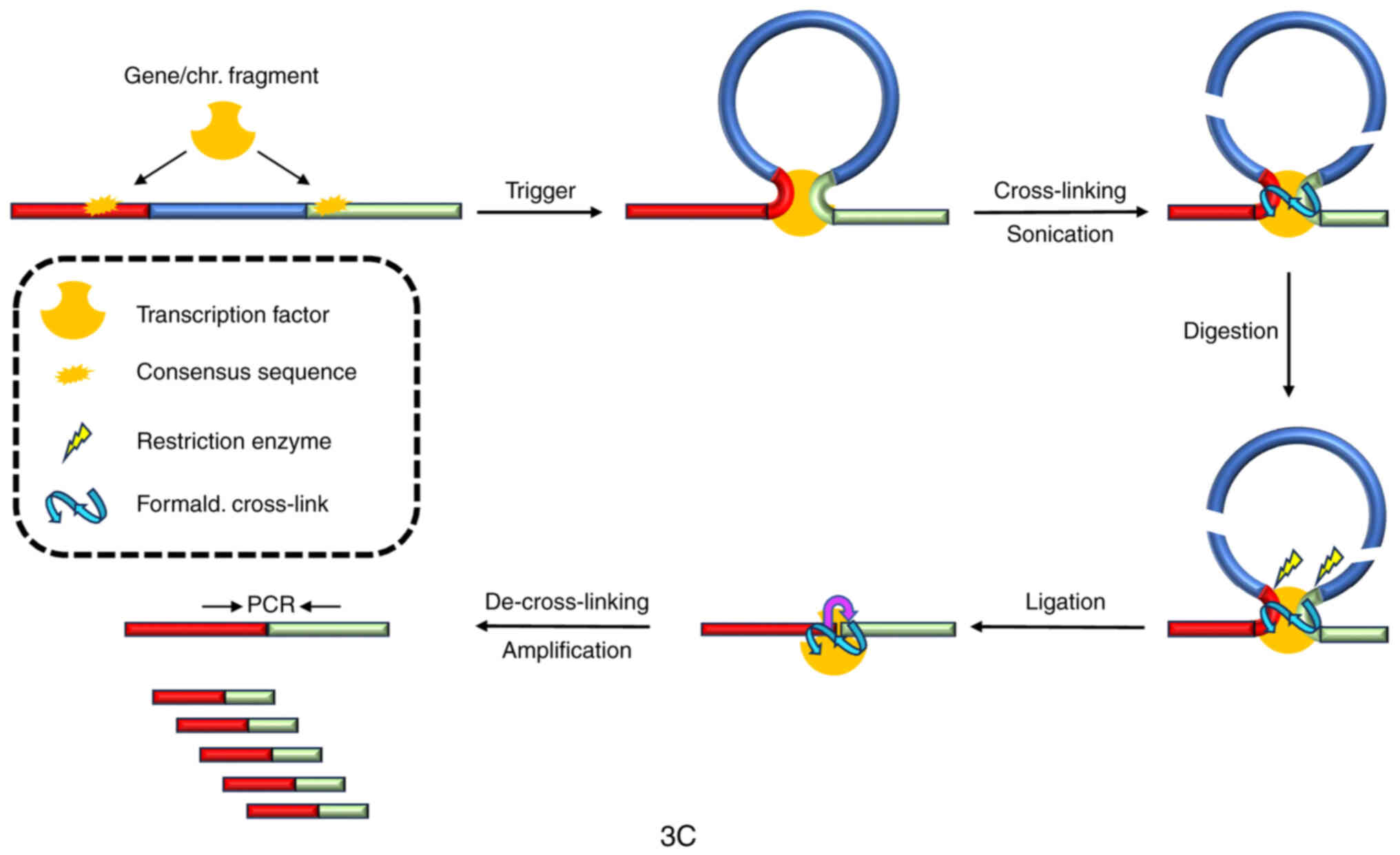
differentiation and in cells with high C value. Mol Biol Cell.
17:2910–2920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Laat W and Grosveld F: Spatial
organization of gene expression: The active chromatin hub.
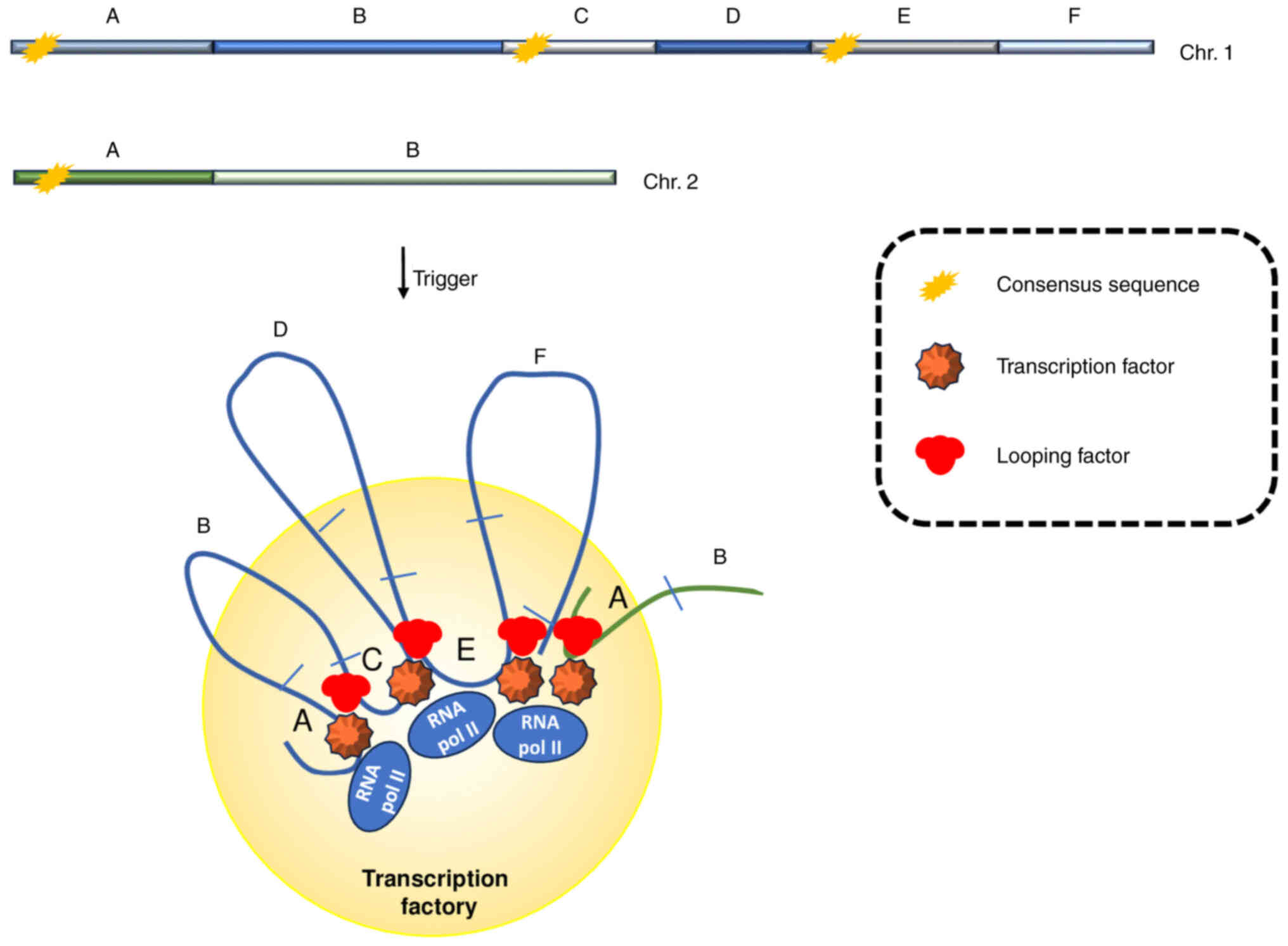
Chromosome Res. 11:447–459. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
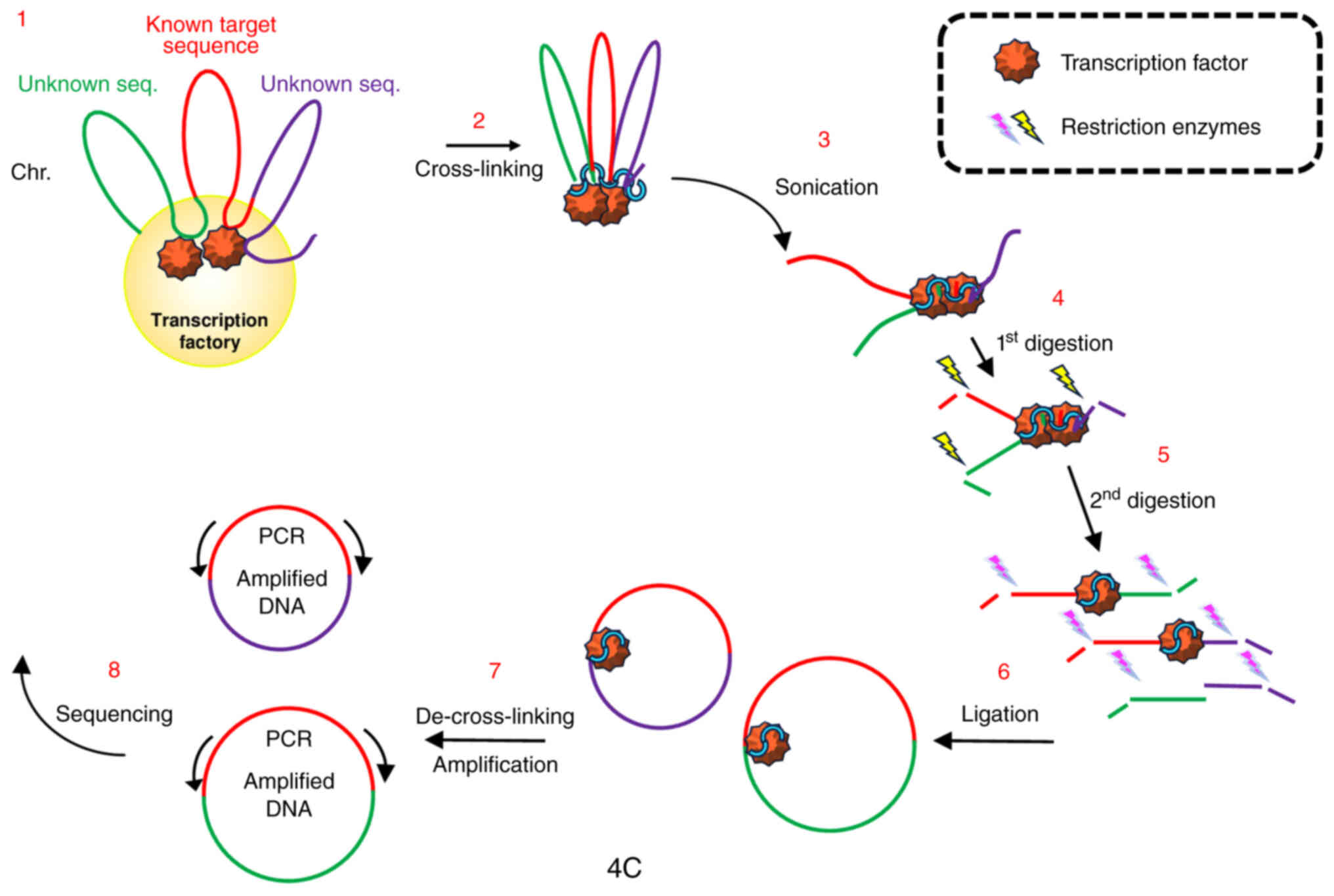
10
|
Papantonis A and Cook PR: Transcription
factories: Genome organization and gene regulation. Chem Rev.
13:8683–8705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Osborne CS, Chakalova L, Brown KE, Carter
D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W
and Fraser P: Active genes dynamically colocalize to shared sites
of ongoing transcription. Nat Genet. 36:1065–1071. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Faro-Trindade I and Cook PR: Transcription
factories: Structures conserved during differentiation and
evolution. Biochem Soc Trans. 34:1133–1137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Larkin JD, Cook PR and Papantonis A:
Dynamic reconfiguration of long human genes during one
transcription cycle. Mol Cell Biol. 32:2738–2747. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mahy NL, Perry PE and Bickmore WA: Gene
density and transcription influence the localization of chromatin
outside of chromosome territories detectable by FISH. J Cell Biol.
159:753–763. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chambeyron S and Bickmore WA: Chromatin
decondensation and nuclear reorganization of the HoxB locus upon
induction of transcription. Genes Dev. 18:1119–1130. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krivega I and Dean A: Enhancer and
promoter interactions-long distance calls. Curr Opin Genet Dev.
22:1–7. 2012. View Article : Google Scholar
|
|
17
|
Stadhouders R, Thongjuea S, Andrieu-Soler
C, Palstra RJ, Bryne JC, van den Heuvel A, Stevens M, de Boer E,
Kockx C, van der Sloot A, et al: Dynamic long-range chromatin
interactions control Myb proto-oncogene transcription during
erythroid development. EMBO J. 31:986–999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kagey MH, Newman JJ, Bilodeau S, Zhan Y,
Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine
SS, et al: Mediator and cohesion connect gene expression and
chromatin architecture. Nature. 467:430–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Q, Yu M, Tirado-Magallanes M, Li B,
Kong L, Guo M, Tan ZH, Lee S, Chai L, Numata A, et al: ZNF143
mediates CTCF-bound promoter-enhancer loops required for murine
hematopoietic stem and progenitor cell function. Nat Commun.
12:432021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weintraub AS, Li CH, Zamudio AV, Sigova
AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL,
et al: YY1 is a structural regulator of enhancer-promoter loops.
Cell. 171:1573–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiao W, Chen Y, Song H, Li D, Mei H, Yang
F, Fang E, Wang X, Huang K, Zheng L and Tong Q: HPSE enhancer RNA
promotes cancer progression through driving chromatin looping and
regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 36:2728–2745.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang
X, Sun T, Sweeney CJ, Lee GS, Chen S, et al: Enhancer RNAs
participate in androgen receptor-driven looping that selectively
enhances gene activation. Proc Natl Acad Sci USA. 111:7319–7324.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan SH, Leong WZ, Ngoc PCT, Tan TK,
Bertulfo FC, Lim MC, An O, Li Z, Yeoh AEJ, Fullwood MJ, et al: The
enhancer RNA ARIEL activates the oncogenic transcriptional program
in T-cell acute lymphoblastic leukemia. Blood. 134:239–251. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He X, Khan AU, Cheng H, Pappas DL Jr,
Hampsey M and Moore CL: Functional interactions between the
transcription and the mRNA 3′ end processing machineries mediated
by Ssu72 and Sub1. Genes Dev. 17:1030–1042. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng W, Lee J, Wang H, Miller J, Reik A,
Gregory PD, Dean A and Blobel GA: Controlling long-range genomic
interactions at a native locus by targeted tethering of a looping
factor. Cell. 149:1233–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chubb JR and Bickmore WA: Considering
nuclear compartmentalization in the light of nuclear dynamics.
Cell. 112:403–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chubb JR, Boyle S, Perry P and Bickmore
WA: Chromatin motion is constrained by association with nuclear
compartments in human cells. Curr Biol. 12:439–445. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meshorer E, Yellajoshula D, George E,
Scambler PJ, Brown DT and Misteli T: Hyper-dynamic plasticity of
chromatin proteins in pluripotent embryonic stem cells. Dev Cell.
10:105–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Turner BM: Open chromatin and
hypertranscription in embryonic stem cells. Cell Stem Cell.
2:408–410. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang HH, Hemberg M, Barahona M, Ingber DE
and Huang S: Transcriptome-wide noise controls lineage choice in
mammalian progenitor cells. Nature. 453:544–547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guelen L, Pagie L, Brasset E, Meuleman W,
Faza MB, Talhout W, Eussen BH, de Klein A, Wesels L, de Laat W and
van Steensel B: Domain organization of human chromosomes revealed
by mapping of nuclear lamina interactions. Nature. 453:948–951.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Solovei I, Kreysing M, Lanctôt C, Kösem S,
Peichl L, Cremer T, Guck J and Joffe B: Nuclear architecture of rod
photoreceptor cells adapts to vision in mammalian evolution. Cell.
137:356–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nitzsche A, Paszkowski-Rogacz M, Matarese
F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L,
Huebner N, Mann M, et al: RAD21 cooperates with pluripotency
transcription factors in the maintenance of embryonic stem cell
identity. PLoS One. 6:e194702011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bertolini JA, Favaro R, Zhu Y, Pagin M,
Ngan CY, Wong CH, Tjong H, Vermunt MW, Martynoga B, Barone C, et
al: Mapping the global chromatin connectivity network for Sox2
function in neural stem cell maintenance. Cell Stem Cell.
24:462–476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Drissen R, Palstra RJ, Gillemans N,
Splinter E, Grosveld F, Philipsen S and de Laat W: The active
spatial organization of the beta-globin locus requires the
transcription factor EKLF. Genes Dev. 18:2485–2490. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Magli A, Baik J, Pota P, Cordero CO, Kwak
IY, Garry DJ, Love PE, Dynlacht BD and Perlingeiro RCR: Pax3
cooperates with Ldb1 to direct local chromosome architecture during
myogenic lineage specification. Nat Commun. 10:23162019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dall'Agnese A, Caputo L, Nicoletti C, di
Iulio J, Schmitt A, Gatto S, Diao Y, Ye Z, Forcato M, Perera R, et
al: Transcription factor-directed re-wiring of chromatin
architecture for somatic cell nuclear reprogramming toward
trans-differentiation. Mol Cell. 76:453–472. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Johanson TM, Lun ATL, Coughlan HD, Tan T,
Smyth GK, Nutt SL and Allan RS: Transcription-factor-mediated
supervision of global genome architecture maintains B cell
identity. Nat Immunol. 19:1257–1264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sołtys K and Ozyhar A: Transcription
regulators and membrane less organelles challenges to investigate
them. Int J Mol Sci. 22:127582021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shrinivas K, Sabari BR, Coffey EL, Klein
IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young
RA and Chakraborty AK: Enhancer features that drive formation of
transcriptional condensates. Mol Cell. 75:549–561.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pederson T: The nucleolus. Cold Spring
Harbor Perspect Biol. 3:a0003682011. View Article : Google Scholar
|
|
42
|
Gondor A and Ohlsson R: Chromosome
crosstalk in three dimensions. Nature. 461:212–217. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pujadas E and Feinberg AP: Regulated noise
in the epigenetic landscape of development and disease. Cell.
148:1123–1131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Osborne CS, Chakalova L, Mitchell JA,
Horton A, Wood AL, Bolland DJ, Corcoran AE and Fraser P: Myc
dynamically and preferentially relocates to a transcription factory
occupied by Igh. PLoS Biol. 5:e1922007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Misteli T: Higher-order genome
organization in human disease. Cold Spring Harb Perspect Biol.
2:a0007942010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev V and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wijchers PJ and de Laat W: Genome
organization influences partner selection for chromosomal
rearrangements. Trends Genet. 27:63–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
De S and Michor F: DNA replication timing
and long-range DNA interactions predict mutational landscapes of
cancer genomes. Nat Biotechnol. 29:1103–1108. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fudenberg G, Getz G, Meyerson M and Mirny
LA: High order chromatin architecture shapes the landscape of
chromosomal alterations in cancer. Nat Biotechnol. 29:1109–1113.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kouzarides T: Chromatin modifications and
their functions. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peters AH, Kubicek S, Mechtler K,
O'Sullivan RJ, Derijck AAH, Perez-Burgos L, Kohlmaier A, Opravil S,
Tachibana M, Shinkai Y, et al: Partitioning and plasticity of
repressive histone methylation states in mammalian chromatin. Mol
Cell. 12:1577–1589. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rice JC, Briggs SD, Ueberheide B, Barber
CM, Shabanowitz J, Hunt DF, Shinkai Y and Allis CD: Histone
methyltransferases direct different degrees of methylation to
define distinct chromatin domains. Mol Cell. 12:1591–1598. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Closs PAC, Christensen J, Agger K and
Helin K: Erasing the methyl mark: Histone demethylases at the
center of cellular differentiation and disease. Genes Dev.
22:1115–1140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Perillo B, Ombra MN, Bertoni A, Cuozzo C,
Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C and
Avvedimento EV: DNA oxidation as triggered by H3K9me2 demethylation
drives estrogen-induced gene expression. Science. 319:202–206.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Papantonis A and Cook PR: Genome
architecture and the role of transcription. Curr Opin Cell Biol.
22:271–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dekker J, Rippe K, Dekker M and Kleckner
N: Capturing chromosome conformation. Science. 295:1306–1311. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Simonis M, Kooren J and de Laat W: An
evaluation of 3C-based methods to capture DNA interactions. Nat
Methods. 4:895–901. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Murrell A, Heeson S and Reik W:
Interaction between differentially methylated regions partitions
the imprinted genes Igf2 and H19 into parent-specific chromatin
loops. Nat Genet. 36:889–893. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gheldof N, Smith EM, Tabuchi TM, Koch CM,
Dunham I, Stamatoyannopoulos JA and Dekker J: Cell-type-specific
long-range looping interactions identify distant regulatory
elements of the CFTR gene. Nucleic Acids Res. 38:4325–4336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dekker J, Marti-Renom MA and Mirny LA:
Exploring the three-dimensional organization of genomes:
Interpreting chromatin interaction data. Nat Rev Genet. 14:390–403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
O'Sullivan JM, Tan-Wong SM, Morillon A,
Lee B, Coles J, Mellor J and Proudfoot NJ: Gene loops juxtapose
promoters and terminators in yeast. Nat Genet. 36:1014–1018. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bentley DL: Rules of engagement:
Co-transcriptional recruitment of pre-mRNA processing factors. Curr
Opin Cell Biol. 17:251–256. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Déjardin J and Kingston RE: Purification
of proteins associated with specific genomic loci. Cell.
136:175–186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Abbondanza C, De Rosa C, Ombra MN, Aceto
F, Medici N, Altucci L, Moncharmont B, Puca GA, Porcellini A,
Avvedimento EV and Perillo B: Highlighting chromosome loops in
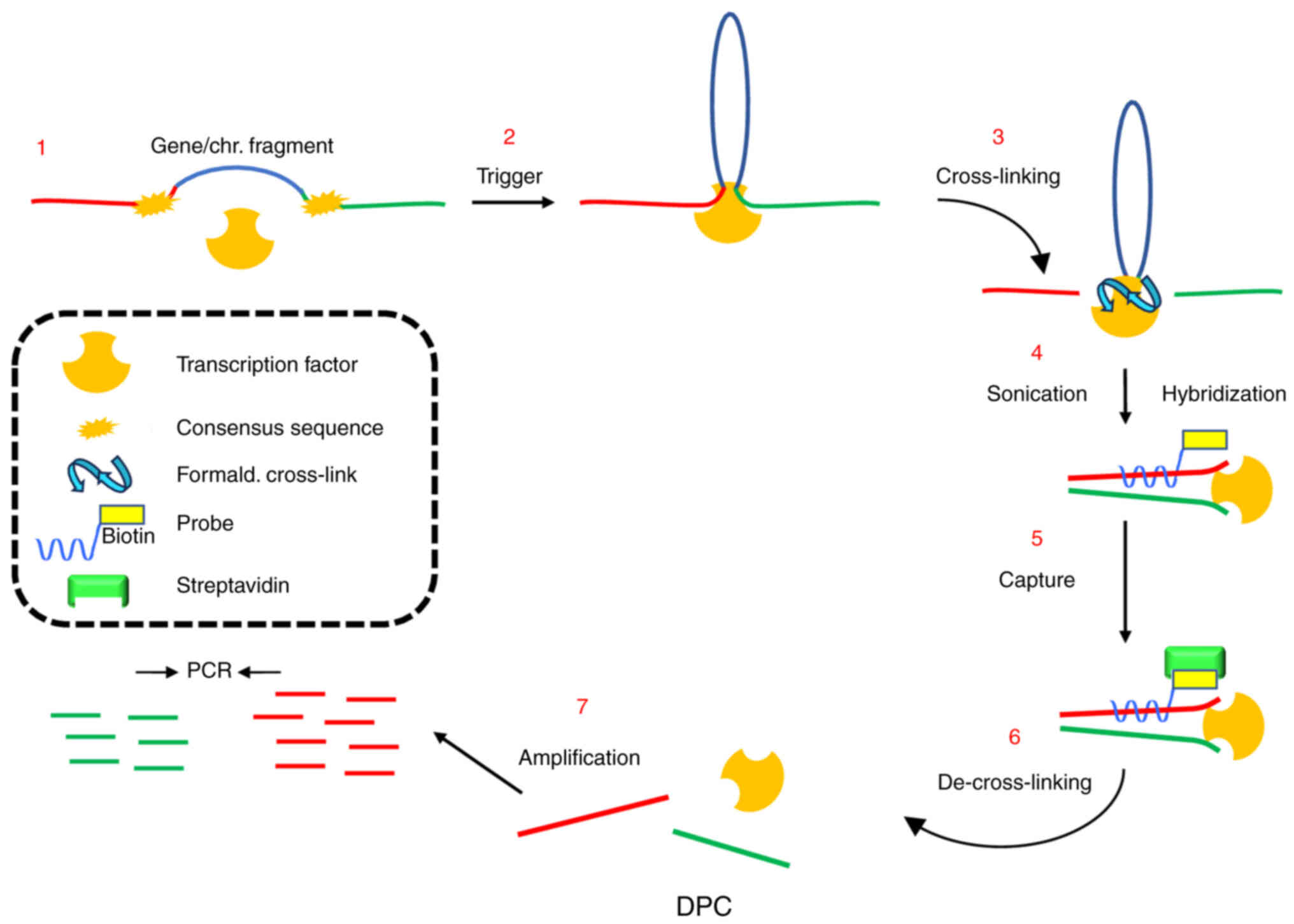
DNA-picked chromatin (DPC). Epigenetics. 6:979–986. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bolzer A, Kreth G, Solovei I, Koehler D,
Sarakoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR and
Cremer T: Three-dimensional maps of all chromosomes in human male
fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e1572005.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Han J, Zhang Z and Wang K: 3C and 3C-based
techniques: The powerful tools for spatial genome organization
deciphering. Mol Cytogen. 9:11–21. 2018.
|
|
69
|
Simonis M, Klous P, Splinter E, Moshkin Y,
Willemsen R, de Wit E, van Steensel B and de Laat W: Nuclear
organization of active and inactive chromatin domains uncovered by
chromosome conformation capture-on-chip (4C). Nat Genet.
38:1348–1354. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wurtele H and Chartrand P: Genome-wide
scanning of HoxB1-associated loci in mouse ES cells using an
open-ended chromosome conformation capture methodology. Chromosome
Res. 14:477–495. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhao Z, Tavoosidana G, Sjölinder M, Göndör
A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et
al: Circular chromosome conformation capture (4C) uncovers
extensive networks of epigenetically regulated intra- and
interchromosomal interactions. Nat Genet. 38:1341–1347. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
van de Werken HJ, Landan G, Holwerda SJ,
Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y,
Bouwman BAM, et al: Robust 4C-seq data analysis to screen for
regulatory DNA interactions. Nat Methods. 9:969–972. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Symmons O, Pan L, Remeseiro S, Aktas T,
Klein F, Huber W and Spitz F: The Shh topological domain
facilitates the action of remote enhancers reducing the effects of
genomic distances. Dev Cell. 5:529–543. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tomasetto C, Rockel N, Mattei MG, Fujita R
and Rio MC: The gene encoding the human spasmolytic protein
(SML1/hSP) is in 21q physically linked to the homologous breast
cancer marker gene BCEI/pS2. Genomics. 13:1328–1330. 1992.
View Article : Google Scholar : PubMed/NCBI
|